Abstract
γ glutamyl transferase (GGT) and albumin (ALB) are two markers of liver function. These two proteins have been associated with non-alcoholic fatty liver disease and cardiovascular disease. The objective of this study was to explore the genetic factors that influence variation in the plasma levels of GGT and ALB and to evaluate their genetic correlations with cardiovascular risk factors. Baboons from the Southwest National Primate Research Center at the Southwest Foundation for Biomedical Research, San Antonio, TX were used as an animal model. The baboons were fed a standard monkey chow diet ad libitum. Fasting plasma concentrations of GGT, ALB, triglycerides, total cholesterol and LDL cholesterol were measured in 350 pedigreed adult baboons by standard assay procedures. A maximum likelihood based variance decomposition approach implemented in the computer program SOLAR was used to conduct genetic analyses. The heritabilities of GGT (h2=0.55; p< 0.0001) and ALB (h2=0.42; p< 0.01) were significant. No statistically significant associations were found between GGT and the cardiovascular related phenotypes. Genetic correlations between ALB and total cholesterol, LDL cholesterol and triglycerides were significant. A QTL (LOD = 2.8) for GGT plasma levels was identified on the baboon homologue of human chromosome 22 between markers D22S304 and D22S280. A QTL (LOD =2.3) near marker D10S1432 was detected on the baboon homologue of human chromosome 10 for ALB. These results imply that variations in the plasma levels of GGT and ALB are under significant genetic regulation and that a common genetic component influences ALB and cardiovascular risk factor phenotypes.
Keywords: NAFLD, obesity, genome scan, atherosclerosis, oxidative stress
Introduction
The leading cause of an abnormal liver function test in overweight and obese individuals is non-alcoholic fatty liver disease (NAFLD) (1). This hepatic pathology encompasses simple steatosis, non-alcoholic steatohepatitis (NASH), fibrosis and cirrhosis (2). Many factors can lead to NAFLD. NAFLD affects approximately 14%-25% of the world's population. The prevalence is higher in the obese and type 2 diabetics (3). Patients with NAFLD might be at a higher risk for cardiovascular disease than those without this hepatic condition (4). Epidemiological studies have shown a strong positive relationship between markers of NAFLD such as liver enzymes and prevalence of cardiovascular disease (CVD) (5,-7). The focus of this paper is on two indicators of liver function, γ glutamyl transferase (GGT) and albumin (ALB), both of which have also been implicated in the pathogenesis of atherosclerosis. It is known that the variation in the plasma level of these liver related proteins is genetically influenced. To explore these genetic factors and to evaluate the relationship of these liver markers with cardiovascular risk phenotypes we have chosen the baboon as an animal model.
The pedigreed baboons from the colony maintained by the Southwest National Primate Research Center are an excellent model for this study. Gene and protein sequence identity is conserved between baboons and humans and both are physiologically and developmentally similar. Baboons have proven useful for genetic studies related to atherosclerosis (8), insulin-resistance (9), obesity (10) and liver gene expression (11). Although all the baboons share similar diet and housing conditions, 10% become obese spontaneously and 4% become hyperglycemic (12). Further, during weight gain fat is deposited primarily in the abdominal area of baboons. It is known that visceral adiposity is strongly linked to several metabolic diseases such as cardiovascular ailments, type 2 diabetes and fatty liver (13) and in obese and insulin resistant baboons from this colony levels of liver enzymes have been found to be altered (14).
It is significant to determine the genetic factors that influence variation in circulating levels of GGT and ALB because to understand the pathogenesis of a disease all of the components contributing to the pathological state have to be identified. Each of the mechanisms involved in the development of a disease will not be fully understood if a component is ignored. Therefore, findings of this study will assist in future hypotheses aimed at identifying the relative contribution of various components to the pathogenesis of liver dysfunction. Additionally, if common set of genes are found to regulate variation in levels of liver proteins and cardiovascular disease risk factors these experimental results can be used to develop future studies that will identify and explore those genes. Therefore, the purpose of this study is to identify chromosomal regions containing genes that affect the variation in the plasma levels of GGT and albumin and to determine whether genes contributing to variation in these hepatic proteins also influence variation in known cardiovascular disease risk factors as well.
METHODS
Animals
The study population consisted of 350 non-inbred (12) ( 254 females, 96 males) pedigreed baboons (Papio hamadrayas) from the colony maintained at the Southwest National Primate Research Center located at the Southwest Foundation for Biomedical Research (SFBR) in San Antonio, TX, USA. These animals consist primarily of olive baboons, but also include yellow baboons and olive-yellow hybrids. The baboons are gang housed and fed a standard monkey chow diet ad libitum (Harlan Teklad 15% monkey diet, 8715, Indianapolis, IN).
Sampling and Analyses
Blood samples were drawn after an overnight fast (12 hours) from animals sedated with ketamine. Heparin tubes were used to collect samples for analysis of GGT, ALB, triglycerides, total cholesterol and LDL cholesterol. The samples were subjected to centrifugation at 2000 × g for 10 minutes and the resultant plasma was stored in aliquots at –80°C for future analysis. The animals were weighed on a calibrated electronic scale (GSE, Chicago, IL). The Institutional Animal Care and Use Committee of the SFBR approved all procedures.
Triglycerides, total cholesterol, LDL cholesterol, GGT and ALB were analyzed by standard laboratory techniques using the Alfa Wasserman ACE clinical chemistry instrument (West Cladwell, NJ). Samples whose replicates deviated > 5% were reanalyzed.
Genotyping
The animals in this study had previously been genotyped at more than 400 highly polymorphic microsatellite marker loci for the construction of a whole genome linkage map with an average marker density of10cM (11, 15). We made use of these maps and identity-by-descent coefficients estimated from the genotype data in our analyses
Statistical genetic methods
The maximum likelihood variance decomposition methods implemented in SOLAR (16) were used to perform the statistical genetic analyses reported in this paper. We used this method to partition the phenotypic variance of the quantitative traits studied into additive genetic and non-genetic (environmental) components. From this decomposition, we estimated the proportion of the variance due to the additive effects of genes – i.e., the heritability (h2).
We further decomposed the additive genetic variance for each trait into a component for individual loci and a residual (polygenic) component and performed multipoint whole genome linkage screens to test for quantitative loci (QTLs). Using the baboon marker map information and the multipoint IBD matrices estimated from the genotype data, we tested the hypothesis of linkage at 1 cM intervals across the baboon genome. Essentially, these tests consisted of comparing the likelihood of a restricted model for the trait in which the variance due to a QTL is constrained to zero (no linkage, null hypothesis) to an unrestricted model in which the QTL-specific variance is freely estimated. Twice the difference of the log likelihoods was asymptomatically distributed as ½: ½ mixture of chi-square variable, with one degree of freedom and a point mass at zero (17). The difference between the two log10 likelihoods yields a LOD score, which measures the support for the hypothesis of linkage over that of “no linkage” at a given chromosomal location. Our threshold for significant evidence of linkage was LOD =2.69, and for suggestive evidence of linkage was LOD=1.46. We obtained these genome-wide significance thresholds using a modification of an approach suggested by Feingold et al. (18) to control for the overall false positive rate in our whole genome linkage screens of a single phenotype. Our approach takes into account the finite marker density in the linkage map utilized in the multipoint QTL screens and the mean recombination rate for these pedigreed baboons.
An extension of the univariate model was used for bivariate genetic analyses. The bivariate phenotype is a result of the animal's phenotypic values, population means, the additive genetic estimates and environmental effects. This model was used to calculate the genetic and environmental variance-covariance matrices, in addition to genetic and environmental correlations. Both univariate and bivariate genetic analyses were conducted using the Sequential Oligogenic Linkage Analysis Routinues (SOLAR) computer program (16). Age, sex, age squared and their interactions were included as covariates for the analyses All the traits were inverse normalised for the analyses. According to this step, observations are ranked and replaced by expected value for that rank from a standard normal distribution. Software program PEDSYS computed the group means and ranges of male and female baboons.
RESULTS
Information describing the number of relative pairs represented in the sample for these analyses is provided in Table 1. Descriptive statistics of the phenotypes according to sex are given in Table 2. Male baboons were younger and had higher body weights than females due to the sexual dimorphism in these animals (19). Female baboons had higher levels of plasma total cholesterol, LDL cholesterol and triglycerides. The activity of GGT was elevated in male baboons, but no difference between the sexes was observed for ALB.
TABLE 1.
Relative pairs within the baboon population
| Relationship | Number |
|---|---|
| Parent-offspring | 44 |
| Siblings | 152 |
| Avuncular | 32 |
| Half-siblings | 2119 |
| Half avuncular | 331 |
| Half 1st cousins | 33 |
| Half 1st cousins, 1 rem | 15 |
| Half siblings & 1st cousins | 3 |
| Half siblings & half 1st cousins | 30 |
| Half siblings & half avuncular | 5 |
TABLE 2.
Descriptive statistics of baboons
| Phenotype | Males/females * | Range |
|---|---|---|
| Number | 96/254 | |
| Age (yrs) | 12.90 (0.4)/15.24 (0.3) | 6.9 - 31 |
| Weight (kg) | 31.90 (0.6)/19.54 (0.3) | 12 - 48 |
| Total cholesterol (mg/dl) | 91.09 (2.6)/ 121.14 (2.6) | 54 - 239 |
| LDL cholestrol (mg/dl) | 31.53 (1.7)/53.52 (1.8) | 6 - 135 |
| Triglycerides (mg/dl) | 46.03 (1.8)/65.20 (2.0) | 19 - 152 |
| γ glutamyl transferase (U/L) | 46.41 (1.5)/43.43 (0.7) | 2 - 148 |
| Albumin (g/dl) | 4.4 (0.05)/4.3 (0.03) | 2.2 - 6 |
Mean (S.E.M)
Table 3 shows the heritabilities of liver function markers and cardiovascular risk factors analyzed as part of this study. All were significantly heritable with total cholesterol exhibiting the greatest hertitability. Table 4 gives the genetic and phenotypic correlations between plasma GGT, albumin and plasma lipids. No statistically significant genetic association was found between GGT and the cardiovascular related phenotypes. However, relationship between plasma albumin and lipids was negative. Significant phenotypic correlation between GGT and ALB ((rho p= 0.25; p < 0.0009) and LDL cholesterol was found in this sample
TABLE 3.
Heritabilities of liver function markers and cardiovascular risk factors
| Phenotype | Heritability ± s.e.m | p |
|---|---|---|
| Total cholesterol | 0.68 ± 0.10 | < 0.01 |
| LDL cholesterol | 0.50 ± 0.10 | < 0.01 |
| Triglycerides | 0.34 ± 0.11 | < 0.01 |
| γglutamyl transferase | 0.55 ± 0.13 | <0.0001 |
| Albumin | 0.42 ± 0.15 | < 0.01 |
TABLE 4.
Genetic (ρG) correlations between plasma concentration of gamma glutamyl transferase, albumin and cardiovascular risk factors
| Gammaglutamyl Transferase | Albumin | |||||||
|---|---|---|---|---|---|---|---|---|
| Phenotype | ρGenetic | p | ρPhenotype | p | ρGenetic | p | ρPhenotype | p |
| Total cholesterol | 0.18 | 0.39 | 0.04 | 0.09 | −0.60 | 0.01 | −0.09 | 0.20 |
| LDL cholesterol | 0.26 | 0.35 | 0.15 | 0.03 | −0.67 | 0.01 | −0.09 | 0.36 |
| Triglycerides | −0.41 | 0.19 | −0.03 | 0.13 | −0.86 | <0.01 | −0.09 | 0.44 |
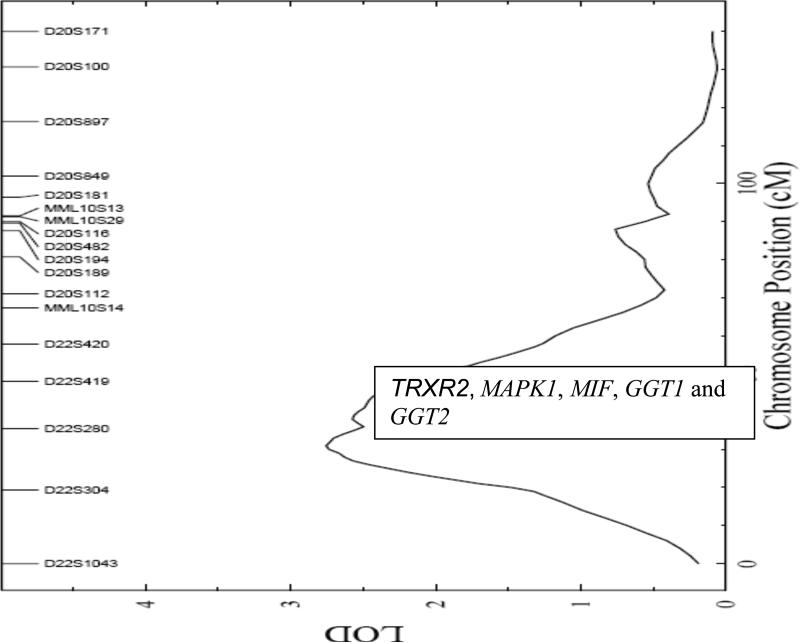
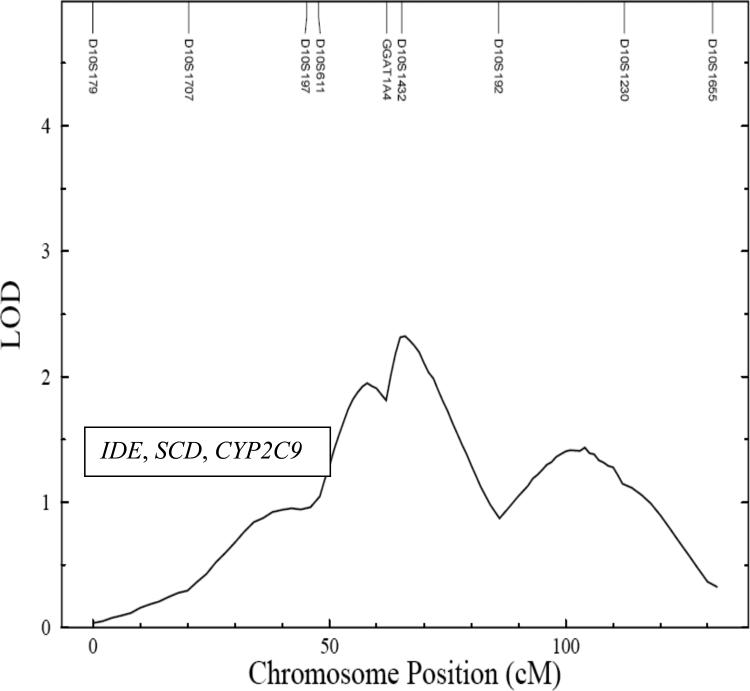
The linkage signal for plasma GGT is displayed in Figure 1. The strongest QTL for GGT was detected between markers D22S304 and D22S280 on the baboon homologue of human chromosome 22, at approximately 30 cM with a LOD score of 2.8. The maximum LOD score detected in a genome wide linkage scan for ALB was detected on the baboon homologue of human chromosome 10, with a LOD score of 2.3 at 66 cM near marker D10S1432 (Figure 2).
FIGURE 1.
Map depicting LOD scores (X axis) and marker distances (Y axis) for plasma gamma glutamyl transferase on chromosome 22
FIGURE 2.
Map depicting LOD scores (X axis) and marker distances (Y axis) for plasma albumin on chromosome 10
DISCUSSION
This study has demonstrated that the variation in the plasma levels of GGT and ALB is influenced by genetic factors. The heritability estimates of these liver function markers in baboons are similar to those found in humans. Heritability estimates of GGT were 52% in Australian men and women (20) and 62% in Danish twins (21), as compared to 55% found in this study. For ALB the additive genetic effect in adult men and women was 36%, which is lower than the value calculated for baboons in this paper (22). This is the first study to publish the QTLs for GGT and ALB in baboons and there is no information on QTLs for these proteins yet published in humans.
Even though phenotypic correlations between ALB and cardiovascular risk factors have been reported (23- 25) the evidence of shared genetic effects (i.e. pleiotropy) between them has been detected for the first time in this paper. The associations found here imply that overlapping, but not identical set of genes regulate their circulating concentrations.
Plasma GGT is a well-established and sensitive marker of hepatic dysfunction. Plasma GGT is a well-established and sensitive marker of hepatic dysfunction. The circulating concentration of GGT is elevated in CVD and NAFLD patients (26 - 30), implying that it might be a link between the two diseases. Yet, in this study, we did not detect any significant genetic relationship between GGT and cardiovascular risk factors. However, lack of a genetic correlation in this experimental model does not rule out the potential for a common set of genes that might operate under a different experimental design or in an atherogenic environment.
In contrast, a significant negative genetic correlation between ALB and cardiovascular risk factors was observed in this study. Traditionally, plasma ALB has been considered a good biomarker of protein malnutrition but recently this protein has been associated with heart disease and was found to be an independent indicator of mortality in patients with NAFLD (23, 31). Since the liver manufactures ALB, damage to this organ reduces the blood levels of this protein (32). Lower levels of ALB raises the concentration of unbound free fatty acids that consequently increases the amount of lipid parameters in the blood (33).
Insulin resistance and oxidative stress are hypothesized to be the link between fatty liver abnormalities and heart disease (34). Another plausible mechanism associating these two disorders could be abnormal lipoprotein metabolism in individuals with steatotic liver, i.e. elevated plasma concentrations of triglycerides and low-density lipoprotein cholesterol (LDL) (35). The high prevalence of cardiovascular disease in NAFLD patients could be due to insulin resistance that results in dyslipidemia (36).
For GGT, potential positional candidate genes (based upon the homologous region in humans) that are present within the one LOD-support interval of the maximum linkage signal on chromosome 22 include selenoprotein Z (TRXR2) (37), mitogen- activated protein kinase 1 (MAPK1) (38), macrophage migration inhibitory factor (MIF) (39), gamma glutamyltransferase 1 (GGT1) and gamma glutamyltransferase 2 (GGT2) (40). The most promising positional candidate genes within this QTL are the two structural genes that produce the two isoforms (GGT1 and GGT2) of GGT. The genes of interest within the one LOD support interval of the ALB QTL on chromosome 10 are insulin degrading enzyme (IDE) (41), stearoyl –CoA desaturase (SCD) (42), cytochorme P450 subfamily IIC, and polypeptide 9 (CYP2C9) (43). All of these genes are related to oxidative stress.
In conclusion, this study indicates that genetic factors significantly influence the variation in circulating levels of GGT and ALB and a common set of genes appear to regulate plasma levels of ALB and cardiovascular related risk factors in baboons.
GRANTS
This investigation was conducted in part in facilities constructed with support from the Research Facilities Improvement Program under grant number C06 RR014578, C06 RR013556, C06 RR015456, C06 RR017515, and with support from NIH grants PO1 HL028972, and P51 RR013986, as well as research support from the Kronkosky Foundation.
REFERENCES
- 1.Ekstedt M, Franzen LE, Mathiesen UL, Thorelius L, Holmqvist M, Bodemar G, Kechagias S. Long-term follow-up of patients with NAFLD and elevated liver enzymes. Hepatology. 2006;44:865–873. doi: 10.1002/hep.21327. [DOI] [PubMed] [Google Scholar]
- 2.Liou I, Kowdley KV. Natural history of nonalcoholic steatohepatitis. J Clin Gastroenterol. 2006;40:S11–16. doi: 10.1097/01.mcg.0000168644.23697.31. [DOI] [PubMed] [Google Scholar]
- 3.Adams LA, Angulo P. Recent concepts in non-alcoholic fatty liver disease. Diabet Med. 2005;22:1129–1133. doi: 10.1111/j.1464-5491.2005.01748.x. [DOI] [PubMed] [Google Scholar]
- 4.Targher G. Non-alcoholic fatty liver disease, the metabolic syndrome and the risk of cardiovascular disease: the plot thickens. Diabet Med. 2007;24:1–6. doi: 10.1111/j.1464-5491.2007.02025.x. [DOI] [PubMed] [Google Scholar]
- 5.Schindhelm RK, Dekker JM, Nijpels G, Bouter LM, Stehouwer CD, Heine RJ, Diamant M. Alanine aminotransferase predicts coronary heart disease events: A 10-year follow-up of the Hoorn Study. Atherosclerosis. 2007;191:391–396. doi: 10.1016/j.atherosclerosis.2006.04.006. [DOI] [PubMed] [Google Scholar]
- 6.Targher G, Bertolini L, Poli F, Rodella S, Scala L, Tessari R, Zenari L, Falezza G. Nonalcoholic fatty liver disease and risk of future cardiovascular events among type 2 diabetic patients. Diabetes. 2005;54:3541–6. doi: 10.2337/diabetes.54.12.3541. [DOI] [PubMed] [Google Scholar]
- 7.Devers MC, Campbell S, Shaw J, Zimmet P, Simmons D. Should liver function tests be included in definitions of metabolic syndrome? Evidence from the association between liver function tests, components of metabolic syndrome and prevalent cardiovascular disease. Diabet Med. 2008;25:523–9. doi: 10.1111/j.1464-5491.2008.02408.x. [DOI] [PubMed] [Google Scholar]
- 8.Mahaney MC, Blangero J, Rainwater DL, Mott GE, Comuzzie AG, MacCluer JW, Vandeberg JL. Pleiotropy and genotype by diet interaction in a baboon model for atherosclerosis: a multivariate quantitative genetic analysis of HDL subfractions in two dietary environments. Arterioscler Thromb Vasc Biol. 1999;19:1134–1341. doi: 10.1161/01.atv.19.4.1134. [DOI] [PubMed] [Google Scholar]
- 9.Cai G, Cole SA, Tejero ME, Proffitt JM, Freeland-Graves JH, Blangero J, Comuzzie AG. Pleiotropic effects of genes for insulin resistance on adiposity in baboons. Obes Res. 2004;12:1766–1772. doi: 10.1038/oby.2004.219. [DOI] [PubMed] [Google Scholar]
- 10.Tejero ME, Cole SA, Cai G, Peebles KW, Freeland-Graves JH, Cox LA, Mahaney MC, Rogers J, VandeBerg JL, Blangero J, Comuzzie AG. Genome-wide scan of resistin mRNA expression in omental adipose tissue of baboons. Int J Obes. 2005;29:406–412. doi: 10.1038/sj.ijo.0802699. [DOI] [PubMed] [Google Scholar]
- 11.Cox LA, Mahaney MC, Vandeberg JL, Rogers J. A second-generation genetic linkage map of the baboon (Papio hamadryas) genome. Genomics. 2006;88:274–281. doi: 10.1016/j.ygeno.2006.03.020. [DOI] [PubMed] [Google Scholar]
- 12.Comuzzie AG, Cole SA, Martin L, Carey KD, Mahaney MC, Blangero J, VandeBerg JL. The baboon as a nonhuman primate model for the study of the genetics of obesity. Obes Res. 2003;11:75–80. doi: 10.1038/oby.2003.12. [DOI] [PubMed] [Google Scholar]
- 13.Licata G, Argano C, Di Chiara T, Parrinello G, Scaglione R. Obesity: a main factor of metabolic syndrome? Panminerva Med. 2006;48:77–85. [PubMed] [Google Scholar]
- 14.Bose T, Lopez-Alvarenga JC, Dick E, Tejero ME, Freeland-Graves JH, Cole SA, Comuzzie AG. Presence of non-alcoholic fatty liver in Baboons (Abstract). Am J Primatol. 2006;68:41. [Google Scholar]
- 15.Rogers J, Mahaney MC, Witte SM, Nair S, Newman D, Wedel S, Rodriguez LA, Rice KS, Slifer SH, Perelygin A, Slifer M, Palladino-Negro P, Newman T, Chambers K, Joslyn G, Parry P, Morin PA. A genetic linkage map of the baboon (papio hamadryas) based on human microsatellite polymorphisms. Genomics. 2000;67:237–247. doi: 10.1006/geno.2000.6245. [DOI] [PubMed] [Google Scholar]
- 16.Almasy L, Blangero J. Mulitpoint quantitative-trait linkage analysis in general pedigrees. Am J Hum Genet. 1998;62:1198–1121. doi: 10.1086/301844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Self S, Liang K. Asymptotic properties of maximum likelihood-ratio tests under nonstandard conditions. J Am Stat Assoc. 1987;82:605–610. [Google Scholar]
- 18.Feingold E, Brown PO, Siegmund D. Gaussian models for genetic linkage analysis using complete high-resolution maps of identity by descent. Am J Hum Genet. 1993;53:234–51. [PMC free article] [PubMed] [Google Scholar]
- 19.Tejero ME, Freeland-Graves JH, Proffitt JM, Peebles KW, Cai G, Cole SA, Comuzzie AG. Adiponectin but not resistin is associated with insulin resistance-related phenotypes in baboons. Obes Res. 2004;12:871–877. doi: 10.1038/oby.2004.105. [DOI] [PubMed] [Google Scholar]
- 20.Whitfield JB, Zhu G, Nestler JE, Heath AC, Martin NG. Genetic covariation between serum gamma-glutamyltransferase activity and cardiovascular risk factors. Clin Chem. 2002;48:1426–1431. [PubMed] [Google Scholar]
- 21.Bathum L, Petersen HC, Rosholm JU, Hyltoft Petersen P, Vaupel J, Christensen K. Evidence for a substantial genetic influence on biochemical liver function tests: results from a population-based Danish twin study. Clin Chem. 2001;47:81–87. [PubMed] [Google Scholar]
- 22.Pankow JS, Folsom AR, Cushman M, Borecki IB, Hopkins PN, Eckfeldt JH, Tracy RP. Familial and genetic determinants of systemic markers of inflammation: the NHLBI family heart study. Atherosclerosis. 2001;154:681–689. doi: 10.1016/s0021-9150(00)00586-4. [DOI] [PubMed] [Google Scholar]
- 23.Schalk BW, Visser M, Bremmer MA, Penninx BW, Bouter LM, Deeg DJ. Change of serum albumin and risk of cardiovascular disease and all-cause mortality: Longitudinal Aging Study Amsterdam. Am J Epidemiol. 2006;164:969–977. doi: 10.1093/aje/kwj312. [DOI] [PubMed] [Google Scholar]
- 24.Beddhu S, Kaysen GA, Yan G, Sarnak M, Agodoa L, Ornt D, Cheung AK, HEMO Study Group Association of serum albumin and atherosclerosis in chronic hemodialysis patients. Am J Kidney Dis. 2002;40:721–727. doi: 10.1053/ajkd.2002.35679. [DOI] [PubMed] [Google Scholar]
- 25.Krijgsman B, Papadakis JA, Ganotakis ES, Mikhailidis DP, Hamilton G. The effect of peripheral vascular disease on the serum levels of natural anti-oxidants: bilirubin and albumin. Int Angiol. 2002;21:44–52. [PubMed] [Google Scholar]
- 26.Meisinger C, Doring A, Schneider A, Lowel H. Serum gamma-glutamyltransferase is a predictor of incident coronary events in apparently healthy men from the general population. Atherosclerosis. 2006;189:297–302. doi: 10.1016/j.atherosclerosis.2006.01.010. [DOI] [PubMed] [Google Scholar]
- 27.Ruttmann E, Brant LJ, Concin H, Diem G, Rapp K, Ulmer H. Gamma-glutamyltransferase as a risk factor for cardiovascular disease mortality: an epidemiological investigation in a cohort of 163,944 Austrian adults. Circulation. 2005;112:2130–2137. doi: 10.1161/CIRCULATIONAHA.105.552547. [DOI] [PubMed] [Google Scholar]
- 28.Lee DS, Evans JC, Robins SJ, Wilson PW, Albano I, Fox CS, Wang TJ, Benjamin EJ, D'Agostino RB, Vasan RS. Gamma glutamyl transferase and metabolic syndrome, cardiovascular disease, and mortality risk: the Framingham Heart Study. Arterioscler Thromb Vasc Biol. 2007;27:127–33. doi: 10.1161/01.ATV.0000251993.20372.40. [DOI] [PubMed] [Google Scholar]
- 29.Strasak AM, Kelleher CC, Klenk J, Brant LJ, Ruttmann E, Rapp K, Concin H, Diem G, Pfeiffer KP, Ulmer H, Vorarlberg Health Monitoring and Promotion Program Study Group Longitudinal change in serum gamma-glutamyltransferase and cardiovascular disease mortality: a prospective population-based study in 76,113 Austrian adults. Arterioscler Thromb Vasc Biol. 2008;28:1857–65. doi: 10.1161/ATVBAHA.108.170597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hozawa A, Okamura T, Kadowaki T, Murakami Y, Nakamura K, Hayakawa T, Kita Y, Nakamura Y, Okayama A, Ueshima H, NIPPON DATA90 Research Group gamma-Glutamyltransferase predicts cardiovascular death among Japanese women. Atherosclerosis. 2007;194:498–504. doi: 10.1016/j.atherosclerosis.2006.08.058. [DOI] [PubMed] [Google Scholar]
- 31.Younossi ZM, Gramlich T, Matteoni CA, Boparai N, McCullough AJ. Nonalcoholic fatty liver disease in patients with type 2 diabetes. Clin Gastroenterol Hepatol. 2004;2:262–5. doi: 10.1016/s1542-3565(04)00014-x. [DOI] [PubMed] [Google Scholar]
- 32.Tietge UJ, Boker KH, Manns MP, Bahr MJ. Elevated circulating adiponectin levels in liver cirrhosis are associated with reduced liver function and altered hepatic hemodynamics. Am J Physiol Endocrinol Metab. 2004;287:E82–89. doi: 10.1152/ajpendo.00494.2003. [DOI] [PubMed] [Google Scholar]
- 33.Kvilekval K, Lin J, Cheng W, Abumrad N. Fatty acids as determinants of triglyceride and cholesteryl ester synthesis by isolated hepatocytes: kinetics as a function of various fatty acids. J Lipid Res. 1994;35:1786–94. [PubMed] [Google Scholar]
- 34.Palomo I, Alarcon M, Moore-Carrasco R, Argiles JM. Hemostasis alterations in metabolic syndrome. Int J Mol Med. 2006;18:969–974. [PubMed] [Google Scholar]
- 35.Targher G, Arcaro G. Non-alcoholic fatty liver disease and increased risk of cardiovascular disease. Atherosclerosis. 2007;191:235–240. doi: 10.1016/j.atherosclerosis.2006.08.021. [DOI] [PubMed] [Google Scholar]
- 36.Ortega E, Koska J, Salbe AD, Tataranni PA, Bunt JC. Serum gamma-glutamyl transpeptidase is a determinant of insulin resistance independently of adiposity in Pima Indian children. J Clin Endocrinol Metab. 2006;9:1419–1422. doi: 10.1210/jc.2005-1783. [DOI] [PubMed] [Google Scholar]
- 37.Sun QA, Wu Y, Zappacosta F, Jeang KT, Lee BJ, Hatfield DL, Gladyshev VN. Redox regulation of cell signaling by selenocysteine in mammalian thioredoxin reductases. J Biol Chem. 1999;274:24522–24530. doi: 10.1074/jbc.274.35.24522. [DOI] [PubMed] [Google Scholar]
- 38.Li L, Wysk M, Gonzalez FA, Davis RJ. Genomic loci of human mitogen-activated protein kinases. Oncogene. 1994;9:647–649. [PubMed] [Google Scholar]
- 39.Bozza M, Kolakowski LF, Jr, Jenkins NA, Gilbert DJ, Copeland NG, David JR, Gerard C. Structural characterization and chromosomal location of the mouse macrophage migration inhibitory factor gene and pseudogenes. Genomics. 1995;27:412–419. doi: 10.1006/geno.1995.1071. [DOI] [PubMed] [Google Scholar]
- 40.Figlewicz DA, Delattre O, Guellaen G, Krizus A, Thomas G, Zucman J, Rouleau GA. Mapping of human gamma-glutamyl transpeptidase genes on chromosome 22 and other human autosomes. Genomics. 1993;17:299–305. doi: 10.1006/geno.1993.1325. [DOI] [PubMed] [Google Scholar]
- 41.Espinosa R, 3rd, Lemons RS, Perlman RK, Kuo WL, Rosner MR, Le Beau MM. Localization of the gene encoding insulin-degrading enzyme to human chromosome 10, bands q23-q25. Cytogenet Cell Genet. 1991;57:184–186. doi: 10.1159/000133142. [DOI] [PubMed] [Google Scholar]
- 42.Zhang L, Ge L, Parimoo S, Stenn K, Prouty SM. Human stearoyl-CoA desaturase: alternative transcripts generated from a single gene by usage of tandem polyadenylation sites. Biochem. J. 1999;340:255–264. [PMC free article] [PubMed] [Google Scholar]
- 43.Gray IC, Nobile C, Muresu R, Ford S, Spurr NK. A 2.4-megabase physical map spanning the CYP2C gene cluster on chromosome 10q24. Genomics. 1995;28:328–332. doi: 10.1006/geno.1995.1149. [DOI] [PubMed] [Google Scholar]