Abstract
Misregulation of REL, a nuclear factor-κB family transcription factor, has been implicated in several human lymphoid malignancies. REL has a conserved N-terminal DNA-binding/dimerization domain called the Rel homology domain (RHD) and a C-terminal transactivation domain (TAD). Here, we define the sequences (amino acids (aa) 323–422) between the RHD and TAD as an REL inhibitory domain (RID) because deletion of these sequences increases both REL transactivation and DNA binding. Furthermore, we have characterized two REL mRNA splice variants that encode proteins with alterations near RID: one lacking exon 9 sequences (aa 308–330; RELΔ9) and one with an exonized Alu fragment insertion of 32 aa after aa 307 (REL + Alu). Deletion of RID or exon 9-encoded sequences increases transactivation by GAL4–REL by approximately threefold. Moreover, deletion of RID or exon 9 sequences increases transactivation by full-length REL from certain κB site-containing promoters and increases DNA binding by REL. Deletion of RID does not affect REL's ability to transform chicken spleen cells. Reverse transcriptase-polymerase chain reaction analysis of mRNA from both primary lymphoma samples and several transformed tissue culture cell lines indicates that the RELΔ9 splice variant is preferentially expressed in lymphoma, suggesting that the REL transcript lacking exon 9 could serve as a marker for certain types of lymphoid tumors.
Keywords: c-Rel, NF-κB, alternative splicing, lymphoma, transactivation, inhibition
Introduction
Nuclear factor (NF)-κB transcription factors are involved in the control of several key biological processes, including cell growth, immune response and apoptosis (Hayden and Ghosh, 2008). In mammals, there are five NF-κB transcription factors, which can form homo- or heterodimers with other NF-κB members, and their nuclear localization is prevented by association with the inhibitor IκB. Signals that activate NF-κB do so via activation of the IκB kinase, which then phosphorylates IκB that leads to its proteolytic degradation. The liberated NF-κB complex can then enter the nucleus and activate target genes.
The NF-κB family member REL (human c-Rel) is a 587 amino-acid (aa) protein. REL contains an approximately 300 aa conserved N-terminal domain called the Rel homology domain (RHD), which is necessary for dimerization, DNA binding, nuclear localization and IκB interaction (Hayden and Ghosh, 2008). The C-terminal half of REL contains a transactivation domain (TAD) comprised of at least two subdomains, I (aa 424–490; TADI) and II (aa 518–587; TADII), which are required for REL to fully activate gene transcription (Martin et al., 2001; Starczynowski et al., 2003). Nevertheless, deletion of either TADI or -II enhances REL's ability to transform chicken lymphoid cells in vitro while reducing its transactivating ability (Starczynowski et al., 2003). The function of the residues (306–423) between RHD and REL TAD is unclear. However, deletion of aa 309–421 can increase transactivation by a GAL4–REL fusion protein in Jurkat T cells (Martin et al., 2001).
c-Rel is expressed at every stage of B-cell development, but its expression is highest in immature/mature B cells and lowest in pre-B cells (Grumont and Gerondakis, 1994; Liou et al., 1994). c-Rel-knockout mice develop normally, but show defects in B-cell proliferation and survival after treatment with some mitogens (Köntgen et al., 1995). These B-cell defects are due to a block in cell-cycle progression from G1 to S phase and due to increased apoptosis because of decreased expression of c-Rel target genes (Grumont et al., 1999).
c-Rel has also been implicated in a number of animal and human lymphoid cell malignancies. The retroviral homologue v-Rel causes rapidly fatal lymphoid cell tumors in young birds, and overexpression of v-Rel or chicken, mouse or human c-Rel can transform chicken B cells in vitro (Gilmore et al., 2004). Moreover, the human REL gene is amplified in several types of B-cell lymphomas, including Hodgkin's lymphomas, diffuse large B-cell lymphomas (DLBCLs) and follicular lymphomas (Gilmore et al., 2004).
In this report, we have identified a region between the RHD and the TAD of REL that affects transcriptional activation and DNA binding. Furthermore, we show that one alternatively spliced variant of REL (RELΔ9) with an alteration in this region is specifically expressed at higher levels in certain lymphoma cell lines and tumors.
Results
A deletion of 99 residues defines a REL inhibitory domain for transactivation
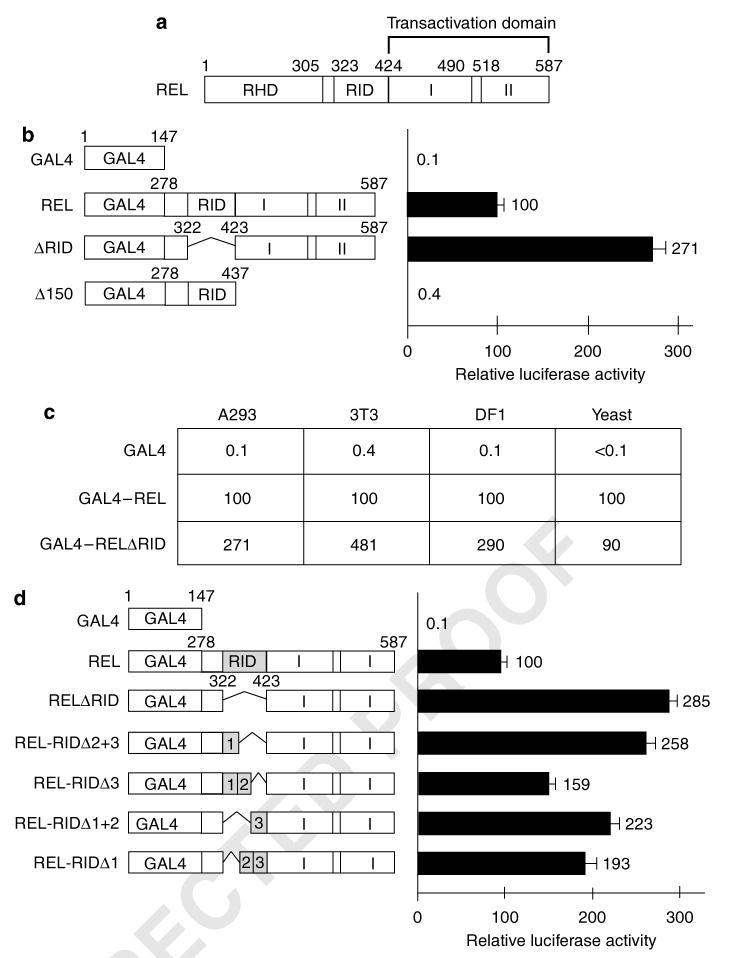
The function of REL sequences between the end of RHD (aa 305) and the start of the two transactivation subdomains (aa 424) has not been defined (Figure 1a). To identify a function for these residues, we first determined the effect of deletion of most of these residues (aa 323–422) on REL's transactivating ability. Expression plasmids were constructed for GAL4–REL fusion proteins containing intact REL C-terminal sequences (from aa 278 to aa 587; GAL4–REL), C-terminal sequences deleted from both TADs (aa 278–437; GAL4–RELΔ150) or C-terminal sequences lacking the residues (aa 323–422) between RHD and TADI (GAL4–RELΔ323–422). Transactivation by these fusion proteins was measured in A293 cells using a GAL4 site luciferase reporter plasmid (Figure 1b). GAL4–REL activated transcription ∼1000-fold more potently than GAL4 alone, whereas GAL4Δ150 had essentially no transactivating ability. The GAL4–REL fusion protein lacking residues 323–422 (GAL4–RELΔ323–422) activated transcription approximately threefold higher than wild-type GAL4–REL. Similar results were obtained in mouse 3T3 cells and chicken DF1 fibroblasts (Figure 1c). In contrast, in yeast cells, GAL4–REL and GAL4–RELΔ323–422 activated transcription to similar extents (Figure 1c). These results demonstrate that residues 323–422 lack inherent transactivating ability, and that their presence can inhibit the transactivating ability of the REL C-terminal TAD (aa 423–587) in mammalian and avian cells, but not in yeast cells. Thus, REL residues 323–422 constitute a REL inhibitory domain (RID) for transactivation in vertebrate cells.
Figure 1.
Deletion of REL residues 323–422 enhances transactivation by REL in vertebrate cells. (a) The general structure of REL is shown at the top (RHD, Rel homology domain; RID, REL inhibitory domain; I and II, transactivation subdomains I and II). (b and d) GAL4 fusion proteins containing the indicated REL sequences were analysed for their abilities to activate transcription from a GAL4 site-containing luciferase reporter in A293 cells. Values are relative to those seen with GAL4–REL (100) and are the averages of three experiments performed in triplicate. Error bars indicate s.e. (c) The indicated GAL4 fusion proteins were analysed for their abilities to activate transcription from a GAL4 site-containing reporter in human (A293), mouse (3T3), chicken (DF1) and S. cerevisiae (yeast) cells. Values are relative to GAL4–REL (100). Experiments were performed three times in triplicate in A293 cells and S. cerevisiae and two times in triplicate in mouse 3T3 cells and chicken DF1 cells.
In an effort to delineate a smaller domain within RID that could mediate inhibition of REL transactivation, 33- or 66 aa deletions within RID were created. These REL mutants were then tested for their ability to activate transcription as GAL4 fusion proteins in A293 cells (Figure 1d). As above, deletion of the entire RID (aa 323–422) increased transactivation by GAL4–REL by approximately threefold. Deletion of discreet subregions within RID resulted in increased transactivation by GAL4–REL, with increases ranging from ∼1.6- to 2.6-fold. There was no obvious pattern as to how the deletions affected REL transactivation, suggesting that any disruption in this region of REL increases transactivation by GAL4–REL.
Replacement of RID with β-galactosidase sequences decreases transactivation by GAL4–REL
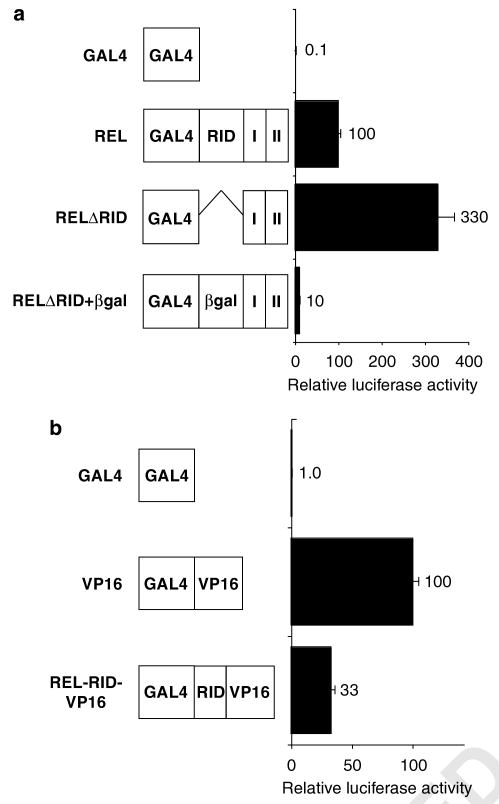
We next investigated whether the inhibitory effect of RID on REL transactivation could be mimicked by other sequences. That is, we replaced the 99 aa that constitute the RID domain with 99 aa from the bacterial protein β-galactosidase (βgal), and measured GAL4-mediated transactivation in A293 cells (Figure 2a). GAL4–REL (containing the RID sequences) transactivated at approximately 30% the level of GAL4–RELΔRID. GAL4–RELΔRID+βgal transactivated at a level approximately 3% that of GAL4–RELΔRID. Therefore, substitution of βgal sequences for RID can effect an even larger decrease in GAL4–REL transactivation than seen with RID itself.
Figure 2.
Replacement of RID sequences with non-REL sequences does not enhance GAL4–REL transactivation. (a and b) GAL4 fusion proteins containing the indicated sequences were analysed for their abilities to activate transcription from a GAL4 site reporter plasmid in A293 cells (RHD, Rel homology domain; RID, REL inhibitory domain; I and II, transactivation subdomains I and II). Values are relative to GAL4–REL (100) and are the averages of three experiments performed in triplicate. Error bars indicate s.e. βgal, β-galactosidase aa 10–106 plus two linker aa; VP16, transactivation domain II from the HSV-1 VP16 protein.
We also wanted to determine whether RID could inhibit the activity of a heterologous TAD. Therefore, we inserted RID into a GAL4–VP16 fusion protein containing a portion of the Herpes simplex virus-1 VP16 TAD (GAL4-RID-VP16). The transactivation abilities of GAL4–VP16 and GAL4-RID-VP16 were then compared in reporter assays in A293 cells. Insertion of RID reduced GAL4–VP16-directed transactivation by approximately threefold (Figure 2b). Therefore, the presence of RID reduces transactivation by GAL4–REL and GAL4–VP16 to approximately the same extent.
RELΔRID differs from wild-type REL in its ability to transactivate κB site-containing promoters
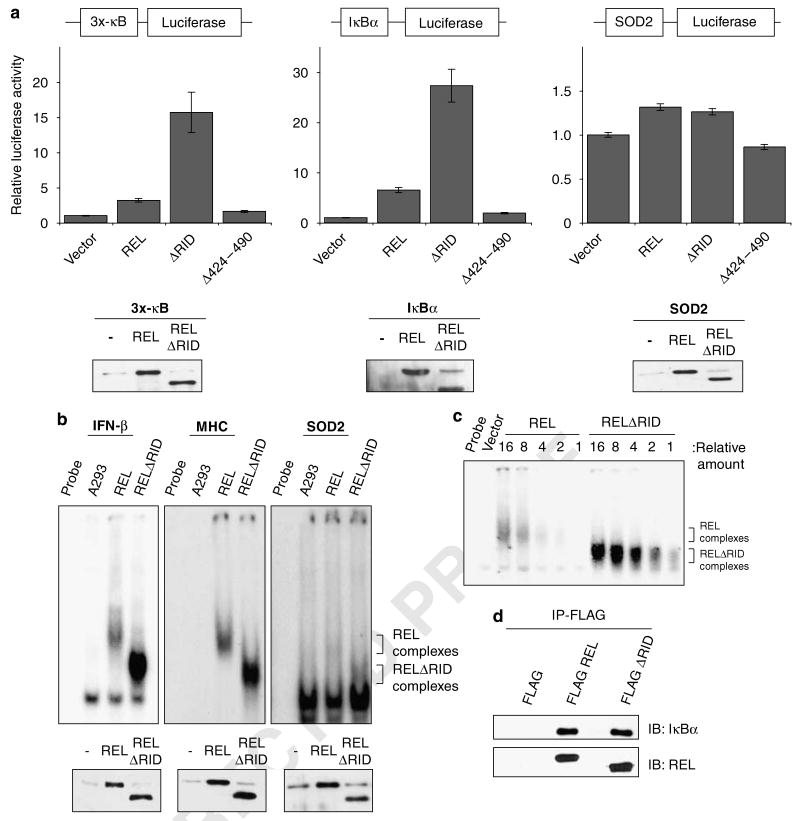
To determine whether RID also influences full-length REL's ability to activate transcription from κB site-containing promoters, we co-transfected A293 cells with expression plasmids for wild-type REL, RELΔ424–490 (deletion of TADI) or RELΔRID and different κB site luciferase reporter gene plasmids. REL's transactivating ability was measured from three κB site luciferase reporter plasmids: a reporter plasmid (3x-κB) containing a minimal c-fos promoter downstream of three κB sites from the MHC-I promoter (Mitchell and Sugden, 1995), a reporter with five κB sites from the chicken ikba promoter (Schatzle et al., 1995); and a κB site-containing reporter that includes 3.3kb of the human manganese superoxide dismutase (SOD2) promoter (two κB sites) and 0.4 kb of the SOD2 intronic enhancer that includes one κB site (Abid et al., 2004). RELΔRID activated transcription ∼fourfold more effectively than wild-type REL from the 3x-κB and IκBα promoter reporter plasmids (Figure 3a). In contrast, REL and RELΔRID both activated transcription to approximately the same extent from the SOD2 promoter (Figure 3a). As a control, we measured the ability of RELΔ424–490 to activate these reporters, as this mutant displays promoter-specific transactivation differences compared with wild-type REL (Starczynowski et al., 2005). Consistent with previous results, RELΔ424–490 activated at levels approximately equal to REL from the 3x-κB reporter and less than REL from both the IκBα and SOD2 reporters (Starczynowski et al., 2005). Anti-REL western blotting showed that REL and RELΔRID were expressed at approximately equal levels in all reporter assays (Figure 3a). Thus, RELΔRID has an enhanced ability to activate transcription from two of three κB site-containing promoters examined, suggesting that the presence of RID also reduces transactivation by full-length REL at some promoters.
Figure 3.
Deletion of REL inhibitory domain (RID) enhances κB site transcription activation and DNA binding by REL. (a) Reporter gene assays with the indicated κB site-containing luciferase cassettes were performed. Co-transfections with the indicated expression and reporter plasmids were performed in A293 cells. Luciferase activities are relative to the value obtained with the pcDNA 3.1 vector alone (1.0). Values are the averages of three independent experiments performed in triplicate. Error bars indicate s.e. In the bottom panels, extracts from triplicate samples used in the reporter assays were analysed by anti-REL western blotting. (b) The ability of REL and RELDRID to bind the indicated κB site-containing probes was measured in electrophoretic mobility shift assays (EMSAs). A293 cells were transfected with expression plasmids for vector alone, REL or RELΔRID. Whole-cell extracts were incubated with the indicated κB site probe. REL and RELΔRID complexes are indicated. Protein levels of REL and RELΔRID were analysed by anti-REL western blotting (bottom panel). (c) An EMSA was used to compare the abilities of REL and RELΔRID to bind a κB site-containing probe. A293 cell extracts containing REL or RELΔRID were titrated as indicated at the top and incubated with a radiolabeled IFN-β κB site probe. REL and RELΔRID complexes are indicated. (d) A293 cells were transiently transfected with the indicated FLAG–REL fusion proteins. Extracts were immunoprecipitated using an anti-FLAG antibody conjugated to agarose beads. The beads were then boiled in SDS sample buffer, proteins were separated by SDS-polyacrylamide gel electrophoresis and samples were subjected to western blotting with anti-REL or anti-IκBα antiserum.
Deletion of RID enhances REL's ability to bind a κB site probe
We next compared the abilities of REL and RELΔRID to bind DNA. Vectors expressing either wild-type REL or RELΔRID were transiently transfected into A293 cells. Whole-cell extracts were first subjected to anti-REL western blotting to ensure equal transfection efficiencies. Extracts were then analysed by electrophoretic mobility shift assays (EMSAs) using three different κB site probes (from the IFN-β and MHC promoters and intron 2 of SOD2). When approximately equal amounts of transfected REL and RELΔRID protein were analysed in an EMSA, RELΔRID bound to all three κB site probes more avidly than REL (Figure 3b). Of note, binding of REL and RELΔRID to the SOD2 κB probe was very weak, perhaps explaining why REL and RELΔRID only poorly activate the SOD2 reporter (Figure 3a). A comparison of DNA-binding activity using twofold serial dilutions of the cell extracts indicated that RELΔRID binds the IFN-β κB site probe with about fourfold greater affinity than wild-type REL (Figure 3c).
Deletion of RID does not affect binding of REL to IκBα
To determine whether the increased transactivation and DNA binding by RELΔRID are due to decreased inhibition by IκBα, we compared the ability of REL and RELΔRID to bind IκBα. Vectors expressing either FLAG–REL or FLAG-RELΔRID were transiently transfected into A293 cells. Extracts were subjected to immunoprecipitation with anti-FLAG antibody, and the immunoprecipitates were analysed by anti-IκBα western blotting (Figure 3d). The membrane was also stripped and probed with anti-REL antibody to ensure equal transfection efficiencies. The amounts of IκBα that were immunoprecipitated with REL and RELΔRID were essentially the same, indicating that the increases in transactivation and DNA binding by RELΔRID are not due to decreased binding of IκBα.
Deletion of RID does not affect REL-mediated transformation of chicken spleen cells
Deletion of REL transactivation subdomain I (RELΔ424–490) enhances REL-mediated transformation of chicken spleen cells (Starczynowski et al., 2003). To determine whether the deletion of RID affects REL-mediated transformation, we infected primary chicken spleen cells with spleen necrosis virus vectors for expression of wild-type REL, RELΔ424–490 and RELΔRID, and measured their ability to transform these cells using a liquid outgrowth assay (Figures 4a and b). Consistent with previous results, RELΔ424–490 showed an enhanced ability to transform chicken spleen cells as compared with wild-type REL. In contrast, deletion of RID sequences did not statistically affect REL-mediated transformation. Anti-REL western blotting of transformed spleen cell extracts confirmed that all REL proteins were expressed at approximately equal levels (Figure 4c).
Figure 4.
Deletion of REL inhibitory domain (RID) sequences does not affect REL-mediated transformation of chicken spleen cells. (a) Retroviral expression vectors for wild-type REL, RELΔ424–490 and RELΔRID were electroporated into primary chicken spleen cells, and transformation was determined by the ability to induce sustained growth in liquid media. Values are the average number of days for the cultures to become overgrown with transformed cells; values in parenthesis are the number of cultures that became transformed over the total number of plates in the given assay. Shown are the results of four independent experiments. (b) The relative days to transform were calculated by dividing the number of days it took for a liquid culture to become transformed (within each of the four spleen cell assays) by the average number of days for that particular assay's wild-type REL cultures to become transformed. These values were then averaged and are relative to REL (100); REL, n = 21; RELΔRID, n = 24; RELΔ424–490, n = 12. Error bars indicate s.e. Plates that did not become transformed were not included in these calculations. (c) Anti-REL western blot of extracts from chicken spleen cells transformed by the indicated proteins.
Identification of alternatively spliced REL mRNAs
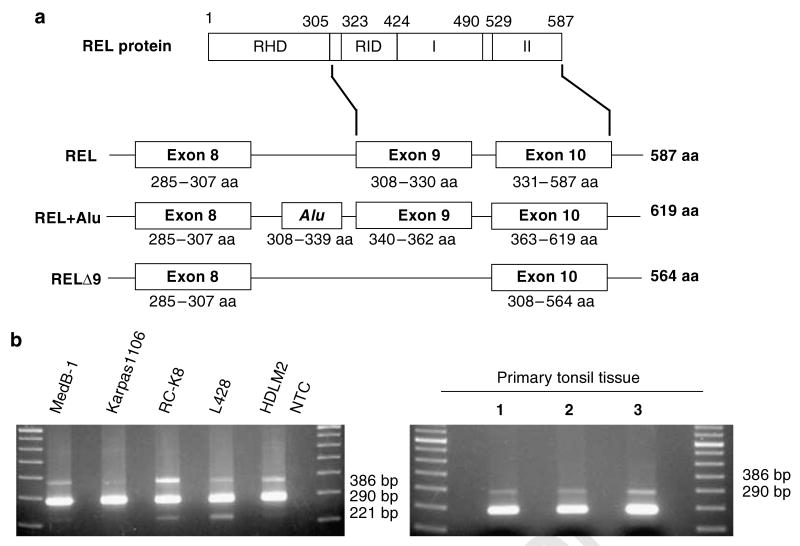
Human c-rel was first cloned as two REL-containing cDNAs from a cDNA library derived from the Daudi human lymphoma cell line (Brownell et al., 1989). One cDNA encoded ‘wild-type’ REL (587 aa), whereas the second cDNA contained a 96 base (32 aa) Alu insertion, such that it encoded a 619 aa protein. The Alu sequences were inserted between REL exons 8 and 9 (to insert 32 aa between REL aa 307 and 308) (Figure 5a).
Figure 5.
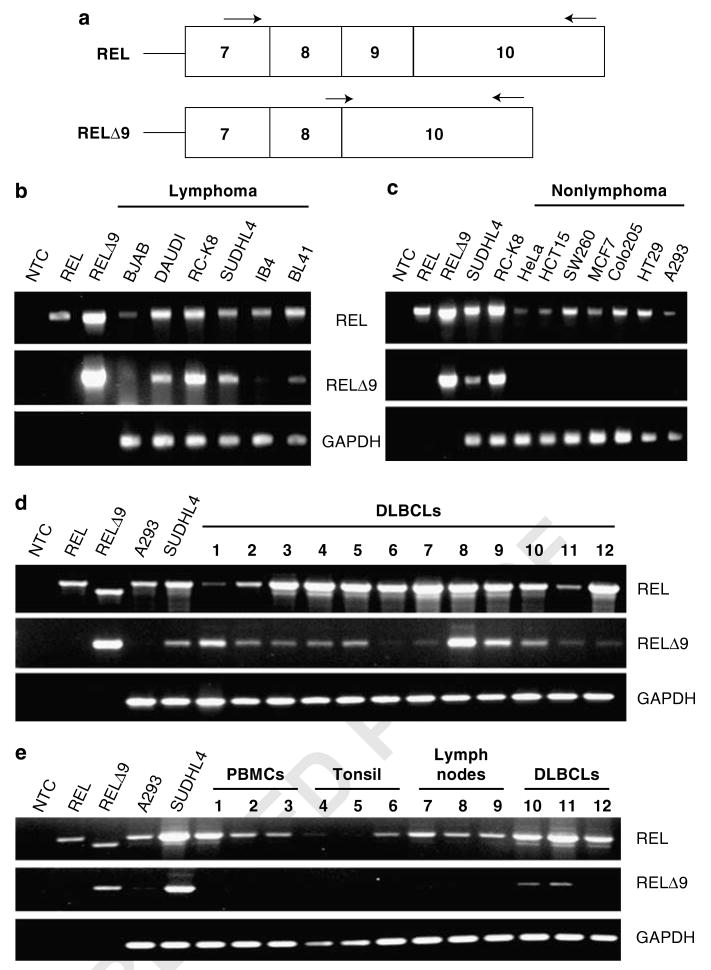
Alternatively spliced forms of REL in human lymphoma cell lines and in primary lymphocytes. (a) Schematic of the wild-type REL protein at the top with diagrams of the exons used to generate the three REL mRNAs found in human lymphoma cell lines. Exon architecture and amino acids encoded by each isoform are indicated (RHD, Rel homology domain; RID, REL inhibitory domain; I and II, transactivation subdomains I and II). (b) PCR amplification was performed on cDNA isolated from the indicated lymphoma cell lines and from three samples of non-neoplastic tonsil tissues. The sizes of the PCR products are indicated.
Because of the proximity of the Alu insertion to RID sequences, we designed primers that bracketed RID and used reverse transcriptase-polymerase chain reaction (RT–PCR) to determine whether there were other REL cDNAs containing insertions or deletions in this region. cDNA obtained from several human lymphoma cell lines and primary lymphocytes were analysed by RT–PCR. As expected, in all cell types, we saw a prominent 290 bp fragment corresponding to wild-type REL and a less abundant 386 bp fragment corresponding to the REL transcript containing the 96 bp Alu insertion. A third minor RT–PCR product of 221 bp was also observed, which did not correspond to any known REL transcript. The 221 bp RT–PCR product was found in some lymphoma cell line cDNAs (MedB-1, RC-K8, L428), but not in cDNA isolated from primary lymphocytes (Figure 5b). DNA sequence analysis showed that the 221 bp cDNA lacked REL exon 9 and was predicted to encode a 564 aa protein missing aa 308–330 (Figure 5a).
REL splice variants differ in their transactivation ability
Because the two REL splice variants (RELΔ9 and REL + Alu) encode REL proteins with alterations in the region near RID, we sought to determine whether the REL proteins encoded by these splice variants also had changes in transactivation and DNA binding.
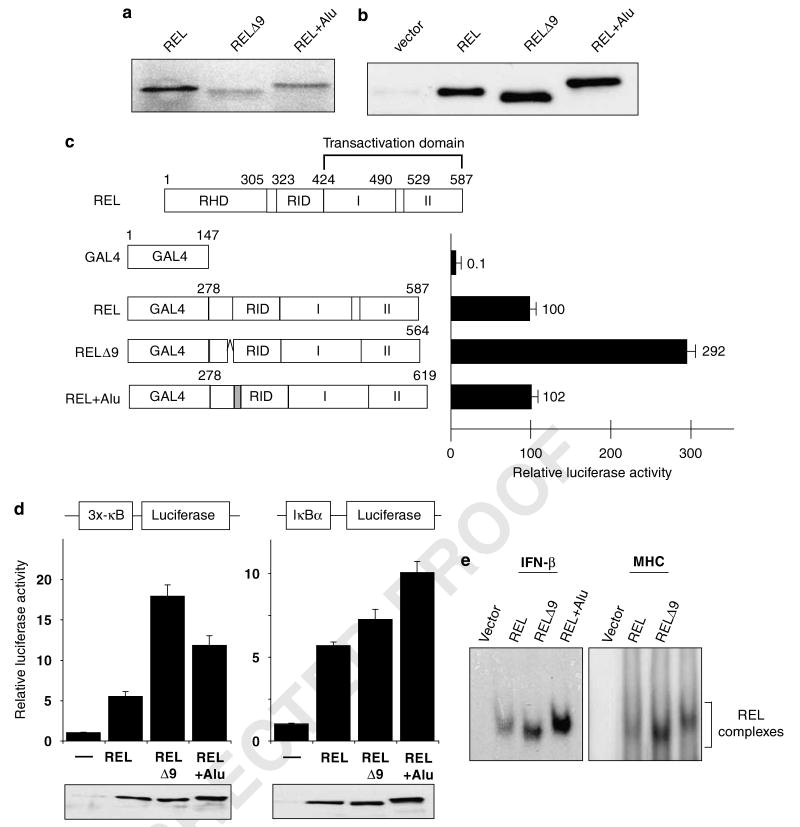
To ensure that the cloned REL splice variants could be translated into the appropriately sized proteins, we isolated full-length cDNAs for all three REL mRNAs (wild-type, Δ9, +Alu) from Karpas1106 cells and analysed each cDNA by both in vitro transcription/translation and in vivo expression after transfection into A293 cells. In both cases, each cDNA encoded a protein of the expected size (Figures 6a and b). The RELΔ9 splice variant lacking 23 aa migrated to a distance corresponding to a MW approximately 2–3 kDa lesser than REL on a 7.5% SDS-polyacrylamide gel, whereas REL + Alu, with an extra 32 aa, migrated to a distance corresponding to a MW approximately 3–4 kDa greater.
Figure 6.
REL isoforms differentially affect REL-mediated transcriptional activation and κB site DNA binding. (a) pcDNA3.1 plasmids containing the indicated REL sequences were subjected to in vitro translation in the presence of [35S]methionine. Proteins were separated by SDS-polyacrylamide gel and detected by phosphorimaging. (b) A293 cells were transiently transfected with pcDNA 3.1 expression vectors containing the indicated REL cDNAs. The cell extracts were subjected to anti-REL western blotting. (c) Shown at top is a schematic of full-length REL. GAL4 fusion proteins containing the indicated REL C-terminal sequences were analysed for their ability to activate transcription from a GAL4 site-containing luciferase reporter locus in A293 cells, as described for Figure 1. Values are relative to that seen with GAL4–REL (100). (d) Reporter gene assays with the indicated κB site-containing luciferase cassettes were performed in A293 cells as described for Figure 3a. Luciferase activities are relative to pcDNA (1.0), and values are the averages of three independent experiments performed in triplicate. In the bottom panels, extracts from triplicate samples used in the reporter assays were analysed by anti-REL western blotting. (e) An electrophoretic mobility shift assay was performed with whole-cell extracts from A293 cells transiently transfected with expression vectors containing each of the three REL splice variants. The κB site-containing probe used is shown at the top. The REL complexes are indicated. RHD, Rel homology domain; RID, REL inhibitory domain; I and II, transactivation subdomains I and II.
To determine whether there was an effect of addition of Alu sequences or removal of exon 9 sequences on intrinsic REL transactivation, we first created expression plasmids for GAL–REL, GAL4–RELΔ9 and GAL4–REL + Alu. All GAL4–REL fusions activated transcription well above control (GAL4 alone) levels in A293 cells. Similar to what is seen with deletion of residues 323–422 (that is, RID), GAL4–RELΔ9 activated transcription approximately threefold higher than wild-type REL (Figure 6c). Addition of Alu sequences did not significantly affect transactivation by GAL–REL.
We next measured the ability of the RELΔ9 and REL + Alu to activate transcription as full-length REL proteins from different κB site-containing reporter plasmids in A293 cells. RELΔ9 activated transcription from the 3x-κB reporter at a level approximately three times higher than wild-type REL, whereas REL + Alu showed a twofold increase in transactivation as compared with wild-type REL (Figure 6d). With the IκBα promoter, we detected only a slight increase in transactivation by RELΔ9 and an approximately twofold increase with REL + Alu, as compared with wild-type REL (Figure 6d). Anti-REL western blotting of A293 cell extracts from the reporter assays confirmed that all REL splice variants were expressed at approximately equal levels (Figure 6d).
It is not clear why RELΔ9 increases transactivation from the two reporter plasmids to different extents, but it could be due to the different contexts of the κB sites within the 3x-κB and IκBα promoters. Similarly, for unknown reasons, the REL + Alu protein differs in its ability to transactivate as compared with wild-type REL in the context of a GAL4 fusion protein (Figure 6c) versus as a full-length protein (Figure 6d).
RELΔ9 and REL + Alu have enhanced DNA binding
Given that the alternatively spliced forms of REL, RELΔ9 and REL + Alu, can have increased transactivation potential as full-length REL proteins, we examined whether these proteins also had increased DNA-binding abilities as compared with wild-type REL. A293 cells were transfected with expression plasmids for full-length REL, RELΔ9, REL + Alu or with vector alone, and extracts were then subjected to EMSA analysis using the IFN-β and MHC κB site probes. With both κB site probes, the REL isoforms RELΔ9 and REL + Alu bound the probe more efficiently than wild-type REL (Figure 6e).
The RELΔ9 splice variant shows increased expression in both primary lymphomas and lymphoma cell lines
Because our preliminary experiments showed that RELΔ9 mRNA was expressed at higher levels in some lymphoma cell lines as compared with primary lymphocytes (Figure 5b), we further investigated the expression levels of RELΔ9 using cDNA from a panel of human lymphoma cell lines and nonlymphoma tumor cell lines (Figures 7b and c). Detection of wild-type REL mRNA was done using a forward PCR primer complementary to REL exon 7 sequences and a reverse primer complementary to sequences within REL exon 10 (Figure 7a). This primer pair amplified products of 919- and 850 bp corresponding to REL and RELΔ9, respectively, from plasmid cDNA. To quantify wild-type REL cDNA, we used a PCR program with low cycle number, a condition in which the less abundant RELΔ9 transcripts were not detected. To examine the expression levels of the RELΔ9 splice variant, we designed a specific forward primer containing 3′-sequences complementary to sequences only present in mRNA coding for the RELΔ9 isoform. That is, the primer contained sequences from both the end of REL exon 8 and the beginning of REL exon 10, and thus could only anneal to sequences present in RELΔ9 cDNA (Figure 7a). By pairing this RELΔ9-specific forward primer with the same exon 10 reverse primer used to amplify wild-type REL cDNA, we specifically amplified a 702bp product corresponding to RELΔ9 cDNA (Figures 7b and c). RELΔ9 mRNA was expressed at detectable levels in most lymphoma-derived cell lines (excluding BJAB) (Figure 7b), but was not detected in a panel of nonlymphoma human tumor cell lines, even though wild-type REL mRNA was easily detected in the nonlymphoma tumor cell lines (Figure 7c).
Figure 7.
The RELΔ9 isoform shows increased expression in lymphoma cell lines. (a) General structure of the relevant section of the REL cDNAs. Arrows indicate approximate positions of RT–PCR primers used. (b) RT–PCR detection of mRNAs for the REL and RELΔ9 transcripts in a panel of lymphoma cell lines. (c) RT–PCR detection of the REL and RELΔ9 transcripts in a panel of nonlymphoma human cell lines: HeLa, cervical cancer; HCT15, colon carcinoma; SW260, colon carcinoma; MCF7, breast cancer; Colo205, colorectal cancer; HT29, colon adenocarcinoma; and A293, adenovirus-transformed embryonic kidney. (d) RT–PCR detection of the REL and RELΔ9 transcripts in a panel of primary human diffuse large B-cell lymphoma (DLBCL) samples. (e) Comparison of RT–PCR of the REL and RELΔ9 transcripts from DLBCLs (samples 8, 9 and 12, as in d) with primary human, non-neoplastic samples from peripheral mononuclear blood cells (PMBCs), tonsil and lymph nodes. The REL-specific forward primer is in exon 7, the RELΔ9-specific forward primer spans the exon 8/exon 10 boundary and the reverse primer for amplification of both REL and RELΔ9 anneals within exon 10. PCRs of SUDHL4 and RC-K8 cDNAs are used as positive controls for amplification of the RELΔ9 splice variant cDNA. NTC, no template control. PCRs of pcDNA–REL and pcDNA–RELΔRID plasmids were used as controls for product size and primer specificity. GAPDH is included as a control for cDNA quantity.
To determine whether the RELΔ9 splice variant was also expressed in primary tumor samples, RT–PCR analysis was performed on 12 DLBCL samples (Figure 7d). Expression of REL and RELΔ9 was detected in all DLBCLs and at levels comparable to the levels seen in the SUDHL4 lymphoma cell line. Of note, the levels of RELΔ9 mRNA do not always correlate with the levels of wild-type REL mRNA in these DLBCL samples.
Next, we compared the expression of the RELΔ9 splice variant in primary DLBCL samples to primary lymphoid tissues. RT–PCR was performed on two high (no. 8 and no. 9) and one low (no. 12) RELΔ9-expressing DLBCL samples and on nine non-neoplastic samples (peripheral mononuclear blood cells, tonsil and lymph nodes) (Figure 7e). Expression of RELΔ9 was detected in two of these three primary DLBCL samples, but in none of the non-neoplastic samples.
Discussion
In this report, we define REL aa 323–422 as an RID that can reduce both REL-mediated transactivation and DNA binding. We have also characterized two REL splice variants that have alterations near RID, and these splice variant-encoded REL proteins show increased transactivation and DNA binding. One REL splice variant, RELΔ9 (lacking exon 9 sequences), is specifically expressed at higher levels in both lymphoma cell lines and primary human DLBCL samples as compared with normal lymphoid tissues and nonlymphoid tumor cell lines.
Alternative splicing can alter various protein properties, including sites of post-translational modification, stability, DNA- and protein-binding properties, sub-cellular localization and enzymatic activities (Stamm et al., 2005). For example, several alternatively spliced transcripts of the c-Myb transcription factor show different transcriptional activities (O'Rourke and Ness, 2008). Alternative splicing of several genes has been implicated in a number of human diseases and cancers (Stoilov et al., 2002). Alternatively spliced variants of tumor suppressor p53 show differential expression between normal and human breast cancer tissue, causing altered expression of p53 target genes (Bourdon et al., 2005). Two splice variants of HDM2, an inhibitor of p53, were found to be expressed only in Hodgkin's lymphoma cell lines and not in non-neoplastic primary lymphocytes (Sánchez-Aguilera et al., 2006). In addition, two isoforms of transcription factor FoxP1 are expressed in a subset of DLBCLs (Brown et al., 2007). Moreover, all mammalian NF-κB transcription factor family members undergo alternative splicing (reviewed in Leeman and Gilmore, 2008).
A plausible model to account for RID inhibition of transactivation and DNA binding by REL is that RID sequences act as an intramolecular hinge, facilitating a direct or indirect interaction between the RHD and the C-terminal TADs. Upon interaction of these domains, REL loses affinity for DNA resulting in decreased transactivation by REL. A similar model is used to explain regulation of transactivation and DNA binding by the NF-κB family member RelA (that is, p65). In the transcriptionally inactive cytoplasmic state, HDAC1 mediates an interaction between the RelA RHD and C-terminal TAD (Zhong et al., 2002). Upon phosphorylation of RelA by PKAc, HDAC1 is released, freeing the C-terminal TAD to interact with the co-activator CBP/p300 and activate transcription (Zhong et al., 1998). A similar model could explain why deletion or alterations of sequences between the RHD and TAD of REL results in increased DNA binding and transactivation. If the intervening RID sequences serve as a hinge to facilitate an interaction between the REL RHD and C-terminal TADs, then alterations of the hinge region in REL (as in RELΔRID or RELΔ9) could make an inhibitory intramolecular interaction less favorable. Consistent with this model, p300 has been shown to enhance REL transactivation in co-transfection experiments (Yu et al., 2004). In addition, deletion of RID sequences results in an approximately threefold increase in transactivation as a GAL4 fusion in a number of vertebrate cell lines, and yet has no effect on GAL4–REL transactivation in yeast cells, suggesting that yeast cells lack a factor that is required for mediating the effects of RID in vertebrate cells.
The enhanced reduction in REL transactivation after replacement of RID with βgal sequences suggests that RID sequences provide a specific structural scaffold that allows the REL TAD to activate transcription within an appropriate range. Perhaps the substituted βgal sequences provide a more favorable environment for an RHD and C-terminal TAD interaction. The crystal structure of REL has only been solved for the RHD, therefore making a direct structural comparison of the RID and βgal sequences impossible (Huang et al., 2001). Web-based bioinformatic prediction of RID and βgal sequences indicates that βgal sequences contain less secondary structural motifs than RID, suggesting that βgal sequences would be more flexible and thus more capable of permitting an intramolecular REL interaction.
Mutations within the REL C-terminal TAD that alter transactivation potential have been shown to enhance REL-mediated transformation (Starczynowski et al., 2003, 2005, 2007). Our data show that deletion of RID sequences does not enhance REL-mediated transformation. Given the close proximity and similar effects of RID and exon 9 deletions on REL activity, we believe it is likely that RELΔ9 would not exhibit enhanced transforming activity. Because previous data have shown that REL C-terminal deletions that decrease transactivation result in increased transforming ability (Starczynowski et al., 2003), it makes sense that RID or exon 9 deletions, which increase transactivation, would not enhance REL-mediated transforming activity. As such, we believe that it is unlikely that the RELΔ9 isoform encodes a lymphoma cell-specific oncogenic variant of REL. Nevertheless, it is possible that REL–RELΔ9 heterodimers alter transactivation from certain κB site-containing promoters and in this way contribute to the oncogenic state in human lymphoma cells.
Gene amplification is the most common alteration in REL that is found in human lymphomas. Increased REL copy number can also lead to increased REL mRNA expression, and, in some cases, may thereby also lead to increased RELΔ9 mRNA. As such, one can speculate that increased expression of the RELΔ9 protein isoform, with its altered DNA binding and transactivation properties, may enhance the expression of a subset of NF-κB target genes in certain lymphomas. Furthermore, the lymphoma cell-specific expression of the RELΔ9 isoform may serve as a marker for certain types of human lymphoma.
Materials and methods
Cells and spleen cell transformation assays
For growth and sources of cell lines, see Supplementary Material. Chicken spleen cell transformation was measured in a liquid outgrowth assay as previously described (Starczynowski et al., 2003; Gilmore et al., 2004).
Plasmids
Recombinant DNA manipulations were carried out using standard procedures (Sambrook et al., 1989). GAL4 expression vectors were constructed by subcloning REL C-terminal sequences starting at aa 278 into pSG424 (Starczynowski et al., 2003). Yeast GAL4-REL expression vectors were constructed in pGBT9 (Epinat et al., 2000). The full-length REL alternatively spliced cDNAs were obtained from Karpas1106 cells and were subcloned into the pcDNA 3.1(−) expression vector (Invitrogen, Carlsbad, CA, USA). For spleen cell transformation assays, REL cDNAs were subcloned into spleen necrosis virus vector JD214BS + (Gilmore et al., 2004). GAL4 and κB site luciferase reporter plasmids were described previously (Starczynowski et al., 2005). A complete list of all primers and subclones used to generate plasmids can be found at http://www.nf-kb.org.
Cell transfection and luciferase reporter assays
Transfections for reporter assays using RID deletion mutants in A293 and mouse 3T3 cells using Superfect and in DF1 cells using DMSO/polybrene were carried out as described previously (Kalaitzidis and Gilmore, 2002; Starczynowski et al., 2007). Transfections using the Superfect reagent were performed according to the manufacturer's recommendations (Qiagen, Valencia, CA, USA). Transfections of A293 cells for EMSAs or reporter assays with REL splice variant isoforms were performed using PEI (polyethylenimine; Polysciences Inc., Warrington, PA, USA). A293 cells were seeded to be 40-60% confluent on the day of transfection. On the day of transfection, cells were incubated with DNA/PEI at a ratio of 3:1 in serum-free media (300 μl for a 60 mm plate) for 15min at room temperature. Following incubation, 4.7ml of DMEM/10% FBS was added to the DNA mixture, and this mixture was added to cells. Twenty-four hours later, the media was replaced with 5ml of fresh DMEM/10% FBS. The following day, the cells were harvested.
After lysing the cells, luciferase activity was measured using the Luciferase Assay System according to the manufacturer's recommendations (Promega, Madison, WI, USA). Luciferase values were normalized to βgal levels in all assays, as described previously (Kalaitzidis et al., 2002).
Yeast reporter gene assays were carried out as previously described (Epinat et al., 2000).
Western blotting, immunoprecipitation and in vitro translation
Western blotting was performed on whole-cell extracts prepared in Promega lysis buffer, SDS-sample buffer or AT buffer, as previously described (Kalaitzidis et al., 2002). The REL antibody used in western blotting was raised against the C-terminal epitope NEQLSDSFPYEFFQV and was a kind gift from Nancy Rice (Kalaitzidis and Gilmore, 2002).
For co-immunoprecipitations, A293 cells were transfected with pcDNA3–FLAG, FLAG–REL or FLAG–RELΔRID, as described above. FLAG fusion proteins were immunoprecipitated with anti-FLAG M2-conjugated agarose beads (Sigma-Aldrich, St Louis, MO, USA). Immunoprecipitated protein extracts were analysed for the presence of REL by western blotting using anti-REL antibody or anti-IκBα antiserum (1:500) (SC-371; Santa Cruz Biotechnology, Santa Cruz, CA, USA).
In vitro transcription/translation-coupled reactions were performed using pcDNA vectors in TNT wheat germ extract according to the manufacturer's recommendations (Promega) in the presence of [35S]methionine. REL isoforms were visualized by phosphorimaging.
Electrophoretic mobility shift assay
Using A293 whole-cell extracts prepared in AT buffer, EMSAs were performed as previously described (Kalaitzidis et al., 2002). DNA-binding reactions contained 40 000 c.p.m. of 32P-end-labeled κB probe from the IFN-β promoter (5′-TCGAGAGGTCGGGAAATTCCCCCCCG-3′), the MHC promoter (5′-TCGAGAGGTTGGGGATTCCCCACCCG-3′) or the second intron of SOD2 (5′-TCGAGAGGTCGGGAATACCCCCCCCG-3′), as well as 10μg of cell extract. Samples were incubated at room temperature for 30min. DNA–protein complexes were resolved on 5% polyacrylamide gels and autoradiography was performed.
Reverse transcriptase-PCR
Total RNA from cells was extracted using TRIzol (Invitrogen). Two microgram of RNA was reverse-transcribed using M-MLV reverse transcriptase (Promega) and cDNA was resuspended in 40 μl of RNase-free water. PCR was then performed with 2μl of cDNA, using the conditions and primers described in Supplemental Material.
Tissue sample preparation
Frozen tissue samples were obtained from the Institute of Pathology (University of Ulm, Ulm, Germany). The material was made anonymous to comply with the German law for usage of archival tissue for clinical research (Deutsches Artzeblatt, 2003) Total RNA was isolated using the RNeasy kit (Qiagen, Hilden, Germany) and treated with RNase-free DNase (Qiagen). Reverse transcription was performed using Superscript II reverse transcriptase (Invitrogen), according to manufacturer's instructions. The PCR conditions used to amplify REL, RELD9 and GAPDH from the primary samples were as described in Supplementary Material.
Supplementary Material
Acknowledgments
We thank Mike Garbati, Melanie Herscovitch, Daniel Starczynowski and Francis Wolenski for helpful discussions, and Michaela Buck, Karola Dorsch, Iwona Nerbase and Ingo Melzner for technical assistance. This work was supported by NIH Grant CA47763 (TDG) and a grant from Deutsche Krebshilfe (108060; TFB).
Footnotes
Supplementary Information accompanies the paper on the Oncogene website (http://www.nature.com/onc)
References
- Abid MR, Schoots IG, Spokes KC, Wu SQ, Mawhinney C, Aird WC. Vascular endothelial growth factor-mediated induction of manganese superoxide dismutase occurs through redox-dependent regulation of forkhead and IκB/NF-κB. J Biol Chem. 2004;279:44030–44038. doi: 10.1074/jbc.M408285200. [DOI] [PubMed] [Google Scholar]
- Bourdon JC, Fernandes K, Murray-Zmijewski F, Liu G, Diot A, Xirodimas DP, et al. p53 isoforms can regulate p53 transcriptional activity. Genes Dev. 2005;19:2122–2137. doi: 10.1101/gad.1339905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown PJ, Ashe SL, Leich E, Burek C, Barrans S, Fenton JA, et al. Potentially oncogenic B-cell activation induced smaller isoforms of FOXP1 are highly expressed in the activated B-cell-like subtype of DLBCL. Blood. 2007;111:2816–2824. doi: 10.1182/blood-2007-09-115113. [DOI] [PubMed] [Google Scholar]
- Brownell E, Mittereder N, Rice NR. A human rel proto-oncogene cDNA containing an Alu fragment as a potential coding exon. Oncogene. 1989;4:935–942. [PubMed] [Google Scholar]
- Deutsches Artzeblatt. 2003;100(23):A 1632. [Google Scholar]
- Epinat JC, Kazandjian D, Harkness DD, Petros S, Dave J, White DW, et al. Mutant envelope residues confer a transactivation function onto N-terminal sequences of the v-Rel oncoprotein. Oncogene. 2000;19:599–607. doi: 10.1038/sj.onc.1203376. [DOI] [PubMed] [Google Scholar]
- Gilmore TD, Kalaitzidis D, Liang MC, Starczynowski DT. The c-Rel transcription factor and B-cell proliferation: a deal with the devil. Oncogene. 2004;23:2275–2286. doi: 10.1038/sj.onc.1207410. [DOI] [PubMed] [Google Scholar]
- Grumont RJ, Gerondakis S. The subunit composition of NF-κB complexes changes during B-cell development. Cell Growth Differ. 1994;5:1321–1331. [PubMed] [Google Scholar]
- Grumont RJ, Rourke IJ, Gerondakis S. Rel-dependent induction of A1 transcription is required to protect B cells from antigen receptor ligation-induced apoptosis. Genes Dev. 1999;13:400–411. doi: 10.1101/gad.13.4.400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayden MS, Ghosh S. Shared principles in NF-κB signaling. Cell. 2008;132:344–362. doi: 10.1016/j.cell.2008.01.020. [DOI] [PubMed] [Google Scholar]
- Huang DB, Chen YQ, Ruetsche M, Phelps CB, Ghosh G. X-ray crystal structure of proto-oncogene product c-Rel bound to the CD28 response element of IL-2. Structure. 2001;9:669–678. doi: 10.1016/s0969-2126(01)00635-9. [DOI] [PubMed] [Google Scholar]
- Kalaitzidis D, Davis RE, Rosenwald A, Staudt LM, Gilmore TD. The human B-cell lymphoma cell line RC-K8 has multiple genetic alterations that dysregulate the Rel/NF-κB signal transduction pathway. Oncogene. 2002;21:8759–8768. doi: 10.1038/sj.onc.1206033. [DOI] [PubMed] [Google Scholar]
- Kalaitzidis D, Gilmore TD. Genomic organization and expression of the rearranged REL proto-oncogene in the human B-cell lymphoma cell line RC-K8. Genes Chromosomes Cancer. 2002;34:129–135. doi: 10.1002/gcc.10051. [DOI] [PubMed] [Google Scholar]
- Köntgen F, Grumont RJ, Strasser A, Metcalf D, Li R, Tarlinton D, et al. Mice lacking the c-rel proto-oncogene exhibit defects in lymphocyte proliferation, humoral immunity, and interleukin-2 expression. Genes Dev. 1995;9:1965–1977. doi: 10.1101/gad.9.16.1965. [DOI] [PubMed] [Google Scholar]
- Leeman JR, Gilmore TD. Alternative splicing in the NF-κB signaling pathway. Gene. 2008 doi: 10.1016/j.gene.2008.07.015. (in press) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liou HC, Sha WC, Scott ML, Baltimore D. Sequential induction of NF-κB/Rel family proteins during B-cell terminal differentiation. Mol Cell Biol. 1994;14:5349–5359. doi: 10.1128/mcb.14.8.5349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin AG, San-Antonio B, Fresno M. Regulation of NF-κB transactivation. Implication of phosphatidylinositol 3-kinase and protein kinase C-ζ in c-Rel activation by tumor necrosis factor α. J Biol Chem. 2001;276:15840–15849. doi: 10.1074/jbc.M011313200. [DOI] [PubMed] [Google Scholar]
- Mitchell T, Sugden B. Stimulation of NF-κB-mediated transcription by mutant derivatives of the latent membrane protein of Epstein–Barr virus. J Virol. 1995;69:2968–2976. doi: 10.1128/jvi.69.5.2968-2976.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sambrook J, Fritsch EF, Maniatis T. Molecular Cloning: A Laboratory Manual. Cold Spring Harbor Press; Cold Spring Harbor, NY: 1989. [Google Scholar]
- Sánchez-Aguilera A, García JF, Sánchez-Beato M, Piris MA. Hodgkin's lymphoma cells express alternatively spliced forms of HDM2 with multiple effects on cell cycle control. Oncogene. 2006;25:2565–2574. doi: 10.1038/sj.onc.1209282. [DOI] [PubMed] [Google Scholar]
- Schatzle JD, Kralova J, Bose HR., Jr Avian IκBα is transcriptionally induced by c-Rel and v-Rel with different kinetics. J Virol. 1995;69:5383–5390. doi: 10.1128/jvi.69.9.5383-5390.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stamm S, Ben-Ari S, Rafalska I, Tang Y, Zhang Z, Toiber D, et al. Function of alternative splicing. Gene. 2005;344:1–20. doi: 10.1016/j.gene.2004.10.022. [DOI] [PubMed] [Google Scholar]
- Starczynowski DT, Reynolds JG, Gilmore TD. Deletion of either C-terminal transactivation subdomain enhances the in vitro transforming activity of human transcription factor REL in chicken spleen cells. Oncogene. 2003;22:6928–6936. doi: 10.1038/sj.onc.1206801. [DOI] [PubMed] [Google Scholar]
- Starczynowski DT, Reynolds JG, Gilmore TD. Mutations of tumor necrosis factor α-responsive serine residues within the C-terminal transactivation domain of human transcription factor REL enhance its in vitro transforming ability. Oncogene. 2005;24:7355–7368. doi: 10.1038/sj.onc.1208902. [DOI] [PubMed] [Google Scholar]
- Starczynowski DT, Trautmann H, Pott C, Harder L, Arnold N, Africa JA, et al. Mutation of an IKK phosphorylation site within the transactivation domain of REL in two patients with B-cell lymphoma enhances REL's in vitro transforming activity. Oncogene. 2007;26:2685–2694. doi: 10.1038/sj.onc.1210089. [DOI] [PubMed] [Google Scholar]
- Stoilov P, Meshorer E, Gencheva M, Glick D, Soreq H, Stamm S. Defects in pre-mRNA processing as causes of and predisposition to diseases. DNA Cell Biol. 2002;21:803–818. doi: 10.1089/104454902320908450. [DOI] [PubMed] [Google Scholar]
- Yu SH, Chiang WC, Shih HM, Wu KJ. Stimulation of c-Rel transcriptional activity by PKA catalytic subunit β. J Mol Med. 2004;82:621–628. doi: 10.1007/s00109-004-0559-7. [DOI] [PubMed] [Google Scholar]
- Zhong H, May MJ, Jimi E, Ghosh S. The phosphorylation status of nuclear NF-κB determines its association with CBP/p300 or HDAC-1. Mol Cell. 2002;9:625–636. doi: 10.1016/s1097-2765(02)00477-x. [DOI] [PubMed] [Google Scholar]
- Zhong H, Voll RE, Ghosh S. Phosphorylation of NF-κB p65 by PKA stimulates transcriptional activity by promoting a novel bivalent interaction with the coactivator CBP/p300. Mol Cell. 1998;1:661–671. doi: 10.1016/s1097-2765(00)80066-0. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.