Abstract
Mix-related homeodomain proteins are involved in endoderm formation in the early vertebrate embryo. We used a yeast two-hybrid screen to identify proteins that interact with Mix.3/mixer to regulate endoderm induction. We demonstrate that cyclin dependent kinase 9 (CDK9) interacts with the carboxyl terminal domain of Mix.3. CDK9 is the catalytic subunit of the PTEF-b transcription elongation complex that phosphorylates the C-terminal domain of RNA polymerase II to promote efficient elongation of nascent transcripts. Using whole embryo transcription reporter and animal pole explant assays, we show that Mix.3 activity is regulated by CDK9/cyclin complexes. Co-expression of cyclin T2 and cyclin K had different effects on Mix.3 transcriptional activity and endoderm induction. Our data suggests that binding of CDK9, and the recruitment of different cyclin partners, can modulate the endoderm inducing activity of Mix.3 during embryonic development.
Keywords: Mix.3, mixer, CDK9, cyclin T2, cyclin K, P-TEFb, endoderm, Xenopus laevis
Introduction
The molecular mechanisms of vertebrate endoderm formation remain largely unknown. Several classes of transcription factors have been implicated with the early events of endoderm specification. These include the GATA-binding factors (such as GATA-4, -5 and -6), the high mobility group (HMG) proteins (such as Sox17), and the Pax-like homeodomain proteins (such as Mix.3/mixer) (Alexander and Stainier, 1999; Shivdasani, 2002). The role of the Mix proteins in endoderm development has been demonstrated in several vertebrate model systems. Mix.3/mixer was first identified in the frog, Xenopus laevis, using expression cloning strategies (Henry and Melton, 1998; Mead et al., 1998). Ectopic expression of Mix.3 alone in animal pole explants results in a cell fate change from ectoderm to endoderm (Doherty et al., 2006; Henry and Melton, 1998; Mead et al., 1998). In zebrafish, the bonnie and clyde mutation results in a failure of definitive endoderm formation. Positional cloning of the bonnie and clyde mutation demonstrated that the affected gene was the zebrafish Mix.3/mixer gene (Kikuchi et al., 2000). A single Mix-like gene has been identified in both mouse and human (Pearce and Evans, 1999; Robb et al., 2000). Targeted deletion of the murine Mix-like gene results embryonic lethality at day 8.5 and null animals die without forming a heart tube or a gut tube. Mixl1−/− embryos have complex defects in axial mesendodermal structures and display increased expression of Brachyury and Nodal (Hart et al., 2002). Expression of the mouse Mix-like protein (mMix/Mml) directly induces endoderm gene expression in Xenopus animal pole explants (Mohn et al., 2003). Morpholino knockdown of Mix.3/mixer protein in the Xenopus embryo results in gastrulation and tailbud abnormalities and down-regulation of general endoderm genes, including endodermin, GATA-5 and Sox17, and the anterior endoderm gene cerberus (Kofron et al., 2004). In addition to the decreased expression of specific endoderm target genes, Mix.3/mixer-depleted Xenopus embryos have elevated expression of several mesodermal genes including eomesodermin and Fgf8 indicating that Mix.3/mixer functions in the early embryo as both a transcription activator and repressor. Expansion of the expression domain of several mesodermal genes following Mix.3 depletion suggested that, in addition to a role in endoderm formation, Mix.3 may play an important role in controlling the amount of mesoderm formed from vegetal cells in the early embryo.
To better understand the role of Mix.3 in endoderm development, we performed a yeast two-hybrid assay to identify proteins that interact with Mix.3. We have demonstrated that the cdc2-like cyclin-dependent kinase 9 (CDK9) directly interacts with Mix.3 and may be a regulator of Mix.3 function. CDK9 was first identified on the basis of its similarity to cell cycle regulated cyclin-dependent kinases (Garriga and Grana, 2004; Pines, 1994). Unlike other CDKs that have important roles in regulating the cell cycle, CDK9 kinase activity is fairly constant throughout the cell cycle (Grana et al., 1994; Garriga et al., 2003). CDK9 was shown to be a component of the positive transcription elongation factor b (P-TEFb) complex that associates with the carboxyl-terminal repeat domain (CTD) of RNA polymerase II (RNAPII) (Zhu et al., 1997). The P-TEFb complex, comprised of CDK9 and cyclin T (or cyclin K), induces hyper-phosphorylation of RNAPII CTD resulting in productive elongation of nascent transcripts (Napolitano et al., 2002; Price, 2000). The transcription initiation complex only efficiently associates with the non-phosphorylated form of RNAPII (Lania et al., 1999). CDK9 phosphorylates the CTD of RNAPII after the formation of the transcription initiation complex and results in processive transcriptional elongation. Subsequent rounds of transcription initiation are thought to be regulated, in part, by the availability of the hypo-phosphorylated form of the RNAPII enzyme. The BRCT (BRCA1 C-terminus)-containing phosphatase Fcp1 associates with the general transcription factor TFIIF, dephosphorylates the CTD of RNAPII, and restores a pool of the non-phosphorylated form of the polymerase for recruitment to nascent transcription complexes (Archambault et al., 1998; Cho et al., 1999; Kobor et al., 1999; Licciardo et al., 2001). Thus, the phosphorylation, and dephosphorylation, of the RNAPII CTD plays an important role in the regulation of transcriptional activation and RNA elongation (Majello and Napolitano, 2001). While the P-TEFb complex has a positive effect on the elongation of transcription, other complexes negatively regulate this process. Two mutliprotein complexes have been identified that inhibit transcription elongation; DSIF (DRB (5,6-dichloro-1-β-D-ribofuranosylbenzimidazole) Inducing Sensitivity Factor) and NELF (Negative Elongation Factor) (Wada et al., 1998a; Yamaguchi et al., 1999). DSIF and NELF associate with hypo-phosphorylated RNAPII in the transcription initiation complex and block productive elongation of the nascent transcript. Recruitment of the P-TEFb complex leads to phosphorylation of RNAPII CTD, SPT5 (TFIIH, a subunit of DSIF (Wada et al., 1998a)) and RD (a subunit of NELF (Yamaguchi et al., 1999)), and results in dissociation of the elongation inhibitory complexes, and allows processive elongation (Wada et al., 1998b). CDK9 complexes have also been shown to interact with (and phosphorylate) transcription factors such as HIV Tat (Wei et al., 1998; Zhu et al., 1997), MyoD (Simone et al., 2002b), retinoblastoma protein (pRb) (De Luca et al., 1997; Grana et al., 1994; Simone et al., 2002a), aryl hydrocarbon receptor (Tian et al., 2003), and c-Myc (Eberhardy and Farnham, 2001; Eberhardy and Farnham, 2002). Transcription factors can thus recruit the P-TEFb complex to specific gene loci to mediate elongation of the nascent transcript. For example, RelA binds P-TEFb and recruits the elongation-promoting complex to the IL-8 promoter. Stimulation of IL-8, a NF-κB-dependent gene, by TNF-α signaling results in recruitment of RelA, CDK9 and cyclin T1 to NF-κB binding sites in the IL-8 promoter and leads to hyper-phosphorylation of the CTD of RNAPII (Barboric et al., 2001).
Like other cyclin dependent kinases, CDK9 associates with multiple cyclins. CDK9 forms complexes with cyclin T1, T2a, T2b (Peng et al., 1998; Wei et al., 1998) and the unique cyclin K (Fu et al., 1999). Each of these CDK9-associated cyclins shares similarity to cyclin C. Cyclin T2a and T2b are derived from alternate splice forms of a single gene. Each of the CDK9:cyclin complexes has been shown to activate transcription in a variety of assays (reviewed in (Sano and Schneider, 2003)). In contrast to complexes with the T cyclins, the CDK9:cyclin K complex can only activate transcription when associated with RNA but not DNA (Lin et al., 2002). PTEFb activity is required for transcription of a broad range of RNA pol II-dependent genes and this function is conserved over wide evolutionary boundaries. For example, chemical inhibition of CDK9 activity in mammalian cells with flavopiridol resulted in a marked (70%) decrease in RNA pol II transcription, suggesting that transcription elongation mediated by CTD phosphorylation by CDK9 positively regulates a wide range of RNA pol II-dependent genes (Chao and Price, 2001). Likewise, knockdown of CDK9 in C. elegans with siRNAs indicated a broad requirement for PTEFb activity during embryonic gene expression (Shim et al., 2002).
In this study, we have used a yeast two-hybrid assay and identified CDK9 as a binding partner with Mix.3. The interaction of these two proteins was confirmed and the interaction domain in Mix.3 was identified by co-immunoprecipitation experiments. We used an in silico approach to identify Xenopus laevis orthologs of CDK9 cyclin partners, cyclin T2 and K. In transient reporter assays, we show that the transcriptional activity of Mix.3 is modulated differentially by CDK9:cyclin T2 and CDK9:cyclin K complexes. For example, Mix.3 activates a heterologous reporter contain three tandem Mix-binding sites cloned upstream of a minimal TATA-box. Addition of CDK9:cyclin T2 represses the ability of Mix.3 to transactivate this reporter. In contrast, CDK9:cyclin K does not repress the activity of Mix.3 on this reporter. Using mutants of Mix.3 and CDK9, we have demonstrated that the repression of Mix.3 in this transient reporter system is dependent on the Mix.3:CDK9 interaction. Using animal pole explants assays (animal caps), we have demonstrated that CDK9:cyclin T2 can repress the ability of Mix.3 to induce endoderm in animal cap assays. Cyclin K did not repress Mix.3-mediated induction of endoderm markers. Using both early and late markers for endoderm differentiation, we have shown that, as with the repression of the transient Mix.3 reporter, the repression of endoderm induction in animal cap explants is dependent on the CDK9-interaction domain in the carboxyl terminal of Mix.3 and also dependent on CDK9 kinase activity. These data suggest a role for CDK9 complexes in the regulation of Mix.3 endoderm induction in early development.
Results
Yeast two-hybrid screen and in vitro co-immunoprecipitations
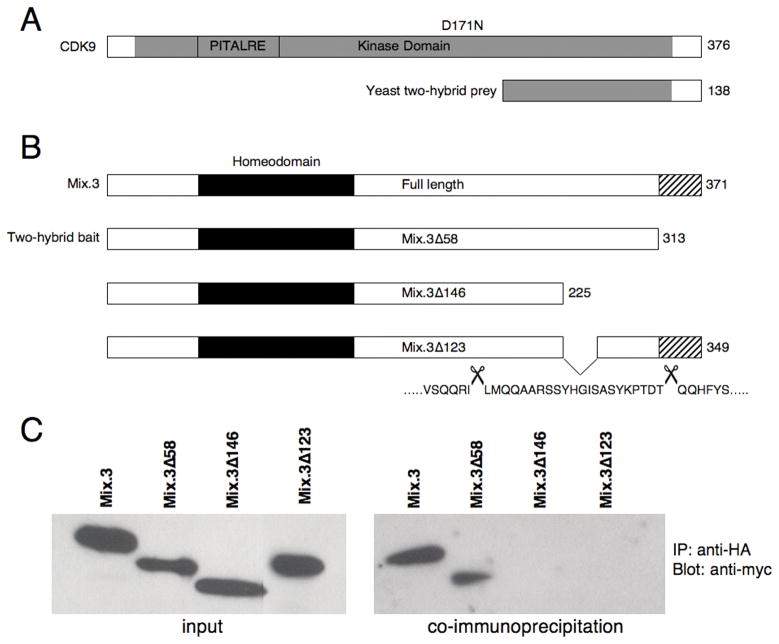
We, and others, have shown that Mix.3 has potent endoderm inducing activity. We reasoned that the activity of this homeodomain-containing transcription factor must be precisely regulated during early development. We used a yeast two-hybrid screen to identify proteins that interact with, and potentially regulate, the activity of Mix.3. We were unable to use the full-length Mix.3 protein as a “bait” in the two-hybrid system due to the presence of a potent activation domain that mapped to the last 58 amino acids at the C-terminus of Mix.3 (data not shown). Indeed, any potential “bait” construct that contained the C-terminal 58 amino acid sequence had strong auto-activation and thus could not be used to screen the yeast two-hybrid library (data not shown). Our earlier studies to identify the domains of Mix.3 that are required for endoderm inducing activity had demonstrated that this C-terminal region, as well as the homeodomain, are required for endoderm induction (Doherty et al., 2006; see Figure 1B). We used the truncated Mix.3 mutant M3Δ58 (Figure 1B), that lacks the C-terminal 58 amino acid sequence, as a bait in our two-hybrid screen. One hundred and twenty-eight positive clones were identified from a screen of 5 × 105 independent clones in the yeast two-hybrid assay. The positive clones were re-plated on four-drop out (4DO) media with X-gal to verify the interactions (data not shown). DNA was isolated from the positive clones, retransformed into competent JM109 E. coli, and plated on ampicillin-containing plates to select for the prey-containing plasmids (pGADT7 vector). Sequence analysis revealed two independent clones sharing sequence identity with the carboxyl terminal of Xenopus laevis CDK9 (amino acids 238–376 of xCDK9; Figure 1A). A full-length cDNA encoding Xenopus CDK9 was obtained from the IMAGE consortium (Open Biosystems, AL) and subcloned into the pGADT7 vector to provide a HA-tag for co-immunoprecipitation studies.
Figure 1.
(A) Schematic representation of CDK9 showing the partial clone recovered from the yeast two-hybrid screen (labeled yeast two-hybrid prey). The grey shading represents the predicted kinase domain (not to scale). The PITALRE sequence, a PSTAIRE-like motif found in CDC2 and related kinases, is boxed. The Mix.3 interaction domain of CDK9 is located in the carboxyl terminal 138 amino acids. The predicted amino acid length of each construct is shown on the right. (B) Schematic diagram of the Mix.3 constructs used in this study (not to scale). The homeodomain is depicted by the black box and the C-terminal 58 amino acid domain is depicted by the stripped box. The C-terminal 58 amino acid domain and the homeodomain are both required for the endoderm inducing activity of Mix.3 (see text for details). The bait construct used for the two-hybrid screen lacked the last 58 amino acids at the carboxyl terminus (Mix.3Δ58). Deletion of 146 amino acids from the carboxyl terminus of Mix.3 ablated the interaction with CDK9 (Mix.3Δ146). The CDK9 interaction domain was further mapped to twenty-two amino acids in the carboxyl terminal domain of Mix.3 (Mix.3Δ123). The numbers on the right of the boxes indicate the predicted amino acid length of each construct. The sequence deleted to generate Mix.3Δ123 is shown beneath the box and is flanked by scissors. (C) In vitro co-immunoprecipitaion assays. Full-length HA-tagged CDK9 and myc-tagged Mix.3 constructs were in vitro transcribed and translated. Immunoprecipitations were performed with an anti-HA antibody and western blots were probed with an anti-myc tag antibody. Full-length Mix.3, and the truncated Mix.3 protein used to screen the yeast two-hybrid library, interact with CDK9. Deletion of 146 amino acids from the carboxyl terminal of Mix.3 ablates the interaction with CDK. The interaction domain further mapped to twenty-two amino acids (227–249; Mix.3Δ123) in the carboxyl terminus of Mix.3.
Co-immunoprecipitation of full-length CDK9 (HA-tagged) and full-length Mix.3 (myc-tagged; pGBKT7 vector) confirmed the interaction of the two proteins (Figure 1). Interaction was also detected by co-immunoprecipitation with CDK9 and the truncated Mix.3 construct (Mix.3Δ58) that was used as the bait in the yeast two-hybrid screen. Deletion analysis of Mix.3 revealed that the CDK9 binding site resides in the carboxyl terminal domain of the Mix.3 protein. Introduction of a stop codon at position 226 in Mix.3 results in truncation of the last 146 amino acids (Figure 1B). This truncated protein did not interact with CDK9 in the co-immunoprecipitation assay (Figure 1C). Further analysis revealed that the CDK9 interaction domain was restricted to a twenty-two amino acid sequence in the carboxyl terminus of Mix.3 (amino acids 227–249; construct Mix.3Δ123, Figure 1 and data not shown).
Isolation and developmental expression of Xenopus CDK9, cyclin T2 and cyclin K
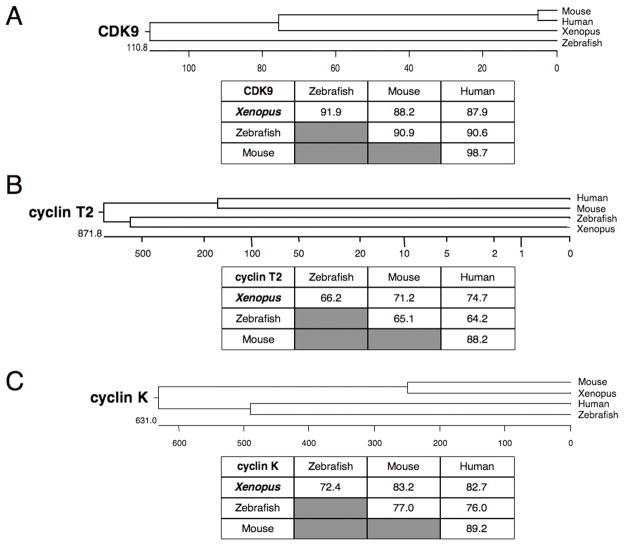
Full-length clones encoding Xenopus laevis CDK9 and its cyclin partners, cyclin T2 and cyclin K, were obtained from the IMAGE consortium full-length sequence collection (Open Biosystems, AL). Xenopus CDK9 is highly homologous to other vertebrate CDK9 proteins and shares nearly 90% amino acid identity with the zebrafish, mouse and human proteins (Figure 2A). Xenopus cyclin K shares more than 70% identity with the vertebrate homologs (Figure 2C). Only a single clone encoding a full-length cyclin T was identified in the Xenopus EST databases. This clone shares more than 60% amino acid sequence identity with vertebrate cyclin T2 proteins (Figure 2B). Partial-length clones for Xenopus laevis cyclin T1 were present in the EST databases indicating that the frog, like other vertebrate species has two cyclin T genes.
Figure 2.
Phylogenetic analysis of vertebrate CDK9, cyclin T2 and cyclin K. Phylogenetic trees and tables of amino acid sequence identity are shown for (A) CDK9, (B) cyclin T2 and (C) cyclin K. Accession numbers used to generate this data are: human CDK9, NP_001252; human cyclin T2, NP_001232; human cyclin K, AAF82290; mouse CDK9, BAC40824; mouse cyclin T2, XP_129395; mouse cyclin K, AAH27297; zebrafish CDK9, AAP47016; zebrafish cyclin T2, AAH44378; zebrafish cyclin K, XP_697908.
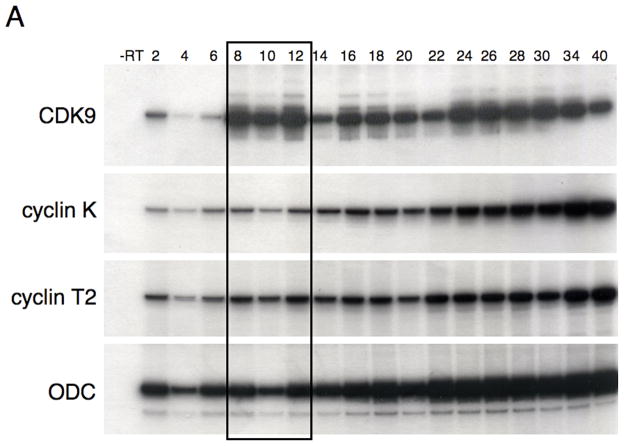
The expression profiles of Xenopus CDK9, cyclin K and cyclin T2 during early embryonic development were examined by reverse-transcription PCR assays. All three genes are present as maternal transcripts and are expressed at all stages of development that were tested (Figure 3A). In contrast, Mix.3 transcripts are detected for a narrow window of development, from the onset of zygotic transcription at the mid-blastula transition to the end of gastrulation (Henry and Melton, 1998; Mead et al., 1998). Thus, the timing of CDK9 expression, and its cyclin partners, coincides with that of Mix.3.
Figure 3.
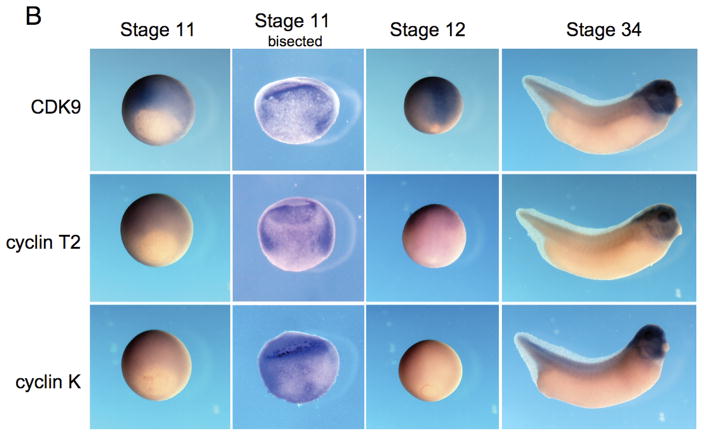
Expression of CDK9, cyclin T2 and cyclin K during Xenopus development. (A) Reverse transcription PCR analysis of the developmental expression of CDK9 and its cyclin partners T and K. RNA isolated from embryos at the indicated stages was used as template for reverse transcription followed by PCR amplification of the target genes with specific oligonucleotide primers. CDK9, cyclin T2 and cyclin K are present as maternal transcripts and are expressed throughout early embryonic development. Mix.3 is expressed during late blastula and gastrula stages (boxed). (B) Whole mount in situ hybridization for CDK9, cyclin T and cyclin K during Xenopus development. Expression of CDK9, and its cyclin partners, is detected throughout the embryo at early gastrula stages. Stage 11 embryos were sliced in half (Stage 11 bisected) to reveal the staining pattern inside the embryo. Each of the genes is expressed broadly throughout the mesendoderm during gastrula stages. During gastrulation the expression of CDK9 remains high while the levels of cyclin T2 and, more profoundly, cyclin K decline. At tailbud stage, high levels of expression of CDK9, cyclin T2 and cyclin K are detected in the head with low-level expression detected throughout the body.
Whole mount in situ hybridization using anti-sense riboprobes revealed that CDK9, cyclin T2 and cyclin K have similar and overlapping expression patterns during embryonic development. CDK9, and its cyclin partners, are widely expressed in the early to mid-gastrula (stage 11; Figure 3B). Bisection of in situ stained embryos revealed that CDK9, cyclin T2 and cyclin K are expressed throughout the developing mesendoderm at early gastrula stage. While the expression of CDK9 remains high during late gastrula stages (Stage 12), the expression of cyclin T2, and more markedly cyclin K, decline at this stage. At stage 34, CDK9, cyclin T2 and cyclin K are highly expressed in the brain and eyes. Expression is also detected throughout the head and is prominent in the hyal and branchial arches. Low-level expression of each gene is detected throughout the body. Thus, the spatial expression of CDK9, and its cyclin partners, overlaps that of Mix.3 (Henry and Melton, 1998; Mead et al., 1998).
Differential activity of CDK9:cyclin T2 and CDK9:cyclin K in reporter assays
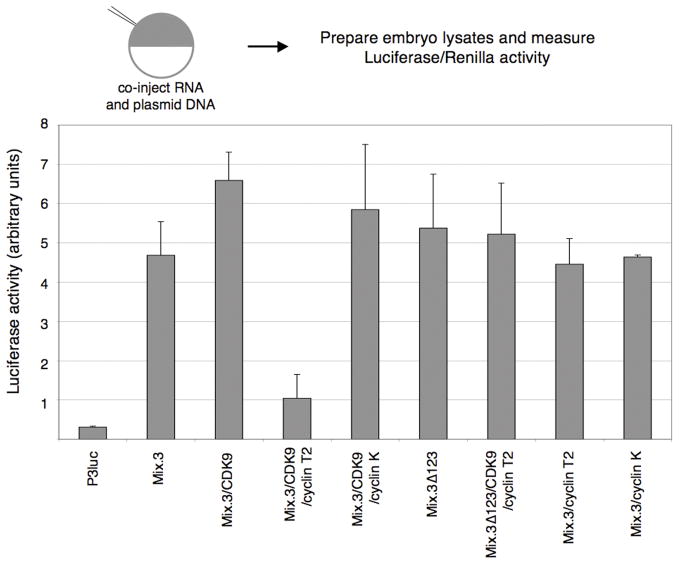
CDK9:cyclin complexes are thought to promote transcriptional elongation by phosphorylation of the CTD of RNAPII. We used two transient whole embryo reporter assays to examine the effect of CDK:cyclin complexes on Mix.3 transcriptional activity. Embryos were injected at the one cell stage with a plasmid containing a reporter and combinations of mRNA encoding Mix.3, CDK9 and cyclin T2 or cyclin K. The reporter assays were normalized by co-injection of a TK-Renilla reporter that served as a control for the micro-injection based assay. We used two reporter constructs, one that would be activated by Mix.3 (P3luc; three tandem Mix-binding sites upstream of a thymidine kinase (TK) promoter TATA-box) and one that is repressed by Mix.3 (Xbraluc; 2.1 kb of the Xenopus brachyury (Xbra) promoter driving luciferase expression; (Latinkic et al., 1997)). In the activation assay, using the P3luc reporter, Mix.3 stimulated expression of luciferase approximately six-fold above the reporter alone (Figure 4). Co-injection of mRNA encoding CDK9 resulted in a modest increase in the luciferase activity. Surprisingly, co-injection of Mix.3, CDK9 and cyclin T2 resulted in a dramatic decrease in reporter activity (approximately five-fold lower than the co-injection of Mix.3 with CDK9). In contrast, co-expression of Mix.3 and CDK9 with cyclin K did not result in decreased activation of the P3luc reporter. This indicates that cyclin K and cyclin T2 have different activities in CDK9-associated complexes with Mix.3. To test whether the activity of cyclin T2 in this assay was a direct effect of its interaction with a Mix.3:CDK9 complex, a mutant form of Mix.3 lacking the CDK9 interaction domain was injected with the reporter construct. Deletion of the CDK9 interaction domain (Mix.3Δ123) alone had no effect on activation activity of Mix.3 on the P3luc reporter (Figure 4). Co-injection of Mix.3Δ123 and CDK9 with cyclin T2 had no effect on Mix.3’s ability to transactivate the reporter construct (Figure 4). This indicates that the ability of cyclin T2 to repress Mix.3 transactivation activity is dependent on the formation of a Mix.3:CDK9 and cyclin T2 complex and not due to an indirect effect of cyclin T2 alone.
Figure 4.
Transient whole embryo reporter assays for Mix.3 trancriptional activation activity. Embryos were injected at the one-cell stage with a Mix-reporter construct (100 pg; P3luc = three tandem Mix-binding sites cloned upstream of a minimal TATA-box driving luciferase expression) and combinations of synthetic mRNA encoding Mix.3, a deletion mutant of Mix.3 that lacks the CDK9 interaction domain (M3Δ123), CDK9, cyclin T2 and cyclin K. The injection cocktail also included a thymidine kinase promoter-Renilla plasmid (TK-Renilla; 50 pg) as an internal control. Luciferase and Renilla activities were measured in three pools of ten embryos each at stage 11. CDK9/cyclin T2, but not CDK9/cyclin K, co-injection blocks Mix.3 transactivation of the P3luc reporter. The inhibition of Mix.3 activity on the P3luc reporter is dependent on the interaction of Mix.3 with CDK9 as deletion of the CDK9-interaction domain from the carboxyl terminus of Mix.3 alleviated the inhibition of CDK9/cyclin T2 (sample Mix.3D123/CDK9/cyclinT2). Mix.3/Cyclin T2, without added CDK9, showed no significant change in P3luc activity relative to Mix.3 mRNA injection alone.
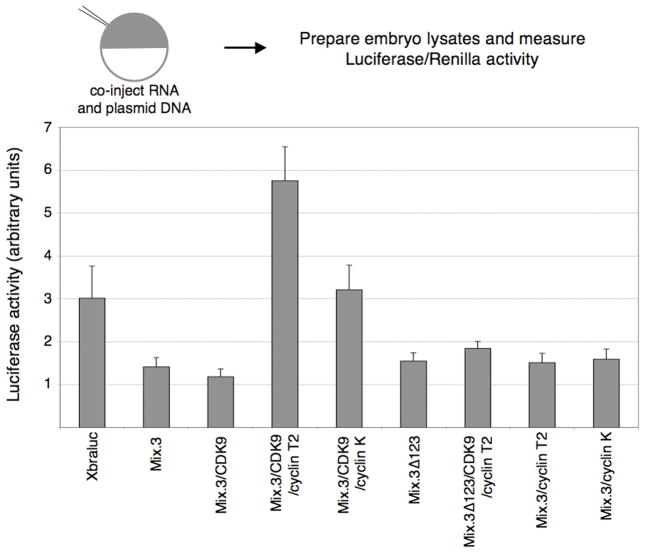
Mix proteins have been reported to act as both transcriptional activators and repressors (Latinkic and Smith, 1999; Latinkic et al., 1997; Lerchner et al., 2000). To determine the effect of the CDK9:cyclin complexes on the transcriptional repression activity of Mix.3, we used a Xenopus brachyury (Xbra) promoter luciferase reporter construct (Xbraluc). Co-expression of Mix.3 with the Xbra reporter resulted a two-fold reduction in reporter activity (Figure 5). Co-expression of CDK9 with either cyclin T2 or cyclin K resulted in attenuation of the Mix.3 repression activity. Co-injection of cyclin K resulted in levels of transcriptional activity similar to the reporter alone. Co-expression with cyclin T2, however, resulted in a two-fold activation of the Xbra reporter construct. To test whether this transactivation activity was dependent on the interaction of Mix.3 with CDK9/cyclin T2, we co-expressed the CDK9-interaction domain mutant (Mix.3Δ123) with CDK9 and cyclin T2. Deletion of the CDK9-interaction domain from the carboxyl terminus of Mix.3 had no effect on the repression activity of Mix.3 when injected alone. Co-injection of Mix.3Δ123 with CDK9 and cyclin T2 resulted in repression of the Xbra reporter to similar levels as wild type Mix.3 (Figure 5). Thus, the inhibition of Mix.3 repression on the Xbra promoter reporter by cyclin T2 is dependent on the interaction of Mix.3 with CDK9.
Figure 5.
Transient whole embryo reporter assays for Mix.3 transcriptional repression activity. Ectopic expression of Mix.3 inhibits the activity of a Xenopus brachyury promoter driving expression of luciferase (Xbraluc). Co-injection of CDK9:cyclin T2 reverses the repression activity of Mix.3 on the Xbraluc reporter and results in a two-fold activation of luciferase activity. The ability of CDK9:cyclin T2 to alleviate the repression of Mix.3 is dependent on the CDK9-interaction domain in Mix.3. Deletion of the CDK9-interaction domain does not affect Mix.3’s ability to repress the brachyury promoter but blocks the opposing activity of co-injection of CDK9:cyclin T2. Co-injection of CDK9:cyclin K also resulted in relief of the repression activity of Mix.3 to control levels (Xbraluc alone) but did not further activate the reporter as CDK9:cyclin T2 did. Mix.3/Cyclin T2, without added CDK9, showed no significant change in Xbraluc activity relative to Mix.3 mRNA injection alone.
Ectopic expression of CDK9:cyclin T2 does not relieve the repression of brachyury expression by Mix.3 in vivo
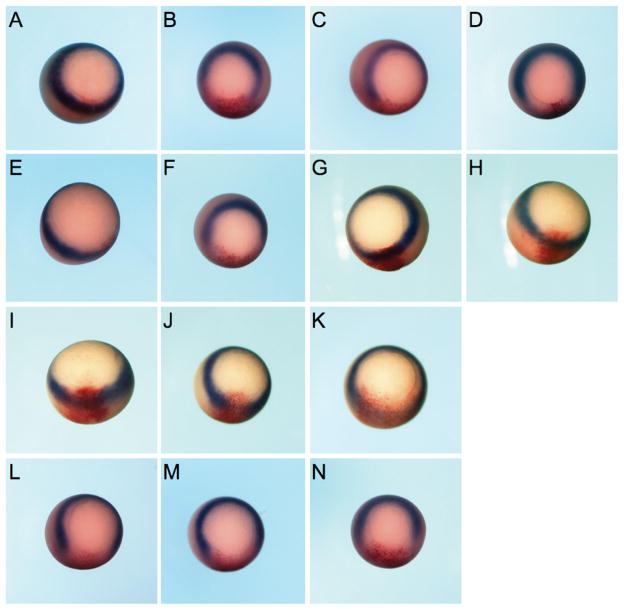
To determine whether the de-repression activity of Mix.3 by CDK9:cyclin T2 occurred in vivo, in situ hybridization studies of the endogenous Xbra gene were performed in albino Xenopus embryos (Figure 6). At early gastrula stages, brachyury is expressed in a ring at the equator of the embryo in the nascent mesoderm (Figure 6A). Injection of Mix.3 mRNA blocked Xbra expression at the site of injection, as determined by co-localization with a co-injected β-galactosidase reporter (Figure 6B, C). Expression of the Mix.3 mutant (Mix.3Δ123) that lacks the CDK9 interaction domain also inhibits Xbra expression indicating that the interaction of Mix.3 and CDK9 is not required for Mix.3 repression of brachyury. Injection of mRNAs encoding cyclin T2 (Figure 6H), cyclin K (Figure 6G) and CDK9 (Figure 6E) alone, or CDK9 and cyclin K (Figure 6I) together, did not disrupt Xbra expression. In the studies above, co-injection of CDK9 and cyclin T2 in transient reporter assays relieved the Mix.3 repression of a Xbraluc reporter in whole embryos (Figure 5). However, when endogenous Xbra expression was assayed by in situ hybridization, CDK9 and cyclin T2 co-injection did not relieve the inhibition of Xbra expression by Mix.3 (Figure 6M). This may be due to the fact that injection of CDK9 and cyclin T2, without added Mix.3, also resulted in modest repression of Xbra expression (Figure 6J and K). This suggests that the CDK9/cyclin T2 complex may also interfere with other components of the Xbra pathway. Furthermore, the differences seen in the in situ hybridization studies and the transient reporter assays may be due to the small (2.1 kb) fragment of the brachyury promoter used to generate the reporter construct may not fully recapitulate activity of the endogenous Xbra gene.
Figure 6.
Endogenous Xenopus brachyury is down-regulated by ectopic expression of Mix.3. Embryos were injected in one blastomere at the two-cell stage with test mRNA together with a β-galactosidase (β-gal) mRNA as a lineage tracer (stained red). In situ hybridization analyses for Xbra expression (stained blue/purple) were performed at early gastrula (Stage 10.5). Representative embryos for each group are shown; the number of embryos examined and the percentage that are the same as the representative are noted below. (A) β-gal alone (100%, n = 61). (B and C) Mix.3. Ectopic expression of Mix.3 results in decreased Xbra expression on the injected side of the embryo (96% have decreased Xbra expression, n = 107/112). (D) M3Δ123. A mutant of Mix.3 that cannot interact with CDK9 maintains Xbra repression activity (100%, n = 21). (E) CDK9. Expression of CDK9 alone does not disrupt Xbra expression in the marginal zone of the embryo (100%, n = 23). (F) Mix.3 + CDK9. CDK9 expression does not block the repression of Xbra expression by Mix.3 (100%, n = 25). (G) Cyclin K (100%, n = 22). (H) Cyclin T2 (100%, n = 23). (I) CDK9 + cyclin K (100%, n = 51). (J and K) CDK9 + cyclin T2. Expression of CDK9 with cyclin T2 disrupts Xbra expression (89%, n = 40/45). (L) Mix.3 + CDK9 + cyclin K (100%, n = 25). (M) Mix.3 + CDK9 + cyclin T2 (100%, n = 44). (N) Mix.3Δ123 + CDK9 + cyclin T2 (98%, n = 46/47).
CDK9:cyclin T2, but not CDK9:cyclin K, blocks Mix.3-directed endoderm induction in animal pole explants and whole embryos
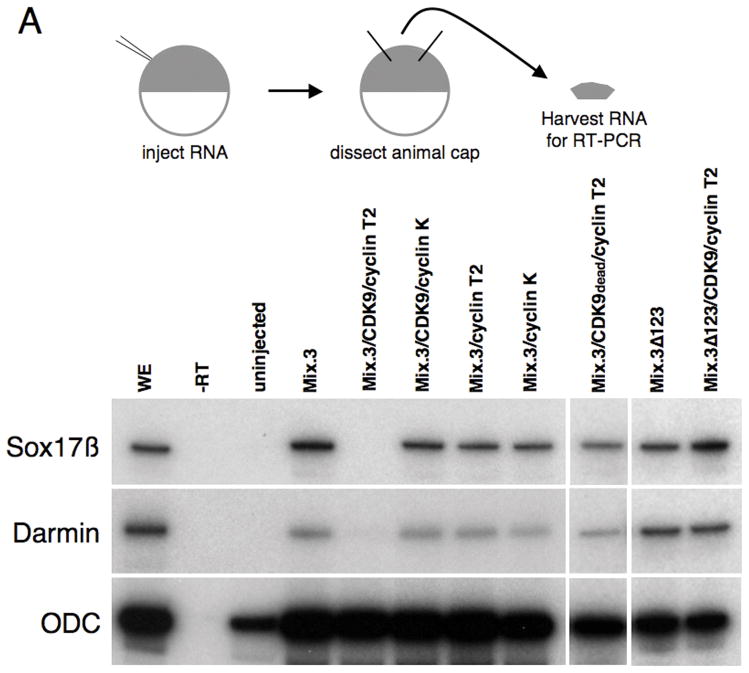
To determine whether the interaction with CDK9 and cyclins affects the endoderm inducing activity of Mix.3, we used animal pole explant assays. Animal caps harvested from embryos injected with Mix.3 mRNA adopt an endodermal cell fate as indicated by the expression of early and late markers of endoderm (Figure 7A, B (Henry and Melton, 1998; Mead et al., 1998)). Two early (Sox17β and Darmin) and two late (Endodermin (edd) and Intestinal Fatty Acid Binding Protein (IFABP)) endoderm markers were used in this study (Hudson et al., 1997; Pera et al., 2003; Sasai et al., 1996; Shi and Hayes, 1994). Similar to results observed with the transient reporter assays using the P3luc construct, co-expression of Mix.3 with CDK9 and cyclin T2 resulted in a decrease in early (Stage 14, Figure 7A) and late (Stage 35, Figure 7B) endodermal marker expression compared to either Mix.3 alone or Mix.3 with CDK9. Co-injection of cyclin K with Mix.3 and CDK9, however, did not result in a decrease in endodermal marker expression. The inhibition of Mix.3-directed endodermal gene expression was dependent on the presence of the CDK9-interaction domain in the carboxyl terminus of Mix.3. The mutant Mix.3 protein that lacked the CDK9 interaction motif (Mix.3Δ123) had similar endoderm inducing activity as the wild type protein (Figure 7A). This indicates that the CDK9-interaction domain of Mix.3 is dispensable for endoderm induction but is required for CDK9 modulation of Mix.3 activity. Co-injection of CDK9 and cyclin T2 with Mix.3Δ123 did not block the endoderm inducing activity of the mutant Mix protein (Figure 7A). To determine whether the suppression of Mix.3 endoderm inducing activity was dependent on the kinase activity of the CDK9 complex, a kinase dead mutant form of CDK9 was co-injected with cyclin T2 and wild type Mix.3. The kinase dead CDK9 (CDK9dead) mutant blocked the suppressive activity of cyclin T2 on early endoderm markers indicating that the CDK9/cyclin T2 complex blocks Mix.3-directed endoderm induction in a kinase-dependent manner. Thus, while the CDK9-interaction domain is dispensable for Mix.3 endoderm inducing activity, it is required for the CDK9:cyclin T2 complex to inhibit Mix.3-directed endoderm induction. Furthermore, the ability of cyclin T2 to attenuate Mix.3’s endoderm inducing activity in animal cap explants is dependent on association with an active CDK9 protein as a kinase dead CDK9 mutant blocked the inhibitory effect. This indicates that the inhibitory effect of cyclin T2 is mediated by the kinase activity of CDK9 and is dependent on the interaction of CDK9 with Mix.3.
Figure 7.
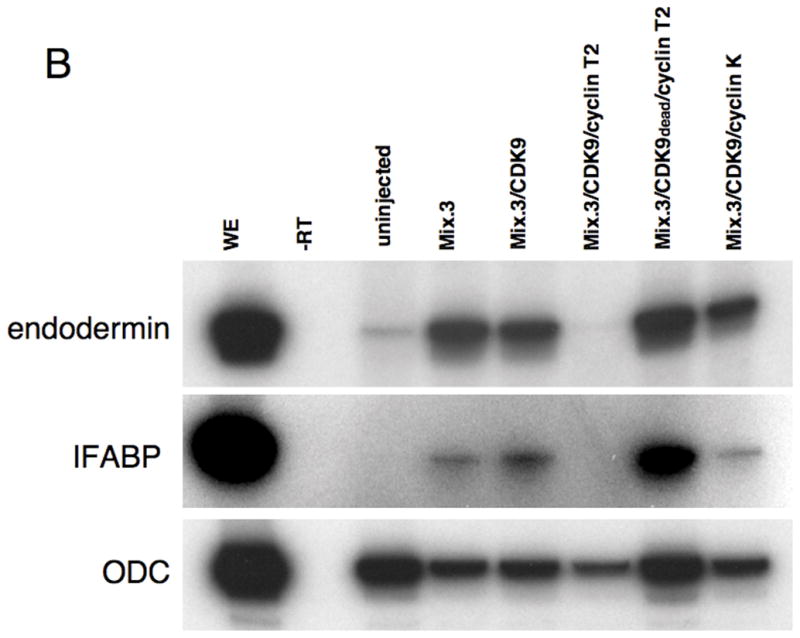
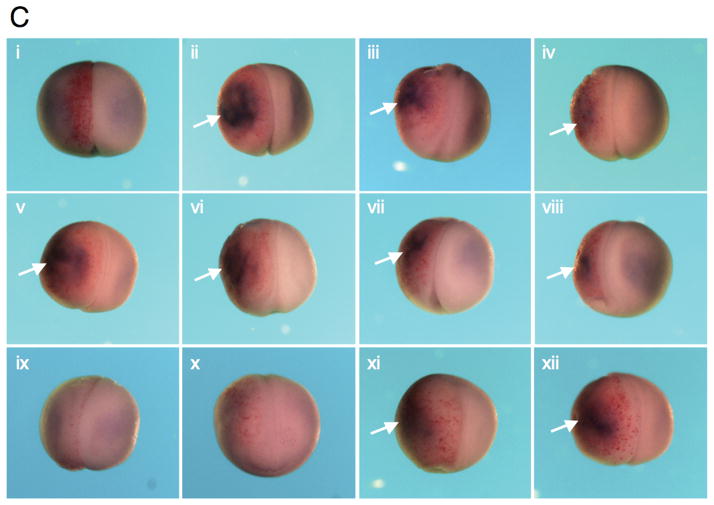
CDK9:cyclin complexes modulate the endoderm inducing activity of Mix.3. RT-PCR analysis for endodermal markers was performed on RNA prepared from animal pole explants dissected from injected embryos. (A) Analysis of early endoderm marker gene expression in animal pole explants. Ectopic expression of Mix.3 induces expression of the early endoderm genes Sox17β and Darmin. Co-injection of CDK9:cyclin T2 blocks Mix.3’s endoderm inducing activity. Deletion of the CDK9-interaction domain from Mix.3 (Mix.3Δ123) eliminates the ability of cyclin T2 to repress Mix.3-mediated endoderm gene expression. The deletion mutant (Mix.3Δ123), when injected alone, induced endoderm markers. The inhibition of Mix.3-directed endoderm induction in animal pole explants requires the kinase activity of CDK9. Co-injection of a kinase-dead mutant of CDK9 (CDK9dead) with cyclin T2 did not block Mix.3 endoderm induction indicating that this is not an inhibitory effect of cyclin T2 alone. Cyclin K co-injected with CDK9 did not interfere with Mix.3 endoderm-inducing activity. (B) RT-PCR analysis of late markers of endoderm differentiation. Animal pole explants were assayed at stage 36 for the late endodermal markers endodermin and intestinal fatty acid binding protein (IFABP). As demonstrated at earlier stages, CDK9:cyclin T2, but not CDK9:cyclin K, can block Mix.3-directed induction of endodermal markers. The inhibition of Mix.3 endoderm by CDK9:cyclin T2 is dependent on the kinase activity of CDK9. (C) The effect of Mix.3, CDK9 and cyclin T2 or K co-injection on endogenous darmin expression was measured by in situ hybridization. Embryos were injected in one blastomere at the two-cell stage with test mRNA together with a β-galactosidase (β-gal) mRNA as a lineage tracer (stained red). In situ hybridization analyses for darmin expression (stained blue/purple) were performed at stage 18. The injected side (stained red) is on the left; endogenous darmin expression is seen on the untreated (right) side of the embryo. Representative embryos for each group are shown; the number of embryos examined and the percentage that are the same as the representative are noted below. (i) β-gal alone (n = 61). (ii) Mix.3 induces ectopic darmin expression (white arrow) in the whole embryo (100% of embryos have ectopic darmin expression, n = 51). (iii) M3Δ123. Deletion of the CDK9 interaction domain has no effect on the ability of Mix.3 to induce ectopic darmin expression (93% displayed ectopic darmin expression, n = 25/27). (iv) Mix.3 + CDK9 induces ectopic darmin expression similar to Mix.3 mRNA alone (98%, n = 45/46). (v) Mix.3 + cyclin T2. Cyclin T2, in the absence of added CDK9, does not block the ability of Mix.3 to induce ectopic darmin expression (100%, n = 27). (vi) Mix.3 + cyclin K. Mix.3 induces ectopic darmin expression in the presence of added cyclin K (100%, n = 20). (vii-viii) Mix.3 + CDK9 + cyclin K. Mix.3 induces ectopic darmin expression in the presence of added CDK9 and cyclin K (96%, n = 47/49). (ix-x) Mix.3 + CDK9 + cyclinT2. Co-injection of cyclin T2 mRNA with Mix.3 + CDK9 blocks ectopic darmin expression (86% showed inhibition of ectopic darmin expression, n = 49/57). (xi-xii) M3Δ123 + CDK9 + cyclin T2. The ability of CDK9 + cyclin T2 to block Mix.3-directed ectopic darmin expression is dependent on the CDK9 interaction domain in the C-terminus of Mix.3 (94%, n = 29/31).
To determine whether CDK9:cyclin T2 was able to attenuate the endoderm inducing activity of Mix.3 in the whole embryo, we used an in situ hybridization assay for darmin expression. Embryos were injected in one blastomere at the two-cell stage with synthetic mRNAs encoding the test proteins together with a lineage tracer (β-galactosidase) and darmin expression was assayed at stage 18. Ectopic expression of Mix.3 lead to increased darmin expression in the progeny of the injected blastomere (Figure 7C, ii). Addition of CDK9 (Figure 7C,iv) or its cyclin partners (cyclin T2, 7C v; cyclin K, 7C vi) alone had no effect on the ability of injected Mix.3 to induce ectopic endoderm. Similar to our findings in the animal pole explants, co-injection of Mix.3 with CDK9 and cyclin K resulted in ectopic darmin expression similar to that of Mix.3 alone (Figure 7C, vii and viii). Co-injection of Mix.3 with CDK9 and cyclin T2, however, resulted in a block in ectopic darmin expression (Figure 7C, ix and x). Interestingly, the endogenous darmin expression appears unchanged by the ectopic expression CDK9:cyclin T2 with Mix.3. As demonstrated in the animal pole explant assays described above, deletion of the CDK9 interaction domain from Mix.3 had no effect on the ability of injected Mix.3 to induce ectopic darmin expression (Figure 7C, iii). The ability of CDK9:cyclin T2 to attenuate Mix.3 endoderm induction was dependent on the presence of the CDK9-interaction domain in the C-terminal region of Mix.3. Co-injection of M3Δ123 with CDK9 and cyclin T2 resulted in similar ectopic darmin expression seen with wild type Mix.3 injection alone (Figure 7C, xi and xii).
In summary, we have shown that the interaction of Mix.3 with CDK9/cyclins alters the transcriptional and endoderm inducing activity of this Pax-like homeodomain protein. CDK9 interacts with both cyclin K and cyclin T2, and each cyclin partner confers different activities on the Mix.3/CDK9/cyclin complex. We have shown that the CDK9:cyclin T2 complex can alter the transcriptional activity of Mix.3 in whole embryo transient reporter assays. CDK9:cyclin T2 can block Mix.3 transactivation on the P3luc reporter and interfere with the repression activity of Mix.3 on the Xbraluc reporter construct. In animal pole explant and whole embryo assays, the CDK9:cyclin T2 complex was able to block Mix.3-directed endoderm induction, and this inhibition was dependent on both the interaction of CDK9 with Mix.3 and the kinase activity of CDK9.
Discussion
Using a yeast two-hybrid approach, we have determined that CDK9 interacts with the Pax-like homeodomain protein Mix.3/mixer. Co-immunoprecipitation pull-down assays were used to confirm the interaction and to map the binding site to a twenty-two amino acid sequence in the carboxyl terminal domain of Mix.3. CDK9 is the catalytic component of the positive transcription elongation factor b (P-TEFb) complex that phosphorylates the carboxyl-terminal repeat domain (CTD) of RNA polymerase II (RNAPII). CDK9 is a cdc2-like kinase that associates with two cyclin partners, cyclin T and cyclin K. The phosphorylation of serine residues in the CTD of RNAPII promotes elongation of the nascent messenger after transcriptional initiation steps have taken place. Thus, the recruitment of CDK9-containing P-TEFb complexes to the transcription machinery provides an important regulatory step in the control of transcriptional activity.
In this study, we show that the interaction of CDK9:cyclin complexes can modulate the endoderm-inducing activity of Mix.3 in transient reporter assays, in animal pole explant assays and in whole embryos. The recruitment of cyclin K and cyclin T2 had different effects on the endoderm-inducing activity of Mix.3. CDK9:cyclin T2, but not CDK9:cyclin K, inhibited Mix.3-directed transcriptional activation of a Mix-luciferase (P3luc) reporter in whole embryo transient assays. The inhibition of Mix.3 transcriptional activity in this assay was dependent on the CDK9-interaction domain of Mix.3. In animal cap assays, CDK9:cyclin T2, but not CDK9:cyclin K, blocked endoderm differentiation by Mix.3. As demonstrated in the transient activation assays, the inhibition of endoderm induction was dependent on the CDK9-interaction domain of Mix.3 and also dependent on the catalytic activity of the kinase. The mechanism of action of CDK9 on Mix.3 activity is unknown. Based on the known role of CDK9 and the PTEFb complex in the elongation of nascent transcripts it is, however, tempting to speculate that Mix.3 recruits CDK9:cyclin K to the transcriptional complex to allow efficient elongation of nascent Mix.3 target genes. Alternatively, the kinase activity of CDK9 may be required for phosphorylation of Mix.3, or other members of the Mix-containing transcriptional complex, to achieve maximal transcriptional activity. Our data indicates that the recruitment of the CDK9:cyclin T2 complex by Mix.3 results in a block in Mix-directed induction of endoderm target genes. CDK9 in complex with cyclin T1, T2a, T2b or K is able to phosphorylate the CTD of RNAPII (Fu et al., 1999; Peng et al., 1998). If each of the cyclin partners can stimulate transcriptional elongation by hyper-phosphorylation of the CTD of RNAPII, why then does the over-expression of cyclin T2 with CDK9 block activation of Mix.3-dependent endoderm gene expression? While the answer to this question remains unsolved, there are, however, other examples of this phenomenon in the literature. For example, Napolitano and co-workers demonstrated that cyclins T1 and T2 had opposing effects on the activation of HIV Tat transcription (Napolitano et al., 1999). The authors show that over-expression of cyclin T2, but not cyclin T1, can inhibit activation of HIV gene expression. Furthermore, the same effect could be observed with over-expression of a deletion mutant of cyclin T2 that contained only the N-terminal (cyclin box) domain, the domain that binds CDK9. Together, their data demonstrates that P-TEFb complexes that contain different cyclin partners can illicit markedly different effects on target gene transactivation. To date, we have been able to identify a single full-length cyclin T cDNA in the Xenopus laevis EST databases that shares highest homology with vertebrate cyclin T2 proteins. The absence of a full-length X. laevis cyclin T1 cDNA clone has precluded analysis of the activity of this cyclin on Mix.3 activity. Recently, however, full-length cDNA clones encoding the Xenopus tropicalis cyclin T1, cyclin T2, cyclin K and CDK9 genes have become available in the IMAGE clone collections, and our future studies will use these clones to determine whether the P-TEFb complex containing cyclin T1 has similar effects on Mix.3-directed transcription as those containing cyclins K and T2.
The potent endoderm inducing activity of Mix.3 must be tightly regulated during early development. Over-expression of Mix.3 results in ectopic endoderm formation and thus the endogenous activity of Mix.3 must be precisely regulated (Doherty et al., 2006; Mead et al., 1998). Our data demonstrate that CDK9:cyclin complexes are involved in the regulation of Mix.3 endoderm inducing activity. Mix proteins have also been implicated in the regulation of mesoderm formation by antagonizing brachyury expression. Knockdown of Mix.3 expression using antisense morpholinos results in expansion of mesoderm markers indicating that Mix proteins play an important role in establishing the boundary of mesoderm and endoderm germ layers during early embryonic development (Kofron et al., 2004). Our data also shows that CDK9:cyclin complexes interact with Mix.3 and can modulate the Mix.3-mediated repression of brachyury. Thus, the interaction of CDK9 with Mix.3, and the recruitment of different cyclin partners, can regulate the transcriptional activation and repression activities of Mix.3 and may provide an important mechanism for regulating the activity of this potent homeodomain protein during early development.
Experimental Procedures
Yeast two hybrid library and screen
A cDNA library was prepared from Xenopus laevis gastrulae (stages 10–12) in the pGADT7 vector using the Matchmaker 3 Gal4 Yeast Two-Hybrid System (Clontech, CA). Baits were constructed by amplifying a Mix.3 cDNA with specific primers, and the resulting PCR products were subcloned as NdeI and SalI fragments into pGBKT7 (Clontech, CA). The bait constructs were transformed into AH109 yeast strain together with pGADT7 empty vector on –Leu;-Trp “two-drop” out media and –Ade;-His;-Leu;-Trp “four-drop” out media to test for auto-transactivation. We chose the pGBKT7-Mix.3Δ58 that had no auto-activation activity as the bait to screen the library. pGBKT7-Mix.3Δ58 was transformed into the AH109 yeast strain, and subsequently, transformed with the cDNA library. At least 5×105 yeast transformants were obtained and were used for screening for Mix.3-interacting proteins according the manufacturers instructions.
Isolation of full length cDNAs
Full length cDNAs for Xenopus laevis CDK9 (IMAGE number 5542442), cyclin T2 (IMAGE number 6318646) and cyclin K (IMAGE number 5542442) were obtained from the IMAGE clone collection (Open Biosystems, AL). The cDNA clones were completely sequenced to verify their identity before use in our assays.
In vitro protein translation and co-immunoprecipitation
Mix.3 with an N-terminal c-myc tag and HA-tagged CDK9 constructs were translated in vitro using the TNT Coupled Reticulocyte Lysate Transcription/Translation System according to the manufacturers instructions (Promega, CA). Immunoprecipitation experiments were performed using the ProFound™ Mammalian HA tag IP/Co-Immunoprecipitation kit according to the manufacturers instructions (Pierce, IL). Co-immunoprecipitated proteins were eluted using 2x non-reducing sample buffer to eliminate interference from co-eluting antibody heavy or light chains. Samples were loaded on pre-cast SDS-PAGE gels (Bio-Rad Laboratories, CA). After electrophoresis, the proteins were transferred onto PVDF membranes for western blot assays using either anti-c myc or anti-HA antibodies.
Whole embryo luciferase assays
A Mix.3 responsive luciferase reporter construct (P3luc) was generated by cloning three tandem Mix.3 DNA binding sites (5′-TAA TCG GAT TAG CCT AAT CGG ATT AGC CTA ATC GGA TTA-3′) upstream of a herpes simplex virus thymidine kinase (TK) minimal promoter (TATA box) in the pGL3-basic luciferase vector (Promega, WI). A 2.1 kb fragment of the Xenopus brachyury promoter was cloned into pGL3-enhancer luciferase vector (Promega, WI). The Renillia luciferase mRNA (pRL-TK, Promega, WI) was used as internal control. Single cell embryos were microinjected with 100 pg of luciferase reporter, 0.6 pg of Renilla mRNA and 1 ng of synthetic test mRNA. Messenger RNA was synthesized using mMessage mMachine transcription kit (Ambion, TX). Embryos were harvested at stage 11 and assayed using the Dual Luciferase Reporter Assay System (Promega, WI). Luciferase activity was measured in a Zylux femtomaster FB12 luminometer. Three sets of ten embryos were assayed for each data point and experiments were repeated at least three times.
Xenopus embryo manipulations
Xenopus laevis embryos were obtained, fertilized and microinjected using standard protocols (Sive et al., 2000). For animal cap assays, embryos were injected at one cell stage and animal pole explants were harvested at stage 8. Animal caps from at least twenty embryos per RNA were cultured for up to five days at 18°C. Total RNA was extracted from caps using RNAeasy Mini Prep kit according to the manufacturers instructions (Qiagen, CA). Reverse transcription was performed using SuperScript II Reverse Transcriptase (Invitrogen, CA). Endodermal markers Darmin and Sox17β were used for RT-PCR analysis. Darmin, primer 1: 5′-AAC ACG CTT GCA GAT GGA AA-3′, primer 2: 5′-GTT AGG GAC TTG CCG AAT GG-3′, XSox17β primer 1: 5′-AAC TCC CAC CAG CAG GCT ACT TTG-3′ primer 2: 5′-TGT CAA TGT CAC TCT CCA GAT GTC C-3′ (Myers et al. 2004). Endoderm specific genes were amplified from animal cap cDNA using HotStar taq DAN polymerase (Qiagen, CA). Ornithine decarboxylase (ODC) was used as a control for RNA recovery. ODC upper: 5′-CAG CTA GCT GTG GTG TGG-3′, ODC lower: 5′-CAA CAT GGA AAC TCA CAC C-3′ (Agius et al., 2000). Trace amounts of α-32P dCTP was included in each reaction to allow detection of the PCR product by autoradiography. Reactions were separated on pre-cast 10% (w/v) polyacrylamide tris-Borate-EDTA gels (Bio-Rad Laboratories, CA), fixed, dried and exposed to X-ray film. Experiments were repeated at least three times and representative data is shown.
Primers used for developmental RT-PCR analysis:
Xcyclin T2, upper: 5′-GGC ATC CAC TTC ATT TCC AG-3′ lower: 5′-GCC AGT GAC GGA GCA TAG AC-3′;
Xcyclin K, P1: 5′-CCA ACA GTC CAA GAA GCC ATC-3′, P2: 5′-AAT TGG TTG TAG ACA TCC CCG-3′;
XCDK9, upper: 5′-AGT TGA CAT GTA CAG CTG CAT ACG-3′, lower: 5′-CCC TAT CAA AAT ACA TAG GTA CAG CAA C-3′
Primers used for Mix.3 and CDK9 mutagenesis using the QuikChange site-directed mutagenesis kit (Stratagene) are listed below. The boldface letters indicate the mutated bases.
Mix.3Δ146 deletion mutant (a stop codon (TAA) was introduced at position 226 to generate a truncated Mix.3 mutant protein.) M3Δ146, upper: 5′-GCT GTT TCC CAG CAG TAA ATC CTG ATG CAG CAG-3′, lower: 5′-CTG CTG CAT CAG GAT TTA CTG CTG GGA AAC AGC-3′.
Mix.3Δ123 deletion mutant (a BamHI site was introduced into the Mix.3 sequence at amino acid position 248–249, digestion of the modified clone with BamHI released an internal fragment and the plasmid was re-ligated to generate the desired in-frame twenty-two amino acid deletion mutant.) M3Δ123, upper: 5′-TCT TAT AAA CCT ACA GGG ATC CAG CAG CAC TTC TAC-3′, lower: 5′-GTA GAA GTG CTG CTG GAT CCC TGT AGG TTT ATA AGA-3′.
CDK9dead (a point mutation was engineered in the active site of the kinase domain to yield a protein that is kinase-deficient; D171N (De Falco et al., 2000)); XCDK9dead, upper: 5′-GTG TTG AAA CTT GCA AAC TTT GGG CTT GCC AGA G-3′, lower: 5′-GAG ACC GTT CGG GTT TCA AAC GTT CAA AGT TGT G-3′
In situ hybridization of whole embryos and Red Gal staining
Whole embryo in situ hybridization was performed using albino embryos as described by Harland (Harland, 1991). Anti-sense probes were generated as described by Kelley et al. (Kelley et al., 1994). Briefly, anti-sense probes were synthesized with digoxigenin-coupled UTP (Roche) and detected with alkaline phosphatase coupled to anti-digoxigenin Fab fragment (Roche) followed by the chromogenic reaction with BM Purple (Roche). β-galactosidase protein was detected as described by Smith and Harland with the modification that 6-chloro-3-indolyl-D-galactoside (RedGal, Research Organics, Inc.) was used in place of X-gal (Smith and Harland, 1991). Following staining, embryos were re-fixed and whole mount in situ hybridizations were carried out according to the method of Harland (Harland, 1991).
Acknowledgments
We thank Drs. Donald Yergeau and Clair Kelley for critical review of the manuscript and members of the MeadLab for helpful discussions. We thank Shelby Benson and Kevin Bergeron for assistance with animal husbandry. DNA sequencing and bioinformatics support was provided by the Harwell Center for Biotechnology and Bioinformatics, a SJCRH shared resource. This work was funded by the American Lebanese and Syrian Associated Charities (ALSAC) and by developmental funds from the Cancer Center (NCI P30 CA21765).
References
- Agius E, Oelgeschlager M, Wessely O, Kemp C, De Robertis EM. Endodermal Nodal-related signals and mesoderm induction in Xenopus. Development. 2000;127:1173–1183. doi: 10.1242/dev.127.6.1173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alexander J, Stainier DY. A molecular pathway leading to endoderm formation in zebrafish. Curr Biol. 1999;9:1147–1157. doi: 10.1016/S0960-9822(00)80016-0. [DOI] [PubMed] [Google Scholar]
- Archambault J, Pan G, Dahmus GK, Cartier M, Marshall N, Zhang S, Dahmus ME, Greenblatt J. FCP1, the RAP74-interacting subunit of a human protein phosphatase that dephosphorylates the carboxyl-terminal domain of RNA polymerase IIO. J Biol Chem. 1998;273:27593–275601. doi: 10.1074/jbc.273.42.27593. [DOI] [PubMed] [Google Scholar]
- Barboric M, Nissen RM, Kanazawa S, Jabrane-Ferrat N, Peterlin BM. NF-kappaB binds P-TEFb to stimulate transcriptional elongation by RNA polymerase II. Mol Cell. 2001;8:327–337. doi: 10.1016/s1097-2765(01)00314-8. [DOI] [PubMed] [Google Scholar]
- Chao SH, Price DH. Flavopiridol inactivates P-TEFb and blocks most RNA polymerase II transcription in vivo. J Biol Chem. 2001;276:31793–31799. doi: 10.1074/jbc.M102306200. [DOI] [PubMed] [Google Scholar]
- Cho H, Kim TK, Mancebo H, Lane WS, Flores O, Reinberg D. A protein phosphatase functions to recycle RNA polymerase II. Genes Dev. 1999;13:1540–1552. doi: 10.1101/gad.13.12.1540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Falco G, Bagella L, Claudio PP, De Luca A, Fu Y, Calabretta B, Sala A, Giordano A. Physical interaction between CDK9 and B-Myb results in suppression of B-Myb gene autoregulation. Oncogene. 2000;19:373–379. doi: 10.1038/sj.onc.1203305. [DOI] [PubMed] [Google Scholar]
- De Luca A, Esposito V, Baldi A, Claudio PP, Fu Y, Caputi M, Pisano MM, Baldi F, Giordano A. CDC2-related kinase PITALRE phosphorylates pRb exclusively on serine and is widely expressed in human tissues. J Cell Physiol. 1997;172:265–273. doi: 10.1002/(SICI)1097-4652(199708)172:2<265::AID-JCP13>3.0.CO;2-8. [DOI] [PubMed] [Google Scholar]
- Doherty JR, Zhu H, Kuliyev E, Mead PE. Determination of the minimal domains of Mix.3/Mixer required for endoderm development. Mech Dev. 2006;123:56–66. doi: 10.1016/j.mod.2005.09.006. [DOI] [PubMed] [Google Scholar]
- Eberhardy SR, Farnham PJ. c-Myc mediates activation of the cad promoter via a post-RNA polymerase II recruitment mechanism. J Biol Chem. 2001;276:48562–48571. doi: 10.1074/jbc.M109014200. [DOI] [PubMed] [Google Scholar]
- Eberhardy SR, Farnham PJ. Myc recruits P-TEFb to mediate the final step in the transcriptional activation of the cad promoter. J Biol Chem. 2002;277:40156–40162. doi: 10.1074/jbc.M207441200. [DOI] [PubMed] [Google Scholar]
- Fu TJ, Peng J, Lee G, Price DH, Flores O. Cyclin K functions as a CDK9 regulatory subunit and participates in RNA polymerase II transcription. J Biol Chem. 1999;274:34527–34530. doi: 10.1074/jbc.274.49.34527. [DOI] [PubMed] [Google Scholar]
- Garriga J, Bhattacharya S, Calbo J, Marshall RM, Truongcao M, Haines DS, Grana X. CDK9 is constitutively expressed throughout the cell cycle, and its steady-state expression is independent of SKP2. Mol Cell Biol. 2003;23:5165–5173. doi: 10.1128/MCB.23.15.5165-5173.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garriga J, Grana X. Cellular control of gene expression by T-type cyclin/CDK9 complexes. Gene. 2004;337:15–23. doi: 10.1016/j.gene.2004.05.007. [DOI] [PubMed] [Google Scholar]
- Grana X, De Luca A, Sang N, Fu Y, Claudio PP, Rosenblatt J, Morgan DO, Giordano A. PITALRE, a nuclear CDC2-related protein kinase that phosphorylates the retinoblastoma protein in vitro. Proc Natl Acad Sci U S A. 1994;91:3834–3838. doi: 10.1073/pnas.91.9.3834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harland RM. In situ hybridization: an improved whole-mount method for Xenopus embryos. Methods Cell Biol. 1991;36:685–695. doi: 10.1016/s0091-679x(08)60307-6. [DOI] [PubMed] [Google Scholar]
- Hart AH, Hartley L, Sourris K, Stadler ES, Li R, Stanley EG, Tam PP, Elefanty AG, Robb L. Mixl1 is required for axial mesendoderm morphogenesis and patterning in the murine embryo. Development. 2002;129:3597–3608. doi: 10.1242/dev.129.15.3597. [DOI] [PubMed] [Google Scholar]
- Henry GL, Melton DA. Mixer, a homeobox gene required for endoderm development. Science. 1998;281:91–96. doi: 10.1126/science.281.5373.91. [DOI] [PubMed] [Google Scholar]
- Hudson C, Clements D, Friday RV, Stott D, Woodland HR. Xsox17alpha and -beta mediate endoderm formation in Xenopus. Cell. 1997;91:397–405. doi: 10.1016/s0092-8674(00)80423-7. [DOI] [PubMed] [Google Scholar]
- Kelley C, Yee K, Harland R, Zon LI. Ventral expression of GATA-1 and GATA-2 in the Xenopus embryo defines induction of hematopoietic mesoderm. Dev Biol. 1994;165:193–205. doi: 10.1006/dbio.1994.1246. [DOI] [PubMed] [Google Scholar]
- Kikuchi Y, Trinh LA, Reiter JF, Alexander J, Yelon D, Stainier DY. The zebrafish bonnie and clyde gene encodes a Mix family homeodomain protein that regulates the generation of endodermal precursors. Genes Dev. 2000;14:1279–1289. [PMC free article] [PubMed] [Google Scholar]
- Kobor MS, Archambault J, Lester W, Holstege FC, Gileadi O, Jansma DB, Jennings EG, Kouyoumdjian F, Davidson AR, Young RA, Greenblatt J. An unusual eukaryotic protein phosphatase required for transcription by RNA polymerase II and CTD dephosphorylation in S. cerevisiae. Mol Cell. 1999;4:55–62. doi: 10.1016/s1097-2765(00)80187-2. [DOI] [PubMed] [Google Scholar]
- Kofron M, Wylie C, Heasman J. The role of Mixer in patterning the early Xenopus embryo. Development. 2004;131:2431–2441. doi: 10.1242/dev.01132. [DOI] [PubMed] [Google Scholar]
- Lania L, Majello B, Napolitano G. Transcriptional control by cell-cycle regulators: a review. J Cell Physiol. 1999;179:134–141. doi: 10.1002/(SICI)1097-4652(199905)179:2<134::AID-JCP3>3.0.CO;2-O. [DOI] [PubMed] [Google Scholar]
- Latinkic BV, Smith JC. Goosecoid and mix.1 repress Brachyury expression and are required for head formation in Xenopus. Development. 1999;126:1769–1779. doi: 10.1242/dev.126.8.1769. [DOI] [PubMed] [Google Scholar]
- Latinkic BV, Umbhauer M, Neal KA, Lerchner W, Smith JC, Cunliffe V. The Xenopus Brachyury promoter is activated by FGF and low concentrations of activin and suppressed by high concentrations of activin and by paired-type homeodomain proteins. Genes Dev. 1997;11:3265–3276. doi: 10.1101/gad.11.23.3265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lerchner W, Latinkic BV, Remacle JE, Huylebroeck D, Smith JC. Region-specific activation of the Xenopus brachyury promoter involves active repression in ectoderm and endoderm: a study using transgenic frog embryos. Development. 2000;127:2729–2739. doi: 10.1242/dev.127.12.2729. [DOI] [PubMed] [Google Scholar]
- Licciardo P, Ruggiero L, Lania L, Majello B. Transcription activation by targeted recruitment of the RNA polymerase II CTD phosphatase FCP1. Nucleic Acids Res. 2001;29:3539–3545. doi: 10.1093/nar/29.17.3539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin X, Taube R, Fujinaga K, Peterlin BM. P-TEFb containing cyclin K and Cdk9 can activate transcription via RNA. J Biol Chem. 2002;277:16873–16878. doi: 10.1074/jbc.M200117200. [DOI] [PubMed] [Google Scholar]
- Majello B, Napolitano G. Control of RNA polymerase II activity by dedicated CTD kinases and phosphatases. Front Biosci. 2001;6:D1358–1368. doi: 10.2741/majello. [DOI] [PubMed] [Google Scholar]
- Mead PE, Zhou Y, Lustig KD, Huber TL, Kirschner MW, Zon LI. Cloning of Mix-related homeodomain proteins using fast retrieval of gel shift activities, (FROGS), a technique for the isolation of DNA-binding proteins. Proc Natl Acad Sci U S A. 1998;95:11251–11256. doi: 10.1073/pnas.95.19.11251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohn D, Chen SW, Dias DC, Weinstein DC, Dyer MA, Sahr K, Ducker CE, Zahradka E, Keller G, Zaret KS, Gudas LJ, Baron MH. Mouse Mix gene is activated early during differentiation of ES and F9 stem cells and induces endoderm in frog embryos. Dev Dyn. 2003;226:446–459. doi: 10.1002/dvdy.10263. [DOI] [PubMed] [Google Scholar]
- Napolitano G, Licciardo P, Gallo P, Majello B, Giordano A, Lania L. The CDK9-associated cyclins T1 and T2 exert opposite effects on HIV-1 Tat activity. Aids. 1999;13:1453–1459. doi: 10.1097/00002030-199908200-00003. [DOI] [PubMed] [Google Scholar]
- Napolitano G, Majello B, Lania L. Role of cyclinT/Cdk9 complex in basal and regulated transcription (review) Int J Oncol. 2002;21:171–177. [PubMed] [Google Scholar]
- Pearce JJ, Evans MJ. Mml, a mouse Mix-like gene expressed in the primitive streak. Mech Dev. 1999;87:189–192. doi: 10.1016/s0925-4773(99)00135-5. [DOI] [PubMed] [Google Scholar]
- Peng J, Marshall NF, Price DH. Identification of a cyclin subunit required for the function of Drosophila P-TEFb. J Biol Chem. 1998;273:13855–13860. doi: 10.1074/jbc.273.22.13855. [DOI] [PubMed] [Google Scholar]
- Pera EM, Martinez SL, Flanagan JJ, Brechner M, Wessely O, De Robertis EM. Darmin is a novel secreted protein expressed during endoderm development in Xenopus. Gene Expr Patterns. 2003;3:147–152. doi: 10.1016/s1567-133x(03)00011-5. [DOI] [PubMed] [Google Scholar]
- Pines J. The cell cycle kinases. Semin Cancer Biol. 1994;5:305–313. [PubMed] [Google Scholar]
- Price DH. P-TEFb, a cyclin-dependent kinase controlling elongation by RNA polymerase II. Mol Cell Biol. 2000;20:2629–2634. doi: 10.1128/mcb.20.8.2629-2634.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robb L, Hartley L, Begley CG, Brodnicki TC, Copeland NG, Gilbert DJ, Jenkins NA, Elefanty AG. Cloning, expression analysis, and chromosomal localization of murine and human homologues of a Xenopus mix gene. Dev Dyn. 2000;219:497–504. doi: 10.1002/1097-0177(2000)9999:9999<::AID-DVDY1070>3.0.CO;2-O. [DOI] [PubMed] [Google Scholar]
- Sano M, Schneider MD. Cyclins that don’t cycle--cyclin T/cyclin-dependent kinase-9 determines cardiac muscle cell size. Cell Cycle. 2003;2:99–104. [PubMed] [Google Scholar]
- Sasai Y, Lu B, Piccolo S, De Robertis EM. Endoderm induction by the organizer-secreted factors chordin and noggin in Xenopus animal caps. Embo J. 1996;15:4547–4555. [PMC free article] [PubMed] [Google Scholar]
- Shi YB, Hayes WP. Thyroid hormone-dependent regulation of the intestinal fatty acid-binding protein gene during amphibian metamorphosis. Dev Biol. 1994;161:48–58. doi: 10.1006/dbio.1994.1006. [DOI] [PubMed] [Google Scholar]
- Shim EY, Walker AK, Shi Y, Blackwell TK. CDK-9/cyclin T (P-TEFb) is required in two postinitiation pathways for transcription in the C. elegans embryo. Genes Dev. 2002;16:2135–2146. doi: 10.1101/gad.999002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shivdasani RA. Molecular regulation of vertebrate early endoderm development. Dev Biol. 2002;249:191–203. doi: 10.1006/dbio.2002.0765. [DOI] [PubMed] [Google Scholar]
- Simone C, Bagella L, Bellan C, Giordano A. Physical interaction between pRb and cdk9/cyclinT2 complex. Oncogene. 2002a;21:4158–4165. doi: 10.1038/sj.onc.1205511. [DOI] [PubMed] [Google Scholar]
- Simone C, Stiegler P, Bagella L, Pucci B, Bellan C, De Falco G, De Luca A, Guanti G, Puri PL, Giordano A. Activation of MyoD-dependent transcription by cdk9/cyclin T2. Oncogene. 2002b;21:4137–4148. doi: 10.1038/sj.onc.1205493. [DOI] [PubMed] [Google Scholar]
- Smith WC, Harland RM. Injected Xwnt-8 RNA acts early in Xenopus embryos to promote formation of a vegetal dorsalizing center. Cell. 1991;67:753–765. doi: 10.1016/0092-8674(91)90070-f. [DOI] [PubMed] [Google Scholar]
- Tian Y, Ke S, Chen M, Sheng T. Interactions between the aryl hydrocarbon receptor and P-TEFb. Sequential recruitment of transcription factors and differential phosphorylation of C-terminal domain of RNA polymerase II at cyp1a1 promoter. J Biol Chem. 2003;278:44041–44048. doi: 10.1074/jbc.M306443200. [DOI] [PubMed] [Google Scholar]
- Wada T, Takagi T, Yamaguchi Y, Ferdous A, Imai T, Hirose S, Sugimoto S, Yano K, Hartzog GA, Winston F, Buratowski S, Handa H. DSIF, a novel transcription elongation factor that regulates RNA polymerase II processivity, is composed of human Spt4 and Spt5 homologs. Genes Dev. 1998a;12:343–356. doi: 10.1101/gad.12.3.343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wada T, Takagi T, Yamaguchi Y, Watanabe D, Handa H. Evidence that P-TEFb alleviates the negative effect of DSIF on RNA polymerase II-dependent transcription in vitro. Embo J. 1998b;17:7395–7403. doi: 10.1093/emboj/17.24.7395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei P, Garber ME, Fang SM, Fischer WH, Jones KA. A novel CDK9-associated C-type cyclin interacts directly with HIV-1 Tat and mediates its high-affinity, loop-specific binding to TAR RNA. Cell. 1998;92:451–462. doi: 10.1016/s0092-8674(00)80939-3. [DOI] [PubMed] [Google Scholar]
- Yamaguchi Y, Takagi T, Wada T, Yano K, Furuya A, Sugimoto S, Hasegawa J, Handa H. NELF, a multisubunit complex containing RD, cooperates with DSIF to repress RNA polymerase II elongation. Cell. 1999;97:41–51. doi: 10.1016/s0092-8674(00)80713-8. [DOI] [PubMed] [Google Scholar]
- Zhu Y, Pe’ery T, Peng J, Ramanathan Y, Marshall N, Marshall T, Amendt B, Mathews MB, Price DH. Transcription elongation factor P-TEFb is required for HIV-1 tat transactivation in vitro. Genes Dev. 1997;11:2622–2632. doi: 10.1101/gad.11.20.2622. [DOI] [PMC free article] [PubMed] [Google Scholar]