Abstract
The ribosome is a complex macromolecular machine that translates the message encoded in the messenger RNA and synthesizes polypeptides by linking the individual amino acids carried by the cognate transfer RNAs (tRNAs). The protein elongation cycle, during which the tRNAs traverse the ribosome in a coordinated manner along a path of more than 100 Å, is facilitated by large-scale rearrangements of the ribosome. These rearrangements go hand in hand with conformational changes of tRNA as well as elongation factors EF-Tu and EF-G – GTPases that catalyze tRNA delivery and translocation, respectively. This review focuses on the structural data related to the dynamics of the ribosomal machinery, which are the basis, in conjunction with existing biochemical, kinetic, and fluorescence resonance energy transfer data, of our knowledge of the decoding and translocation steps of protein elongation.
1. Introduction
The ribosome is a molecular machine responsible for translating the information carried by the messenger RNA (mRNA) into a sequence of amino acids, which are linked to form a polypeptide chain and eventually a protein. Translation is a dynamic process during which the ribosome and all of its functional ligands undergo conformational changes following various induced-fit mechanisms. Characterization of these changes and their functional significance is accomplished by a combination of cryo-electron microscopy (cryo-EM) and single-molecule fluorescence resonance energy transfer (smFRET), drawing from the rich trove of X-ray structures solved in the past decade.
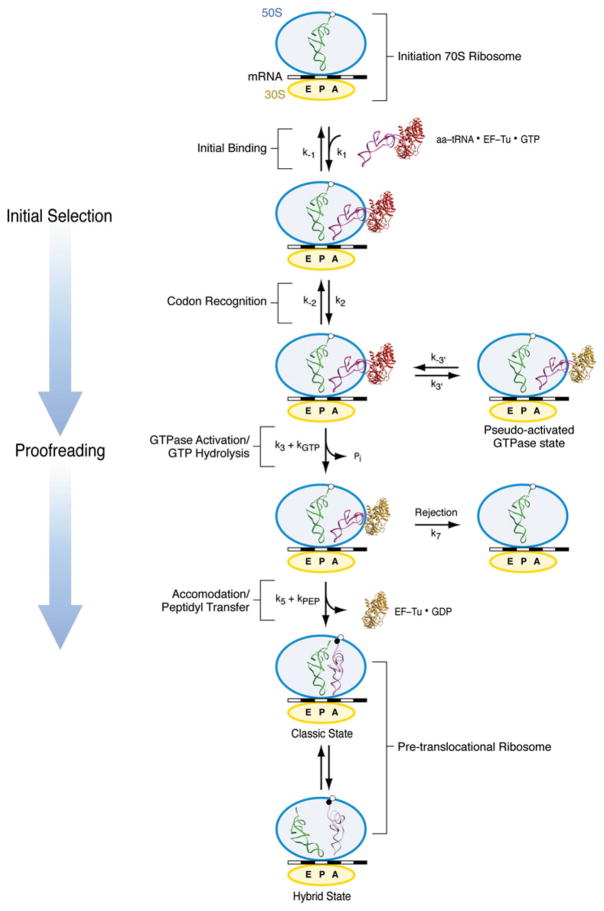
The work cycle during which a new amino acid is added to the growing polypeptide chain, referred to as elongation cycle, is a repetitive multistep process encompassing aminoacyl-tRNA (aa-tRNA) selection, peptide bond formation, and mRNA–tRNA translocation (Fig. 1). This process requires a fine balance between rate and fidelity (Johansson et al. 2008; Zaher & Green, 2009). It has been estimated that proteins are synthesised in vivo at a rate of 15–20 amino acids per second, with an estimated error rate of below ~10−4 (Bouadloun et al. 1983). The ribosome achieves this balance by working in coordination with EF-Tu and EF-G during the aa-tRNA delivery and tRNA translocation steps, respectively.
Fig. 1.
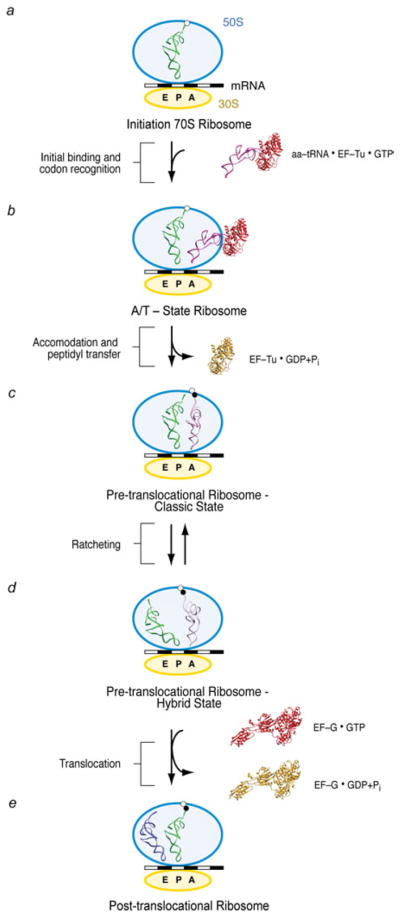
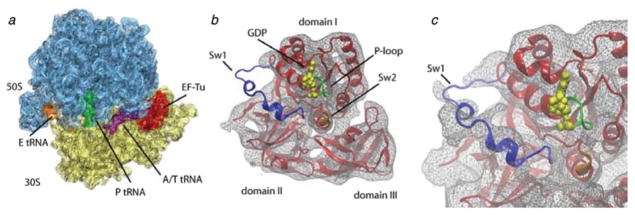
General overview of the protein elongation cycle. Once the initiation ribosome bearing an fMet-tRNAfMet in the P site is formed (a), the first aa-tRNA is incorporated (b). The cognate codon–anticodon match triggers GTP hydrolysis on EF-Tu and EF-Tu release. The accommodation of the aa-tRNA (c) is immediately followed by peptide bond transfer. At this stage, the pre-translocational ribosome samples the hybrid A/P and P/E states, a reconfiguration coupled to the ratchet motion of the 30S subunit (d). In the last step of the protein elongation cycle, the GTPase EF-G catalyzes the complete translocation of the mRNA–(tRNA)2 complex (e), freeing up the A site for the next aa-tRNA.
The ribosome is formed by a small and a large subunit, referred to as 30S and 50S, respectively, in prokaryotes (40S and 60S in eukaryotes). Despite this difference in size, the centrally located functional sites of the ribosome are highly conserved in all domains of life, as the structural variations (due to the presence of additional proteins and the so-called RNA expansion segments) are predominantly located at the periphery of the ribosome. While the small subunit is responsible for the decoding of the mRNA, monitoring the codon–anticodon interaction in the aminoacyl (A) site, the large subunit provides the proper environment (located at the peptidyl transferase center, or PTC) for the transfer of the polypeptide from the P-site tRNA to the aminoacyl group on the A-site tRNA. The mRNA is positioned around the neck of the small subunit such that it displays a 135° kink between adjacent codons (Yusupova et al. 2001), a conformation that enables the simultaneous base-pairing with the tRNAs in the A and P sites.
The following account gives a brief overview of the main steps in the course of the elongation cycle on which this article is focused (see Noller et al. 2002; Ogle & Ramakrishnan, 2005; Ninio, 2006; Frank et al. 2007; Korostelev et al. 2008; Steitz, 2008). In the initiation of protein synthesis, the ribosomal subunits, mRNA, and the initiator tRNA (fMet-tRNAfMet) are brought together, guided by initiation factors, to form an elongation-competent ribosome. Once this initiation complex bearing an fMet-tRNAfMet in the P site is formed (Fig. 1a), the first elongation cycle starts with the incorporation of the first aa-tRNA into the ribosome (Fig. 1b). In the initial binding state, referred to as A/T state, this aa-tRNA is in a ternary complex with the GTPase EF-Tu (eEF1A in eukaryotes) and GTP. When a Watson–Crick codon–anticodon match is recognized by the ribosome, a signal is transmitted to EF-Tu that triggers GTP hydrolysis and thereby causes the dissociation of EF-Tu from the ribosome. The subsequent accommodation of the 3′ acceptor arm of the tRNA in the PTC of the large subunit leads to a rapid peptide bond transfer (Fig. 1c).
After peptide bond formation, the resulting pre-translocational ribosome contains a peptidyl-tRNA in the A site and a deacylated tRNA in the P site. At this stage, the A- and P-site tRNAs sample hybrid configurations, referred to as A/P and P/E, respectively, in which the anticodon stem loops (ASLs) of the tRNAs reside in the A and P sites of the small subunit, whereas the acceptor ends interact with the P and E sites of the large subunit (Fig. 1d). The transition from the classic to the hybrid state of the ribosome (i.e., Fig. 1c, d) is accompanied by a large conformational change, the ratchet-like rotation. Binding of the GTPase EF-G (eEF2 in eukaryotes) catalyzes the translocation of the mRNA–tRNA complex (Fig. 1e). The concerted, precise movement of tRNA and mRNA is essential for translation, as any error in this process would result in the loss of the reading frame. All three steps of the elongation cycle (decoding, peptide bond transfer, and translocation) are repeated until the complete mRNA sequence has been read and a stop codon marking termination is sensed by the small subunit’s decoding center. A comparison of the positions and orientations of the tRNAs in the course of the elongation cycle, as determined from cryo-EM density maps by fitting of the X-ray structure, is shown in Fig. 2.
Fig. 2.
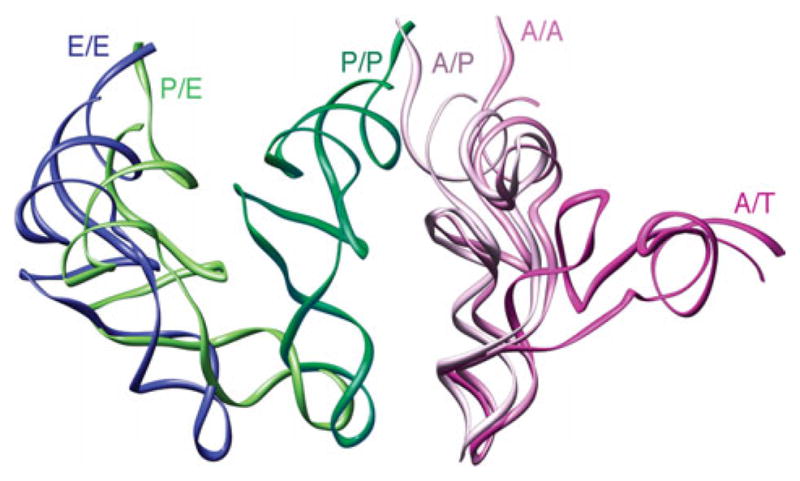
Comparison between the positions and orientations of the tRNAs in the ribosome at different sites. The tRNA positions and conformations (displayed in ribbons) were obtained by fitting of the experimental cryo-EM densities with the X-ray structure of the tRNAs by real-space refinement.
This review will specifically focus on what we have learned thus far about the dynamic interactions among the ribosome, tRNA, and elongation factors in the course of the elongation cycle, especially by means of cryo-EM, but also by another important emerging source of dynamic information, smFRET. Questions about the detailed mechanism of decoding succinctly formulated by Ogle and Ramakrishnan (2005), although still unresolved, appear in a new light thanks to new high-resolution cryo-EM studies (Lebarron et al. 2008; Li et al. 2008; Schuette et al. 2009; Villa et al. 2009a). Advances in the understanding of mRNA–tRNA translocation have been particularly dramatic, showing a remarkable convergence of data from smFRET with dynamic changes visualized by cryo-EM (Agirrezabala et al. 2008; Cornish et al. 2008; Frank et al. 2007; Julian et al. 2008; Kim et al. 2007; see Korostelev et al. 2008).
2. The decoding process: aa-tRNA selection and the A/T state
For the maintenance of fidelity in the decoding of the aa-tRNA, the cognate codon has to be recognized correctly by the anticodon, leading to the incorporation of the tRNA, while incorrect pairings must lead to rejection. The structural basis for decoding is the recognition of the complementary Watson–Crick base-pairing between the codon of the mRNA and the anticodon of the tRNA, through the mechanism described by Ramakrishnan and coworkers (Ogle et al. 2001). The discrimination between cognate and noncognate aa-tRNAs (i.e., more than two mismatches with bases in the codon) can rely on the difference in stability of the codon–anticodon base-pairing as the difference in free energy of binding is sufficiently large. However, the difference in thermodynamic stability between cognate and near-cognate (i.e., just one mismatch with bases in the codon) is too small to be able to account for the accuracy of decoding (Sugimoto et al. 1986; Freier et al. 1986; Meroueh & Chow, 1999; Almlof et al. 2007).
To explain this paradox, a kinetically driven discrimination mechanism has been postulated to be involved in tRNA selection. Accordingly, the recognition of a cognate aa-tRNA and its incorporation into the ribosome is achieved not in one but two steps, initial selection and proofreading, separated by an irreversible step of GTP hydrolysis (Fig. 3) (Hopfield, 1974; Ninio, 1975; Thompson & Stone, 1979; Ruusala et al. 1982). A two-step selection mechanism allows the ribosome to sample the energy landscape twice, resulting in enhanced selectivity.
Fig. 3.
Kinetic scheme for aa-tRNA incorporation. Initial selection starts with a codon-independent, reversible rapid initial binding of the ternary complex (k1, k−1). During codon recognition, the codon–anticodon interaction is still labile and reversible (k2, k−2). Once the cognate codon–anticodon match is recognized, the ternary complex is stabilized and a signal transmitted to GTPase EF-Tu calling for activation (k3) and GTP hydrolysis (k4=kGTP). In the reversible, short life-time, pseudo-activated GTPase state, the cognate ternary complex establishes initial unsuccessful contacts with the ribosome (k3′, k−3′). Once GTP is hydrolyzed, the release of Pi and subsequent conformational change of EF-Tu leads to the dissociation of the factor from the ribosome and the accommodation of the aminoacyl end of the tRNA into the A site of the 50S subunit (k5). Accommodation is followed by rapid peptide bond transfer (k6=kpep). In case of rejection, the aa-tRNA dissociates from the ribosome (k7). After peptide bond transfer, the pre-translocational ribosome alternates between the classic and hybrid states.
Subsequent research has led to a more refined kinetic model. The high level of accuracy does not originate solely from differences in dissociation rates (k−2 and k7 in our diagram), as originally proposed. Instead, pre-steady-state kinetic studies demonstrated that the discrimination between cognate and near-cognate tRNAs relies mainly on differences between the cognate and near-cognate case in GTPase activation (k3) and tRNA accommodation (k5) rates. The rates for GTPase activation (during initial selection) and accommodation (during proofreading) are much higher for the cognate than for the near-cognate species (Pape et al. 1999; Rodnina & Wintermeyer, 2001; Gromadski & Rodnina, 2004; Cochella & Green, 2005; Gromadski et al. 2006). In summary, such a double-checking mechanism during decoding, in which both the initial selection and proofreading steps contribute to a similar extent to the overall selection accuracy, in conjunction with high kinetic efficiency of the cognate species, ensures the high accuracy in tRNA selection that is essential for the faithful expression of the genetic information.
In structural terms, the differences in forward rates between the cognate and near-cognate case are due to an induced-fit mechanism, i.e., differences in the rate-limiting conformational changes in ribosome and tRNA complex generated by the codon–anticodon match. According to this view (see Ogle & Ramakrishnan, 2005), the structural changes required to allow the final incorporation of aa-tRNA are not taking place unless a cognate aa-tRNA is encountered. Important breakthroughs toward understanding the induced-fit mechanism have recently been made by means of X-ray crystallography and cryo-EM. We will first describe the relevant crystal structures of the 30S subunit from Thermus thermophilus. Next, we will discuss published cryo-EM structures of different 70S ribosome-bound kirromycin-stalled ternary complexes (aa-tRNA•EF-Tu•GTP) for both Escherichia coli and T. thermophilus, which are in the postcodon recognition, GTPase-activated state. We will end this section by presenting a model for the decoding mechanism that attempts to integrate the aforementioned structural data with biochemical, kinetic, and smFRET data.
2.1 X-ray crystallography of the 30S subunit: interactions at the decoding site
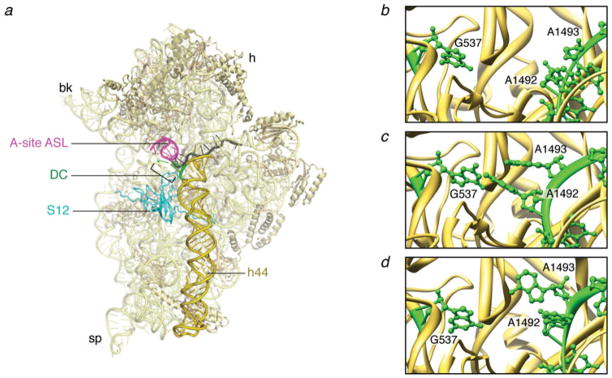
Before the emergence of X-ray data, different experimental approaches already pointed out the importance of three universally conserved bases of the small subunit 16S rRNA, G530, A1492, and A1493, which were found to be involved in the interaction with the incoming aa-tRNA (Bowman et al. 1971; Senior & Holland, 1971; Noller & Chaires, 1972; Moazed & Noller, 1986; Fourmy et al. 1996; Yoshizawa et al. 1999; Noller, 2006). However, a more conclusive understanding regarding the interactions at the decoding site was not achieved until high-resolution X-ray structures of the 30S subunit were obtained (Ogle et al. 2001, 2002). Structures of the 30S subunits obtained from crystals of T. thermophilus soaked with ASLs and mRNA showed that the nucleotides G530, A1492, and A1493 effectively monitor the geometry of the codon–anticodon helix: binding of cognate ASL in the A site induces bases A1492 and A1493 to flip out of the internal loop of helix 44 and base G530 to rotate from a syn to an anti conformation (Fig. 4b, c). These data showed that A-minor contacts are formed between the three bases A1492, A1493, and G530 and the minor groove of the short helix formed by cognate codon–anticodon interaction. These contacts are not stabilized unless the minor groove has Watson–Crick base-pairing geometry.
Fig. 4.
The interactions at the decoding center. (a) Crystal structure of the 30S subunit (PDB code: 2J00). (b) Close-up of the decoding center in the absence of mRNA (PDB code: 1J5E). (c) Close-up of the decoding center in the presence of cognate mRNA (PDB code: 1IBM). (d) Close-up of the decoding center in the presence of a near-cognate mRNA (PDB code: 1N34). The structures are displayed in Ribbons. Labels and landmarks: sp, spur; bk, beak; h, head; S12, protein S12; DC, decoding center; h44, helix 44; A-tRNA/ASL, A-site tRNA ASL.
Unlike the first two base pairs, the geometry of the third (wobble) base pair between the codon and the anticodon was found not to be closely monitored by the ribosome, explaining why noncanonical base pairs are often permitted at the third position. Hand in hand with the action of the monitoring bases goes a more global reconfiguration of head and shoulder domains, referred to as domain closure. During this reconfiguration, the shoulder domain (comprised of S12 and the G530 loop from h18) connects to h44 at the decoding center through the codon–anticodon helix.
The response of the subunit to the binding of a near-cognate ASL is strikingly different from that in the cognate situation (Ogle et al. 2002). Here, bases G530 and A1492 do not change their original positions, and only A1493 is partially flipped out upon near-cognate ASL binding (Fig. 4d). In addition, domain closure is not observed in the near-cognate case.
2.2 Incorporation of the ternary complex to the ribosome and the nature and origin of the distorted A/T tRNA conformation
While crystallography of the 30S subunit thus provided very detailed information on the interactions of mRNA and the ASL of tRNA at the decoding site, the structural dynamics of the aa-tRNA incorporation into the ribosome remained largely obscure. Cryo-EM has provided essential insights into the tRNA selection process, by describing one of the intermediates during this process, the 70S ribosome bound with the cognate Phe-tRNAPhe•EFTu•GDP ternary complex (Stark et al. 2002; Valle et al. 2002, 2003a). Stalling was achieved by using kirromycin, an antibiotic that impedes the selection mechanism immediately following GTP hydrolysis by EF-Tu, but before dissociation of the aminoacylated acceptor end of the tRNA from the protein. Kirromycin binds directly to EF-Tu at the interface of its three domains (Parmeggiani & Nissen, 2006). The ternary complex aa-tRNA•EFTu•GTP with cognate aa-tRNA binds tightly to the ribosome in the presence of the antibiotic, becoming trapped in the so-called kirromycin-stalled A/T state. The analysis of this complex is of particular relevance here because the antibiotic stalls the factor in the GTPase-activated configuration but before the conformational changes leading to EF-Tu release (Rodnina et al. 1995).
The cryo-EM maps (Fig. 5) revealed that the aa-tRNA molecule adopts a bent, twisted conformation when in the A/T site of the ribosome. The location of the kink between the anticodon- and D-stem loop regions was narrowed down to positions 44, 45, and 26 (Valle et al. 2003a). This distortion was suggested to be a key element of the cognate versus near-cognate aa-tRNA selection mechanism, as it could set the threshold for triggering the GTPase activity of EF-Tu and thus hold the key to kinetic discrimination (Yarus et al. 2003).
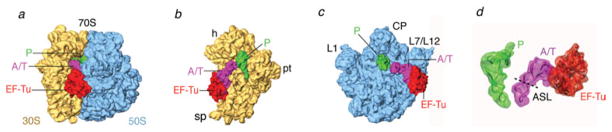
Fig. 5.
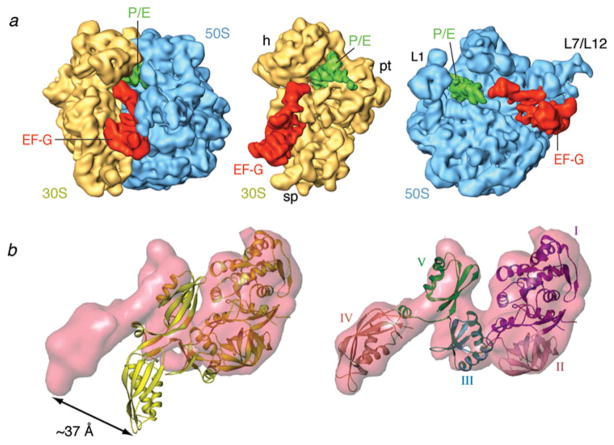
Cryo-EM reconstruction of ribosome bearing mRNA and a cognate ternary complex stalled with kirromycin after GTP hydrolysis. Cryo-EM map of the 70S•fMet-tRNAfMet•Trp-tRNATrp•EF-TuGDP•kir complex (X. Agirrezabala, J. Lei, R. F. Ortiz-Meoz, R. Green, & J. Frank, unpublished results). Cryo-EM densities are shown for the small (yellow) and large (blue) subunits, A- and P-site tRNAs (magenta and green, respectively), and EF-Tu (red). (a) Side view of 70S ribosome, (b) interface view of 30S subunit, (c) interface view of 50S subunit, (d) arrangement of P-site tRNA and ternary complex. Labels and landmarks: sp, spur; pt, platform; h, head; CP, central protuberance; L1, L1 stalk; L7/L12, L7/L12 stalk.
From this analysis, it became evident that Phe-tRNAPhe (and probably all aa-tRNAs; see below), as a flexible molecule with intrinsic instability, is tailored for dynamic interactions with the ribosome from the initial binding, through the selection and proofreading phases, to the final accommodation into the A site. According to the manual expert-guided fitting used by Valle et al. (2003a), later confirmed by molecular dynamics flexible fitting (MDFF) of a higher-resolution map of the same complex (Trabuco et al. 2008; Villa et al. 2009a), accommodation involves the relaxation of the A/T tRNA distortion localized at positions 44, 45, and 26, coupled to a ~45° untwisting rotation around the ASL.
The entire process of cognate aa-tRNA selection can be described in terms of the actions of a molecular spring (Valle et al. 2003a): from a relaxed, low-energy conformation (i.e., before its interaction with the ribosome), to a high-energy conformation (the distorted, A/T conformation), and then back to the relaxed conformation (as it is accommodated into the ribosome’s A site). However, as the cryo-EM ribosome results were from a single type of aa-tRNA (namely Phe-tRNAPhe) in the A/T state, the question remained whether the observed deformation and the implied molecular spring mechanism are a universal feature of aa-tRNAs or merely a feature due to the intrinsic properties of the Phe-tRNAPhe studied.
To address this question, two additional ribosome complexes stalled during the aa-tRNA selection process, bearing Trp-tRNATrp and Leu-tRNALeu, were recently analyzed by cryo-EM (Li et al. 2008). Examination of these new structures strongly suggests that the overall mechanism of initial aa-tRNA selection by the programmed ribosome is the same for all species of aa-tRNA during the selection process, as virtually identical structural perturbations of the tRNA are observed in all three complexes, representing two out of the three existing classes of aa-tRNA. The high preservation of this high-energy conformation is perhaps surprising, considering the differences in sequence, size, amino acid the tRNA is charged with or considering the presence of chemically different nucleoside modifications at homologous positions. Particularly interesting is Leu-tRNALeuI, whose variable stem-loop is unusually long (14 nucleotides) but is apparently not used as a discriminating element in the selection.
The results also show identically positioned anticodons and 3′ CCA-ends in the three tRNAs, and suggested that the slight differences in the conformations of EF-Tu, the GAC, the D and T loops of the aa-tRNAs reflect structural adjustments to ensure that the positioning of anticodons and CCA-ends at the respective functional sites of the ribosome are conserved. Indeed, these small variations may be related to the different kinetic rates obtained for these three distinct aa-tRNAs during the decoding process (Gromadski & Rodnina, 2004; Cochella & Green, 2005; M. Ehrenberg, personal communication).
Using flexible fitting of X-ray coordinates into the cryo-EM maps of the three separate 70S•EF-Tu-tRNA•GTP ternary complexes, Li et al. (2008) further suggested that protein S12 may play a direct role in establishing a communication path between codon–anticodon interaction site on the 30S ribosomal subunit and activation of EF-Tu-dependent GTP hydrolysis on the 50S ribosomal subunit side (Fig. 6). The pivotal role of S12 in tRNA selection was already suggested by Ogle and Ramakrishnan (2005). The resulting atomic models showed that nucleotide 1491 of h44 interacts directly with Lys43 of protein S12 in each of the complexes. In addition, amino acids 78–80 of S12 are in close contact with nt 69 of the aa-tRNA, a position near EF-Tu (Fig. 6a). These common features allowed Li and coworkers (2008) to define a bonding network that connects the anticodon, the two sites of S12, and the CCA arm. The contact of S12 to the CCA arm of the aa-tRNA, which is present in the EF-Tu-bound state but absent after the aa-tRNA has been accommodated into the A site (Fig. 6b), could be essential for the propagation of the signal that is required to be conveyed from the 30S subunit decoding site to the GTPase EF-Tu.
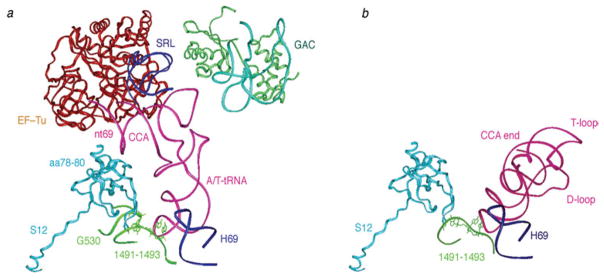
Fig. 6.
Interactions between ribosome and ternary complex and between ribosome and A-site tRNA, inferred by real-space refinement of cryo-EM density maps. (a) Structural elements located around A/T-tRNA in the fitting model of the 70S ribosomal complex bearing Phe-tRNAPhe. tRNA (pink), EF-Tu (red), SRL (blue), L11(green), rRNA in the GAC (light blue), S12 (orange), H69 (dark blue), fragments of h44 and h18 (dark green). (b) Interactions of tRNA in the A site, from X-ray structure (PDB code: 1GIX), with its surrounding elements from X-ray structure (PDB code: 2AVY): S12 (orange); h44 fragment (green); H69 (blue). Protein S12 in PDB files 2AVY and 1GIX were aligned. This figure was altered for the purposes of this article. Data adapted from Li et al. (2008), copyright (2008) Nature Publishing Group.
In two recent articles, Villa et al. (2009a) and Schuette et al. (2009) have confirmed the existence of the aforementioned pronounced conformational distortion for Phe-tRNAPhe bound to the ribosome in the A/T state. While Villa et al., employing a novel MDFF procedure (Trabuco et al. 2008), obtained an atomic model closely resembling the one described by Valle et al. (2003a), already outlined above, Schuette et al. (2009) used rigid-body fitting and obtained a model in which the main conformational change is localized in the elbow region. It is unlikely that these differences originate in the difference in species – E. coli versus T. thermophilus, respectively – since both the sequences and functional states of the ribosomes closely match. Most likely, the difference in tRNA structure inferred is the result of the limited resolution of the maps and the different fitting methods used.
One of the unresolved questions is the origin of the conformational change from the known structure of tRNA within the unbound ternary complex to its structure within the ribosome-bound complex. Valle et al. (2003a) and Frank et al. (2005) proposed that the change in conformation precedes and actually enables probing codon–anticodon interaction and that it is triggered by a contact with the ribosome, while Schuette et al. (2009) postulate that there is no ribosome-induced conformational change before codon–anticodon interaction and that a nearly correct steric engagement in the initial approach might result from conformational fluctuations of the tRNA. Valle et al.’s original proposal, involving helix 69 of the 23S rRNA as pivotal contact, was considerably weakened by the discovery that ribosomes with helix 69 deleted still translate with virtually unchanged fidelity (Ali et al. 2006; R. Green, personal communication). On the other hand, the hypothesis spelled out by Schuette et al. that the pronounced steric difficulties in codon–anticodon interaction might be overcome as a result of chance fluctuations occurring with sufficient frequency commensurate with physiological translation rates, has not been closely examined.
A study addressing this question by MD simulations (Villa et al. 2009b) has now shown that aa-tRNA in the context of the ternary complex has a distinct dynamic behavior with a pronounced mode of bending in the region identified by Valle et al. (2003a), toward a conformation that facilitates codon–anticodon contact. Thus, Schuette et al.’s hypothesis of spontaneous deformation toward a geometry that permits codon–anticodon interaction has gained plausibility, and the present results confirm the localization of the instability around positions 44, 45, and 26, in line with Yarus’ earlier mutation results (Smith & Yarus, 1989a, 1989b; Schultz & Yarus, 1994a, b).
2.3 The mechanism of GTPase activation
The guanine nucleotide binding proteins, or GTPases, are molecular switches that alternate between active and inactive states. Activation involves the exchange of GDP by GTP, a reversible process mediated by nucleotide exchange factors. In the case of EF-Tu (eEF1A in eukaryotes), while EF-Ts (eEF-1Bα in eukaryotes) is responsible for exchanging GDP for GTP, the GTPase activity is controlled by the ribosome. It has been demonstrated that the low intrinsic GTPase activity of EF-Tu is enhanced 105-fold by the ribosome (Pape et al. 1998).
Several structures of EF-Tu have been solved in different binding states by X-ray crystallography (Abel et al. 1996; Berchtold et al. 1993; Kjeldgaard et al. 1993; La Cour et al. 1985; Polekhina et al. 1996; Song et al. 1999), in the presence of different antibiotics (reviewed by Parmeggiani & Nissen, 2006), as well as in complexes with tRNAs (Nissen et al. 1995, 1999; Kristensen, O., Nissen, P. & Nyborg, J., unpublished data; PDB: 1OB2). These structures show that EF-Tu is composed of three domains: nucleotide-binding domain I (GTPase or G domain), which is highly conserved among all GTPases (Bourne et al. 1991; Hilgenfeld, 1995), and domains II and III. While the acceptor arm of the tRNA interacts primarily with domain III, the 3′ CCA-end resides in the crevice between domains I and II. Upon GTP hydrolysis, the major variability in domain I is localized in the two conserved loops, Switch I (residues 40–62, E. coli numbering) and Switch II (residues 80–100). This rearrangement of the switch regions causes a drastic reorientation of domain I with respect to domains II and III in the GDP state, which leads to the disruption of the tRNA binding surface as the affinity to the aa-tRNA is greatly diminished (Dell et al. 1990).
For several GTPases, the mechanism of GTP hydrolysis is well known. In the case of many heterotrimeric GTPases, conserved arginine and glutamine residues which are located, respectively, in the switch regions I and II, are known to stabilize the transition state for the hydrolysis (Vetter & Wittinghofer, 2001). EF-Tu presents the conserved arginine residue (position 58) and a histidine residue (position 84) in a position homologous to that of the catalytic glutamine. Indeed, several studies have brought evidence for the importance of residues Arg58 and His84 in the GTPase activity of EF-Tu (Cool & Parmeggiani, 1991; Knudsen et al. 1995; Scarano et al. 1995; Zeidler et al. 1995; Daviter et al. 2003).
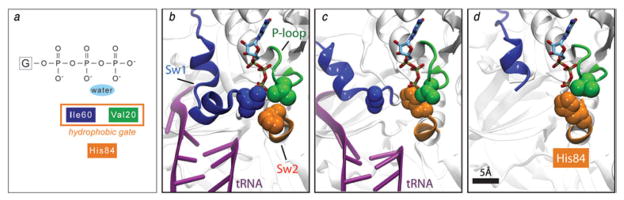
During GTP hydrolysis, the gamma-phosphate is directly attacked by a water molecule. In many GDP- and GTP-bound EF-Tu structures the nucleophilic water is protected by a hydrophobic gate formed by the side-chains of Ile60 and Val20. Neither His84 nor Arg58 is able to interact with the water to stabilize the transition state. In contrast, in one of the structures, the GDP-aurodox-bound form (Vogeley et al. 2001), the hydrophobic gate is open and His84 is oriented toward the nucleotide-binding site, suggesting a mechanism for activation. However, the fact that GTPase activation by aurodox may proceed by a distinct path; that the ternary complex was not bound to the ribosome and that aurodox-promoted GTP hydrolysis rates are relatively low, the significance of these results was limited (Daviter et al. 2003).
Recent results obtained by MDFF (Trabuco et al. 2008) of a high-resolution cryo-EM density map have demonstrated that the changes induced by the ribosome are indeed similar to those described in the aurodox-bound form of free EF-Tu (Villa et al. 2009a). The experimental density map provided sufficient resolution (6·7 Å) to visualize the conformational changes that facilitate the hydrolysis (Fig. 7). The structure shows that the sarcin–ricin loop (SRL; nucleotides 2646–2674 of the 23S rRNA), specifically nucleotide 2660, interacts with His19, thus stabilizing the P-loop in which Val20 is located. The SRL is a highly conserved component of 23S rRNA and is known to be a crucial binding site of both EF-Tu and EF-G (Hausner et al. 1987; Moazed et al. 1998). In contrast, switch I, where Ile60 is located, shows a conformation not observed in any of the crystal structures, and is seen to interact with h8 and h14 of 16S rRNA in the ribosome-bound form. Thus, one of the wings of the hydrophobic gate is indeed pulled away upon binding of the ribosome (Fig. 8). In addition, a conformational change in switch II was discerned, likely originating at the 3′ CCA-end of the tRNA. This change positions His84 closer to the nucleotide, to a final distance of ~4 Å. The repositioning would allow the residue to act as a general base activating the water, thereby triggering the hydrolysis of GTP. Results obtained by Schuette et al. (2009) for T. thermophilus are virtually identical in the regions of the switches and led to the same model of GTPase activation as proposed by Villa et al. (2009a), with the only difference being in the structure inferred for A/T tRNA by fitting, as already mentioned above.
Fig. 7.
GTP hydrolysis on EF-Tu. (a) Cryo-EM map of the 70S•fMet-tRNAfMet•Phe-tRNAPhe•EF-TuGDP•kir complex. Cryo-EM densities are shown for the small (yellow) and large (blue) subunits, A-, P-, and E-site tRNAs (magenta green and orange, respectively), and EF-Tu (red) as transparent surface, along with the atomic model (shown in cartoon representation) obtained by flexible fitting using MDFF. (b) Switch regions in the MDFF-generated atomic model of ribosome-bound EF-Tu: Switch I (Sw1, blue), Switch II (Sw2, orange), and P-loop (green). (c) Close-up of the GTPase domain of EF-Tu. The cryo-EM density map is displayed at a lower threshold compared to that in panel (b) (1·4σ versus 2·2σ) to make the Switch I density visible. Data reproduced from Villa et al. (2009a), copyright (2009) National Academy of Sciences.
Fig. 8.
Hydrophobic gate in EF-Tu. (a) Schematic depiction of the hydrophobic gate formed by residues Ile60 and Val20, which prevents His84 from activating the water molecule and catalyzing the hydrolysis of GTP. (b) Close-up of the gate of the X-ray structure of the ternary complex bound to kirromycin in the presence of GDPNP (PDB code: 1OB2). (c) Close-up of the ribosome-bound ternary complex model obtained through MDFF. The movement of Switch I opens the gate on the Ile60 side; the acceptor stem of the tRNA moves closer to Switch II, and the side-chain of His84 is repositioned toward the nucleotide, resulting in catalysis of GTP hydrolysis. (d) Close-up of the hydrophobic gate in the open position as seen in the crystal structure of the aurodox-bound ternary complex in the presence of GDP (PDB code: 1HA3). The conformation of the gate is very similar to that shown in panel (c). A GTP molecule is shown in all panels for comparison purposes. Data reproduced from Villa et al. (2009a), copyright (2009) National Academy of Sciences.
2.4 The decoding process: an integrated model for aa-tRNA selection
Kinetic (Pape et al. 1998, 1999; Rodnina & Wintermeyer, 2001; Gromadski & Rodnina, 2004; Cochella & Green, 2005; Gromadski et al. 2006; Cochella et al. 2007) and smFRET studies (Blanchard et al. 2004a; Lee et al. 2007; Gonzalez et al. 2007) of the decoding process have defined the following steps in the decoding process: reversible initial binding, codon recognition, GTPase activation, GTP hydrolysis and, finally, accommodation leading to peptidyl transfer.
During the initial binding, the aa-tRNA•EF-Tu•GTP ternary complex binds with the ribo-some, an event apparently mediated by the tentacle-like L7/L12 proteins protruding from the L10 stalk of the large subunit (Diaconu et al. 2005; Helgstrand et al. 2007). In this binding event, the anticodon of the aa-tRNA reversibly interacts with the decoding site in the small subunit for initial codon recognition (Moazed & Noller, 1989a). Kinetic studies have shown that upon this initial binding of the ternary complex to the ribosome, the conformation of the aa-tRNA changes in a characteristic way regardless of whether a cognate or a near-cognate tRNA is encountered (Rodnina et al. 1994). These conformational changes were localized in the D-loop of the tRNA, where the proflavin used to follow the reaction was placed.
The absence of structural data at this point precludes a definite description of this initial conformation, yet it is likely distinct from the conformation observed by cryo-EM for a cognate tRNA bound in the A/T site before (Frank et al. 2005) and after GTP hydrolysis (Valle et al. 2003a; Li et al. 2008; Villa et al. 2009a; Schuette et al. 2009). At this stage, the ternary complex is bound in a labile manner to the ribosome (Rodnina et al. 1996). Upon cognate codon recognition, the conformational distortion of the tRNA is magnified and the ternary complex stabilized on the ribosome. The GTPase-associated center (GAC) of the large subunit moves inward, toward the intersubunit space (Valle et al. 2003a). The incoming tRNA presents the final distorted conformation observed by cryo-EM, a disposition that leads to GTPase activation (Stark et al. 2002; Valle et al. 2002, 2003a; Li et al. 2008).
As the initial binding, codon recognition and GTPase activation are separate steps (Rodnina et al. 1994, 1995, 1996), the deformation seen in the kirromycin-stalled ternary complexes is not likely the one acquired during the initial binding of the ternary complex. As this initial conformation is known to be independent of the cognate or near-cognate nature of the incoming tRNA, it is reasonable to assume that only if the correct decoding interactions are formed (Ogle et al. 2001), the tRNA is stabilized sufficiently to acquire the final distorted A/T conformation observed in the cognate forms. Noncognate ternary complexes are efficiently excluded in this initial selection step, probably because the absence of enough stabilizing contacts in the decoding center makes the transition to the final A/T form highly unfavorable.
Upon successful codon recognition, the interaction between the ternary complex and the ribosome induces conformational changes in both the 30S subunit and the aa-tRNA (induced fit; Pape et al. 1999), thus stabilizing the transition state for the ternary complex. Mutational analysis has shown that these changes in the tRNA and small subunit are partially independent of one another (Cochella et al. 2007). Besides interacting with nucleotides 1492–1493 of h44 and 530 in h18 (Ogle et al. 2001), H69, and GAC, the ternary complex makes contacts with S12 and the SRL in H95. Proper geometry fostering these interactions is ensured by the distorted conformation of the aa-tRNA (Li et al. 2008). Although ternary complexes bound with ribosomes programmed with near-cognate codons have never been visualized to date, smFRET data suggest that the near-cognate binding implies not only a different conformation and positioning of RNA, but also the formation of incorrect contacts between the ternary complex and the ribosome (Blanchard et al. 2004a; Gonzalez et al. 2007; Lee et al. 2007). In fact, the recognition of the cognate tRNA is known to enhance EF-Tu-catalyzed GTP hydrolysis rates by several orders of magnitude compared to the near-cognate substrate (Gromadski & Rodnina, 2004).
During this initial selection, a ternary complex with near-cognate tRNA dissociates from the ribosome more rapidly, and the GTPase activation of EF-Tu occurs more slowly, than for the cognate tRNA. Together, these features ensure a high probability for the acceptance of the cognate, but not a near-cognate, ternary complex. The codon–anticodon match triggers an activation signal that is received by the G domain of EF-Tu, leading to the activated GTPase state of the factor (Rodnina et al. 1995). This postulated signal propagation, from the 30S subunit decoding site to EF-Tu, likely occurs through the tRNA with mediation of protein S12 (Li et al. 2008; Ogle & Ramakrishnan, 2005). The importance of S12 during the decoding process is unequivocal as demonstrated by mutagenesis analysis (Sharma et al. 2007). The distorted A/T disposition described by cryo-EM studies would also play a central role, as this conformation facilitates the simultaneous binding of the anticodon to the decoding center and S12 and the acceptor arm to EF-Tu, GAC, and SRL (Li et al. 2008). Kinetic studies have shown that a mutant tRNATrp form (the Hirsh suppressor mutant, which carries a single substitution, G24A, in its D-loop) results in elevated levels of miscoding by accelerating the GTPase activation and accommodation rates (Cochella & Green, 2005), providing strong evidence for the active role of the tRNA itself in the decoding process. The fact that an intact tRNA is required to trigger the hydrolysis of GTP supports this hypothesis (Piepenburg et al. 2000).
During the next steep, termed proofreading, the inorganic phosphate is released as EF-Tu changes its conformation. In the GTP bound state, EF-Tu shows three compactly arranged domains. Upon signal transmission, conformational changes in the conserved GTPase switch regions trigger the hydrolysis of GTP (Daviter et al. 2003; Schuette et al. 2009; Villa et al. 2009a). The release of the gamma-phosphate leads to a structural rearrangement of the three EF-Tu domains. This step marks the irreversibility of the process. Once EF-Tu has departed from the ribosome, the 3′ CCA-end of the tRNA is accommodated in the A site, leading to rapid peptide transfer.
As in the case of initial selection, an induced-fit mechanism likely promotes the increased accommodation rates observed for the cognate species. Indeed, although constituting kinetically distinct steps, they likely follow the same mechanism. The accommodation movement implicates a rotation of the tRNA body pivoting around the ASL. We argue, much in line with previous suggestions (Ogle & Ramakrishan, 2005), that the more fixed configuration of the cognate distorted ASL due to the correct (i.e., with properly aligned ASL) interactions with the decoding center, coupled with the narrowing of the space available for the tRNA to reach the A site due to the domain closure, sterically facilitates the rotation and movement of the incoming aa-tRNA, and therefore, lead to higher accommodation rates for the cognate species. In contrast, the occasional near-cognate tRNA, which may have “survived” the initial selection, is likely to dissociate from the ribosome due to the partial mismatch that leads to an unstable anchoring of the ASL in the decoding center.
2.5 Peptidyl transfer
As described in the previous section, during the process of tRNA incorporation, the aa-tRNA is brought to the ribosome as part of the ternary complex. At this stage, the anticodon of the incoming aa-tRNA interacts with the codon in the decoding site, and the 3′ CCA-aminoacyl end is bound to EF-Tu. After the conformational change of EF-Tu induced by the hydrolysis of GTP, the factor dissociates from the aminoacyl end of the aa-tRNA and the ribosome. Once EF-Tu is released, the tRNA is free to accommodate in the A site, positioning its 3′ CCA-end (specifically nucleotide C76, covalently bound with the amino acid) in contact with the PTC of the 50S subunit. As soon as the final positioning has taken place, the ribosome catalyzes the peptide bond formation between the α-amino group of the incoming amino acid and the terminal carboxyl group of the peptide chain attached to the P-site tRNA (fMet-tRNA in the first round of the protein elongation cycle). This formation of the peptide bond is known to be very rapid, as it follows the process of accommodation almost instantaneously (Pape et al. 1998; for more details, see Beringer & Rodnina, 2007, and references therein).
Although there is as yet no contribution by cryo-EM due to resolution limitations, structural studies by X-ray crystallography of several species have shed light on the structure of the PTC and the catalytic mechanism of the peptide bond formation on the ribosome (Nissen et al. 2000a; Yusupov et al. 2001; Harms et al. 2001; Hansen et al. 2002; Bashan et al. 2003; Schmeing et al. 2002, 2005a, 2005b; Schuwirth et al. 2005; Selmer et al. 2006; Korostelev et al. 2006). These studies have demonstrated that the PTC is composed of RNA (specifically, domain V of 23S rRNA) and that its central core and active residues are mostly preserved in all kingdoms. In fact, early experiments employing largely protein-free 50S subunits already suggested that the 23S ribosomal RNA is in charge of the peptidyl transferase reaction, evidencing that the ribosome is an RNA-based enzyme (Noller et al. 1992). The role of protein components such as L16 or L27, residing in the vicinity of the PTC and shown to contact the tRNA substrates in T. thermophilus (Selmer et al. 2006; Voorhees et al. 2009) still remains unclear. L27, for example, is known to directly interfere in the peptidyl transferase activity of Escherichia coli ribosomes (Maguire et al. 2005), but the fact that L27 is not conserved in all organism (the archaeon Haloarcula marismortui is an example lacking a homolog of L27) may suggest that it is not part of the evolutionarily conserved machinery. In fact, 50S subunit prereaction state crystal structures from the very Haloarcula marismortui, obtained using substrate and transition state analogs, have served as the basis for the proposal of a model that follows an induced-fit mechanism involving 23S RNA nucleotides 2583–2585 and 2506 (E. coli numbering).
In this model, the repositioning of these nucleotides would make the P-site aa/peptidyl-tRNA C-terminal group accessible to the nucleophilic attack by the aminoacyl group of the incoming A-site aa-tRNA (Schmeing et al. 2005b). Recent X-ray analysis have demonstrated that the PTC is indeed very similar in the 50S subunit and in the intact 70S ribosome (Simonovic & Steitz, 2008; Voorhees et al. 2009), confirming the relevance of the results and conclusions obtained with isolated large subunits (Nissen et al. 2000a; Schmeing et al. 2002, 2005a, 2005b) and supporting kinetic studies that showed that the 50S subunit has the intrinsic potential of catalyzing the peptide bond at equal rates without any requirement from the small subunit when a full-length tRNA is used as the P-site substrate (Wohlgemuth et al. 2006).
3. The process of mRNA–tRNA translocation
3.1 EF-G-mediated translocation: movement of tRNAs from A and P to P and E sites
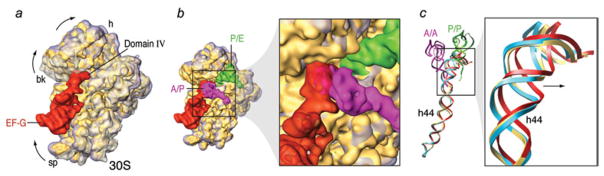
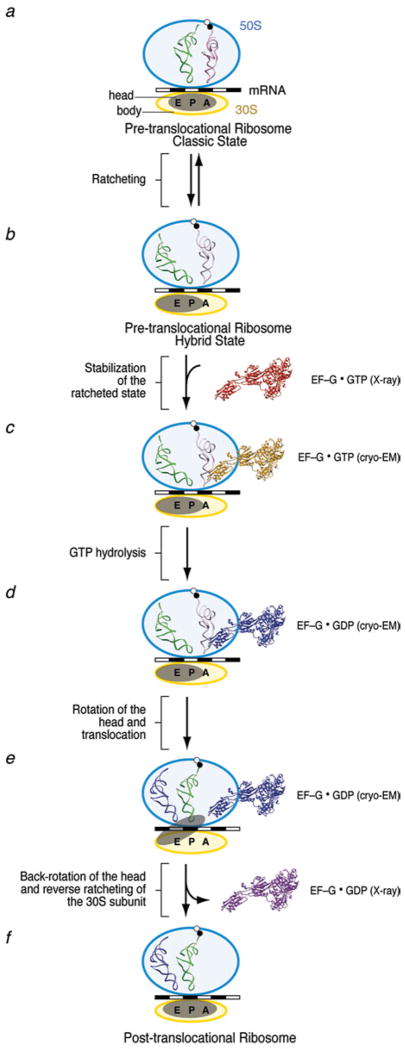
As soon as the incoming aa-tRNA has been accommodated and the large subunit has catalyzed peptide bond formation, the translocation of the mRNA–(tRNA)2 complex from the A and P sites to the P and E sites, respectively, must occur such that the A site is vacated for another aa-tRNA to be incorporated. Translocation normally requires the action of the GTPase EF-G (or eEF2 in eukaryotes). However, the finding that under certain experimental conditions, translocation can occur spontaneously (i.e., in the absence of the factor), albeit with much reduced rate (Pestka, 1968, 1969; Gavrilova & Spirin, 1971) suggested that the capacity for this essential process is intrinsic to the ribosome, closely tied to its architecture. Early on Spirin (1968) speculated, on the basis of the composition of the ribosome from two subunits, that translocation might entail a relative movement of the subunits. Such a movement was indeed observed by cryo-EM (Frank & Agrawal, 2000; Valle et al. 2003b; Fig. 9), a discovery that spawned new avenues of research and motivated numerous studies since. Due to the absence of any X-ray structure of complexes formed by the ribosome and EF-G, the bulk of what we know now about the structural underpinnings of the translocation process has come from cryo-EM (with the help of interpretations requiring the X-ray data), smFRET, cross-linking, modification, and protection assays. In the course of these studies, our understanding of the role of EF-G and GTP hydrolysis has been thoroughly revised. The cryo-EM studies (Agrawal et al. 1998, 1999; Frank & Agrawal, 2000; Stark et al. 2000) showed EF-G interacting with both subunits in multiple contacts. Its position on the ribosome and the constellation of the binding sites were strikingly similar to those earlier obtained for the ternary complex, underlining Nissen’s molecular mimicry paradigm that was based on a comparison of X-ray structures of the unbound molecules (Nissen et al. 2000b). EF-G in its GDP form was found to mimic EF-Tu in its GTP form. Following this paradigm, domains III, IV, and V jointly mimic the tRNA; specifically, domain IV of EF-G takes the place of the anticodon arm. Indeed, the tip of domain IV of EF-G is seen in the cryo-EM map to reach into the decoding center where it occupies the place of the anticodon. In a complex stalled before GTP hydrolysis by adding GDPNP, or after GTP hydrolysis by fusidic acid, the interaction of EF-G with the ribosome was seen to result in large conformational changes both in the ribosome and in EF-G (Agrawal et al. 1998; Stark et al. 2000; Frank & Agrawal, 2000). The bound EF-G exhibited a conformation quite unlike the X-ray structures of EF-G in the GDP or GTP states, which in turn differed from each other insignificantly (discussed in Section 3.5).
Fig. 9.
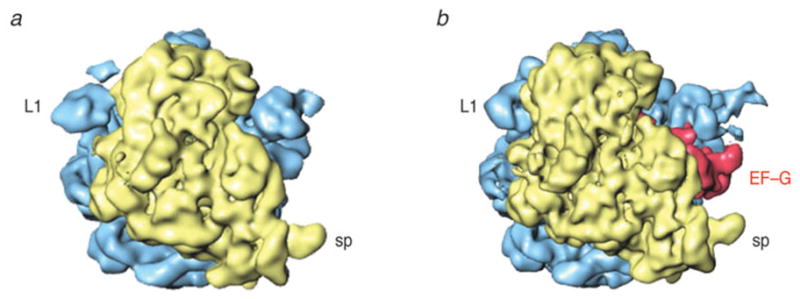
Ratchet motion of the E. coli ribosome observed with binding of EF-G. (a) Normal (MSI) conformation; (b) Conformation (MSII) after ratchet-like rotation of the 30S subunit, with EF-G (red) bound. Labels and landmarks: sp (spur), L1 (L1 stalk). This figure was altered for the purposes of this article. Data adapted from Valle et al. (2003b), copyright (2003) Cell Press.
The finding that even the binding of nonhydrolyzable GTP analogs (e.g., GDPNP) promotes translocation led to the idea that EF-G presents its active form when bound to GTP, and in its inactive form upon GTP hydrolysis (Kaziro, 1978). In this scenario, GTP hydrolysis would only be required for EF-G’s “recycling,” generating conformational changes in EF-G that would result in a reduction of affinity to the ribosome and subsequent dissociation. However, later fast kinetic experiments challenged these proposals, as they demonstrated that GTP hydrolysis actually precedes mRNA–(tRNA)2 translocation and, in addition, causes conformational changes in the ribosome itself that precede – and limit the rates of – subsequent translocation and Pi release from EF-G–GDP-Pi (Rodnina et al. 1997; Peske et al. 2000; Savelsbergh et al. 2003; Wilden et al. 2006). In the experiment by Savelsbergh et al. (2003), for instance, four different fluorescence techniques were used to monitor fluorescence changes produced by GTP hydrolysis on EF-G, by Pi release, by proflavin attached to the elbow of tRNA, and by fluorescin attached to mRNA during translocation.
The confusing picture presented by these results can be reconciled by looking at translocation as a process involving two major motions (Frank & Agrawal, 2000; Frank et al. 2007; Taylor et al. 2007): one is the translocation of the mRNA–(tRNA)2 relative to the large subunit, the other the translocation of the mRNA–tRNAs relative to the small subunit. The first motion is linked to the counterclockwise “ratchet” motion of the small subunit with respect to the large subunit, while the second is linked to the reversal of that motion, back into the original position. In this second step – aided by the movement of the small subunit head to be detailed below – the mRNA moves up by one codon, essentially keeping its place in a coordinate system affixed to the large subunit. Thus, since the reverse rotation of the small subunit follows GTP hydrolysis, translocation of mRNA with respect to the small subunit, which is the action actually measured in the fluorescence quenching of a probe attached to mRNA, also follows GTP hydrolysis. In contrast, tRNA movement on the large subunit is prompted by the ratchet motion, which itself precedes GTP hydrolysis as it is triggered by the binding of EF-G even in the absence of GTP, or even occurs spontaneously under certain conditions.
3.2 The hybrid state of tRNA binding, and its relationship to ribosome ratcheting
Translocation cannot be understood without taking account of the so-called hybrid state of the ribosome. Following transfer of the peptide bond, the ribosome is in the pre-translocational state, where it contains a deacylated tRNA in the P site and a peptidyl-tRNA in the A site. Chemical protection experiments demonstrated that after peptidyl transfer, the interactions between ribosome and acceptor stems of the A- and P-site tRNAs are remodeled to form the hybrid state of tRNA binding (Moazed & Noller, 1989b), confirming early suggestions by Bretscher (1968). In this new configuration, the ASLs of the tRNAs reside in the A and P sites of the small subunit, whereas the acceptor ends interact with the P and E sites of the large subunit. Moazed and Noller already observed that the hybrid state may be sampled in the absence of EF-G when the Mg2+ concentration is sufficiently low, a fact whose relevance has come to be appreciated in recent studies.
The understanding of the hybrid state and its structural basis has been significantly advanced through a combination of studies by cryo-EM, X-ray crystallography, and, more recently, smFRET. As pointed out before, cryo-EM brought first evidence of a large conformational change of the ribosome accompanying translocation (Frank & Agrawal, 2000; Stark et al. 2000), termed ratchet-like motion. Observed when EF-G was bound either in the presence of fusidic acid or GDPNP, the small subunit is seen to rotate with respect to the large subunit in a counterclockwise direction. Initially spelled out as a hypothesis (Frank & Agrawal, 2000), there has been increasing evidence indicating that the transition from the classic to the hybrid state and the ratchet motion are coupled. Implied in the observation of the ratchet motion is the idea that the reverse, clockwise motion of the small subunit will contribute toward the translocation of mRNA–(tRNA)2 relative to the decoding center, which brings the ribosome into the post-translocational state, with the next codon exposed at the A site (Frank & Agrawal, 2000).
For the purpose of this review, it is advantageous to use terminology that was introduced after the discovery of the ratchet motion: we distinguish two macrostates (or global states; see Fei et al. 2008) of the ribosome in which it has pronouncedly different conformations: Macrostate I (MAI; nonratcheted, classic) and II (MSII; ratcheted, a precondition for the hybrid state). Thus, we will use the terms ratchet motion and MSI→MSII transition or, respectively, reverse ratchet motion and MSII→MSI transition interchangedly.
Valle et al. (2003b) observed that as a rule, the transition MSI→MSII is never observed unless the P-site tRNA is deacylated. This observation is concordant with a result of Zavialov and Ehrenberg (2003) that the acylation state of P-site tRNA controls the GTPase activity of EF-G. Since during elongation a deacylated P-site tRNA occurs ater peptide bond formation and is a characteristic of the pre-translocational ribosome, this rule reflects the existence of a locking mechanism whose rationale, in the framework of the elongation cycle, is to ensure maintenance of MSI, the conformation favouring classic tRNA binding, unless a vital condition for translocation has been met, namely that the P-site tRNA is deacylated and the peptide is linked to the A-site tRNA. Upon unlocking, the state MSII can be visited, a state that apparently creates an environment favouring the hybrid A/P and P/E positions of tRNA.
Thus, we need to see unlocking as a necessary, but not sufficient condition for the ribosome to go from MSI to into a stable MSII configuration – for this stabilization, the action of EF-G is needed under the conditions of Valle et al.’s (2003b) experiments. A tRNA in the hybrid P/E position was first visualized by cryo-EM in EF-G-bound ribosome complexes with a single deacylated tRNA in the P site, in the presence of GDPNP (Valle et al. 2003b). The interpretation of this finding was initially that EF-G is required to catalyze the MSI→MSII transition, a prerequisite for the transition of tRNA to the A/P and P/E hybrid positions, until several lines of study indicated that after peptidyl transfer, in the absence of EF-G, the ribosome fluctuates between the classic and hybrid configurations. (As we will see, an even more radical departure from the conventional picture occurred later on, relegating EF-G’s role from a catalyst of the MSI→MSII transition to a mere ligand whose true substrate is MSII.)
It must be pointed out that the evidence from the smFRET studies (Blanchard et al. 2004b; Kim et al. 2007; Fei et al. 2008) for fluctuations from classic to hybrid positions of the tRNAs is indirect since it is based on the measurement of a distance between fluorophores placed on two moving components – the elbows of tRNAs in the A and P site, or, in the case of Fei et al. on L1 and the P-site tRNA – without a reference probe on the body of the ribosome itself. Still, structural data from X-ray crystallography and cryo-EM did lend plausibility to their interpretations.
Independently, there were measurements reflecting on the transition MSI→MSII of the ribosome itself. In an elegant experiment, Ermolenko et al. (2007a) placed a donor on the large subunit and two acceptors on adjacent large-subunit proteins and showed by bulk FRET measurements that the ribosome fluctuates between MSI and MSII in the course of elongation in the absence of EF-G. Subsequent smFRET studies by Cornish et al. (2008) and Marshall et al. (2008) confirmed these findings, and allowed a more detailed characterization of lifetimes in the two states. Conclusive evidence of the link between the structural MSI→MSII transition and translocation came from Noller’s group. Translocation, it was found, cannot take place if ribosomal proteins belonging to the two subunits and facing each other across the intersubunit space are cross-linked; in other words, unless they are free to rotate (Horan & Noller, 2007).
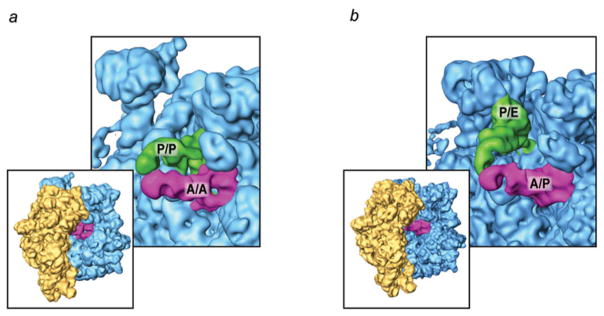
The study by Kim et al. (2007) brought in a new aspect – the magnesium ion dependence of observed interconversions between classic and hybrid states of the pre-translocational ribosome in the absence of EF-G – which put new light on earlier results obtained by Moazed and Noller (1989b) and cryo-EM (Valle et al. 2003b). High Mg2+ concentrations, typically used in cryo-EM and X-ray crystallography, was seen to shift the equilibrium to the classical state, while low Mg2+ (<5 mM), closer to physiological levels, shifted it toward the hybrid state. Following this lead, Agirrezabala et al. (2008) studied a pre-translocational complex at 3·5 mM Mg2+ with cryo-EM and found two subpopulations, representing classic (i.e., MSI with tRNAs in classic positions) and hybrid state (i.e., MSII with tRNAs in hybrid position) in a pure form (Fig. 10). Failure to find a population in any other conformation indicated that the lifetime of the two macrostate conformations would dwarf the lifetime of any transition states, if present. Similar results were obtained by Julian et al. (2008).
Fig. 10.
Spontaneous ratchet motion: classic and hybrid ribosome configurations. (a) Classic A/A and P/P tRNA positions, with ribosome in MSI conformation; (b) hybrid A/P and P/E tRNA positions, with ribosome in MSII conformation.
Interestingly, Agirrezabala et al. (2008) and Julian et al. (2008) also found that the E site was empty in the MSI complex, in contrast to earlier results (Valle et al. 2003b) in which it was seen occupied by a tRNA without a round of translocation; i.e., apparently by a tRNA diffusing in “backward” from the solvent. This comparison suggested an explanation for the Mg2+ dependence of the MSI→MSII transition, and – by implication – of the classic hybrid transition, as a result of the Mg2+ dependence of promiscuous E-site tRNA binding, which could obstruct the efficient transition of the P-site tRNA to the P/E configuration (Agirrezabala et al. 2008). If the explanation were correct, it would imply that the reconfiguration of the ribosome–tRNA system from the classic to the hybrid state, along with the ratchet motion, is affected by the presence of the E-site tRNA as was previously suggested in the case of A-site tRNA binding (Rheinberger & Nierhaus, 1986).
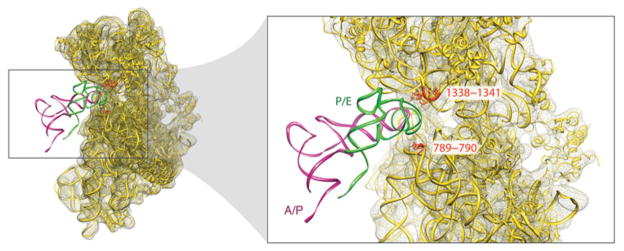
3.3 Anatomy of the MSI→MSII transition
Atomic models obtained from the cryo-EM maps by flexible fitting show that the reconfiguration of the tRNAs is influenced by S13, L1, L5, H68, H69, and H38, as these components change their relative positions during the ratcheting (Fig. 11). This reconfiguration would allow different environments “seen” by the tRNA to be created. In the transition from the classic to the hybrid state, the trajectories of the tRNAs originating at the classical A/A and P/P sites are quite different. While the elbow (formed by D and T loops) of the P-site tRNA is displaced by ~40 Å in the P/P to P/E transition, the elbow of the A-site tRNA remains virtually fixed as it goes from A/A to A/P. Thus, with the exception of the CCA-end, the interaction of the tRNA with the ribosome remains basically unaltered between classic A/A and hybrid A/P tRNA configurations. The maps suggest that it is likely the lateral displacement of H69, in conjunction with the conformational change of H38 accompanying the ratcheting of the small subunit, that promotes the pivoting of the A-site tRNA around its anticodon and the reorientation of the elbow region. This rearrangement allows the CCA-end to reach the 50S subunit’s active site in the A/P hybrid configuration, explaining the puromicyn reactivity of the A site-bound peptidyl-tRNA (Semenkov et al. 1992; Sharma et al. 2004). On the other hand, it is clear that the P/E configuration of the tRNA is coupled with the rearrangement of the L1 stalk. Indeed, this interaction is likely to assist the transition. The interaction of L1 protein with the P/E-tRNA in the hybrid state is facilitated by the large displacement of H76 (i.e., the base of the stalk) with respect to the classic state, toward the intersubunit space. The essential role of L1 protein promoting the P/E tRNA configuration is unequivocal, as deletion of L1 has been shown to stabilize the classical state (Munro et al. 2007). Very recently, smFRET experiments using fluorophores attached to L1 and L33 have allowed to identify three different L1 positions, open, half-closed, and fully closed, relative to the body of the large subunit (Cornish et al. 2009). Thus, it is likely that different L1 conformational states are linked to the configuration of the tRNA during its transition from the P to the E site.
Fig. 11.
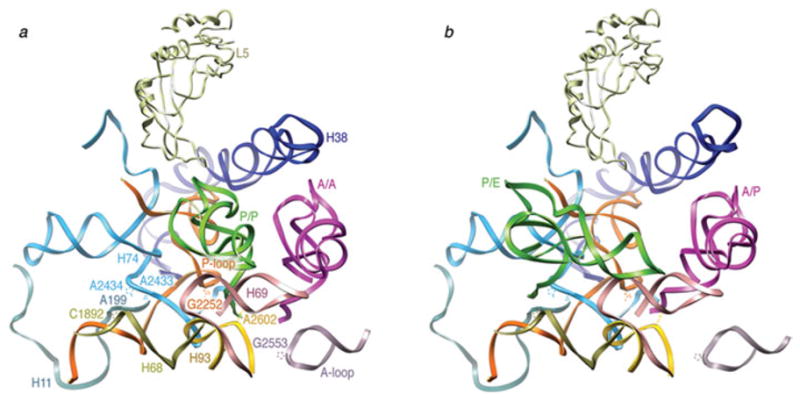
tRNA environment in the large subunit. (a) Close-up of the 50S subunit showing the classic tRNA configuration. (b) Close-up of the 50S subunit showing the hybrid tRNA configuration. Protein L5, as well as 23S rRNA fragments 197–228 (H11), 1189–1905 (H68), 2586–2608 (H93), 2547–2564 (A-loop), 1906–1934 (H69), 2227–2286 (P-loop), and 2372–2451 (H74) are shown. Residues C1892 (H68), A2602 (H93), G2252 (P-loop), A2433 and A2434 (H74), and G2553 (A-loop) are highlighted. The fitting was done by real space refinement. Data reproduced from Agirrezabala et al. (2008), copyright (2008) Cell Press.
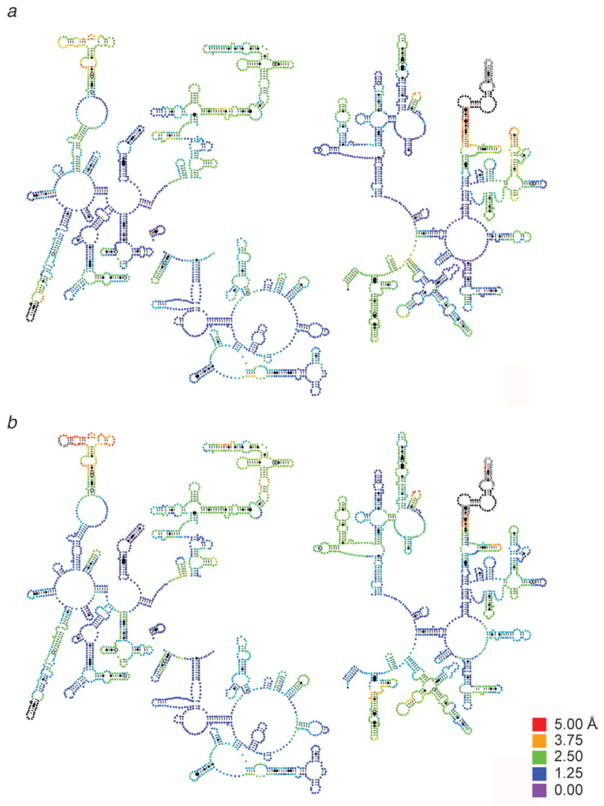
Remarkably, the maps obtained by Agirrezabala et al. (2008) provide structural evidence for the ability of the ribosome to undergo spontaneous factor-free MSI→MSII transition, as an intrinsic characteristic of the pre-translocational ribosome. Another indication of the intrinsic character of this capacity, accumulated over the time of several studies, is the observation of very similar changes in conformation in response to the binding of several ribosomal factors (elongation factor G (Valle et al. 2003b), IF2 (Allen et al. 2005), class II release factor RF3 (Klaholz et al. 2004; Gao et al. 2007), recycling factor RRF (Gao et al. 2005; see compilation in Frank et al. 2007 and gallery in Fig. 12). In Fig. 13, a detailed analysis at molecular level is shown, in which the extent of factor-related ratchet motions seen with the binding of EF-G and RF3 is depicted on the structural models of 16S and 23S rRNA. For further illumination, the displacement of each of the residues was mapped onto the secondary structure of the rRNA using the RNA visualization tool ColoRNA (Lebarron et al. 2007). This analysis indicates that while overall the displacements for 23S RNA are very similar in the cases of EF-G and RF3, there are differences apparently related to specific requirements of the translocation and termination processes (Fig. 14; see Gao et al. 2009 for more details).
Fig. 12.
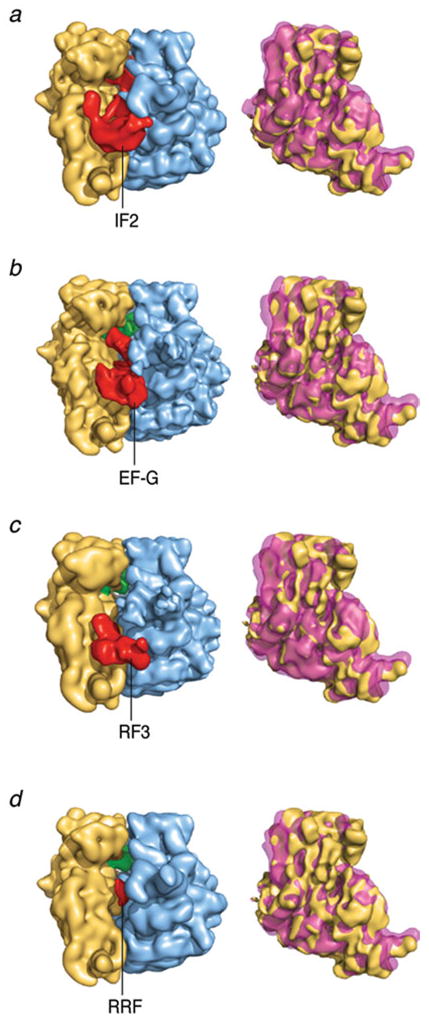
Ratchet motion induced by binding of ribosomal factors. (left) Cryo-EM density maps of 70S ribosome with factors bound; right column: density maps of the 30S subunit overlaid in MSI and MSII positions. This figure was altered for the purposes of this article. Data adapted from Frank et al. (2007), copyright (2007) National Academy of Sciences.
Fig. 13.
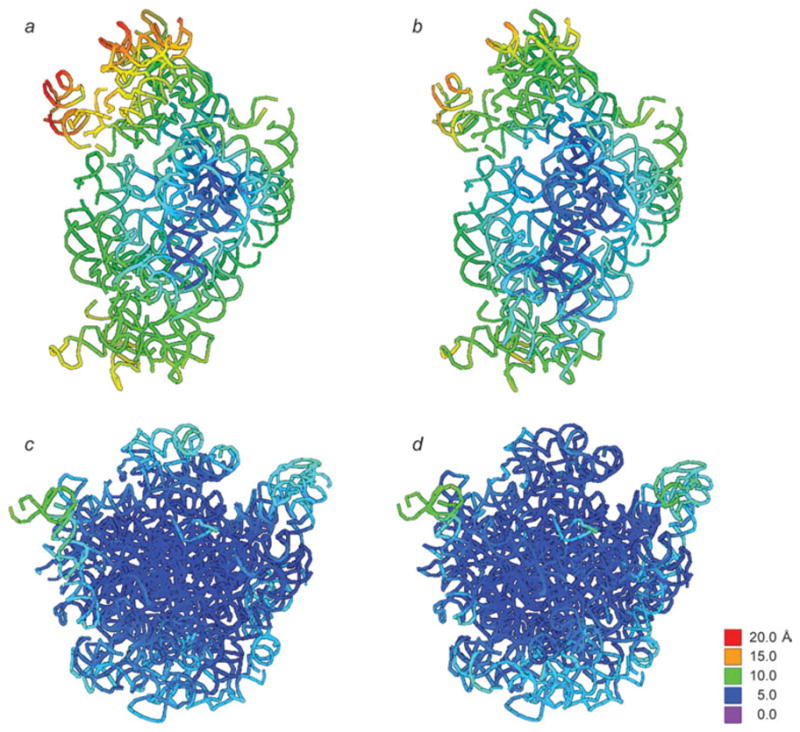
Extent of ratchet-related motions induced by the binding of EF-G and RF3. Color mapping was used to display the factor-induced displacements of rRNA residues on the structure of the unbound ribosome (i.e., MSI). (a, c) 16S and 23S rRNA responding to binding of EF-G; (b, d) 16S and 23S rRNA responding to binding of RF3. Data reproduced from Gao et al. 2009, copyright (2009) Springer Publishing.
Fig. 14.
16S rRNA secondary structure mapping of ratchet-related motions induced by the binding of EF-G and RF3. Displacements of residues upon factor binding are mapped onto the initial (MSI) secondary structure following a color-coding scheme. Displacements above 1·25 Å (blue) are significant. This display focuses on movements up to 5 Å – displacements larger than 5 Å are all displayed in black. Data reproduced from Gao et al. (2009), copyright (2009) Springer Publishing.
The architectural features of the ribosome giving rise to its preference for ratchet-like motions have been summarized earlier (Frank et al. 2007), namely the concentration of stronger rRNA–rRNA intersubunit bridges in the center, versus weak, elastic bridges in the periphery favours a mode of motion that entails a rotation around an axis normal to the plane separating the subunits. Indeed, it has been shown by normal mode analysis of the X-ray structure (Tama et al. 2003; Wang et al. 2004) and of the EM density map itself (Wriggers et al. 2000) that the ratchet-like rotation of the small versus the large subunit (along with the experimentally observed swivelling motion of the L1 stalk, important for P/P→P/E tRNA transport and E-site tRNA ejection) is the ribosome’s favourite mode of internal motion.
3.4 Do intermediate steps exist during the classic-to-hybrid state reconfiguration?
It is well known that specific Watson–Crick pairing interactions between the universally conserved P- and A-loops of 23S rRNA and the 3′ CCA-ends of tRNAs (nucleotides 74 and 75) dictate whether the tRNAs’ CCA-ends are bound to the large subunit’s A or P site. Mutations in the dipeptidyl-tRNA disrupt pairing with the P loop of wt ribosomes, promoting the classic A/A configuration. Similarly, substitutions in the P-loop (nucleotides G2252 and G2251) prevent the wt tRNAs in the classic A/A state from reaching the hybrid A/P configuration. Conversely, mutations in the A-loop (nucleotide G2553) and in the deacylated P-site tRNA promote the hybrid A/P and P/E positions, respectively (Samaha et al. 1995; Kim & Green, 1999; Dorner et al. 2006).
Recently, wild-type as well as these specifically mutated pre-translocational ribosomes have been further characterized using high spatial- and time resolution smFRET techniques (Munro et al. 2007). Remarkably, a sequential translocation trend was deduced from the experiments, as the movement of the deacylated tRNA to its P/E hybrid configuration was observed to be uncoupled from the subsequent movement of the A-site tRNA (schematized in Fig. 15). Indeed, the transition of the deacylated P-site tRNA into the hybrid configuration was shown to be rate-limiting in the equilibrium between classical and hybrid state. In contrast, in the cryo-EM data, a single hybrid-state configuration, characterized as A/P and P/E, was reported (Agirrezabala et al. 2008; Julian et al. 2008). Similarly, smFRET data from other groups (Fei et al. 2008; Cornish et al. 2008) do not indicate the presence of hybrid-state intermediates. Although these results do not exclude the existence of the P/E–A/A intermediate in every case, they do suggest that this intermediate, if it existed, cannot be very stable and that its lifetime must be shorter than the time resolution of some of the experimental approaches.
Fig. 15.
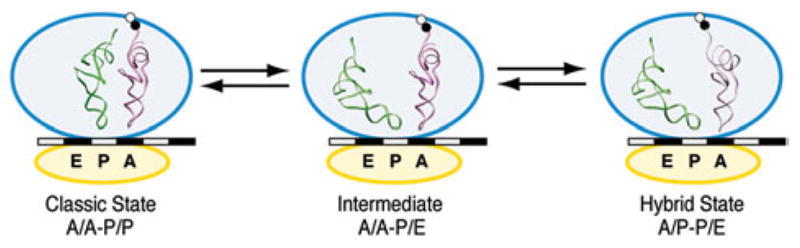
A possible intermediate during tRNA hybrid reconfiguration. Single-molecule FRET data appear to indicate the existence of an A/A–P/E intermediate, which equilibrates with the classic A/A–P/P and the fully hybrid A/P–P/E configuration.
3.5 Conformational changes of EF-G upon its binding to the ribosome
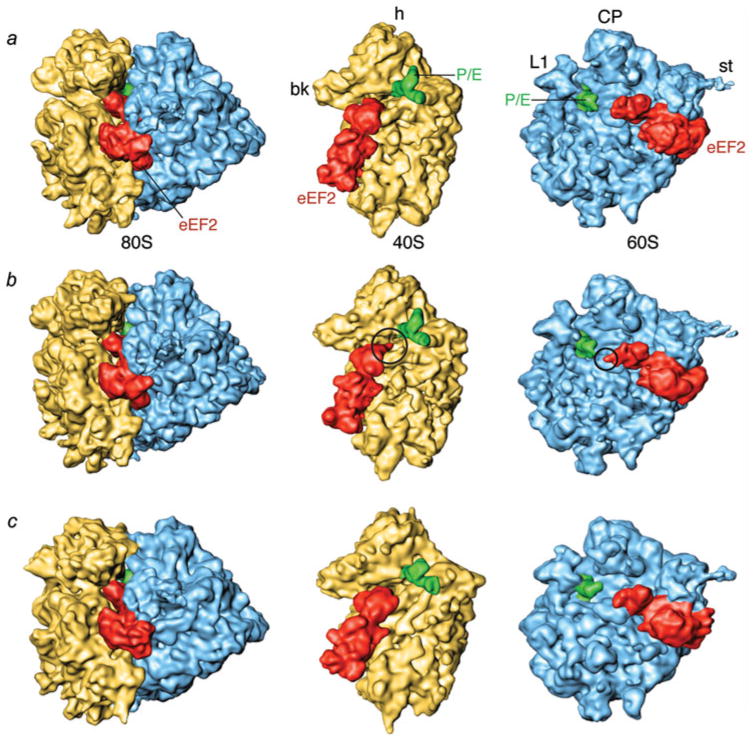
Several crystal structures of EF-G (Aevarsson et al. 1994; Czworkowski et al. 1994; Al-Karadaghi et al. 1996; Hansson et al. 2005), as well as of its eukaryotic counterpart eEF2 (Jorgensen et al. 2003, 2004, 2005) in solution have been solved in different states, showing little structural variations in each of its domains (domains I-V and G′). However, a comparison of these X-ray structures with cryo-EM reconstructions of ribosome-bound EF-G (Agrawal et al. 1998, 1999; Stark et al. 2000; Valle et al. 2003b) and eEF2 forms (Gomez-Lorenzo et al. 2000; Spahn et al. 2004) showed that the binding to the ribosome causes large changes in the factor. A remarkable exception is the crystal structure of EF-G-2, a homolog of EF-G, which was recently found to be crystallized in a conformation related to the ribosome-bound conformation (Connell et al. 2007).
The ribosome-bound factor shows three interaction sites with the large subunit, all located on the base of L7/L12 stalk, and two sites on the small subunit, in the vicinity of S12, as well as the decoding center region, respectively (Fig. 16). The conformational changes upon ribosome binding can be described as joint hinge-like motion of domains III, IV, and V with respect to domains I, II, and G’ (Agrawal et al. 1998; Valle et al. 2003b; Fig. 16b). This rearrangement moves the tip of domain IV by ~40 Å from its original position. Domain IV contacts h44 and the head of the small subunit in the vicinity of the decoding site. In addition to the conformational change of the factor, the reconstructions of EF-G-bound ribosomes demonstrate that the small subunit is ratcheted when the factor is present, and h44 is shifted toward the P site by ~8 Å (van Loock et al. 2000).
Fig. 16.
Cryo-EM reconstruction of EF-GGDPNP bound ribosomes. (a) Cryo-EM densities are shown for the small (yellow) and large (blue) subunits, P/E tRNA (green), and EF-G (red). (b) Manually fitted EF-GGDP structure (PDB code: 1FNM) in the cryo-EM density for EF-GGDPNP (semitransparent red). Relative movements between domains were allowed during the docking procedure. Domains I and II do not require any relative movement compared to their position in the EF-G•GDP crystal structure and they were fitted as one piece. In contrast, domains III, IV, and V shift and rotate jointly, moving the tip of domain IV by approximately 37 Å (see arrow). Labels and landmarks: sp, spur; pt, platform; h, head; CP, central protuberance; L1, L1 stalk; L7/L12, L7/L12 stalk. This figure was altered for the purposes of this article. Data adapted from Valle et al. (2003b), copyright (2003) Cell Press.
Further insight into the translocation has been recently obtained by cryo-EM reconstructions of eEF2 and ADP-ribosylated (ADPR) eEF2 bound to 80S ribosomes in the presence of GDP-sordarin and nonhydrolyzable GTP analog GDPNP (Taylor et al. 2007; Fig. 17). The ADP-ribosylated form was used to amplify and follow the movement of the tip of eEF2’s domain IV and led to the discovery of a further conformational change of domain IV upon GTP hydrolysis. As shown in Fig. 18, domains III and V are seen moving away from switch II in domain I. In addition, domain III rotates by 5° relative to domain I and domains IV and V jointly by 9° relative to domain II.
Fig. 17.
Cryo-EM reconstruction of eEF-2 bound 80S ribosomes. Cryo-EM densities are shown for the small (yellow) and large (blue) subunits, P/E tRNA (green), and eEF2 (red). (a) Cryo-EM map of 80S•eEF2•GDPNP, (b) 80S•ADPR-eEF2•GDPNP, (c) 80S•ADPR-eEF2•GDP•sordarin. Density attributed to the ADPR moiety is circled in (b). Labels and landmarks: bk, beak; h, head; st, stalk; CP, central protuberance; L1, L1 stalk. This figure was altered for the purposes of this article. Data adapted from Taylor et al. (2007), copyright (2007) Nature Publishing Group.
Fig. 18.
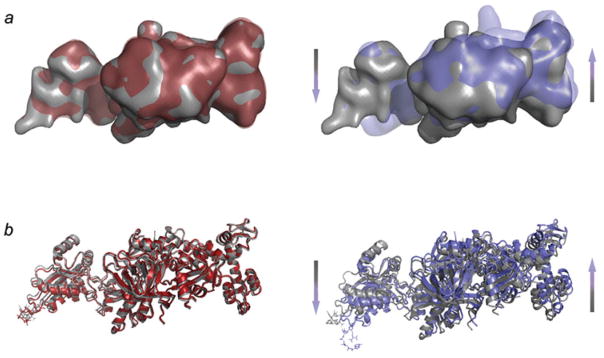
Conformational changes in ADPR-eEF2 as a direct result of GTP hydrolysis. (a) Density attributed to ADPR-eEF2 from the 80S•ADPR-eEF2•GDPNP map (red) is virtually superimposable with that from the 80S•ADPR-eEF2•GDPNP•sordarin map (gray), a map equivalent to the 80S•ADPR-eEF2•GDPNP shown in Fig. 18b (see Taylor et al. 2007 for details). Conversely, superimposition of density attributed to ADPR-eEF2 from the 80S•ADPR-eEF2•GDP•sordarin map (blue) with that from the 80S•ADPR-eEF2•GDPNP•sordarin map (gray) demonstrates significant conformational changes in the factor that are due exclusively to GTP hydrolysis. (b) The atomic models from the 80S•ADPR-eEF2•GDPNP (red) and 80S•ADPReEF2•GDPNP•sordarin (gray) complexes are very similar, whereas a comparison of the 80S•ADPR-eEF2•GDP•sordarin (blue) and 80S•ADPR-eEF2•GDPNP•sordarin (gray) atomic models shows evidence of rearrangements due to GTP hydrolysis. The arrows indicate the direction and magnitude of the conformational changes of the factor. All atomic models in this figure were obtained by flexible fitting using real-space refinement. Data reproduced from Taylor et al. (2007), copyright (2007) Nature Publishing Group.
These rearrangements, which result in a shift of the tip of domain IV by 6 Å toward the decoding center, were suggested to promote disruption of the connection of the codon–anti-codon complex with h44. This event potentially leads to tRNA anticodon movement from the A to the P site of the small subunit. The changes observed in EF-G/eEF2 by means of cryo-EM (ribosome bound forms) and X-ray crystallography (in solution) are summarized in Fig. 19 (see legend for details).
Fig. 19.
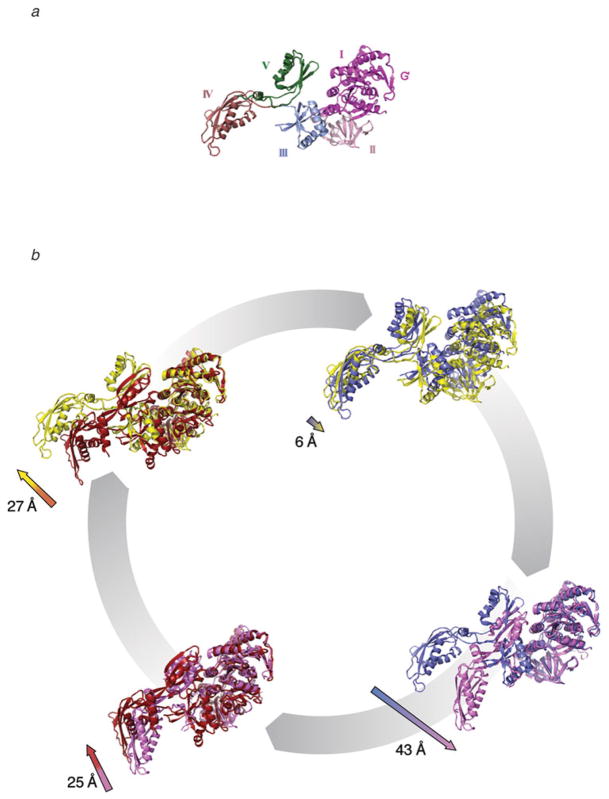
Comparison of structural changes observed in EF-G. (a) The conformational changes observed in EF-G involve a hinge-like movement of the C-terminal domains (III, IV, and V) with respect to the N-terminal domains (I, II, and G′). (b) The conformations observed in EF-G/eEF2 by means of cryo-EM (ribosome-bound forms) and X-ray crystallography (in solution) can be summarized as follows: EF-G in complex with GTP in solution (red), before interacting with the ribosome, as seen in the crystal structure of the EF-G analog EF-G-2 (PDB code: 1WDT). When EF-G•GTP binds to the ribosome, the conformational change of EF-G causes a shift in the tip of domain IV (structure show in yellow). EF-G in the new conformation likely stabilizes the ribosome in the ratcheted conformation. This EF-G•GTP•ribosome conformation was determined by cryo-EM using the nonhydrolyzable GTP analog GDPNP (PDB code: 1PN6). The conformational changes in ribosome-bound EF-G, upon GTP hydrolysis, are likely similar to those described for eEF2•80S complexes in Taylor et al. (2007). A comparison of the complexes before (yellow) and after GTP hydrolysis (blue, PDB code: 2P8Y) reveals small magnitude shifts in domains I, II, and G′ toward the GAC of the ribosome, a reorganization of the Switch I loop, and an ~6-Å shift in domain IV toward the decoding center of the ribosome. This movement is thought to sever the connection between the decoding center in the body of the small subunit and the A-site mRNA–tRNA duplex bound to the head of the small subunit, so that movement of the mRNA–tRNA complex can occur via a head rotation of the small subunit. Conformational changes of EF-G•GDP from the ribosome-bound conformation (blue), determined by cryo-EM (PDB code: 2P8Y), to the solution structure (pink), determined by X-ray crystallography (PDB code: 1EFG), include a movement at the tip of domain IV by ~43 Å. This conformational change in EF-G likely contributes to its dissociation from the ribosome. A comparison of the GDP-bound conformation (pink) of EF-G with the GTP-bound conformation (red) reveals a shift at the tip of domain IV of the factor by ~25 Å. The presence of GTP also reveals an ordered Switch I loop and a different conformation of the Switch II loop. Altogether, these rearrangements likely are the basis of a much higher affinity of the pre-translocational ribosome for the GTP-bound conformation of EF-G. This figure was altered for the purposes of this article. Data adapted from Frank et al. (2007), copyright (2007) National Academy of Sciences.
3.6 Implications of the hybrid configuration for EF-G-based tRNA translocation
Several studies have shown that the hybrid-state configuration of the ribosome makes energetic contributions to EF-G-dependent translocation (Semenkov et al. 2000; Dorner et al. 2006). These experiments demonstrated that the hybrid-state ribosomal complex is the preferred substrate for EF-G, as the maximal rates of translocation were measured in complexes where tRNAs adopt the hybrid configuration. The structural basis for this increased kinetic efficiency was clarified when cryo-EM reconstructions of classic and hybrid-state pre-translocational ribosomes (Agirrezabala et al. 2008) were compared with those of the EF-G•GDPNP-bound ribosome (Valle et al. 2003b).
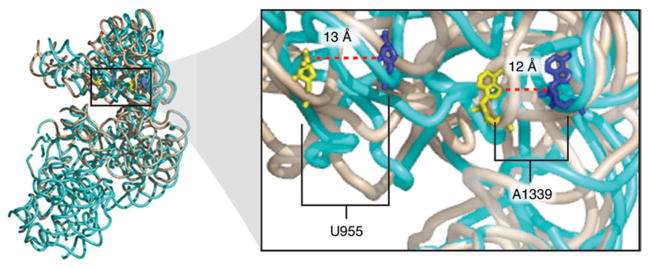
A superimposition of the 30S subunits from the two complexes indicates that the movement of the small subunit in the hybrid-state ribosome goes in the same direction as the one observed in the presence of EF-G (Fig. 20). Although the spontaneous MSI→MSII transition of the ribosome is less extensive than the one finally stabilized by the action of EF-G, it is nevertheless sufficient to position the tRNAs in the hybrid configuration. From the superimposition is also evident that the tip of domain IV of EF-G overlaps with h34 in the classic 30S subunit configuration. As the small subunit is already ratcheted in the hybrid-state ribosome, it no longer presents steric constraints in the vicinity of the 30S head that would interfere with the positioning of EF-G’s domain IV tip in the vicinity of the 30S subunit’s decoding center. Along these lines, FRET data have shown that raising Mg2+ concentrations from 3·5 to 15 mM increases the lifetime of the classic-state fivefold while it leaves the lifetime of the hybrid state unchanged (Kim et al. 2007), so it is tempting to suggest that at physiological conditions (1–2 mM; Cromie et al. 2006), the equilibrium may be shifted well toward the kinetically more efficient hybrid/ratcheted configuration.
Fig. 20.
Kinetic efficiency of the hybrid configuration. (a) Superimposition of aligned 30S subunits from hybrid (solid yellow), classic (transparent gray), and EF-G-bound (transparent purple) ribosomes. The binding position of EF-G (red) is shown for illustration purposes. (b) Close-up of the superimposition of hybrid (solid yellow) and EF-G-bound (transparent purple) ribosomes in the presence of EF-G•GDPNP and hybrid tRNAs. (c) Superimposed atomic models of 16S rRNA h44 for the classic state ribosome (blue), the hybrid state ribosome (yellow) and the EF-GGDPNP bound ribosome (red). The lower portion of h44 has been kept fixed in the fitting to show the displacement of the decoding region (top). The arrow points in the direction of the movement. A/A and P/P tRNAs are shown for clarity. This figure was altered for the purposes of this article. Data adapted from Agirrezabala et al. (2008), copyright (2008) Cell Press.
On the other hand, domain IV of EF-G contacts h44 and the head of the small subunit upon initial binding to the ribosome, shifting the tip of h44 toward the P site by ~8 Å (van Loock et al. 2000). As this motion is caused by the direct action of EF-G and not by the ratcheting of the subunit (Fig. 20c), it is clear that the simultaneous stable binding of EF-G and A/P hybrid tRNA is not sterically feasible.
3.7 Involvement of the head rotation in the translocation process
As discussed above, the binding of EF-G in its GTP form is accompanied by a bending movement of the tip of h44 toward the P site by ~8 Å, coupled with the stabilization of the ratcheted position of the small subunit and concomitant transition to the hybrid tRNA configuration. This movement is required to fully accommodate domain IV of EF-G in the ribosomal A site, but it is not sufficient to complete the translocation. The full translocation step requires a movement of ~20 Å, which is the distance from the small subunit’s A to its P site.
A mechanism by which the remaining distance can be covered has been recently proposed. As described before, by comparing eEF2 bound 80S ribosome structures before and after GTP hydrolysis, it was shown that further changes in domain IV of the factor occur upon hydrolysis (Taylor et al. 2007). These changes were proposed to result in the decoupling of the interaction between the decoding center and the mRNA–tRNA complex in the A-site tRNA (bound to the subunit’s head), allowing a rotation of the head with respect to the body of the small subunit (Spahn et al. 2004). This rotation, which has been shown to go around the neck and in the direction toward the E site, parallels the trajectory of the tRNAs though the ribosome and can account for the 10–12 Å that separates the A and P sites (Fig. 21).
Fig. 21.
Head rotation of the small subunit. Superimposition of atomic models (obtained by real-space refinement) of the bodies of the small subunit of the pre-translocational (tan, PDB code: 1K5X) and the ratcheted, eEF2-bound (blue, PDB code: 1S1H) 80S ribosomes, by explicit least-squares fitting. Highlighted residues U955 and A1339 are in regions known to interact with ribosome-bound A- and P-site tRNAs, respectively. This figure was altered for the purposes of this article. Data adapted from Taylor et al. (2007), copyright (2007) Nature Publishing Group.
The implications of such a rotation of the small subunit head for tRNA movement have been also noted based on the observation of different orientations of the head with respect to the body of the subunit in crystal structures of vacant E. coli 70S ribosomes (Schuwirth et al. 2005). Indeed, a comparison of the X-ray structures shows that while the path for the A to P transition is quite unobstructed, nucleotides G1338 to U1341 and nucleotide A790 located in between the P and E sites at the head and shoulder of the small subunit, respectively, form a gate that alternates between a “close” and “open” position as a function of this head rotation. This path is occluded in the pre-translocational ribosome in the hybrid tRNA configuration (Fig. 22). However, upon factor binding and GTP hydrolysis, the head movement opens the gate, creating the ~26-Å space that is required for the P-site ASL to translocate to the E site (Taylor et al. 2007).
Fig. 22.
The A1338-790 ridge in the hybrid ribosome. The ASL of the hybrid P/E tRNA interacts with the small subunit head, near the G1338-U1341 ridge, as well as with the small subunit body, via the A790 loop of the 18S rRNA. Before the head of the small subunit rotates, the distance between A790 in the body and A1339 in the head of the small subunit is ~18 Å.
3.8 The basis of the translocation of mRNA–(tRNA)2 by the ribosome: a model
The conclusions from structural observations by cryo-EM, in conjunction with previous X-ray, biochemical, kinetic, and FRET data, can be summarized in a three-step model, schematized in Fig. 23. In this model, EF-G is not required to drive the process but rather to make it more efficient, since the labile architecture and dynamic nature of the ribosome and the chemical potential of the aa-tRNAs are likely sufficient to drive the protein elongation forward. GTP hydrolysis is known to precede translocation; however, it is not absolutely essential, as translocation, albeit at low rates, is promoted even by nonhydrolyzable GTP analogs (Belitsina et al. 1975; Inoue-Yukosawa et al. 1974). Indeed, as mentioned before, early experiments already showed that in vitro spontaneous translocation may proceed, although very slowly, in the absence of EF-G and GTP under certain experimental conditions (Pestka, 1968, 1969; Gavrilova & Spirin, 1971), indicating that translocation is a process based on an inherent property of the ribosome.
Fig. 23.
A model for EF-G based translocation. Once the incoming aa-tRNA is incorporated and the peptide transferred, the pre-translocational ribosomes starts fluctuating between (a) the classic (MSI) and (b) the hybrid tRNA configuration (MSII), a rearrangement coupled to the ratcheting of the small subunit. (c) The binding of EF-G stabilizes the ratcheted, MSII conformation of the ribosome. (d) Hydrolysis of GTP promotes the shift of domain IV of EF-G, which in turn detaches the mRNA–tRNA complex from the decoding center and allows rotation of the head. (e) The head rotation translocates the hybrid tRNAs to the P and E sites. (f) Translocation is completed by back-rotation of the head and reverse-ratcheting of the entire small subunit as EF-G in its GDP form is released from the ribosome.
In our model, the first energetic barrier (Fig. 23a, b) corresponds to the reversible ratcheting movement of the 30S subunit and the repositioning of the CCA-ends of both tRNAs from the A and P sites to the P and E sites, respectively, leading to the configuration of the ribosome in its hybrid state (Fig. 23b). It is the unlocking of the ribosome (i.e., the consequence of the deacylation of the P/P-site tRNA; Valle et al. 2003b; Zavialov & Ehrenberg, 2003) that unleashes the inherent capacity of the ribosome to ratchet from MSI to MSII and in this process reconfigures the tRNA positions. Modifications or actions that lead to the hybrid configuration, such as mutations in the ribosome and tRNA (Dorner et al. 2006; Kim & Green, 1999; Samaha et al. 1995) or the binding of some antibiotics such as sparsomycin (Fredrick & Noller, 2003) and viomycin (Ermolenko et al. 2007b; Pan et al. 2007) can be explained as a result of their capacity to lower the first energy barrier.
Structural as well as FRET data have demonstrated that translocation is a spontaneous rearrangement facilitated by the dynamic nature of the ribosome. During the reconfiguration of tRNA, sequential domino-like events are likely to occur, in which the P-site tRNA movement precedes the A-site movement (Munro et al. 2007; Pan et al. 2006, 2007; Walker et al. 2008). The fact that proteins L5 and S13, involved in forming two of the ribosomal intersubunit bridges and known to play an important role in the ratchet-like rearrangement of the ribosome, interact with the P-site tRNA (Gao et al. 2003; Valle et al. 2003b; Selmer et al. 2006; Frank et al. 2007), implicates a close relation between the ratchet motion, the P-site tRNA adopting the hybrid configuration, and an inward movement of the L1 stalk. Experimental evidence for the decoupling between P- and A-site tRNA movements (Munro et al. 2007) and the partial coupling between ratchet motion and assumption of hybrid positions of the tRNAs indicate that the energy barrier has a complex shape with several minima.
Once the P-site tRNA has moved into its hybrid position, and thus removed the steric constraint in the 50S P-site, the A-site tRNA is able to adopt the hybrid configuration, as well. However, in this domino-like sequence of events, the initial move by the P-site tRNA cannot take place unless the E site is empty. The presence of a deacylated tRNA already placed in the E site will increase the initial barrier, as it logically will obstruct an efficient transition of the P-site tRNA to the P/E configuration and the stabilization of the contacts between the 3′ CCA-end and the ribosomal exit site (Lill et al. 1989; Feinberg & Joseph, 2001; Virumae et al. 2002).
The second step in the model is the partial translocation of the mRNA–tRNA complex involving the GTPase EF-G. During its initial binding to the ribosome, EF-G (still in the GTP form) changes its conformation and extends domain IV toward the 30S decoding site (Fig. 23b–c). This rearrangement of EF-G induces the movement of the tip of h44 that displaces the A-site ASL associated to it by approximately 8–10 Å along the translocation trajectory (van Loock et al. 2000). In addition, the ratcheted position (macrostate II) of the small subunit is stabilized as reported by smFRET (Spiegel et al. 2007).
In the third step, EF-G, upon hydrolysis of GTP (Fig. 23c–d), facilitates the translocation of the anticodon part of the tRNA from the A to the P site, coupled with the movement of the mRNA. The anticodon loop of the A-site tRNA is essential for EF-G dependent translocation (Joseph & Noller, 1998). On the other hand, residue His 583 (dipthamide in the eukaryotic case), located at the tip of domain IV, is known to be crucial for maintaining proper rates of translocation (Savelsbergh et al. 2000). Together, these data indicate an intimate interaction between domain IV of EF-G and the ASL of A-site tRNA for translocation. Cryo-EM data suggest (Taylor et al. 2007) that domain IV of eEF2 actually disrupts the binding network between the codon–anticodon moiety and the decoding center region upon GTP hydrolysis, an event that would allow the rotation of the head of the small subunit (Fig. 23d, e). As the head of the small subunit rotates, the distance between A790 in the body and A1339 in the head of the small subunit increases from ~18 to ~26 Å (Taylor et al. 2007), allowing the passage of the ASL from the P to the E site.
In contrast to the P-site hybrid tRNA, the hybrid A/P-site tRNA shows a tighter association with the body of the 30S rather than with the head in the cryo-EM reconstructions of pre-translocational ribosomes (Agirrezabala et al. 2008; Julian et al. 2008). This observation would suggest that the P-site tRNA, through the mRNA, actually pulls the A-site tRNA along with the head rotation. However, early experiments already demonstrated that the underlying mechanism acts directly on both tRNAs, rather than indirectly via mRNA, as EF-G-dependent tRNA translocation was detected even in the absence of mRNA (Belitsina et al. 1981). An interaction between the rotating head and the A-site tRNA would explain these results. Indeed, nucleotides 955 and 956 (h30 of 16S rRNA) in the head of the small subunit (see Fig. 21) are positioned close enough to interact with the hybrid A/P ASL around positions 40–41. This contact has also been observed at atomic resolution in X-ray structures of classic-state pre-translocational ribosomes (Yusupov et al. 2001). If this interpretation is correct, it would explain the link between the rotating head and the A-site tRNA, with immediate consequences for an understanding of previous results regarding the requirements for EF-G-dependent translocation (Joseph & Noller, 1998). These experiments showed that a complete tRNA in the P site and a 15-nucleotide ASL analog are the minimal requirements for translocation. The additional anchor point between the head and ASL in the A site may be necessary to efficiently “drag” the mRNA forward with respect to the ribosome by the space of three nucleotides so that the next codon can be presented in the A site.
The final, necessary disruption of the contact between the head region and tRNAs is likely promoted by a combination of back-ratcheting of the small subunit and back-rotation of the head (Fig. 23e, f) that likely occurs after dissociation of EF-G. Therefore, as the hybrid tRNAs maintain their contacts with the head throughout the rotation, this rearrangement of the head (understood as the rate-limiting conformational change described by kinetic studies), would finally translocate the tRNA–mRNA complex relative to the decoding center by the remaining 10- to 12-Å distance that separates the different ribosomal binding sites (Taylor et al. 2007).
4. Concluding remarks
To infer the mechanism by which translation is accomplished, a detailed three-dimensional knowledge of each of the elements involved in every functional state of the process is required, as well as the way they are dynamically engaged. Notwithstanding the enormous success of X-ray crystallography in the determination of several structures, including high-resolution structures of 70S ribosomes complexed with tRNAs or release factors (Schuwirth et al. 2005; Korostelev et al. 2006; Selmer et al. 2006; Laurberg et al. 2008; Weixlbaumer et al. 2008), the vast majority of the structural knowledge about how the ribosome interacts with translation factors during the protein elongation cycle has been provided by means of cryo-EM. As the density maps improve in resolution – thanks to improved protocols for sample preparation, image acquisition and processing, automated collection of large amounts of data, and to sophisticated methods of fitting atomic structures into the experimental density maps – the description of the molecular events during ribosomal action by cryo-EM is becoming increasingly detailed and accurate. As we have seen, smFRET is capable of adding the time dimension, in a perfect complement to cryo-EM. Fluorescent probes strategically placed on the ribosome and its functional ligands make it possible to observe the stochastic fluctuations between functional states and relate them to a structural frame of reference. These combined data now allow us to integrate genetic and biochemical results collected over half a century of ribosome research into mechanistic models that shed light on the molecular events during protein synthesis.
The results on ribosomal dynamics coming from cryo-EM and smFRET, when interpreted within the framework of existing X-ray data, depict a molecular machine of vast complexity, relying in its function on conformational signaling and allosteric processes that interconnect remote parts of the large structure. Liljas’s (2004) and a recent article by Spirin (in press) contain an appreciation of how far we have advanced in our understanding of the structural aspects of translation, but also of the challenges that lie ahead: how to advance to a full kinetic model of translation that does justice to the thermodynamics of this multistep process.
Acknowledgments
We thank Derek Taylor and Wen Li for discussions of the manuscript and Michael Watters and Lila Iino-Rubenstein for assistance with the preparation of the illustrations. Supported by HHMI and NIH grants R01 GM55440 and R37 GM29169.
References
- Abel KM, Yoder MD, Hilgenfeld R, Jurnak F. An α to β conformational switch in EF-Tu. Structure. 1996;4:1153–1159. doi: 10.1016/s0969-2126(96)00123-2. [DOI] [PubMed] [Google Scholar]
- Aevarsson A, Brazhnikov E, Garber M, Zheltonosava J, Chirgadze Y, Al-karadaghi S, Svensson LA, Liljas A. Three-dimensional structure of the ribosomal translocase: elongation factor G from Thermus thermophilus. EMBO Journal. 1994;13:3669–3677. doi: 10.1002/j.1460-2075.1994.tb06676.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Agirrezabala X, Lei J, Brunelle JL, Ortiz-Meoz RF, Green R, Frank J. Visualization of the hybrid state of binding promoted by spontaneous ratcheting of the ribosome. Molecular Cell. 2008;32:190–197. doi: 10.1016/j.molcel.2008.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Agrawal RK, Penczek P, Grassucci RA, Frank J. Visualization of elongation factor G on the Escherichia coli 70S ribosome: the mechanism of translocation. Proceedings of the National Academy of Sciences USA. 1998;95:6134–6138. doi: 10.1073/pnas.95.11.6134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Agrawal RK, Heagle AB, Penczek P, Grassucci R, Frank J. EF-G-dependent GTP hydrolysis induces translocation accompanied by large conformational changes in the 70S ribosome. Nature Structural Biology. 1999;6:643–647. doi: 10.1038/10695. [DOI] [PubMed] [Google Scholar]
- Ali IK, Lancaster L, Feinberg J, Joseph S, Noller HF. Deletion of a central ribosomal intersubunit RNA bridge. Molecular Cell. 2006;23:865–874. doi: 10.1016/j.molcel.2006.08.011. [DOI] [PubMed] [Google Scholar]
- Al-Karadaghi S, Aevarsson A, Garber M, Zheltonova J, Liljas A. The structure of elongation factor G in complex with GDP: conformational flexibility and nucleotide exchange. Structure. 1996;4:555–565. doi: 10.1016/s0969-2126(96)00061-5. [DOI] [PubMed] [Google Scholar]
- Allen GS, Zavialov A, Gursky R, Ehrenberg M, Frank J. The cryo-EM structure of a translation initiation complex from Escherichia coli. Cell. 2005;121:703–712. doi: 10.1016/j.cell.2005.03.023. [DOI] [PubMed] [Google Scholar]
- Almlof M, Ander M, Aqvist J. Energetics of codon–anticodon recognition on the small ribosomal subunit. Biochemistry. 2007;46:200–209. doi: 10.1021/bi061713i. [DOI] [PubMed] [Google Scholar]
- Bashan A, Agmon I, Zarivach R, Schluenzen F, Harms J, Berisio R, Bartels H, Franceschi F, Auerbach T, Hansen HA, Kossoy E, Kessler M, Yonath A. Structural basis of the ribosomal machinery for peptide bond formation, translocation, and nascent chain progression. Molecular Cell. 2003;11:91–102. doi: 10.1016/s1097-2765(03)00009-1. [DOI] [PubMed] [Google Scholar]
- Belitsina NV, Glukhova MA, Spirin AS. Translocation in ribosomes by attachment-detachment of elongation factor G without GTP cleavage: evidence from a column-bound ribosome system. FEBS Letters. 1975;54:35–38. doi: 10.1016/0014-5793(75)81062-3. [DOI] [PubMed] [Google Scholar]
- Belitsina NV, Tnalina GZ, Spirin AS. Template-free ribosomal synthesis of polylysine from lysyl-tRNA. FEBS Letters. 1981;131:289–292. doi: 10.1016/0014-5793(81)80387-0. [DOI] [PubMed] [Google Scholar]
- Berchtold H, Reshetnikova L, Reiser CO, Schirmer NK, Sprinzl M, Hilgenfeld R. Crystal structure of active elongation factor Tu reveals major domain rearrangements. Nature. 1993;365:126–132. doi: 10.1038/365126a0. [DOI] [PubMed] [Google Scholar]
- Beringer M, Rodnina MV. The ribosomal peptidyl transferase. Molecular Cell. 2007;26:311–321. doi: 10.1016/j.molcel.2007.03.015. [DOI] [PubMed] [Google Scholar]
- Blanchard SC, Gonzalez RL, Kim HD, Chu S, Puglisi JD. tRNA selection and kinetic proofreading in translation. Nature Structural and Molecular Biology. 2004a;11:1008–1014. doi: 10.1038/nsmb831. [DOI] [PubMed] [Google Scholar]
- Blanchard SC, Kim HD, Gonzalez RL, Puglisi JD, Chu S. tRNA dynamics on the ribosome during translation. Proceedings of the National Academy of Sciences USA. 2004b;101:12893–12898. doi: 10.1073/pnas.0403884101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouadloun F, Donner D, Kurland CG. Codon-specific missense errors in vivo. EMBO Journal. 1983;2:1351–1356. doi: 10.1002/j.1460-2075.1983.tb01591.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bourne HR, Sanders DA, Mc Cormick F. The GTPase superfamily: conserved structure and molecular mechanism. Nature. 1991;349:117–127. doi: 10.1038/349117a0. [DOI] [PubMed] [Google Scholar]
- Bowman CM, Dahlberg JE, Ikemura T, Konisky J, Nomura M. Specific inactivation of 16S ribosomal RNA induced by colicin E3 in vivo. Proceedings of the National Academy of Sciences USA. 1971;68:964–968. doi: 10.1073/pnas.68.5.964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bretscher MS. Translocation in protein synthesis: a hybrid structure model. Nature. 1968;218:675–677. doi: 10.1038/218675a0. [DOI] [PubMed] [Google Scholar]
- Cochella L, Green R. An active role for tRNA in decoding beyond codon: anticodon pairing. Science. 2005;308:1178–1180. doi: 10.1126/science.1111408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cochella L, Brunelle JL, Green R. Mutational analysis reveals two independent molecular requirements during transfer RNA selection on the. Nature Structural and Molecular Biology. 2007;14:30–36. doi: 10.1038/nsmb1183. [DOI] [PubMed] [Google Scholar]
- Connell SR, Takemoto C, Wilson DN, Wang H, Muryama K, Terada T, Shirouzu M, Rost M, Schuler M, Giesebrecht J, Dabrowski M, Mielke T, Fucini P, Yokoyama S, Spahn CM. Structural basis for interaction of the ribosome with the switch regions of GTP-bound elongation factors. Molecular Cell. 2007;25:751–764. doi: 10.1016/j.molcel.2007.01.027. [DOI] [PubMed] [Google Scholar]
- Cool RH, Parmeggiani A. Substitution of histidine-84 and the GTPase mechanism of elongation factor Tu. Biochemistry. 1991;30:362–366. doi: 10.1021/bi00216a008. [DOI] [PubMed] [Google Scholar]
- Cornish PV, Ermolenko DN, Noller HF, Ha T. Spontaneous intersubunit rotation in single ribosomes. Molecular Cell. 2008;30:578–588. doi: 10.1016/j.molcel.2008.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cornish PV, Ermolenko DN, Staple DW, Hoang L, Hickerson RP, Noller HF, Ha T. Following movement of the L1 stalk between three functional states in single ribosomes. Proceedings of the National Academy of Sciences USA. 2009;106:2571–2576. doi: 10.1073/pnas.0813180106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cromie MJ, Shi Y, Latifi T, Groisman EA. An RNA sensor for intracellular Mg2+ Cell. 2006;125:71–84. doi: 10.1016/j.cell.2006.01.043. [DOI] [PubMed] [Google Scholar]
- Czworkowski J, Wang J, Steitz TA, Moore PB. The crystal structure of elongation factor G complexed with GDP, at 2·7 Å resolution. EMBO Journal. 1994;13:3661–3668. doi: 10.1002/j.1460-2075.1994.tb06675.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daviter T, Wieden HJ, Rodnina MV. Essential role of histidine 84 in elongation factor Tu for the chemical step of GTP hydrolysis on the ribosome. Journal of Molecular Biology. 2003;332:689–699. doi: 10.1016/s0022-2836(03)00947-1. [DOI] [PubMed] [Google Scholar]
- Dell VA, Miller DL, Jonhson AE. Effects of nucleotide- and aurodox-induced changes in elongation factor Tu conformation upon its interactions with aminoacyl transfer RNA. A fluorescence study. Biochemistry. 1990;29:1757–1763. doi: 10.1021/bi00459a014. [DOI] [PubMed] [Google Scholar]
- Diaconu M, Kothe U, Schlunzen F, Fischer N, Harms JM, Tonevitsky AG, Stark H, Rodnina MV, Wahl MC. Structural basis for the function of the ribosomal L7/12 stalk in factor binding and GTPase activation. Cell. 2005;121:991–1004. doi: 10.1016/j.cell.2005.04.015. [DOI] [PubMed] [Google Scholar]
- Dorner S, Brunelle JL, Sharma D, Green R. The hybrid state of tRNA binding is an authentic translation elongation intermediate. Nature Structural and Molecular Biology. 2006;13:234–241. doi: 10.1038/nsmb1060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ermolenko DN, Majumdar ZK, Hickerson RP, Spiegel PC, Clegg RM, Noller HF. Observation of intersubunit movement of the ribosome in solution using FRET. Journal of Molecular Biology. 2007a;370:530–540. doi: 10.1016/j.jmb.2007.04.042. [DOI] [PubMed] [Google Scholar]
- Ermolenko DN, Spiegel PC, Majumdar ZK, Hickerson RP, Clegg RM, Noller HF. The antibiotic viomycin traps the ribosome in an intermediate state of translocation. Nature Structural and Molecular Biology. 2007b;14:493–497. doi: 10.1038/nsmb1243. [DOI] [PubMed] [Google Scholar]
- Fei J, Kosuri P, Mac Dougall DD, Gonzalez RL. Coupling of ribosomal L1 stalk and tRNA dynamics during translation elongation. Molecular Cell. 2008;30:348–359. doi: 10.1016/j.molcel.2008.03.012. [DOI] [PubMed] [Google Scholar]
- Feinberg JS, Joseph S. Identification of molecular interactions between P-site tRNA and the ribosome essential for translocation. Proceedings of the National Academy of Sciences USA. 2001;98:11120–11125. doi: 10.1073/pnas.211184098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fourmy D, Recht MI, Blanchard SC, Puglisi JD. Structure of the A site of Escherichia coli 16S ribosomal RNA complexed with an aminoglycoside antibiotic. Science. 1996;274:1367–1371. doi: 10.1126/science.274.5291.1367. [DOI] [PubMed] [Google Scholar]
- Frank J, Agrawal RK. A ratchet-like intersubunit reorganization of the ribosome during translocation. Nature. 2000;406:318–322. doi: 10.1038/35018597. [DOI] [PubMed] [Google Scholar]
- Frank J, Sengupta J, Gao H, Li W, Valle M, Zavialov A, Ehrenberg M. The role of tRNA as a molecular spring in decoding, accommodation, and peptidyl transfer. FEBS Letters. 2005;579:959–962. doi: 10.1016/j.febslet.2004.10.105. [DOI] [PubMed] [Google Scholar]
- Frank J, Gao H, Sengupta J, Gao N, Taylor DJ. The process of mRNA–tRNA translocation. Proceedings of the National Academy of Sciences USA. 2007;104:19671–19678. doi: 10.1073/pnas.0708517104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fredrick K, Noller HF. Catalysis of ribosomal translocation by sparsomycin. Science. 2003;300:1159–1162. doi: 10.1126/science.1084571. [DOI] [PubMed] [Google Scholar]
- Freier SM, Kierzek R, Jaeger JA, Sugimoto N, Caruthers MH, Neilson T, Turner DH. Improved free-energy parameters for predictions of RNA duplex stability. Proceedings of the National Academy of Sciences of Sciences of the United States of America. 1986;83:9373–9377. doi: 10.1073/pnas.83.24.9373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao H, Sengupta J, Valle M, Korostelev A, Eswar N, Stagg SM, Van Roey P, Agrawal RK, Harvey SC, Sali A, Chapman MS, Frank J. Study of the structural dynamics of the E. coli 70S ribosome using real-space refinement. Cell. 2003;113:789–801. doi: 10.1016/s0092-8674(03)00427-6. [DOI] [PubMed] [Google Scholar]
- Gao N, Zavialov AV, Li W, Sengupta J, Valle M, Gursky RP, Ehrenberg M, Frank J. Mechanism for the disassembly of the post-termination complex inferred from cryo-EM studies. Molecular Cell. 2005;18:663–674. doi: 10.1016/j.molcel.2005.05.005. [DOI] [PubMed] [Google Scholar]
- Gao H, Zhou Z, Rawat U, Huang C, Bouakaz L, Wang C, Cheng Z, Liu Y, Zavialov A, Gursky R, Sanyal S, Ehrenberg M, Frank J, Song H. RF3 induces ribosomal conformational changes responsible for dissociation of class I release factors. Cell. 2007;129:929–941. doi: 10.1016/j.cell.2007.03.050. [DOI] [PubMed] [Google Scholar]
- Gao H, Lebarron J, Frank J. Ribosomal dynamics: intrinsic instability of a molecular machine. In: Walter NG, Woodson SA, Batey RT, editors. Non-Protein Coding RNAs. Berlin: Springer- Verlag; 2009. pp. 303–316. [Google Scholar]
- Gavrilova LP, Spirin AS. Stimulation of “non-enzymic” translocation in ribosomes by p-chloromercuribenzoate. FEBS Letters. 1971;17:324–326. doi: 10.1016/0014-5793(71)80177-1. [DOI] [PubMed] [Google Scholar]
- Gomez-Lorenzo MG, Spahn CM, Agrawal RK, Grassucci RA, Penczek P, Chakraburtty K, Ballesta JP, Lavandera JL, Garcia-Bustos JF, Frank J. Three-dimensional cryo-electron microscopy localization of EF2 in the Saccharomyces cerevisiae 80S ribosome at 17·5 Å resolution. EMBO Journal. 2000;19:2710–2718. doi: 10.1093/emboj/19.11.2710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez RL, Chu S, Puglisi JD. Thiostrepton inhibition of tRNA delivery to the ribosome. RNA. 2007;13:2091–2097. doi: 10.1261/rna.499407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gromadski KB, Rodnina MV. Kinetic determinants of high-fidelity tRNA discrimination on the ribosome. Molecular Cell. 2004;13:191–200. doi: 10.1016/s1097-2765(04)00005-x. [DOI] [PubMed] [Google Scholar]
- Gromadski KB, Daviter T, Rodnina MV. A uniform response to mismatches in codon–anticodon complexes ensures ribosomal fidelity. Molecular Cell. 2006;21:369–377. doi: 10.1016/j.molcel.2005.12.018. [DOI] [PubMed] [Google Scholar]
- Hansen JL, Ippolito JA, Ban N, Nissen P, Moore PB, Steitz TA. The structures of four macrolide antibiotics bound to the large ribosomal subunit. Molecular Cell. 2002;10:117–128. doi: 10.1016/s1097-2765(02)00570-1. [DOI] [PubMed] [Google Scholar]
- Hansson S, Singh R, Gudkov AT, Liljas A, Logan DT. Crystal structure of a mutant elongation factor G trapped with a GTP analogue. FEBS Letters. 2005;579:4492–4497. doi: 10.1016/j.febslet.2005.07.016. [DOI] [PubMed] [Google Scholar]
- Harms J, Schluenzen F, Zarivach R, Bashan A, Gat S, Agmon I, Bartels H, Franceschi F, Yonath A. High resolution structure of the large ribosomal subunit from a mesophilic eubacterium. Cell. 2001;107:679–688. doi: 10.1016/s0092-8674(01)00546-3. [DOI] [PubMed] [Google Scholar]
- Hausner TP, Atmadja J, Nierhaus KH. Evidence that the G2661 region of 23S rRNA is located at the ribosomal binding sites of both elongation factors. Biochimie. 1987;69:911–923. doi: 10.1016/0300-9084(87)90225-2. [DOI] [PubMed] [Google Scholar]
- Helgstrand M, Mandava CS, Mulder FA, Liljas A, Sanyal S, Akke M. The ribosomal stalk binds to translation factors IF2, EF-Tu, EF-G and RF3 via a conserved region of the L12 C-terminal domain. Journal of Molecular Biology. 2007;365:468–479. doi: 10.1016/j.jmb.2006.10.025. [DOI] [PubMed] [Google Scholar]
- Hilgenfeld R. Regulatory GTPases. Current Opinion in Structural Biology. 1995;5:810–817. doi: 10.1016/0959-440x(95)80015-8. [DOI] [PubMed] [Google Scholar]
- Hopfield JJ. Kinetic proofreading: a new mechanism for reducing errors in biosynthetic processes requiring high specificity. Proceedings of the National Academy of Sciences USA. 1974;71:4135–4139. doi: 10.1073/pnas.71.10.4135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horan LH, Noller HF. Intersubunit movement is required for ribosomal translocation. Proceedings of the National Academy of Sciences USA. 2007;104:4881–4885. doi: 10.1073/pnas.0700762104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inoue-Yokosawa N, Ishikawa C, Kaziro Y. The role of guanosine triphosphate in translocation reaction catalyzed by elongation factor G. Journal of Biological Chemistry. 1974;249:4321–4323. [PubMed] [Google Scholar]
- Johansson M, Lovmar M, Ehrenberg M. Rate and accuracy of bacterial protein synthesis revisited. Current Opinion in Microbiology. 2008;11:141–147. doi: 10.1016/j.mib.2008.02.015. [DOI] [PubMed] [Google Scholar]
- Jorgensen R, Ortiz PA, Carr-Schmid A, Nissen P, Kinzy TG, Andersen GR. Two crystal structures demonstrate large conformational changes in the eukaryotic ribosomal translocase. Nature Structural Biology. 2003;10:379–385. doi: 10.1038/nsb923. [DOI] [PubMed] [Google Scholar]
- Jorgensen R, Yates SP, Teal DJ, Nilsson J, Prentice GA, Merrill AR, Andersen GR. Crystal structure of ADP-ribosylated ribosomal translocase from Saccharomyces cerevisiae. Journal of Biological Chemistry. 2004;279:45919–45925. doi: 10.1074/jbc.M406218200. [DOI] [PubMed] [Google Scholar]
- Jorgensen R, Merrill AR, Yates SP, Marquez VE, Schawan AL, Boesen T, Andersen GR. Exotoxin A–eEF2 complex structure indicates ADP ribosylation by ribosome mimicry. Nature. 2005;436:979–984. doi: 10.1038/nature03871. [DOI] [PubMed] [Google Scholar]
- Joseph S, Noller HF. EF-G-catalyzed translocation of anticodon stem-loop analogs of transfer RNA in the ribosome. EMBO Journal. 1998;17:3478–3483. doi: 10.1093/emboj/17.12.3478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Julian P, Konevega AL, Scheres SH, Lazaro M, Gil D, Wintemeyer W, Rodnina MV, Valle M. Structure of ratcheted ribosomes with tRNAs in hybrid states. Proceedings of the National Academy of Sciences USA. 2008;104:4881–4885. doi: 10.1073/pnas.0809587105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaziro Y. The role of guanosine 5′-triphosphate in polypeptide chain elongation. Biochimica et Biophysica Acta. 1978;505:95–127. doi: 10.1016/0304-4173(78)90009-5. [DOI] [PubMed] [Google Scholar]
- Kim DF, Green R. Base-pairing between 23S rRNA and tRNA in the ribosomal A site. Molecular Cell. 1999;4:859–864. doi: 10.1016/s1097-2765(00)80395-0. [DOI] [PubMed] [Google Scholar]
- Kim HD, Puglisi JD, Chu S. Fluctuations of transfer RNAs between classical and hybrid states. Biophysical Journal. 2007;93:3575–3582. doi: 10.1529/biophysj.107.109884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kjeldgaard M, Nissen P, Thirup S, Nyborg J. The crystal structure of elongation factor EF-Tu from Thermus aquaticus in the GTP conformation. Structure. 1993;1:35–50. doi: 10.1016/0969-2126(93)90007-4. [DOI] [PubMed] [Google Scholar]
- Klaholz BP, Myasnikov AG, Van Heel M. Visualization of release factor 3 on the ribosome during termination of protein synthesis. Nature. 2004;427:862–865. doi: 10.1038/nature02332. [DOI] [PubMed] [Google Scholar]
- Knudsen CR, Clark BF. Site-directed mutagenesis of Arg58 and Asp86 of elongation factor Tu from Escherichia coli: effects on the GTPase reaction and aminoacyl-tRNA binding. Protein Engineering. 1995;8:1267–1273. doi: 10.1093/protein/8.12.1267. [DOI] [PubMed] [Google Scholar]
- Korostelev A, Trakhanov S, Laurberg M, Noller HF. Crystal structure of a 70S ribosome–tRNA complex reveals functional interactions and rearrangements. Cell. 2006;126:1065–1077. doi: 10.1016/j.cell.2006.08.032. [DOI] [PubMed] [Google Scholar]
- Korostelev A, Ermolenko DN, Noller HF. Structural dynamics of the ribosome. Current Opinion in Chemical Biology. 2008;12:674–683. doi: 10.1016/j.cbpa.2008.08.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- La Cour TF, Nyborg J, Thirup S, Clark BF. Structural details of the binding of guanosine diphosphate to elongation factor Tu from E. coli as studied by X-ray crystallography. EMBO Journal. 1985;4:2385–2388. doi: 10.1002/j.1460-2075.1985.tb03943.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laurberg M, Asahara H, Korostelev A, Zhu J, Trakhanov S, Noller HF. Structural basis for translation termination on the 70S ribosome. Nature. 2008;454:852–857. doi: 10.1038/nature07115. [DOI] [PubMed] [Google Scholar]
- Lebarron J, Mitra K, Frank J. Displaying 3D data on RNA secondary structures: ColoRNA. Journal of Structural Biology. 2007;157:262–270. doi: 10.1016/j.jsb.2006.08.018. [DOI] [PubMed] [Google Scholar]
- Lebarron J, Grassucci RA, Shaikh TR, Baxter WT, Sengupta J, Frank J. Exploration of parameters in cryo-EM leading to an improved density map of the E. coli ribosome. Journal of Structural Biology. 2008;164:24–32. doi: 10.1016/j.jsb.2008.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee TH, Blanchard SC, Kim HD, Puglisi JD, Chu S. The role of fluctuations in tRNA selection by the ribosome. Proceedings of the National Academy of Sciences USA. 2007;104:13661–13665. doi: 10.1073/pnas.0705988104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W, Agirrezabala X, Lei J, Bouakaz L, Brunelle JL, Ortiz-Meoz RF, Green R, Sanyal S, Ehrenberg M, Frank J. Recognition of aminoacyl-tRNA: a common molecular mechanism revealed by cryo-EM. EMBO Journal. 2008;27:3322–3331. doi: 10.1038/emboj.2008.243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liljas A. Structural Aspects of Protein Synthesis. Hackensack, NJ: World Scientific Publishing; 2004. [Google Scholar]
- Lill R, Robertson JM, Wintermeyer W. Binding of the 3′ terminus of tRNA to 23S rRNA in the ribosomal exit site actively promotes translocation. EMBO Journal. 1989;8:3933–3938. doi: 10.1002/j.1460-2075.1989.tb08574.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maguire BA, Beniaminov AD, Ramu H, Mankin AS, Zimmermann RA. A protein component at the heart of an RNA machine: the importance of protein L27 for the function of the bacterial ribosome. Molecular Cell. 2005;20:427–435. doi: 10.1016/j.molcel.2005.09.009. [DOI] [PubMed] [Google Scholar]
- Marshall RA, Dorywalska M, Puglisi JD. Irreversible chemical steps control intersubunit dynamics during translation. Proceedings of the National Academy of Sciences USA. 2008;105:15364–15369. doi: 10.1073/pnas.0805299105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meroueh M, Chow CS. Thermodynamics of RNA hairpins containing single internal mismatches. Nucleic Acids Research. 1999;27:1118–1125. doi: 10.1093/nar/27.4.1118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moazed D, Noller HF. Transfer RNA shields specific nucleotides in 16S ribosomal RNA from attack by chemical probes. Cell. 1986;47:985–994. doi: 10.1016/0092-8674(86)90813-5. [DOI] [PubMed] [Google Scholar]
- Moazed D, Robertson JM, Noller HF. Interaction of elongation factors EF-G and EF-Tu with a conserved loop in 23S RNA. Nature. 1988;180:362–364. doi: 10.1038/334362a0. [DOI] [PubMed] [Google Scholar]
- Moazed D, Noller HF. Interaction of tRNA with 23S rRNA in the ribosomal A, P, and E sites. Cell. 1989a;4:585–597. doi: 10.1016/0092-8674(89)90128-1. [DOI] [PubMed] [Google Scholar]
- Moazed D, Noller HF. Intermediate states in the movement of transfer RNA in the ribosome. Nature. 1989b;342:142–148. doi: 10.1038/342142a0. [DOI] [PubMed] [Google Scholar]
- Munro JB, Altman RB, O’connor N, Blanchard SC. Identification of two distinct hybrid state intermediates on the ribosome. Molecular Cell. 2007;25:505–517. doi: 10.1016/j.molcel.2007.01.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ninio J. Kinetic amplification of enzyme discrimination. Biochimie. 1975;57:587–595. doi: 10.1016/s0300-9084(75)80139-8. [DOI] [PubMed] [Google Scholar]
- Ninio J. Multiple stages in codon–anticodon recognition: double-trigger mechanisms and geometric constraints. Biochimie. 2006;88:963–992. doi: 10.1016/j.biochi.2006.06.002. [DOI] [PubMed] [Google Scholar]
- Nissen P, Kjeldgaard M, Thirup S, Polekhina G, Reshetnikova L, Clark BF, Nyborg J. Crystal structure of the ternary complex of Phe-tRNAPhe, EF-Tu and a GTP analog. Science. 1995;270:1464–1472. doi: 10.1126/science.270.5241.1464. [DOI] [PubMed] [Google Scholar]
- Nissen P, Thirup S, Kjeldgaard M, Nyborg J. The crystal structure of Cys-tRNACys-EF-Tu-GDPNP reveals general and specific features in the ternary complex and in the tRNA. Structure. 1999;15:143–156. doi: 10.1016/s0969-2126(99)80021-5. [DOI] [PubMed] [Google Scholar]
- Nissen P, Hansen J, Ban N, Moore PB, Steitz TA. The structural basis of ribosome activity in peptide bond synthesis. Science. 2000a;289:920–930. doi: 10.1126/science.289.5481.920. [DOI] [PubMed] [Google Scholar]
- Nissen P, Kjeldgaard M, Nyborg J. Macromolecular mimicry. EMBO Journal. 2000b;19:489–495. doi: 10.1093/emboj/19.4.489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noller HF, Chaires JB. Functional modification of 16S ribosomal RNA by kethoxal. Proceedings of the National Academy of Sciences USA. 1972;69:3113–3118. doi: 10.1073/pnas.69.11.3115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noller HF, Hoffarth V, Zimniak L. Unusual resistance of peptidyl transferase to protein extraction procedures. Science. 1992;256:1416–1419. doi: 10.1126/science.1604315. [DOI] [PubMed] [Google Scholar]
- Noller HF, Yusupov MM, Yusupova GZ, Baucom A, Cate JHD. Translocation of tRNA during protein synthesis. FEBS Letters. 2002;514:11–16. doi: 10.1016/s0014-5793(02)02327-x. [DOI] [PubMed] [Google Scholar]
- Noller HF. Biochemical characterization of the ribosomal decoding site. Biochimie. 2006;88:935–941. doi: 10.1016/j.biochi.2006.04.006. [DOI] [PubMed] [Google Scholar]
- Ogle JM, Brodersen DE, Clemons WM, Jr, Tarry MJ, Carter AP, Ramakrishnan V. Recognition of cognate transfer RNA by the 30S ribosomal subunit. Science. 2001;292:897–902. doi: 10.1126/science.1060612. [DOI] [PubMed] [Google Scholar]
- Ogle JM, Murphy FV, IV, Tarry MJ, Ramakrishnan V. Selection of tRNA by the ribosome requires a transition from an open to a closed form. Cell. 2002;111:721–732. doi: 10.1016/s0092-8674(02)01086-3. [DOI] [PubMed] [Google Scholar]
- Ogle JM, Ramakrishnan V. Structural insights into translational fidelity. Annual Reviews in Biochemistry. 2005;74:129–177. doi: 10.1146/annurev.biochem.74.061903.155440. [DOI] [PubMed] [Google Scholar]
- Pan D, Kirillov SV, Zhang CM, Hou YM, Cooperman BS. Rapid ribosomal translocation depends on the conserved 18–55 base pair in the P-site transfer RNA. Nature Structural and Molecular Biology. 2006;13:354–359. doi: 10.1038/nsmb1074. [DOI] [PubMed] [Google Scholar]
- Pan D, Kirillov SV, Cooperman BS. Kinetically competent intermediates in the translocation step of protein synthesis. Molecular Cell. 2007;25:519–529. doi: 10.1016/j.molcel.2007.01.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pape T, Wintermeyer W, Rodnina MV. Complete kinetic mechanism of elongation factor Tu-dependant binding of aminoacyl-tRNA to the A site of E. coli ribosome. EMBO Journal. 1998;17:7490–7497. doi: 10.1093/emboj/17.24.7490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pape T, Wintermeyer W, Rodnina MV. Induced fit in initial selection and proofreading of aminoacyl-tRNA on the ribosome. EMBO Journal. 1999;18:3800–3807. doi: 10.1093/emboj/18.13.3800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parmeggiani A, Nissen P. Elongation factor Tu-targeted antibiotics: four structures, two mechanisms of action. FEBS Letters. 2006;580:4576–4581. doi: 10.1016/j.febslet.2006.07.039. [DOI] [PubMed] [Google Scholar]
- Peske F, Matasovva NB, Savelbergh A, Rodnina MV, Wintermeyer W. Conformationally restricted elongation factor G retains GTPase activity but is inactive in translocation on the ribosome. Molecular Cell. 2000;6:501–505. doi: 10.1016/s1097-2765(00)00049-6. [DOI] [PubMed] [Google Scholar]
- Pestka S. Studies on the formation of trensfer ribonucleic acid-ribosome complexes. V. On the function of a soluble transfer factor in protein synthesis. Proceedings of the National Academy of Sciences USA. 1968;61:726–733. doi: 10.1073/pnas.61.2.726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pestka S. Studies on the formation of transfer ribonucleic acid–ribosome complexes. VI. Oligopeptide synthesis and translocation on ribosomes in the presence and absence of soluble transfer factors. Journal of Biological Chemistry. 1969;244:1533–1539. [PubMed] [Google Scholar]
- Piepenburg O, Pape T, Pleiss JA, Wintermeyer W, Uhlenbeck OC, Rodnina MV. Intact aminoacyl-tRNA is required to trigger GTP hydrolysis by elongation factor Tu on the ribosome. Biochemistry. 2000;39:1734–1738. doi: 10.1021/bi992331y. [DOI] [PubMed] [Google Scholar]
- Polekhina G, Thirup S, Kjeldgaard M, Nissen P, Lippmann C, Nyborg J. Helix unwinding in the effector region of elongation factor EF-Tu-GDP. Structure. 1996;4:1141–1151. doi: 10.1016/s0969-2126(96)00122-0. [DOI] [PubMed] [Google Scholar]
- Rheinberger HJ, Nierhaus KH. Allosteric interactions between the ribosomal transfer RNA-binding sites A and E. Journal of Biological Chemistry. 1986;261:9133–9139. [PubMed] [Google Scholar]
- Rodnina MV, Fricke R, Wintermeyer W. Transient conformational states of aminoacyl-tRNA during ribosome binding catalyzed by elongation factor Tu. Biochemistry. 1994;33:12267–12275. doi: 10.1021/bi00206a033. [DOI] [PubMed] [Google Scholar]
- Rodnina MV, Fricke R, Kuhn L, Wintermeyer W. Codon-dependent conformational change of elongation factor Tu preceding GTP hydrolysis on the ribosome. EMBO Journal. 1995;14:2613–2619. doi: 10.1002/j.1460-2075.1995.tb07259.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodnina MV, Pape T, Fricke R, Kuhn L, Wintermeyer W. Initial binding of the elongation factor Tu.GTP.aminoacyl–tRNA complex preceding codon recognition on the ribosome. Journal of Biological Chemistry. 1996;27:646–652. doi: 10.1074/jbc.271.2.646. [DOI] [PubMed] [Google Scholar]
- Rodnina MV, Savelsbergh A, Katutin VI, Wintermeyer W. Hydrolysis of GTP by elongation factor G drives tRNA movement on the ribosome. Nature. 1997;385:37–41. doi: 10.1038/385037a0. [DOI] [PubMed] [Google Scholar]
- Rodnina MV, Wintermeyer W. Fidelity of aminoacyl-tRNA selection on the ribosome: kinetic and structural mechanism. Annual Reviews in Biochemistry. 2001;70:415–435. doi: 10.1146/annurev.biochem.70.1.415. [DOI] [PubMed] [Google Scholar]
- Ruusala T, Ehrenberg M, Kurland CG. Is there proofreading during polypeptide synthesis? EMBO Journal. 1982;1:741–745. doi: 10.1002/j.1460-2075.1982.tb01240.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samaha RR, Green R, Noller HF. A base pair between tRNA and 23S rRNA in the peptidyl transferase centre of the ribosome. Nature. 1995;377:309–314. doi: 10.1038/377309a0. [DOI] [PubMed] [Google Scholar]
- Savelsbergh A, Matassova NB, Rodnina MV, Wintermeyer W. Role of domains 4 and 5 in elongation factor G functions on the ribosome. Journal of Molecular Biology. 2000;300:951–961. doi: 10.1006/jmbi.2000.3886. [DOI] [PubMed] [Google Scholar]
- Savelsbergh A, Katutin VI, Mohr D, Peske F, Rodnina MV, Wintermeyer W. Role of domains 4 and 5 in elongation factor G functions on the ribosome. Molecular Cell. 2003;11:1517–1523. [Google Scholar]
- Scarano G, Krab IM, Bocchini V, Parmeggiani A. Relevance of histidine-84 in the elongation factor Tu GTPase activity and in poly(Phe) synthesis: its substitution by glutamine and alanine. FEBS Letters. 1995;365:214–218. doi: 10.1016/0014-5793(95)00469-p. [DOI] [PubMed] [Google Scholar]
- Schmeing TM, Seila AC, Hansen JL, Freeborn B, Soukup JK, Scaringe SA, Strobel SA, Moore PB, Steitz TA. A pre-translocational intermediate in protein synthesis observed in crystals of enzymatically active 50S subunits. Nature Structural Biology. 2002;9:225–230. doi: 10.1038/nsb758. [DOI] [PubMed] [Google Scholar]
- Schmeing TM, Huang KS, Kitchen DE, Strobel SA, Steitz TA. Structural insights into the roles of water and the 2′ hydroxyl of the P site tRNA n the peptidyl transferase reaction. Molecular Cell. 2005a;20:437–448. doi: 10.1016/j.molcel.2005.09.006. [DOI] [PubMed] [Google Scholar]
- Schmeing TM, Huang KS, Strobel SA, Steitz TA. An induced-fit mechanism to promote peptide bond formation and exclude hydrolysis of peptidyl- tRNA. Nature. 2005b;438:520–524. doi: 10.1038/nature04152. [DOI] [PubMed] [Google Scholar]
- Schuette JC, Murphy FV, IV, Kelley AC, Weir JR, Giesebrecht J, Connell SR, Loerke J, Mielke T, Zhang W, Penczek PA, Ramakrishnan V, Spahn CM. GTPase activation of elongation factor EF-Tu by the ribosome during decoding. EMBO Journal. 2009;28:755–765. doi: 10.1038/emboj.2009.26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schultz DW, Yarus M. tRNA structure and ribosomal function. I. tRNA nucleotide 27–43 mutations enhance first position wobble. Journal of Molecular Biology. 1994a;235:1381–1394. doi: 10.1006/jmbi.1994.1095. [DOI] [PubMed] [Google Scholar]
- Schultz DW, Yarus M. tRNA structure and ribosomal function. II. Interaction between anticodon helix and other tRNA mutations. Journal of Molecular Biology. 1994b;235:1395–1405. doi: 10.1006/jmbi.1994.1096. [DOI] [PubMed] [Google Scholar]
- Schuwirth BS, Borovinskaya MA, Hau CW, Zhang W, Vila-Sanjuro A, Holton JM, Cate JHD. Structures of the bacterial ribosome at 3·5 Å resolution. Science. 2005;310:827–834. doi: 10.1126/science.1117230. [DOI] [PubMed] [Google Scholar]
- Selmer M, Dunham CM, Murphy FV, IV, Weixlbaumer A, Petry S, Kelley AC, Weir JR, Ramakrishnan V. Structure of the 70S ribosome complexed with mRNA and tRNA. Science. 2006;313:1935–1942. doi: 10.1126/science.1131127. [DOI] [PubMed] [Google Scholar]
- Semenkov YP, Shapkina T, Makhono V, Kirillov S. Puromycin reaction for the A site-bound peptidyl-tRNA. FEBS Letters. 1992;296:207–210. doi: 10.1016/0014-5793(92)80380-y. [DOI] [PubMed] [Google Scholar]
- Semenkov YP, Rodnina MV, Wintermeyer W. Energetic contribution of tRNA hybrid state formation to translocation catalysis on the ribosome. Nature Structural Biology. 2000;7:1027–1031. doi: 10.1038/80938. [DOI] [PubMed] [Google Scholar]
- Senior BW, Holland IB. Effect of colicin E3 upon the 30S ribosomal subunit of Escherichia coli. Proceedings of the National Academy of Sciences USA. 1971;69:959–964. doi: 10.1073/pnas.68.5.959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma D, Southworth DR, Green R. EFG- independent reactivity of a pre-translocation-state ribosome complex with the aminoacyl tRNA substrate puromycin supports an intermediate (hybrid) state of tRNA binding. RNA. 2004;10:102–113. doi: 10.1261/rna.5148704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma D, Cukras AR, Rogers EJ, Southworth DR, Green R. Mutational analysis of S12 protein and implications for the accuracy of decoding by the ribosome. Journal of Molecular Biology. 2007;374:1065–1076. doi: 10.1016/j.jmb.2007.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simonovic M, Steitz TA. Cross-crystal averaging reveals that the structure of the peptidyl-transferase center is the same in the 70S ribosome and the 50S subunit. Proceedings of the National Academy of Sciences USA. 2008;105:500–505. doi: 10.1073/pnas.0711076105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith D, Yarus M. Transfer RNA structure and coding specificity. I. Evidence that a D-arm mutation reduces tRNA dissociation form the ribosome. Journal of Molecular Biology. 1989a;206:489–501. doi: 10.1016/0022-2836(89)90496-8. [DOI] [PubMed] [Google Scholar]
- Smith D, Yarus M. Transfer RNA structure and coding specificity. II. A D-arm tertiary interaction that restricts coding range. Journal of Molecular Biology. 1989b;206:503–511. doi: 10.1016/0022-2836(89)90497-x. [DOI] [PubMed] [Google Scholar]
- Song H, Parsons MR, Rowsell S, Leonard G, Phillips SE. Crystal structure of intact elongation factor EF-Tu from Escherichia coli in GDP conformation at 2·05 Å resolution. Journal of Molecular Biology. 1999;285:1245–1256. doi: 10.1006/jmbi.1998.2387. [DOI] [PubMed] [Google Scholar]
- Spahn CM, Gomez-Lorenzo MG, Grassucci RA, Jorgensen R, Andersen GR, Beckmann R, Penczek PA, Ballesta JP, Frank J. Domain movements of elongation factor eEF2 and the eukaryotic 80S ribosome facilitate tRNA translocation. EMBO Journal. 2004;23:1008–1019. doi: 10.1038/sj.emboj.7600102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spiegel PC, Ermolenko DN, Noller HF. Elongation factor G stabilized the hybrid-state conformation of the 70S ribosome. RNA. 2007;13:1473–1482. doi: 10.1261/rna.601507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spirin AS. How does the ribosome work? A hypothesis based on the two subunit construction of the ribosome. Currents in Modern Biology. 1968;2:115–127. doi: 10.1016/0303-2647(68)90017-8. [DOI] [PubMed] [Google Scholar]
- Spirin AS. The ribosome as a conveying thermal ratchet machine. Journal of Biological Chemistry. 2009;284:21103–21119. doi: 10.1074/jbc.X109.001552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stark H, Rodnina MV, Widen HJ, Van Heel M, Wintermeyer Large-scale movement of elongation factor G and extensive conformational change of the ribosome during translocation. Cell. 2000;100:301–309. doi: 10.1016/s0092-8674(00)80666-2. [DOI] [PubMed] [Google Scholar]
- Stark H, Rodnina MV, Widen HJ, Zemlin F, Wintermeyer W, Van Heel M. Ribosome interaction of aminoacyl-tRNA and elongation factor Tu in the codon-recognition complex. Nature Structural Biology. 2002;9:849–854. doi: 10.1038/nsb859. [DOI] [PubMed] [Google Scholar]
- Steitz TA. A structural understanding of the dynamic ribosome machine. Nature Reviews Molecular Cell Biology. 2008;9:242–253. doi: 10.1038/nrm2352. [DOI] [PubMed] [Google Scholar]
- Sugimoto N, Kierzek R, Freier SM, Turner DH. Energetics of internal GU mismatches in ribooligonucleotide helixes. Biochemistry. 1986;25:5755–5759. doi: 10.1021/bi00367a061. [DOI] [PubMed] [Google Scholar]
- Tama F, Valle M, Frank J, Brooks CL., III Dynamic reorganization of the functionally active ribosome explored by normal mode analysis and cryo-electron microscopy. Proceedings of the National Academy of Sciences USA. 2003;100:9319–9323. doi: 10.1073/pnas.1632476100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor DJ, Nilsson J, Merrill AR, Andersen GR, Nissen P, Frank J. Structures of modified eEF2 80S ribosome complexes reveal the role of GTP hydrolysis in translocation. EMBO Journal. 2007;26:2421–2431. doi: 10.1038/sj.emboj.7601677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson RC, Stone PJ. Proofreading of the codon–anticodon interaction on ribosomes. Proceedings of the National Academy of Sciences USA. 1979;74:198–202. doi: 10.1073/pnas.74.1.198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trabuco LG, Villa E, Mitra K, Frank J, Schulten K. Flexible fitting of atomic structures into electron microscopy maps using molecular dynamics. Structure. 2008;16:673–683. doi: 10.1016/j.str.2008.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valle M, Sengupta J, Swami NK, Grassucci RA, Burkhardt N, Nierhaus KH, Agrawal RK, Frank J. Cryo-EM reveals an active role for aminoacyl-tRNA in the accommodation process. EMBO Journal. 2002;21:3557–3567. doi: 10.1093/emboj/cdf326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valle M, Zavialov A, Li W, Stagg SM, Sengupta J, Nielsen RC, Nissen P, Harvey SC, Ehrenberg M, Frank J. Incorporation of aminoacyl-tRNA into the ribosome as seen by cryo-electron microscopy. Nature Structural Biology. 2003a;10:899–906. doi: 10.1038/nsb1003. [DOI] [PubMed] [Google Scholar]
- Valle M, Zavialov AV, Sengupta J, Rawat U, Ehrenberg M, Frank J. Locking and unlocking of ribosomal motions. Cell. 2003b;114:123–134. doi: 10.1016/s0092-8674(03)00476-8. [DOI] [PubMed] [Google Scholar]
- Van Loock MS, Agrawal RK, Gabashvili IS, Qi L, Frank J, Harvey SC. Movement of the decoding region of the 16S ribosomal RNA accompanies tRNA translocation. Journal of Molecular Biology. 2000;304:507–515. doi: 10.1006/jmbi.2000.4213. [DOI] [PubMed] [Google Scholar]
- Vetter IR, Wittinghofer A. The guanine nucleotide-binding switch in three dimensions. Science. 2001;294:1299–1304. doi: 10.1126/science.1062023. [DOI] [PubMed] [Google Scholar]
- Villa E, Sengupta J, Trabuco LG, Lebarron J, Baxter WT, Shaikh TR, Grassucci RA, Nissen P, Ehrenberg M, Schulten K, Frank J. Ribosome-induced changes in elongation factor Tu conformation control GTP hydrolysis. Proceedings of the National Academy of Sciences USA. 2009a;106:1063–1068. doi: 10.1073/pnas.0811370106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Villa E, Li W, Frank J. tRNA entry into the ribosome during the decoding process. 2009b. In preparation. [Google Scholar]
- Virumae K, Saarma U, Horowitz J, Remme J. Functional importance of the 3′-terminal adenosine of tRNA in ribosomal translation. Journal of Biological Chemistry. 2002;277:24128–24134. doi: 10.1074/jbc.M200393200. [DOI] [PubMed] [Google Scholar]
- Vogeley L, Palm GJ, Mesters JR, Hilgenfeld R. Conformational change of elongation factor Tu (EF-Tu) induced by antibiotic binding. Crystal structure of the complex between EF-Tu.GDP and aurodox. Journal of Biological Chemistry. 2001;276:17149–17155. doi: 10.1074/jbc.M100017200. [DOI] [PubMed] [Google Scholar]
- Voorhees RM, Weixlbaumer A, Loakes D, Kelley AC, Ramakrishnan V. Insights into substrate stabilization from snapshots of the peptidyl transferase center of the intact 70S ribosome. Nature Structural and Molecular Biology. 2009;16:528–533. doi: 10.1038/nsmb.1577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker SE, Shoji S, Pan D, Cooperman BS, Fredrick K. Role of hybrid tRNA-binding states in ribosomal translocation. Proceedings of the National Academy of Sciences USA. 2008;105:9192–9197. doi: 10.1073/pnas.0710146105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Rader AJ, Bahar I, Jernigan RL. Global ribosome motions revealed with elastic network model. Journal of Structural Biology. 2004;147:302–314. doi: 10.1016/j.jsb.2004.01.005. [DOI] [PubMed] [Google Scholar]
- Weixlbaumer A, Jin H, Neubauer C, Voorhees RM, Petry S, Kelley AC, Ramakrishnan V. Insights into translational termination from the structure of RF2 bound to the ribosome. Science. 2008;322:953–956. doi: 10.1126/science.1164840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilden B, Savelsbergh A, Rodnina MV, Wintermeyer W. Role and timing of GTP binding and hydrolysis during EF-G-dependant tRNA translocation on the ribosome. Proceedings of the National Academy of Sciences USA. 2006;103:13670–13675. doi: 10.1073/pnas.0606099103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wohlgemuth I, Beringer M, Rodnina MV. Rapid peptide bond formation on isolate 50S ribosomal subunits. EMBO Reports. 2006;7:699–703. doi: 10.1038/sj.embor.7400732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wriggers W, Agrawal RK, Drew DL, Mccammon A, Frank J. Domain motions of EF-G bound to the 70S ribosome: insights from a hand-shaking between multi-resolution structures. Biophysical Journal. 2000;79:1670–1678. doi: 10.1016/S0006-3495(00)76416-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yarus M, Valle M, Frank J. A twisted tRNA intermediate sets the threshold for decoding. RNA. 2003;9:364–365. doi: 10.1261/rna.2184703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshizawa S, Fourmy D, Puglisi JD. Recognition of the codon–anticodon helix by ribosomal RNA. Science. 1999;285:1722–1725. doi: 10.1126/science.285.5434.1722. [DOI] [PubMed] [Google Scholar]
- Yusupov MM, Yusupova GZ, Baucom A, Lieberman K, Earnest TN, Cate JHD, Noller HF. Crystal structure of the ribosome at 5·5 Å resolution. Science. 2001;292:883–896. doi: 10.1126/science.1060089. [DOI] [PubMed] [Google Scholar]
- Yusupova GZ, Yusupov MM, Cate JHD, Noller HF. The path of messenger RNA through the ribosome. Cell. 2001;106:233–241. doi: 10.1016/s0092-8674(01)00435-4. [DOI] [PubMed] [Google Scholar]
- Zaher HS, Green R. Fidelity at the molecular level: lessons from protein synthesis. Cell. 2009;136:746–762. doi: 10.1016/j.cell.2009.01.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zavialov AV, Ehrenberg M. Peptidyl-tRNA regulates the GTPase activity of translation factors. Cell. 2003;114:113–122. doi: 10.1016/s0092-8674(03)00478-1. [DOI] [PubMed] [Google Scholar]
- Zeidler W, Egle C, Ribeiro S, Wagner A, Katunin V, Kreutzer R, Rodnina M, Wintermeyer W, Sprinzl M. Site-directed mutagenesis of Thermus thermophilus elongation factor Tu. Replacement of His85, Asp81, and Arg300. European Journal of Biochemistry. 1995;229:596–604. doi: 10.1111/j.1432-1033.1995.tb20503.x. [DOI] [PubMed] [Google Scholar]