Abstract
The mammalian target of rapamycin (mTOR) Ser/Thr kinase signals in at least two multiprotein complexes distinguished by their different partners and sensitivities to rapamycin. Acute rapamycin inhibits signaling by mTOR complex 1 (mTORC1) but not mTOR complex 2 (mTORC2), which both promote cell growth, proliferation, and survival. Although mTORC2 regulation remains poorly defined, diverse cellular mitogens activate mTORC1 signaling in a manner that requires sufficient levels of amino acids and cellular energy. Before the identification of distinct mTOR complexes, mTOR was reported to autophosphorylate on Ser-2481 in vivo in a rapamycin- and amino acid-insensitive manner. These results suggested that modulation of mTOR intrinsic catalytic activity does not universally underlie mTOR regulation. Here we re-examine the regulation of mTOR Ser-2481 autophosphorylation (Ser(P)-2481) in vivo by studying mTORC-specific Ser(P)-2481 in mTORC1 and mTORC2, with a primary focus on mTORC1. In contrast to previous work, we find that acute rapamycin and amino acid withdrawal markedly attenuate mTORC1-associated mTOR Ser(P)-2481 in cycling cells. Although insulin stimulates both mTORC1- and mTORC2-associated mTOR Ser(P)-2481 in a phosphatidylinositol 3-kinase-dependent manner, rapamycin acutely inhibits insulin-stimulated mTOR Ser(P)-2481 in mTORC1 but not mTORC2. By interrogating diverse mTORC1 regulatory input, we find that without exception mTORC1-activating signals promote, whereas mTORC1-inhibitory signals decrease mTORC1-associated mTOR Ser(P)-2481. These data suggest that mTORC1- and likely mTORC2-associated mTOR Ser-2481 autophosphorylation directly monitors intrinsic mTORC-specific catalytic activity and reveal that rapamycin inhibits mTORC1 signaling in vivo by reducing mTORC1 catalytic activity.
Keywords: Protein Kinases, Protein Phosphorylation, Serine Threonine Protein Kinase, Signal Transduction, TOR, TOR Complex (TORC)
Introduction
The evolutionarily conserved mammalian target of rapamycin (mTOR)4 protein kinase functions as an environmental sensor, integrating signals from diverse cellular stimuli to control cellular physiology (1–3). mTOR signals in at least two distinct multiprotein complexes distinguished by their partner proteins and differing sensitivities to rapamycin, a clinically employed immunosuppressive drug and allosteric mTOR inhibitor (4, 5). Rapamycin acutely inhibits signaling by mTOR complex 1 (mTORC1) but not mTOR complex 2 (mTORC2) (5). mTORC1 and mTORC2 contain mTOR, mLST8/GβL, and deptor (DEP domain protein that interacts with mTOR) but contain mutually exclusive partners, in particular raptor, which defines mTORC1, and rictor, which defines mTORC2 (1, 5–12). Although acute rapamycin fails to inhibit mTORC2 signaling, chronic rapamycin at high concentrations inhibits the assembly of mTORC2 and, thus, its signaling capacity (13). mTORC1 promotes a plethora of cellular processes including protein synthesis, cell growth/size, cell proliferation, cell survival, and cell metabolism during growth factor, amino acid, and energy sufficiency (2, 14–16). Although the cellular stimuli that regulate mTORC2 remain poorly defined due to its more recent discovery and rapamycin insensitivity, this complex appears to promote cell growth/size, cell proliferation, cell survival, and the organization of the actin cytoskeleton (1, 3, 10, 11, 17).
mTORC1 phosphorylates the ribosomal protein S6 kinase 1 (S6K1), an AGC kinase family member, on its hydrophobic motif site, Thr-389, and the eukaryotic translation initiation factor 4E-binding protein 1 (4EBP1) on several sites (2, 3, 14). Both S6K1 and 4EBP1 possess a TOR signaling motif that interacts with raptor to facilitate substrate delivery to the mTOR kinase (18–21). mTORC1-mediated S6K1 and 4EBP1 phosphorylation coordinately up-regulate cap-dependent translation, cell growth, and cell cycle progression (14, 22, 23). mTORC2 mediates the phosphorylation of the AGC kinase family members Akt (also known as protein kinase B), protein kinase Cα, and SGK1 on their respective hydrophobic motif sites (e.g. Ser-473, Ser-657, and Ser-422, respectively) (11, 24–27).
The insulin pathway represents the most intensively studied mTORC1 regulator to date (1, 28). Insulin/PI3K signaling activates Akt, which phosphorylates both TSC2 and PRAS40 (proline-rich Akt substrate of 40 kDa) to suppress their inhibitory action on mTORC1 (29–33). TSC2 interacts with TSC1 to form a tumor suppressor known as tuberous sclerosis complex (TSC) that functions as an mTORC1 inhibitor (28). Akt-mediated phosphorylation of TSC2 inactivates TSC function and results in strong and constitutive mTORC1 signaling as well as the formation of benign tumors in diverse organs (28, 34). TSC2 acts as a GTPase activating protein toward Rheb, a small G- protein that weakly binds to mTOR and promotes mTORC1 signaling when GTP-bound via an ill-defined mechanism (32, 35–38). Thus, by suppressing TSC, insulin/PI3K/Akt signaling promotes Rheb-mediated activation of mTORC1. During energy stress, AMPK phosphorylates TSC2 on distinct sites, which enhances TSC-mediated mTORC1 inhibition (39). Although the biochemical mechanisms by which amino acids promote mTORC1 signaling remain poorly defined, the Rag family GTPases bind raptor during amino acid sufficiency and induce the translocation of mTORC1 to a subcellular compartment that contains the activator Rheb (40, 41).
Although rapamycin potently inhibits mTORC1-mediated phosphorylation of S6K1 in intact cells, its mechanism of action remains poorly understood. Rapamycin, a bacterially derived, membrane-permeable macrolide, binds to an intracellular protein, FK506-binding protein 12 (FKBP12) (5). The rapamycin-FKBP12 complex directly binds to the mTOR FKBP12-rapamycin binding domain (42, 43), which lies immediately N-terminal to the C-terminal kinase domain, resulting in inhibition of mTORC1 signaling, presumably due to allosteric conformational changes in mTORC1. Although rapamycin induces partial dissociation of mTOR and raptor (44), this mechanism likely fails to account fully for the complete inhibitory effect of rapamycin on S6K1 phosphorylation. Moreover, rapamycin and amino acid withdrawal, although mediating the complete dephosphorylation of S6K1, were reported to have no effect on the autophosphorylation of mTOR Ser-2481 in vivo, a site of mTOR-catalyzed autophosphorylation in vitro (45). These findings suggested that inhibition of mTOR intrinsic catalytic activity cannot explain the mechanism of action of rapamycin or amino acid withdrawal on mTOR-mediated signaling (45). Thus, alternate mechanisms have been postulated (46). Rapamycin binding could theoretically hinder reception of upstream activating signals or impair access of docked substrates to the mTOR catalytic domain without affecting mTOR intrinsic catalytic activity; alternatively, rapamycin binding could lead to activation of a phosphatase that dephosphorylates mTORC1 substrates (47).
Here we re-examine the regulation of mTOR Ser-2481 autophosphorylation (Ser(P)-2481) by interrogating its mTORC-specificity. Contrary to earlier work in which the existence of distinct mTOR complexes with differing sensitivities to rapamycin was not yet known (45), we find that rapamycin strongly reduces mTORC1 but not mTORC2-associated mTOR Ser(P)-2481, consistent with the known rapamycin sensitivity or a lack thereof of each complex, respectively. Insulin promotes mTOR Ser(P)-2481 in a wortmannin-sensitive manner in both mTORC1 and mTORC2, demonstrating a requirement for PI3K in mTORC1 and mTOR2 autophosphorylation. In diverse cellular conditions, the level of mTORC1-associated mTOR Ser(P)-2481 correlates positively with the degree of mTORC1 signaling. Therefore, we propose that mTORC1- and likely mTORC2-associated mTOR Ser(P)-2481 directly monitors mTORC1 and mTORC2 intrinsic catalytic activity. Importantly, our data reveal that rapamycin inhibits mTORC1 signaling in vivo by reducing mTORC1 catalytic activity, thus clarifying its poorly defined mechanism of action. As recent data suggest that mTORC1 may also signal in a rapamycin-resistant manner toward certain substrates (e.g. 4EBP1) (48–50), we have also demonstrated that Torin1, a second generation mTOR catalytic inhibitor that completely inhibits both mTORC1 and mTORC2 signaling, eliminates the residual mTORC1-associated mTOR Ser(P)-2481 that remains upon cellular rapamycin treatment. Torin1 also completely inhibits mTORC2-associated mTOR Ser(P)-2481. These data indicate that complete inhibition of mTORC1 correlates with the virtual absence of mTORC1-associated mTOR Ser-2481 autophosphorylation and further demonstrate that mTOR Ser-2481 autophosphorylation monitors mTORC1 (rapamycin-sensitive and -insensitive) and mTORC2 catalytic activities. As mTORC1 dysregulation contributes to a variety of neoplastic syndromes, rapamycin analogs (rapalogs) (e.g. CCI-779; RAD001) continue to be tested in clinical trials for efficacy against diverse cancers (5); moreover, mTOR catalytic inhibitors hold promise for greater anti-tumor efficacy. Thus, understanding inhibitor action and mTORC1 regulation will likely provide therapeutic benefit.
EXPERIMENTAL PROCEDURES
Materials
Reagents were obtained from the following sources. Protein A-Sepharose CL4B and protein G-Sepharose Fast Flow were from GE Healthcare, CHAPS was from Pierce, Immobilon-P polyvinylidene difluoride membrane (.45 μm) was from Millipore, autoradiography film (HyBlot CL) was from Denville Scientific, reagents for enhanced chemiluminescence (ECL) were from Millipore (Immobilon Western chemiluminescent horseradish peroxidase substrate), and all chemicals were from either Fisher or Sigma.
Commercial Antibodies
AU1, Myc (9E10), and HA (HA.11) antibodies were from Covance. FLAG-M2 and phosphorylated MAPK (Thr-202/Tyr-204) were from Sigma. Donkey anti-rabbit horseradish peroxidase and sheep anti-mouse horseradish peroxidase antibodies were from GE Healthcare. The following antibodies were from Cell Signaling Technology: Ser(P)-2481-mTOR, Thr(P)-389-S6K1 (rabbit monoclonal 108D2), Ser(P)-473-Akt, total Akt, Thr(P)-172-AMPK, total AMPK, total S6, and total TSC1.
Generation of Antibodies to Raptor, mTOR, S6K1, and P-S6
Polyclonal antibodies were generated in rabbits against keyhole limpet hemocyanin-coupled peptides using the Covance custom antibody service. To generate immunogen, peptides and phosphorylated peptides (70% pure; Advanced Peptides, Inc, Boston, MA) were coupled via an N-terminal cysteine to maleimide-activated mariculture keyhole limpet hemocyanin (Pierce). Anti-peptide antibodies were affinity-purified by positive selection on antigen peptide that was coupled to Affi-Gel matrix (Bio-Rad). The S6 phosphopeptide antibody was affinity-purified by positive selection on antigen phosphopeptide followed by negative selection on cognate antigen non-phosphopeptide and irrelevant Ser phosphopeptide (using a phosphoserine Jak2 peptide). The following peptides were used to generate antibodies: 1) raptor (amino acids 885–901, human, CSSSLTNDVAKQPVSRDL), 2) mTOR (amino acids 221–237, rat, CTQREPKEMQKPQWYRHT), 3) S6K1 (C-terminal 17 amino acids; 485–502 of the 70-kDa rat isoform, CKQAFPMISKRPEHLRMNL), 4) P-S6 (amino acids 232–249, CRRL(pS)(pS)LRA(pS)TSK(pS)EE(pS)QK). The following Ser(P)-Jak2 peptide was used for negative selection of P-S6 antibodies on irrelevant Ser phosphopeptide: CSDVQI(pS)PTLRQ.
Plasmids, cDNA Mutagenesis, and Sequencing
The pRK5/Myc-raptor plasmid was obtained from D. Sabatini (Massachusetts Institute of Technology, Boston, MA). pRK5/Myc-mTOR, pRK5/Myc-mTOR-KD, and pRK5/HA-raptor plasmids were obtained from D. Sabatini via Addgene (#1861, #8482, and #8513, respectively). pcDNA3/AU1-mTOR plasmid was originally from R. Abraham (Wyeth, Pearl River, NY). We generated the following mutations in pRK5/Myc-mTOR: S2481A, RR(S2035I), RR(S2035I)/S2481A. We generated the following mutation in pRK5/Myc-mTOR-KD: RR(S2035I). The pKH3/HA-mLST8/GβL and pRK7/FLAG-Rheb plasmids were from J. Blenis (Harvard Medical School, Boston, MA). The following oligonucleotides were used to create point mutations in the rat mTOR cDNA (GenBankTM accession #L37085) using QuikChange II XL (Stratagene). Capital letters indicate mismatch, and the three underlined nucleotides represent the codon mutated: S2035I, forward (5′-ggcctagaagaggccATtcgcttgtactttggg-3′) and reverse (5′-cccaaagtacaagcgaATggcctcttctaggcc-3′; S2481A, forward (5′-tgccagaatccatccatgcCttcattggagatggtttgg-3′) and reverse (5′-ccaaaccatctccaatgaagGcatggatggattctggca-3′). The mutated cDNAs were fully sequenced.
Cell Culture, Drug Treatment, and Transfection
HEK293 cells and immortalized TSC1+/+ and TSC1−/− mouse embryonic fibroblasts (originally from D. Kwiatkowski; Brigham and Women's Hospital, Boston, MA) were cultured in Dulbecco's modified Eagle's medium (DMEM) that contained high glucose (4.5 g/liter), glutamine (584 mg/liter), and sodium pyruvate (110 mg/liter) (Invitrogen) supplemented with 10% fetal bovine serum (FBS) (Hyclone). 3T3-L1 fibroblasts were cultured in DMEM containing 10% calf serum and differentiated into adipocytes by a standard protocol (51, 52). All cells were incubated at 37 °C in a humidified atmosphere containing 5% CO2. All cells were serum-deprived via incubation in DMEM supplemented with 20 mm Hepes (pH 7.2) for ∼20 h. Insulin (100 nm) (Invitrogen), EGF (25 ng/ml) (Invitrogen), or phorbol 12-myristate 13-acetate (PMA) (100 ng/ml) (Sigma) was added to serum-deprived cells for 30 min unless indicated otherwise. For drug treatment, serum-deprived cells were pretreated with rapamycin (20 ng/ml) (Calbiochem) or wortmannin (100 nm) (Upstate/Millipore) for 30 min before the addition of insulin. To effect amino acid deprivation, HEK293 cells were incubated in Dulbecco's PBS containing d-glucose (1 g/liter) and sodium pyruvate (36 mg/liter) (D-PBS/Glc) for 60 min. Cells were stimulated with amino acids by re-feeding with DMEM as a source of amino acids. HEK293 cells on 60-mm plates were transfected using TransIT-LT1 (Mirus) using a total of 4–5 μg of DNA per plate; the specific amounts of experimental plasmid transfected are stated in the legends to Figs. 6C and 7B. Cells were lysed ∼24–48 h post-transfection.
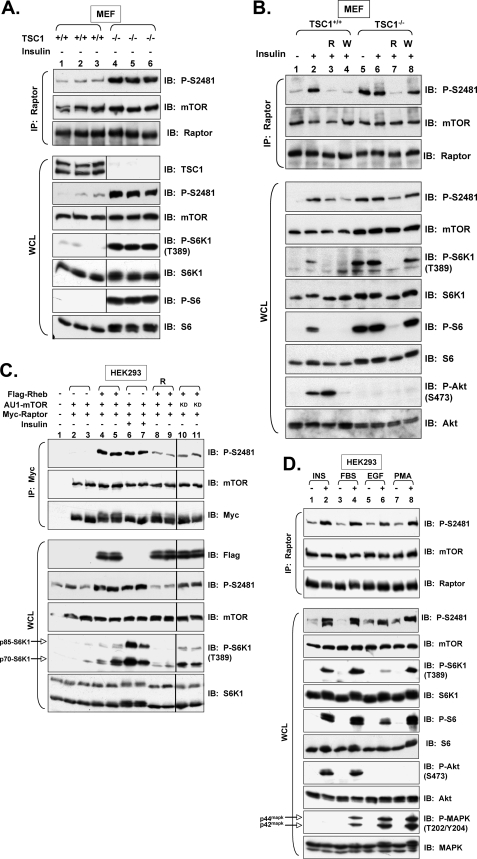
FIGURE 6.
TSC null status, Rheb overexpression, and PI3K/Akt-independent signaling promote mTORC1-associated mTOR Ser-2481 autophosphorylation. A, TSC suppresses mTORC1-associated mTOR Ser(P)-2481. Littermate-matched, 3T3 immortalized MEFs derived from TSC1+/+ or TSC1−/− mice were serum-deprived. Triplicate lysates were immunoprecipitated with anti-raptor antibodies and immunoblotted as indicated. WCL was also immunoblotted directly to confirm the absence of TSC1 and the expected activation of mTORC1 signaling. Note: we find that TSC1−/− fibroblasts express higher levels of total raptor protein when normalized for total protein content; thus, two-thirds of the immunoprecipitate from TSC1−/− cells was loaded relative to TSC1+/+ cells to normalize the amount of raptor between the two cell lines. WCL was loaded similarly. B, TSC1−/− cells exhibit constitutive, rapamycin-sensitive, and wortmannin-resistant mTORC1-associated mTOR Ser(P)-2481. Experiments were similar to A above, except that serum-deprived MEFs from TSC1+/+ or TSC1−/− animals were pretreated with rapamycin (R) or wortmannin (W) and then stimulated with or without insulin. C, Rheb promotes constitutive, mTORC1-associated mTOR Ser(P)-2481 in a rapamycin-sensitive manner that requires mTOR kinase activity. HEK293 cells were co-transfected with Myc-raptor (0.5 μg) and AU1-mTOR (2 μg) in the absence (lanes 1 and 3 and lanes 6 and 7) or presence (lanes 4 and 5 and lanes 8–11) of FLAG-Rheb (3 μg). Rheb-transfected cells were also treated with rapamycin (R) for 24 h (lanes 6-and 7) or were co-transfected with KD Myc-TOR (lanes 10 and 11) rather than wild type mTOR. Cells were serum-deprived and stimulated in the absence (lanes 1–5 and 8–11) or presence (lanes 6 and 7) of insulin. WCL was immunoprecipitated with Myc antibodies to pull down mTORC1 and immunoblotted with the indicated antibodies. WCL was also immunoblotted directly to confirm the expected activation and/or inhibition of mTORC1 signaling by the various treatments. D, EGF and PMA promote mTORC1-associated mTOR Ser(P)-2481 independently of PI3K. HEK293 cells were serum-deprived and stimulated with insulin (INS) (lane 2), 10% FBS (lane 4), EGF (lane 6), or PMA (lane 8) for 30 min. WCL was immunoprecipitated with anti-raptor antibodies and immunoblotted with the indicated antibodies. WCL was also immunoblotted directly to confirm the expected modulation of PI3K (P-Akt (S473)), MAPK (P-MAPK (T202/Y204)I, and mTORC1 (P-S6K1 (T389); P-S6) signaling by the various growth factors/mitogens. Note: p44mapk and p42mapk are also known as ERK1 and -2 (extracellular signal-regulated kinase 1 and 2), respectively.
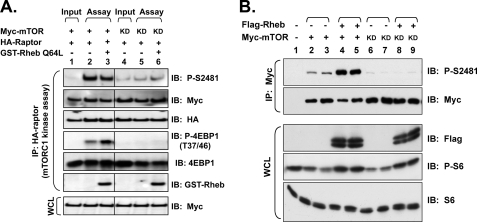
FIGURE 7.
Mechanism of mTOR Ser-2481 autophosphorylation. A, Rheb-GTP provided in vitro does not increase mTORC1-associated mTOR Ser(P)-2481 over basal levels. HEK293 cells on 10-cm plates were co-transfected with HA-raptor (2 μg) together with WT or KD Myc-mTOR alleles (8 μg) and serum-deprived. mTORC1 was immuno-isolated via anti-HA-raptor immunoprecipitation and preincubated with recombinant, GTPase-deficient GST-Rheb (Q64L) protein loaded with GTP. The level of HA-raptor-associated mTOR Ser(P)-2481 after immunoprecipitation but before the addition of GST-Rheb-GTP and initiation of in vitro kinase reactions is shown as Input. In vitro kinase reactions primed with Rheb-GTP were initiated with the addition of recombinant GST-4EBP1 and ATP (Assay). Myc-mTOR autophosphorylation was assessed by immunoblotting IVK reactions with Ser(P)-2481 antibodies, and substrate phosphorylation was assessed with P-Thr-37/46–4EBP1 (P-4EBP1 (T37/46)) antibodies. IVK reactions and WCLs were also immunoblotted with the antibodies indicated. B, mTOR Ser-2481 autophosphorylation occurs in cis rather than in trans. HEK293 cells were co-transfected with Myc-mTOR (1 μg) WT or KD in the absence or presence of FLAG-Rheb (4 μg). Myc-mTOR was immunoprecipitated from cycling cells and immunoblotted with the indicated antibodies. WCLs were also immunoblotted as shown.
Cell Lysis, Immunoprecipitation, and Immunoblotting
Cells were washed 2× with ice-cold PBS (pH 7.4) and collected in ice-cold lysis Buffer A (10 mm KPO4 (pH 7.2), 1 mm EDTA, 5 mm EGTA, 10 mm MgCl2, 50 mm β-glycerophosphate, 1 mm sodium orthovanadate (Na3VO4), 5 μg/ml pepstatin A, 10 μg/ml leupeptin, 40 μg/ml phenylmethylsulfonyl fluoride) containing the detergent CHAPS (0.3%) (to preserve the mTOR/raptor interaction upon lysis (6, 7)) unless indicated otherwise. In some experiments (indicated in legends to supplemental Figs. S2 and S3) cells were lysed in Buffer A containing 0.5% Nonidet P-40, 0.5%Brij35) as detergent. Lysates were spun at 13,200 rpm for 5 min at 4 °C, and the post-nuclear supernatants were collected. A Bradford assay was used to normalize protein levels for immunoprecipitation and immunoblot analyses. For immunoprecipitation, whole cell lysate (WCL) was incubated with antibodies for ∼2 h at 4 °C, incubated with protein A- or G-Sepharose beads for 1.5 h, washed 3 times in lysis buffer, and resuspended in 1× sample buffer (50 mm Tris-HCl (pH 6.8), 10% glycerol, 2% SDS, 2% β-mercaptoethanol, 0.02% bromphenol blue). Samples were heated at 95 °C for 5 min, resolved on SDS-PAGE gels, and transferred to polyvinylidene difluoride membranes using Towbin transfer buffer (25 mm Tris, 192 mm glycine, 10% methanol, 0.02% SDS). Immunoblotting was performed by blocking polyvinylidene difluoride membranes in TBST (40 mm Tris-HCl (pH 7.5), 0.9% NaCl, 0.1% Tween 20) containing 3% nonfat milk and incubating the membranes in TBST with 2% bovine serum albumin containing primary antibodies or secondary horseradish peroxidase-conjugated antibodies. Blots were developed by ECL. mTOR immunoprecipitates were treated with λ-phosphatase as described (53).
m7GTP Cap Pulldown Assay
Whole cell lysates were incubated in 20 μl of m7GTP-Sepharose CL4B (GE Healthcare) for 2 h at 4 °C and then washed twice in the same lysis buffer. Sepharose beads were resuspended in 1× sample buffer with 2% β-mercaptoethanol and resolved on SDS-PAGE.
mTORC1 in Vitro Kinase (IVK) Assays
mTORC1 IVKs were performed basically as described (32, 54). Transfected cells were lysed in ice-cold Buffer C (40 mm Hepes (pH 7.4), 2 mm EDTA, 10 mm pyrophosphate, 10 mm β-glycerophosphate, 40 μg/ml phenylmethylsulfonyl fluoride, 5 μg/ml leupeptin, 10 μg/ml pepstatin A) containing 0.3% CHAPS (to preserve the raptor-mTOR interaction). After HA-raptor immunoprecipitation, immune complexes were washed 3× in Buffer C/CHAPS that contained 150 mm NaCl, washed 2× in Buffer D (25 mm HEPES (pH 7.4), 20 mm KCl), and resuspended in 10 μl of 3× mTOR kinase buffer (75 mm HEPES (pH 7.4), 30 mm MgCl2, 60 mm KC), 5 μl of GST-Rheb loaded with GTPγS (∼150 ng; see below), and 15 μl of distilled H2O. The immune complexes (volume = 30 μl) were preincubated at 30 °C for 5 min and then stored on ice until initiation of kinase assay. To initiate in vitro kinase reactions, 10 μl of IVK Start solution was added (contains 500 μm ATP and 150 ng of GST-4EBP1 in Start Bf (20 mm Hepes (pH 7.4), 10 mm MgCl2, 140 mm KCl)) to immune complexes, mixed, and incubated at 30 °C for 30 min (final (ATP) = 125 μm). IVK reactions were stopped by the addition of 40 μl of 2× sample buffer.
GST-Rheb-E64Q (GTPase-deficient) was isolated from transfected HEK293T cells using immobilized glutathione beads and stored in Rheb Storage Buffer (20 mm HEPES (pH 8), 200 mm NaCl, 5 mm MgCl2) as described (54). GST-Rheb was loaded with GTPγS via incubation with 10 mm EDTA and 0.1 mm GTPγS at 30 °C for 10 min, and the reaction was terminated by the addition of MgCl2 (20 mm).
Image Editing
In Figs. 4C, 6A, 6C, and 7A (indicated by a vertical black line), irrelevant lanes were removed from a scanned autoradiograph, and flanking lanes were juxtaposed using Adobe Photoshop.
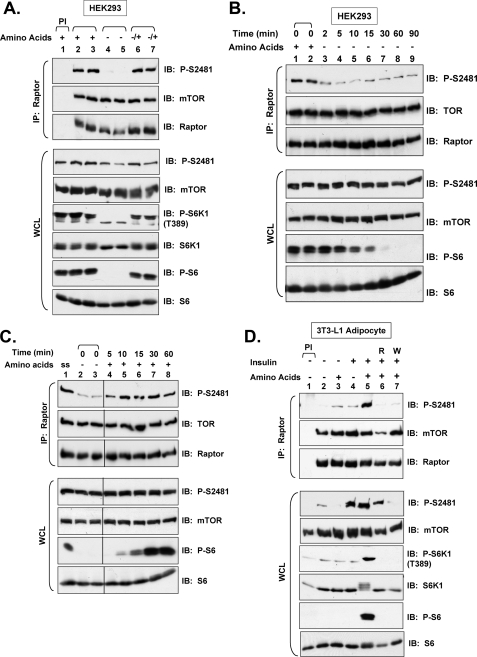
FIGURE 4.
Sufficient levels of amino acids are required for mTORC1-associated mTOR Ser-2481 autophosphorylation. A, amino acid withdrawal mediates the dephosphorylation of mTORC1-associated mTOR Ser(P)-2481. Cycling HEK293 cells cultured in DMEM/FBS (lanes 1–3) were incubated in D-PBS/glucose containing dialyzed 10% FBS (D-PBS/Glc/FBS) for 60 min (lanes 4–7) to effect amino acid deprivation. Amino acid-deprived cells were then re-stimulated with DMEM/FBS for 30 min (lanes 6–7) as a source of amino acids. WCL was immunoprecipitated with anti-raptor antibodies and immunoblotted as indicated (upper panels). WCL was also immunoblotted directly to confirm the expected regulation of mTORC1 signaling (lower panels). B, experiments were similar to A above, except that amino acids were withdrawn for various amounts of time, 2–90 min. C, experiments were similar to A above, except that after amino acid deprivation for 60 min (lanes 2–8), amino acids were added back by refeeding with DMEM/FBS for 5–60 min. Note: ss indicates steady state, whereby cycling cells were cultured in DMEM/FBS before incubation in d-PBS/Glc/FBS. D, both amino acids and insulin are required for mTORC1-associated mTOR Ser(P)-2481. HEK293 cells were serum-deprived (∼20 h) and then amino acid-deprived via incubation in D-PBS/Glc (60 min). Factor-deprived cells were then pretreated with rapamycin (R) (lane 6) or wortmannin (W) (lane 7) and stimulated with amino acids alone (lane 3), insulin alone (lane 4), or both amino acids and insulin (lanes 5–7) for 30 min as indicated. Note: DMEM was used as source of amino acids. WCL was immunoprecipitated with pre-immune (PI) sera or with anti-raptor antibodies and immunoblotted as indicated. WCL was also immunoblotted directly to confirm the expected activation and/or inhibition of mTORC1 signaling by the various treatments.
RESULTS
Rapamycin Inhibits mTORC1 but Not mTORC2-associated mTOR Ser-2481 Autophosphorylation in Response to Insulin/PI3K Signaling
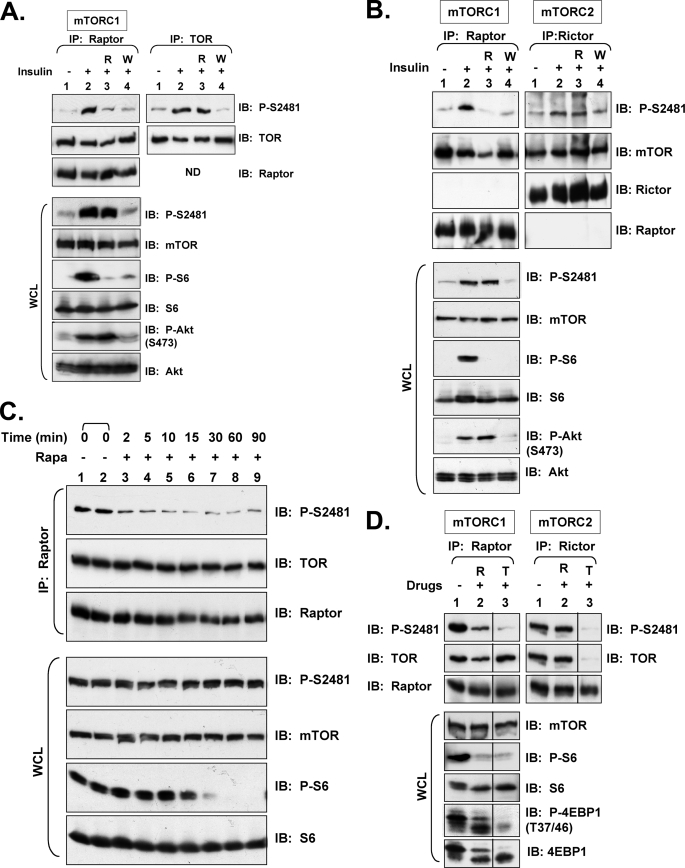
While immunoblotting whole cell lysates derived from 3T3-L1 adipocytes, we were surprised to observe that pretreatment of cells with rapamycin, an mTORC1-specific inhibitor, reduced insulin-stimulated mTOR Ser-2481 autophosphorylation (Ser(P)-2481). As expected, rapamycin inhibited mTORC1 signaling, as measured by analyzing the phosphorylation of S6K1 Thr-389 and the ribosomal protein S6, an S6K1 substrate (Fig. 1A). The inhibition of mTOR Ser(P)-2481 by rapamycin was unexpected, as rapamycin was reported to have no effect on mTOR Ser-2481 autophosphorylation (45). To investigate this observation more carefully, we characterized the mTORC specificity of mTOR Ser(P)-2481 by examining mTOR Ser(P)-2481 in raptor (TORC1) versus rictor (mTORC2) immunoprecipitates in 3T3-L1 adipocytes pretreated with rapamycin or wortmannin (a PI3K inhibitor) (Fig. 1B). Consistent with the results obtained by directly immunoblotting whole cell lysates, mTOR Ser(P)-2481 displayed rapamycin sensitivity in mTORC1 but rapamycin insensitivity in mTORC2, consistent with the well established sensitivity and insensitivity of these two complexes, respectively, to acute rapamycin. Additionally, insulin-stimulated Ser(P)-2481 on total TOR or on mTORC-specific immunoprecipitates displayed wortmannin sensitivity in all cases; as expected, wortmannin inhibited Akt Ser-473 phosphorylation (Fig. 1B). These data demonstrate that in 3T3-L1 adipocytes, rapamycin inhibits mTORC1 but not mTORC2-associated mTOR Ser-2481 autophosphorylation. Moreover, they show that insulin requires PI3K to promote mTOR Ser-2481 autophosphorylation in both mTORC1 and mTORC2. We also confirmed Ser-2481 to be a site of mTOR autophosphorylation in vivo, as Myc-tagged wild type (WT) but not kinase-dead (KD) mTOR reacted with the Ser(P)-2481 antibody, as published previously (45) (Fig. 1C). Note that we confirmed the site- and phospho-specificity of the commercially available mTOR Ser(P)-2481 antibody (supplemental Fig. S1). Ser-2481, which localizes to the mTOR extreme C terminus in a hydrophobic region between the kinase domain and FATC domain, displays conservation in mammals (Homo sapiens, Rattus norvegicus, Mus musculus), chick (Gallus gallus), and zebrafish (Danio rerio) but not in flies (Drosophila melanogaster), yeast (Saccharomyces cerevisiae, Schizosaccharomyces pombe), or plants (Arabidopsis thaliana) (Fig. 1D). Such evolutionary conservation suggests a vertebrate-specific role for mTOR Ser-2481 autophosphorylation.
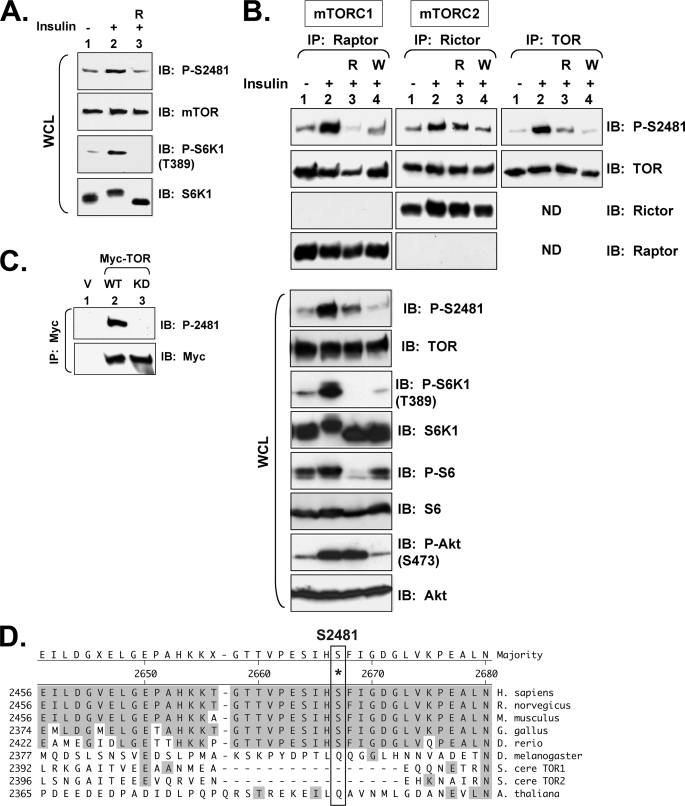
FIGURE 1.
In 3T3-L1 adipocytes, insulin/PI3K signaling promotes rapamycin-sensitive and -resistant mTOR Ser-2481 autophosphorylation in mTORC1 and mTORC2, respectively. A, shown is regulation of total mTOR Ser(P)-2481 in whole cell lysates. Differentiated 3T3-L1 adipocytes were serum-deprived, pretreated with rapamycin (R), and stimulated with insulin (100 nm) for 30 min. After cell lysis, whole cell lysate was resolved on SDS-PAGE and immunoblotted with the indicated antibodies. B, shown is regulation of mTORC1- and mTORC2-associated mTOR Ser(P)-2481. 3T3-L1 adipocytes were serum-deprived, pretreated with rapamycin (R) or wortmannin (W), and stimulated with insulin. WCL was immunoprecipitated with anti-raptor antibodies (left panels) to immuno-isolate mTORC1, with anti-rictor antibodies (middle panels) to immuno-isolate mTORC2, and with anti-mTOR antibodies. Immunoprecipitates were immunoblotted with the indicated antibodies. WCL was also immunoblotted directly (lower panels) with the indicated antibodies to confirm the expected activation and/or inhibition of mTORC1 signaling. Note: the same four lysates (lanes 1–4) were used for raptor, rictor, and mTOR immunoprecipitation as well as for direct immunoblotting. ND, not done. C, shown is confirmation that Ser-2481 is a site of mTOR autophosphorylation. HEK293 cells were transfected with vector control (V), WT (1.0 μg), or KD (1.0 μg) Myc-mTOR plasmids. WCL was immunoprecipitated with Myc antibodies and immunoblotted as indicated. D, shown is alignment of mTOR Ser-2481 from various organisms using the algorithm ClustalW. The Caenorhabditis elegans sequence was omitted because of poor alignment resulting from large regions of non-homology.
As Peterson et al. (45) reported that rapamycin had no effect on Ser-2481 autophosphorylation state in vivo in several cell types (e.g. HEK293; Tag Jurkat cells), we were surprised by our observed rapamycin-mediated inhibition of mTOR Ser(P)-2481 in 3T3-L1 adipocytes. We reasoned that perhaps the relative abundance of rapamycin-insensitive mTORC2 is higher than mTORC1 in certain cell types. Thus, when analyzing total mTOR and disregarding mTORC specificity, an abundant mTORC2-associated mTOR Ser(P)-2481 signal could easily obscure detection of a relatively less abundant, rapamycin-sensitive, mTORC1-associated Ser(P)-2481 signal. We, therefore, examined Ser(P)-2481 on total mTOR as well as on mTORC-specific mTOR in HEK293 cells. Similar to Peterson et al. (45), we found that rapamycin had no effect on mTOR Ser(P)-2481 when whole cell lysates from insulin-stimulated HEK293 were directly immunoblotted (Fig. 2, A–C) or when mTOR was immunoprecipitated (Fig. 2A). We, thus, next investigated the TORC specificity of mTOR Ser(P)-2481 in HEK293 cells. Similar to adipocytes, rapamycin inhibited insulin-stimulated mTOR Ser(P)-2481 on raptor-associated mTOR (mTORC1) (Fig. 2A) but not on rictor-associated mTOR (mTORC2) (Fig. 2B), consistent with the known sensitivity and resistance of these mTORCs, respectively, to acute rapamycin. These data indicate that in HEK293 cells and 3T3-L1 adipocytes, rapamycin indeed inhibits mTORC1-associated mTOR Ser-2481 autophosphorylation. Also similar to adipocytes, insulin increased mTOR Ser(P)-2481 in both raptor (mTORC1) and rictor immunoprecipitates (mTORC2) in a wortmannin-sensitive manner (Fig. 2, A and B). These data indicate that insulin signals via PI3K to promote both mTORC1- and mTORC2-associated mTOR Ser-2481 autophosphorylation. Taken together, our analysis suggests that Peterson et al. (45) failed to observe rapamycin-mediated inhibition of mTOR Ser-2481 autophosphorylation due to the simple fact that their experiments were performed and interpreted before the knowledge that mTOR signals in distinct complexes with differing sensitivities to rapamycin. Thus, a dominant, rapamycin-insensitive, mTORC2-associated Ser(P)-2481 signal obscured their detection of a rapamycin-sensitive, mTORC1-associated Ser(P)-2481 signal in the cell types they analyzed (e.g. HEK293 cells; Tag Jurkat cells). Interestingly, as rapamycin inhibits Ser(P)-2481 on total mTOR in 3T3-L1 adipocytes (Fig. 1, A and B) but not in HEK293 cells (Fig. 2, A–C), the abundance of mTORC1 relative to mTORC2 must be higher in 3T3-L1 adipocytes than in HEK293 cells.
FIGURE 2.
In HEK293 cells, insulin/PI3K signaling promotes rapamycin-sensitive and -resistant mTOR Ser-2481 autophosphorylation in mTORC1 and mTORC2, respectively. A, shown is regulation of mTORC1-associated mTOR Ser(P)-2481. HEK293 cells were serum deprived, pretreated with rapamycin (R) or wortmannin (W), and stimulated with insulin. WCL was immunoprecipitated with anti-raptor antibodies (left panels) to immuno-isolate mTORC1 or anti-mTOR antibodies (right panels). Immunoprecipitates were immunoblotted with the indicated antibodies. WCL was also immunoblotted directly (lower panels) with the indicated antibodies to confirm the expected activation and/or inhibition of mTORC1 signaling. Note: the same four lysates (lanes 1–4) were used for raptor and mTOR immunoprecipitation as well as for WCL immunoblotting. ND, not done. B, shown is regulation of mTORC1- and mTORC2-associated mTOR Ser(P)-2481. Experiments were similar to A above, except WCL was immunoprecipitated with anti-raptor antibodies (left panels) to immuno-isolate mTORC1 or anti-rictor antibodies (right panels) to immuno-isolate mTORC2. C, rapamycin treatment of cycling cells mediates the rapid dephosphorylation of mTORC1-associated mTOR Ser-2481 in mTORC1. HEK293 cells cycling in DMEM/FBS were incubated in the absence (lanes 1 and 2) or presence of rapamycin for the indicated times (2–90 min) (lanes 3–9). WCL was immunoprecipitated with anti-raptor antibodies and immunoblotted with the indicated antibodies. WCL was also immunoblotted directly to confirm the expected inhibition of mTORC1 signaling (lower panels). D, Torin1 eliminates the residual mTORC1-associated mTOR Ser(P)-2481 and P-4EBP1 that remains upon rapamycin treatment. Raptor or rictor was immunoprecipitated from cycling HEK293 cells treated in the absence or presence of rapamycin or Torin1 for 1 h. Immunoprecipitates were immunoblotted for Ser(P)-2481 on raptor- or rictor-associated mTOR. WCLs were also immunoblotted to confirm the expected inhibition of mTORC1 and mTORC2 signaling by these drugs.
As rapamycin inhibits mTORC1-associated mTOR Ser-2481 autophosphorylation and mTORC1-mediated signaling to S6K1, we investigated a functional role for mTOR Ser-2481 autophosphorylation. To study in vivo signaling by exogenously expressed mTOR in the absence of signaling by endogenous mTORC1, we took advantage of a rapamycin-resistant (RR) mutant of Myc-mTOR that contains a point mutation (S2035I) in the FKBP12-rapamycin binding domain, rendering the FKBP12-rapamycin complex unable to bind to and inhibit mTOR as part of mTORC1 (42, 43, 55). Thus, when cells co-transfected with RR-mTOR and HA-S6K1 are cultured in rapamycin, HA-S6K1 phosphorylation occurs via the sole action of the exogenously expressed RR-mTORs. Using a RR-mTOR mutant bearing a phosphorylation site-defective Ala substitution at Ser-2481 (S2481A), we found that mTORC1 containing RR-mTOR-S2481A mediated S6K1 phosphorylation in response to insulin similar to RR-mTOR (supplemental Fig. S2). Additionally, RR-mTOR-S2481A mediated normal insulin-induced dissociation of 4EBP1 from eukaryotic translation initiation factor 4E, a read-out of mTORC1-dependent 4EBP1 phosphorylation (supplemental Fig. S3). Thus, mTOR Ser-2481 autophosphorylation is not required for mTORC1-mediated phosphorylation of S6K1 or 4EBP1. These data are consistent with that of Peterson et al. (45), who concluded that mTOR Ser-2481 autophosphorylation is not required for mTOR-mediated activation of S6K1, as measured by in vitro kinase assay. Additionally, Myc-mTOR-S2481A interacted with HA-raptor and HA-mLST8/GβL similar to wild type mTOR in co-immunoprecipitation assays (supplemental Fig. S4). Thus, mTOR Ser-2481 autophosphorylation is not required for mTORC1 signaling to S6K1 or 4EBP1 or for mTOR interaction with raptor and mLST8/GβL.
As Peterson et al. (45) examined the rapamycin sensitivity of mTOR Ser-2481 autophosphorylation in cycling cells rather than in growth factor-deprived and stimulated cells (as we have done in Figs. 1, A and B, and 2, A and B), we re-examined the rapamycin sensitivity of mTORC1-associated mTOR Ser(P)-2481 in cycling HEK293 cells. We, thus, added rapamycin to cycling cells for various amounts of time (2–90 min) and found that rapamycin promoted rapid mTORC1-associated mTOR Ser-2481 dephosphorylation as early as 2 min post-treatment (Fig. 2C). Similar to Peterson et al. (45), no rapamycin-induced Ser-2481 dephosphorylation was observed when total mTOR was examined in whole cell lysates. As before, failure to observe an effect of rapamycin on total mTOR Ser-2481 autophosphorylation in this context likely results from other rapamycin-insensitive mTORCs (e.g. mTORC2), obscuring detection of rapamycin-sensitive mTOR Ser-2481 autophosphorylation. We additionally determined the kinetics of mTORC1-associated mTOR Ser(P)-2481 in response to insulin (2–120 min stimulation). In HEK293 cells, strong mTORC1-associated mTOR Ser(P)-2481 was observed as early as 5 min after insulin stimulation, before maximal phosphorylation of downstream substrates (e.g. S6), and was maintained for up to 2 h (supplemental Fig. S5).
Recent data suggest that mTORC1 may also signal in a rapamycin-insensitive manner toward certain substrates (e.g. 4EBP1), as rapamycin minimally reduces, whereas mTOR catalytic inhibitors strongly reduce, 4EBP1 phosphorylation (48–50). Consistent with such an idea, we found that residual mTORC1-associated mTOR Ser(P)-2481 signal remained upon rapamycin treatment (see Fig. 2D). We, thus, utilized a novel mTOR catalytic inhibitor, Torin1, to determine whether complete inhibition of mTORC1 catalytic activity correlates with greater reduction in mTORC1-associated mTOR Ser(P)-2481 relative to rapamycin. By treating cycling HEK293 cells in the absence or presence of rapamycin or Torin1, we found that Torin1 more strongly reduced mTORC1-associated mTOR Ser(P)-2481 than did rapamycin (Fig. 2D). These results support the notion that mTORC1 can signal in a rapamycin-insensitive manner. Importantly, they suggest that complete inhibition of mTORC1 correlates with the virtual absence of mTORC1-associated mTOR Ser-2481 autophosphorylation. Additionally, Torin1 also strongly reduced mTORC2-associated mTOR Ser-2481 autophosphorylation, consistent with published data that Torin1 blocks mTORC1 and mTORC2 signaling (48).
Serum Withdrawal and PI3K Inhibition Strongly Reduce mTORC1-associated mTOR Ser-2481 Autophosphorylation
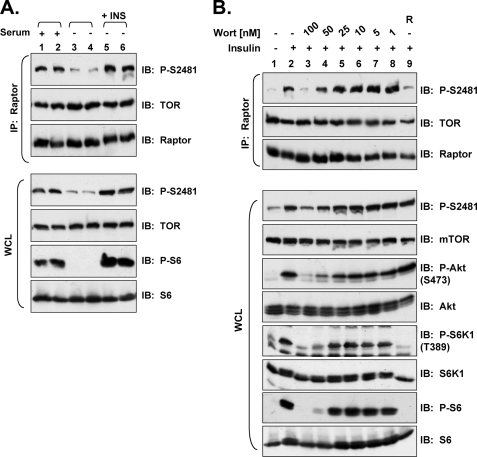
As Peterson et al. (45) found that serum withdrawal minimally decreased (<2-fold) mTOR Ser(P)-2481, we assayed the sensitivity of mTORC1-associated mTOR Ser-2481 autophosphorylation to serum withdrawal. We found that incubation of cycling HEK293 cells in serum-free DMEM for 20 h markedly decreased, whereas insulin re-stimulation increased mTORC1-associated mTOR Ser(P)-2481 and mTORC1 signaling (Fig. 3A). Interestingly, in whole cell lysates, we observed significantly decreased mTOR Ser(P)-2481 in serum-deprived cells (Fig. 3A) (45). It is possible that our HEK293 cell line possesses greater dependence on serum growth factors than the HEK293 line used by Peterson et al. (45). These data together with our finding that insulin increases mTORC2-associated mTOR Ser-2481 autophosphorylation when added to growth factor-deprived cells (Figs. 1 and 2) indicate that serum growth factors modulate mTORC1- and mTORC2-associated mTOR Ser-2481 autophosphorylation.
FIGURE 3.
Serum withdrawal and inhibition of PI3K inhibit mTORC1-associated mTOR Ser(P)-2481 in HEK293 cells. A, withdrawal of serum growth factors mediates the dephosphorylation of mTORC1-associated mTOR Ser-2481. HEK293 cells were cultured in DMEM/FBS (lanes 1–2) or in media lacking serum growth factors for 20 h (lanes 3–6). Serum-deprived cells were then re-stimulated with insulin (INS) for 30 min (lanes 5 and 6). WCL was immunoprecipitated with anti-raptor antibodies and immunoblotted with the indicated antibodies. Note: 1 h before lysis the cycling cells (lanes 1 and 2) were re-fed with fresh media. WCL was also immunoblotted directly to confirm the expected regulation of mTORC1 signaling (lower panels). B, insulin-stimulated, mTORC1-associated mTOR Ser(P)-2481 requires PI3K. HEK293 cells were serum-deprived, pretreated in the absence (lane 2) or presence of various concentrations of wortmannin (100–1 nm) (lanes 3–8), or pretreated with rapamycin (R) (lane 9) for 30 min and then stimulated with insulin for 30 min (lanes 2–9). WCL was immunoprecipitated with anti-raptor antibodies and immunoblotted as indicated (upper panels). WCLs were also immunoblotted directly to confirm the expected activation and/or inhibition of PI3K and mTORC1 signaling by the various treatments (lower panels).
As Peterson et al. (45) found that wortmannin concentrations higher than 100 nm were required to fully inhibit mTOR Ser-2481 autophosphorylation, they concluded that wortmannin most likely inhibits mTOR Ser-2481 autophosphorylation directly by binding to the mTOR kinase domain. We, therefore, examined the wortmannin sensitivity of mTORC1-associated mTOR Ser-2481 autophosphorylation to investigate a regulatory role for PI3K. We thus pretreated serum-deprived HEK293 cells with various doses of wortmannin (1–100 nm) before insulin stimulation and found that 100 nm wortmannin completely inhibited, whereas 50 nm partially inhibited, mTORC1-associated and total mTOR Ser(P)-2481 as well as mTORC1 (as determined by P-S6K1 and P-S6) and PI3K (as determined by P-Akt) signaling (Fig. 3B) (45). As wortmannin concentrations ∼100 nm and lower are generally believed to bind to and inhibit PI3K specifically with minimal inhibition of other PI3K family members (e.g. mTOR) (56), these data identify PI3K as an obligate signaling intermediate in the pathways leading to mTORC1 and mTORC2 (see Figs. 1B and 2, A and B) mTOR Ser-2481 autophosphorylation.
Amino Acids Are Required for mTORC1-associated mTOR Ser- 2481 Autophosphorylation
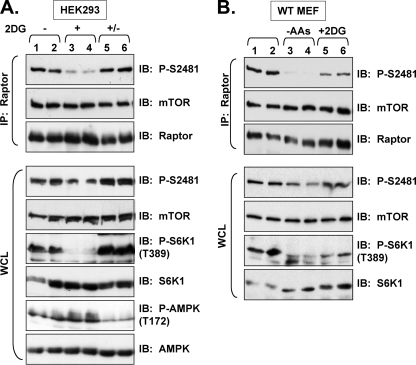
As Peterson et al. (45) found that amino acid withdrawal had no effect on total mTOR Ser-2481 autophosphorylation, we re-analyzed the effect of amino acid levels on mTORC1-associated mTOR Ser(P)-2481. We first deprived cycling HEK293 cells of amino acids via incubation in D-PBS-containing glucose and dialyzed FBS for 60 min. These factor-deprived cells were additionally re-stimulated with amino acids by re-feeding with DMEM/FBS for 30 min. We found that amino acid withdrawal resulted in robust dephosphorylation of mTORC1-associated mTOR Ser-2481 but minimally reduced Ser(P)-2481 on total mTOR; re-addition of amino acids promoted robust re-phosphorylation of mTORC1-associated mTOR Ser-2481 (Fig. 4A). As expected, amino acid levels modulated mTORC1 signaling. Additionally, amino acid withdrawal resulted in dephosphorylation of mTORC1-associated mTOR Ser-2481 in immortalized, wild type mouse embryonic fibroblasts (MEFs) (Fig. 5B). In a similar experiment, we investigated the time course with which amino acid deprivation down-regulates mTORC1-associated mTOR Ser(P)-2481 in HEK293 cells and found that amino acid withdrawal for as little as 2 min led to the dephosphorylation of mTORC1-associated mTOR Ser-2481 (Fig. 4B). Consistent with rapid kinetics of mTOR Ser(P)-2481, re-addition of amino acids to factor-deprived cells for as early as 5 min increased mTORC1-associated mTOR Ser(P)-2481 (Fig. 4C). Last, we examined the requirement for amino acids in insulin-stimulated, mTORC1-associated mTOR Ser(P)-2481 by stimulating serum- and amino acid-deprived HEK293 cells with amino acids alone, insulin alone, or both amino acids and insulin. We found that neither amino acids alone nor insulin alone was sufficient to promote mTORC1-associated mTOR Ser(P)-2481; rather, both were required (Fig. 4D). The addition of both amino acids and insulin promoted mTORC1-associated mTOR Ser(P)-2481 in a rapamycin- and wortmannin-sensitive manner. As expected, the activation of mTORC1 signaling displayed similar regulation (Fig. 4D). Interestingly, when total mTOR Ser(P)-2481 was examined by direct immunoblotting, insulin addition in the absence of amino acids was sufficient to promote mTOR Ser(P)-2481, which presumably reflects amino acid-insensitive but insulin-stimulated TORC2-associated mTOR Ser-2481 autophosphorylation.
FIGURE 5.
Energy stress down-regulates mTORC1-associated mTOR Ser-2481 autophosphorylation. A, energy stress mediates mTORC1-associated mTOR Ser-2481 dephosphorylation. Cycling HEK293 cells were untreated (lanes 1 and 2) or treated with 2DG) (25 mm) for 15 min (lanes 3–6). After 2DG treatment, cells were re-fed with media lacking 2DG and incubated for an additional 30 min (lanes 5 and 6). WCL was immunoprecipitated with anti-raptor antibodies and immunoblotted as indicated. WCL was also immunoblotted directly to confirm the expected activation of AMPK and inhibition of mTORC1 signaling by 2DG. B, in MEFs, energy stress and amino acid withdrawal mediates mTORC1-associated mTOR Ser-2481 dephosphorylation. Cycling wild type MEFs (lanes 1 and 2) were incubated in D-PBS/Glc/FBS for 60 min to effect amino acid (AA) deprivation (lanes 3 and 4) or treated with 2DG (25 mm) for 15 min (lanes 5 and 6). WCL was immunoprecipitated with anti-raptor antibodies and immunoblotted as indicated. WCL was also immunoblotted directly to confirm the expected activation and/or inhibition of mTORC1 signaling by the various treatments.
Energy Stress Down-regulates mTORC1-associated mTOR Ser-2481 Autophosphorylation
As sufficient levels of cellular energy are required for mTORC1 signaling, we investigated whether energy stress modulates mTORC1-associated mTOR Ser(P)-2481. We, therefore, induced energy stress using 2-deoxyglucose (2DG), a glucose analog that blocks cellular glucose utilization, which raises the cellular AMP/ATP ratio, activates AMPK, and down-regulates mTORC1 signaling via a two-pronged mechanism involving AMPK-mediated phosphorylation of TSC2 and raptor (39, 57). Cycling HEK293 cells were, thus, incubated in 2DG for 15 min, which mediated the dephosphorylation of mTORC1-associated mTOR Ser-2481 (Fig. 5A). As expected, 2DG increased AMPK signaling (as measured by increased AMPK P-T172) and decreased mTORC1 signaling. We additionally confirmed that energy stress down-regulates mTORC1-associated mTOR Ser(P)-2481 in immortalized, wild type MEFs (Fig. 5B). These data indicate that in HEK293 cells and MEFs, energy stress down-regulates mTORC1-associated mTOR Ser(P)-2481 and mTORC1 signaling.
TSC Null Status, Rheb Overexpression, and PI3K/Akt-independent Signaling Promote mTORC1-associated mTOR Ser-2481 Autophosphorylation
To identify further the insulin signaling intermediates that modulate mTORC1-associated mTOR Ser(P)-2481, we utilized immortalized MEFs derived from wild type and TSC1−/− mice. Wild type and null MEFs were serum-deprived, and the level of raptor-associated mTOR Ser(P)-2481 was determined. Deletion of TSC1, which conferred the expected up-regulation of mTORC1 signaling, strongly increased mTORC1-associated mTOR Ser(P)-2481 in serum-deprived cells (Fig. 6A). To confirm that the increased mTOR P-2481 in TSC1−/− cells resulted from increased basal phosphorylation rather than increased stoichiometry of phosphorylation, we incubated serum-deprived wild type and TSC1−/− MEFs in the absence and presence of insulin and found that whereas insulin increased raptor-associated mTOR Ser(P)-2481 in wild type MEFs in a rapamycin- and wortmannin-sensitive manner, it failed to increase mTOR Ser(P)-2481 over basal (Fig. 6B). The constitutive mTOR Ser(P)-2481 in the TSC1−/− MEFs displayed sensitivity to rapamycin, consistent with mTORC1 lying downstream of TSC, but displayed wortmannin resistance, consistent with the known uncoupling of PI3K from mTORC1 signaling conferred by TSC inactivation (Fig. 6B).
Moving downstream of TSC in the insulin/PI3K pathway, we asked whether overexpression of Rheb modulates mTORC1-associated mTOR Ser(P)-2481, which strongly promotes mTORC1 signaling in the absence of serum growth factors. We, thus, co-transfected HEK293 cells with AU1-TOR (wild type or KD) and Myc-raptor without or with FLAG-Rheb. The transfected cells were serum-deprived and incubated in the absence or presence of insulin, and the level of mTOR Ser(P)-2481 in Myc-raptor immunoprecipitates was determined. We found that Rheb overexpression strongly increased mTORC1-associated mTOR Ser(P)-2481 in the absence of serum growth factors (lanes 4–5) in a rapamycin-sensitive manner (lanes 8–9) similar to insulin stimulation (lanes 6–7) (Fig. 6C). Consistent with Ser-2481 as a site of autophosphorylation, co-transfection of KD rather than wild type AU1-TOR blunted Rheb-mediated, mTORC1-associated mTOR Ser(P)-2481 (lanes 10–11). Interestingly, rapamycin blunted Ser(P)-2481 on total mTOR in Rheb-overexpressing cells. Although this observation appears to contradict our data and the data of Peterson et al. (45), in which rapamycin fails to reduce Ser(P)-2481 on total mTOR, there is no contradiction when one considers cellular context. As Rheb activates mTORC1 but not mTORC2 signaling (58), the total mTOR Ser(P)-2481 signal in Fig. 6C likely derives predominantly from rapamycin-sensitive mTORC1 with little contribution from rapamycin-insensitive mTORC2, which displays low Ser(P)-2481 in the absence of serum growth factors (Fig. 2B). Taken together, these data demonstrate that insulin signals via the PI3K/TSC/Rheb pathway to promote mTORC1-associated mTOR Ser(P)-2481.
In addition to the canonical insulin/PI3K pathway, the Ras-regulated MAPK pathway activates mTORC1 signaling in a PI3K-independent manner (59–61). We, therefore, asked whether EGF or PMA, mitogens that activate MAPK signaling, modulate raptor-associated mTOR Ser(P)-2481. We found that EGF and PMA as well as FBS increased mTOR Ser(P)-2481 to a degree similar to insulin (Fig. 6D). As expected, these mitogens activated MAPK and mTORC1 signaling, whereas they failed to activate PI3K/Akt signaling (Fig. 6D). Taken together, we find that without exception, mTORC1-activating signals promote, whereas mTORC1-inhibitory signals reduce, mTORC1-associated mTOR Ser(P)-2481, suggesting that mTORC1-associated mTOR Ser-2481 autophosphorylation monitors mTORC1 intrinsic catalytic kinase activity.
Mechanism of mTOR Ser-2481 Autophosphorylation
We next asked whether Rheb-GTP provided in vitro promotes mTORC1-associated mTOR Ser-2481 autophosphorylation, similar to the ability of overexpressed Rheb to do so in intact cells. It is important to note that Sato et al. (62) reported that recombinant Rheb-GTP provided in vitro failed to increase mTOR autophosphorylation, as measured by the incorporation of 32P. We, thus, performed mTORC1 IVK assays by adding recombinant, GTP-loaded GST-Rheb as well as recombinant GST-4EBP1 substrate to HA-raptor-Myc-mTOR complexes immunoprecipitated from serum-deprived cells. Although the addition of GST-Rheb-GTP in vitro promoted mTORC1-mediated phosphorylation of GST-4EBP1 in a kinase-dependent manner, as reported previously (32, 54, 62), GST-Rheb-GTP failed to increase mTOR Ser(P)-2481 over basal levels (Fig. 7A). Importantly, we found that mTORC1 subjected to in vitro kinase assay conditions autophosphorylated measurably in the absence of GST-Rheb-GTP (compare Input to Assay in Fig. 7A), suggesting that extraction of mTORC1 from intact cells causes the loss of regulated Ser-2481 autophosphorylation. Thus, it appears as though Rheb-GTP provided in vitro cannot increase further mTOR autophosphorylation over the high basal autophosphorylation rate.
As mTORC1 may form dimeric or oligomeric complexes (63), we asked whether mTOR Ser-2481 autophosphorylation occurs in cis (within an individual mTORC1) or in trans (between distinct mTORC1s). If mTORC1 autophosphorylation occurs via a bound mTORC1 partner (in trans), we postulated that mTORC1 containing KD Myc-mTOR should oligomerize with mTORC1s containing endogenous WT mTOR, and thus, these active, WT mTORC1s should mediate Ser-2481 autophosphorylation on partnered KD mTORC1s. We, thus, strongly elevated cellular mTORC1 activity by overexpressing FLAG-Rheb together with either WT or KD Myc-mTOR and analyzed mTOR Ser(P)-2481. We found that although FLAG-Rheb increased Ser(P)-2481 on WT Myc-mTOR, it failed to do so on KD Myc-mTOR (Fig. 7B). These data suggest that the mTOR Ser-2481 autophosphorylation mechanism occurs in cis and not in trans.
DISCUSSION
Despite understanding for more than a decade that rapamycin/FKBP12 potently inhibits mTOR-mediated S6K1 phosphorylation by directly binding to the mTOR FKBP12-rapamycin binding domain (42, 43, 64–67), the molecular details of rapamycin-mediated mTOR inhibition remain poorly defined. The earlier inability to observe reduced mTOR Ser(P)-2481 upon rapamycin treatment or amino acid withdrawal suggested that these conditions inhibit mTORC1 signaling by a mechanism other than inhibition of mTOR intrinsic catalytic activity (45). The corollary from such a conclusion, thus, implied that monitoring mTOR Ser-2481 autophosphorylation could not serve as a biomarker for mTORC1 catalytic activity in vivo. Our data alter these conclusions. We find that rapamycin treatment or amino acid withdrawal strongly reduce mTORC1-associated mTOR Ser-2481 autophosphorylation in HEK293 cells, 3T3-L1 adipocytes, and MEFs. Consistent with the established insensitivity of mTORC2 to acute rapamycin treatment, rapamycin fails to reduce mTORC2-associated mTOR Ser-2481 autophosphorylation. In support of our findings, Jacinto et al. (10) reported that recombinant FKBP12 bound to rapamycin inhibited the in vitro autophosphorylation of mTORC1-associated but not mTORC2-associated mTOR, as monitored by 32P incorporation. Thus, rapamycin inhibits mTOR autophosphorylation both in vitro and in vivo. Based on our knowledge today that unique mTOR complexes exist with different sensitivities to rapamycin, it is clear that the presence of a rapamycin-insensitive form of mTOR (e.g. mTORC2) obscured earlier attempts (45) to detect rapamycin- and amino acid-modulated mTOR Ser-2481 autophosphorylation.
We have found that in HEK293 cells and 3T3-L1 adipocytes, insulin promotes both raptor- and rictor-associated mTOR Ser(P)-2481 in a wortmannin-sensitive manner. Thus, insulin signals via PI3K to promote both mTORC1- and mTORC2-associated mTOR Ser-2481 autophosphorylation. Moving downstream of PI3K, we found that TSC suppresses and Rheb promotes raptor-associated mTOR Ser(P)-2481. Thus, the canonical insulin/PI3K/TSC/Rheb pathway promotes mTORC1-associated mTOR Ser-2481 autophosphorylation. As diverse cellular signals regulate mTORC1 signaling, we asked whether insulin/PI3K-independent activation of mTORC1 signaling promotes mTOR Ser-2481 autophosphorylation in mTORC1. Indeed, we found that amino acid withdrawal and energy deprivation reduce, whereas EGF-activated MAPK signaling promotes mTORC1-associated mTOR Ser(P)-2481. In every cellular condition tested, mTORC1-activating signals promoted, whereas mTORC1 inhibitory signals reduced, mTORC1-associated Ser(P)-2481. These data suggest that mTORC1-associated mTOR Ser-2481 autophosphorylation monitors mTORC1 intrinsic catalytic activity in vivo. Additionally, mTORC2-associated mTOR Ser-2481 autophosphorylation likely monitors mTORC2 intrinsic catalytic activity in vivo as well. Our finding of regulated, mTORC1-associated mTOR Ser-2481 autophosphorylation differs from the work of Copp et al. (68), who recently investigated the mTORC specificity of mTOR Ser-2481 autophosphorylation. As they were unable to detect mTORC1-associated mTOR Ser(P)-2481 but readily detected mTORC2-associated mTOR Ser(P)-2481, they concluded that mTOR Ser-2481 autophosphorylation occurs in a TORC-specific manner with mTORC2 but not mTORC1 undergoing Ser-2481 autophosphorylation (68). Our results agree with Copp et al. (68), however, who also found that insulin promotes rictor-associated mTOR Ser(P)-2481. Extending this work, we find that wortmannin inhibits mTORC2-associated mTOR Ser-2481 autophosphorylation, consistent with the report that inhibition of cellular PI3K signaling with wortmannin reduces insulin-stimulated mTORC2 in vitro kinase activity (69). Taken together, the data indicate that insulin signals via PI3K to promote mTORC1- and mTORC2-associated mTOR Ser-2481 autophosphorylation, which serves as a biomarker for mTORC-specific catalytic activity.
Upon growth factor (e.g. insulin; EGF)- and nutrient (e.g. amino acids; glucose)-mediated activation of mTORC1 catalytic activity, we propose that mTOR phosphorylates itself (on Ser-2481) and downstream substrates (e.g. S6K1; 4EBP1). Consistent with this model, mTORC1 containing an mTOR mutant that cannot be phosphorylated on Ser-1261 (S1261A), a novel site of mTOR phosphorylation, exhibits impaired mTOR Ser-2481 autophosphorylation as well as impaired substrate phosphorylation and cell growth (53). Also consistent with this model, Sancak et al. (32) showed that mTORC1-activating signals (e.g. insulin; Rheb) indeed increase mTORC1 in vitro kinase activity toward exogenous substrate. Interestingly, Sato et al. (62) reported recently that recombinant Rheb-GTP failed to increase mTORC1-associated mTOR autophosphorylation in vitro, as measured via 32P incorporation. These data differ from our in vivo results, as we find that cellular Rheb overexpression strikingly increases mTORC1-associated mTOR Ser-2481 autophosphorylation. To investigate this discrepancy, we provided recombinant GST-Rheb loaded with GTP in vitro to HA-raptor-Myc-mTOR complexes isolated from serum-deprived cells. Importantly, the addition of GST-Rheb-GTP promoted mTORC1-mediated phosphorylation of recombinant GST-4EBP1 in a kinase-dependent manner, confirming that our assay worked as expected. GST-Rheb-GTP did not increase mTOR Ser-2481 autophosphorylation over basal levels, however, similar to the data of Sato et al. (62). These data suggest that Rheb-GTP-mediated activation of mTORC1 kinase activity in vitro does not fully recapitulate what occurs in vivo. It is important to note, however, that when mTORC1 was subjected to in vitro kinase assay conditions, mTOR autophosphorylated on Ser-2481 measurably in the absence of GST-Rheb-GTP. These data suggest that mTORC1 extraction from intact cells causes the loss of regulated mTORC1 autophosphorylation. Thus, Rheb-GTP provided in vitro cannot increase further mTOR autophosphorylation over the high basal rate. The data suggest further that Rheb-GTP provided in vitro promotes mTORC1-mediated substrate phosphorylation via a mechanism other than activation of mTORC1 catalytic activity. The recent work of Sato et al. (62) suggests that Rheb-GTP-mediated recruitment of 4EBP1 substrate to mTORC1 may represent such a mechanism (e.g. 4EBP1).
The rapid kinetics with which rapamycin, amino acids, and insulin modulate mTORC1-associated mTOR Ser-2481 autophosphorylation support the notion of mTORC1 as a dynamic, highly responsive sensor that controls cellular physiology based on environmental cues. Autophosphorylation sometimes, but not always, controls kinase function in diverse ways including modulation of catalytic activity, substrate specificity, protein-protein interactions, membrane localization, and proteolysis. To date, functional consequences for site-specific mTOR Ser-2481 autophosphorylation remain unknown. Our work here together with that of Peterson et al. (45) indicates that mTOR Ser-2481 autophosphorylation is not required for mTORC1 signaling to S6K1 or 4EBP1. As mTORC1 may oligomerize (63), we also investigated whether mTOR Ser-2481 autophosphorylation occurs in cis (within an individual mTORC1) or in trans (between distinct, partnered mTORC1s). Our data support a model whereby mTOR Ser-2481 autophosphorylation occurs in cis and not in trans.
The use of mTOR Ser-2481 autophosphorylation as a biomarker to monitor mTORC-specific catalytic activity should greatly simplify assessment of mTORC1 and/or mTORC2 activation state, particularly when screening cells or tissue for inappropriate mTORC signaling in various disease states. Importantly, our data clarify the poorly defined mechanism of action of rapamycin. By analyzing the mTORC-specificity of mTOR Ser-2481 autophosphorylation in vivo, our data reveal that rapamycin (and amino acid withdrawal) indeed inhibits mTORC1 catalytic activity. Our in vivo data combined with the in vitro data of Jacinto et al. (10) support the notion that rapamycin/FKBP12 binding to the mTOR FKBP12-rapamycin binding domain induces an allosteric conformational change that reduces mTORC1 intrinsic catalytic activity. The observation that rapamycin often reduces the strength of the mTOR-raptor interaction (44) supports the idea that rapamycin binding induces a conformational change in mTORC1. It is likely that the cooperative effect of both reduced mTOR-raptor interaction and mTOR catalytic activity contribute to rapamycin-mediated inhibition of mTORC1 signaling. As the mTOR catalytic inhibitor Torin1 eliminates the residual mTOR Ser-2481 autophosphorylation that remains upon cellular rapamycin treatment, our data support the notion that rapamycin does not completely inhibit all mTORC1 function and, thus, may also signal in a rapamycin-insensitive manner.
Supplementary Material
Acknowledgments
We thank all members of the laboratory as well as M. G. Myers, Jr. for critical reading of the manuscript. Thanks to D. Sabatini, J. Blenis, R. Abraham, O. MacDougald, D. Kwiatkowski, and N. Weigel for generously sharing reagents. This work utilized the Cell and Molecular Biology Core(s) of the Michigan Diabetes Research and Training Center, funded by National Institutes of Health Grant NIH5P60 DK20572 (NIDDK) at the University of Michigan.
This work was supported, in whole or in part, by National Institutes of Health Grants R01 DK-078135 (to D. C. F.) and K01 DK-60654 (to G. A. S.). This work was also supported by the American Diabetes Association, the Michigan Diabetes Research and Training Center (to D. C. F.), and the American Heart Association (to G. A. S. and B. E.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1–S5.
- mTOR
- mammalian target of rapamycin
- mTORC1 and mTORC2
- mTOR complex 1 and 2, respectively
- DMEM
- Dulbecco's modified Eagle's medium
- FBS
- fetal bovine serum
- EGF
- epidermal growth factor
- GST
- glutathione S-transferase
- raptor
- regulatory-associated protein of mTOR
- rictor
- rapamycin-insensitive companion of mTOR
- GβL
- G-protein β-subunit-like protein
- mLST8
- mammalian lethal with sec13 protein 8
- PI3K
- phosphatidylinositol 3-kinase
- Rheb
- Ras homolog enriched in brain
- AMPK
- AMP-activated protein kinase
- MAPK
- mitogen-activated protein kinase
- CHAPS
- 3-[(3-cholamidopropyl)-dimethylammonio]-1-propanesulfonate
- PMA
- phorbol 12-myristate 13-acetate
- IVK
- in vitro kinase
- 2DG
- 2-deoxyglucose
- WCL
- whole cell lysate
- IP
- immunoprecipitate
- IB
- immunoblot
- GTPγS
- guanosine 5′-O-(thiotriphosphate)
- WT
- wild type
- RR
- rapamycin-resistant
- S6K1
- S6 kinase 1
- 4EBP1
- 4E-binding protein 1
- TSC
- tuberous sclerosis complex
- FKBP12
- FK506-binding protein 12
- HA
- hemagglutinin
- P-
- phosphorylated
- KD
- kinase dead
- MEF
- mouse embryonic fibroblast.
REFERENCES
- 1.Dunlop E. A., Tee A. R. (2009) Cell. Signal. 21, 827–835 [DOI] [PubMed] [Google Scholar]
- 2.Fingar D. C., Blenis J. (2004) Oncogene 23, 3151–3171 [DOI] [PubMed] [Google Scholar]
- 3.Jacinto E., Lorberg A. (2008) Biochem. J. 410, 19–37 [DOI] [PubMed] [Google Scholar]
- 4.Bhaskar P. T., Hay N. (2007) Dev. Cell 12, 487–502 [DOI] [PubMed] [Google Scholar]
- 5.Guertin D. A., Sabatini D. M. (2009) Sci. Signal. 2, pe24. [DOI] [PubMed] [Google Scholar]
- 6.Hara K., Maruki Y., Long X., Yoshino K., Oshiro N., Hidayat S., Tokunaga C., Avruch J., Yonezawa K. (2002) Cell 110, 177–189 [DOI] [PubMed] [Google Scholar]
- 7.Kim D. H., Sarbassov D. D., Ali S. M., King J. E., Latek R. R., Erdjument-Bromage H., Tempst P., Sabatini D. M. (2002) Cell 110, 163–175 [DOI] [PubMed] [Google Scholar]
- 8.Loewith R., Jacinto E., Wullschleger S., Lorberg A., Crespo J. L., Bonenfant D., Oppliger W., Jenoe P., Hall M. N. (2002) Mol. Cell 10, 457–468 [DOI] [PubMed] [Google Scholar]
- 9.Kim D. H., Sarbassov D. D., Ali S. M., Latek R. R., Guntur K. V., Erdjument-Bromage H., Tempst P., Sabatini D. M. (2003) Mol. Cell 11, 895–904 [DOI] [PubMed] [Google Scholar]
- 10.Jacinto E., Loewith R., Schmidt A., Lin S., Rüegg M. A., Hall A., Hall M. N. (2004) Nat. Cell Biol. 6, 1122–1128 [DOI] [PubMed] [Google Scholar]
- 11.Sarbassov D. D., Ali S. M., Kim D. H., Guertin D. A., Latek R. R., Erdjument-Bromage H., Tempst P., Sabatini D. M. (2004) Curr. Biol. 14, 1296–1302 [DOI] [PubMed] [Google Scholar]
- 12.Peterson T. R., Laplante M., Thoreen C. C., Sancak Y., Kang S. A., Kuehl W. M., Gray N. S., Sabatini D. M. (2009) Cell 137, 873–886 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sarbassov D. D., Ali S. M., Sengupta S., Sheen J. H., Hsu P. P., Bagley A. F., Markhard A. L., Sabatini D. M. (2006) Mol. Cell 22, 159–168 [DOI] [PubMed] [Google Scholar]
- 14.Ma X. M., Blenis J. (2009) Nat. Rev. Mol. Cell Biol. 10, 307–318 [DOI] [PubMed] [Google Scholar]
- 15.Wullschleger S., Loewith R., Hall M. N. (2006) Cell 124, 471–484 [DOI] [PubMed] [Google Scholar]
- 16.Polak P., Hall M. N. (2009) Curr. Opin. Cell Biol. 21, 209–218 [DOI] [PubMed] [Google Scholar]
- 17.Rosner M., Fuchs C., Siegel N., Valli A., Hengstschläger M. (2009) Hum. Mol. Genet. 18, 3298–3310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Choi K. M., McMahon L. P., Lawrence J. C., Jr. (2003) J. Biol. Chem. 278, 19667–19673 [DOI] [PubMed] [Google Scholar]
- 19.Nojima H., Tokunaga C., Eguchi S., Oshiro N., Hidayat S., Yoshino K., Hara K., Tanaka N., Avruch J., Yonezawa K. (2003) J. Biol. Chem. 278, 15461–15464 [DOI] [PubMed] [Google Scholar]
- 20.Schalm S. S., Blenis J. (2002) Curr. Biol. 12, 632–639 [DOI] [PubMed] [Google Scholar]
- 21.Schalm S. S., Fingar D. C., Sabatini D. M., Blenis J. (2003) Curr. Biol. 13, 797–806 [DOI] [PubMed] [Google Scholar]
- 22.Fingar D. C., Richardson C. J., Tee A. R., Cheatham L., Tsou C., Blenis J. (2004) Mol. Cell. Biol. 24, 200–216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fingar D. C., Salama S., Tsou C., Harlow E., Blenis J. (2002) Genes Dev. 16, 1472–1487 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hresko R. C., Mueckler M. (2005) J. Biol. Chem. 280, 40406–40416 [DOI] [PubMed] [Google Scholar]
- 25.Sarbassov D. D., Guertin D. A., Ali S. M., Sabatini D. M. (2005) Science 307, 1098–1101 [DOI] [PubMed] [Google Scholar]
- 26.García-Martínez J. M., Alessi D. R. (2008) Biochem. J. 416, 375–385 [DOI] [PubMed] [Google Scholar]
- 27.Alessi D. R., Pearce L. R., García-Martínez J. M. (2009) Sci. Signal. 2, pe27. [DOI] [PubMed] [Google Scholar]
- 28.Huang J., Manning B. D. (2008) Biochem. J. 412, 179–190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Inoki K., Li Y., Zhu T., Wu J., Guan K. L. (2002) Nat. Cell Biol. 4, 648–657 [DOI] [PubMed] [Google Scholar]
- 30.Manning B. D., Tee A. R., Logsdon M. N., Blenis J., Cantley L. C. (2002) Mol. Cell 10, 151–162 [DOI] [PubMed] [Google Scholar]
- 31.Tee A. R., Fingar D. C., Manning B. D., Kwiatkowski D. J., Cantley L. C., Blenis J. (2002) Proc. Natl. Acad. Sci. U.S.A. 99, 13571–13576 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sancak Y., Thoreen C. C., Peterson T. R., Lindquist R. A., Kang S. A., Spooner E., Carr S. A., Sabatini D. M. (2007) Mol. Cell 25, 903–915 [DOI] [PubMed] [Google Scholar]
- 33.Vander Haar E., Lee S. I., Bandhakavi S., Griffin T. J., Kim D. H. (2007) Nat. Cell Biol. 9, 316–323 [DOI] [PubMed] [Google Scholar]
- 34.Kwiatkowski D. J., Manning B. D. (2005) Hum. Mol. Genet. 14, R251–R258 [DOI] [PubMed] [Google Scholar]
- 35.Garami A., Zwartkruis F. J., Nobukuni T., Joaquin M., Roccio M., Stocker H., Kozma S. C., Hafen E., Bos J. L., Thomas G. (2003) Mol. Cell 11, 1457–1466 [DOI] [PubMed] [Google Scholar]
- 36.Long X., Lin Y., Ortiz-Vega S., Yonezawa K., Avruch J. (2005) Curr. Biol. 15, 702–713 [DOI] [PubMed] [Google Scholar]
- 37.Tee A. R., Manning B. D., Roux P. P., Cantley L. C., Blenis J. (2003) Curr. Biol. 13, 1259–1268 [DOI] [PubMed] [Google Scholar]
- 38.Avruch J., Long X., Lin Y., Ortiz-Vega S., Rapley J., Papageorgiou A., Oshiro N., Kikkawa U. (2009) Biochem. Soc. Trans. 37, 223–226 [DOI] [PubMed] [Google Scholar]
- 39.Inoki K., Zhu T., Guan K. L. (2003) Cell 115, 577–590 [DOI] [PubMed] [Google Scholar]
- 40.Kim E., Goraksha-Hicks P., Li L., Neufeld T. P., Guan K. L. (2008) Nat. Cell Biol. 10, 935–945 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sancak Y., Peterson T. R., Shaul Y. D., Lindquist R. A., Thoreen C. C., Bar-Peled L., Sabatini D. M. (2008) Science 320, 1496–1501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Chen J., Zheng X. F., Brown E. J., Schreiber S. L. (1995) Proc. Natl. Acad. Sci. U.S.A. 92, 4947–4951 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Choi J., Chen J., Schreiber S. L., Clardy J. (1996) Science 273, 239–242 [DOI] [PubMed] [Google Scholar]
- 44.Oshiro N., Yoshino K., Hidayat S., Tokunaga C., Hara K., Eguchi S., Avruch J., Yonezawa K. (2004) Genes Cells 9, 359–366 [DOI] [PubMed] [Google Scholar]
- 45.Peterson R. T., Beal P. A., Comb M. J., Schreiber S. L. (2000) J. Biol. Chem. 275, 7416–7423 [DOI] [PubMed] [Google Scholar]
- 46.Jacinto E. (2008) IUBMB Life 60, 483–496 [DOI] [PubMed] [Google Scholar]
- 47.Peterson R. T., Desai B. N., Hardwick J. S., Schreiber S. L. (1999) Proc. Natl. Acad. Sci. U.S.A. 96, 4438–4442 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Thoreen C. C., Kang S. A., Chang J. W., Liu Q., Zhang J., Gao Y., Reichling L. J., Sim T., Sabatini D. M., Gray N. S. (2009) J. Biol. Chem. 284, 8023–8032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Thoreen C. C., Sabatini D. M. (2009) Autophagy 5, 725–726 [DOI] [PubMed] [Google Scholar]
- 50.Feldman M. E., Apsel B., Uotila A., Loewith R., Knight Z. A., Ruggero D., Shokat K. M. (2009) PLoS Biol. 7, e38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.MacDougald O. A., Cornelius P., Liu R., Lane M. D. (1995) J. Biol. Chem. 270, 647–654 [DOI] [PubMed] [Google Scholar]
- 52.Student A. K., Hsu R. Y., Lane M. D. (1980) J. Biol. Chem. 255, 4745–4750 [PubMed] [Google Scholar]
- 53.Acosta-Jaquez H. A., Keller J. A., Foster K. G., Ekim B., Soliman G. A., Feener E. P., Ballif B. A., Fingar D. C. (2009) Mol. Cell. Biol. 29, 4308–4324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Dunlop E. A., Dodd K. M., Seymour L. A., Tee A. R. (2009) Cell. Signal. 21, 1073–1084 [DOI] [PubMed] [Google Scholar]
- 55.Stan R., McLaughlin M. M., Cafferkey R., Johnson R. K., Rosenberg M., Livi G. P. (1994) J. Biol. Chem. 269, 32027–32030 [PubMed] [Google Scholar]
- 56.Brunn G. J., Williams J., Sabers C., Wiederrecht G., Lawrence J. C., Jr., Abraham R. T. (1996) EMBO J. 15, 5256–5267 [PMC free article] [PubMed] [Google Scholar]
- 57.Gwinn D. M., Shackelford D. B., Egan D. F., Mihaylova M. M., Mery A., Vasquez D. S., Turk B. E., Shaw R. J. (2008) Mol. Cell 30, 214–226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Yang Q., Inoki K., Kim E., Guan K. L. (2006) Proc. Natl. Acad. Sci. U.S.A. 103, 6811–6816 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Ma L., Chen Z., Erdjument-Bromage H., Tempst P., Pandolfi P. P. (2005) Cell 121, 179–193 [DOI] [PubMed] [Google Scholar]
- 60.Roux P. P., Ballif B. A., Anjum R., Gygi S. P., Blenis J. (2004) Proc. Natl. Acad. Sci. U.S.A. 101, 13489–13494 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Tee A. R., Anjum R., Blenis J. (2003) J. Biol. Chem. 278, 37288–37296 [DOI] [PubMed] [Google Scholar]
- 62.Sato T., Nakashima A., Guo L., Tamanoi F. (2009) J. Biol. Chem. 284, 12783–12791 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Wang L., Rhodes C. J., Lawrence J. C., Jr. (2006) J. Biol. Chem. 281, 24293–24303 [DOI] [PubMed] [Google Scholar]
- 64.Chung J., Kuo C. J., Crabtree G. R., Blenis J. (1992) Cell 69, 1227–1236 [DOI] [PubMed] [Google Scholar]
- 65.Price D. J., Grove J. R., Calvo V., Avruch J., Bierer B. E. (1992) Science 257, 973–977 [DOI] [PubMed] [Google Scholar]
- 66.Brown E. J., Beal P. A., Keith C. T., Chen J., Shin T. B., Schreiber S. L. (1995) Nature 377, 441–446 [DOI] [PubMed] [Google Scholar]
- 67.Isotani S., Hara K., Tokunaga C., Inoue H., Avruch J., Yonezawa K. (1999) J. Biol. Chem. 274, 34493–34498 [DOI] [PubMed] [Google Scholar]
- 68.Copp J., Manning G., Hunter T. (2009) Cancer Res. 69, 1821–1827 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Huang J., Dibble C. C., Matsuzaki M., Manning B. D. (2008) Mol. Cell. Biol. 28, 4104–4115 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.