Abstract
Osteopontin (OPN) is a highly modified integrin-binding protein present in most tissues and body fluids where it has been implicated in numerous biological processes. A significant regulation of OPN function is mediated through phosphorylation and proteolytic processing. Proteolytic cleavage by thrombin and matrix metalloproteinases close to the integrin-binding Arg-Gly-Asp sequence modulates the function of OPN and its integrin binding properties. In this study, seven N-terminal OPN fragments originating from proteolytic cleavage have been characterized from human milk. Identification of the cleavage sites revealed that all fragments contained the Arg–Gly–Asp145 sequence and were generated by cleavage of the Leu151–Arg152, Arg152–Ser153, Ser153–Lys154, Lys154–Ser155, Ser155–Lys156, Lys156–Lys157, or Phe158–Arg159 peptide bonds. Six cleavages cannot be ascribed to thrombin or matrix metalloproteinase activity, whereas the cleavage at Arg152–Ser153 matches thrombin specificity for OPN. The principal protease in milk, plasmin, hydrolyzed the same peptide bond as thrombin, but its main cleavage site was identified to be Lys154–Ser155. Another endogenous milk protease, cathepsin D, cleaved the Leu151–Arg152 bond. OPN fragments corresponding to plasmin activity were also identified in urine showing that plasmin cleavage of OPN is not restricted to milk. Plasmin, but not cathepsin D, cleavage of OPN increased cell adhesion mediated by the αVβ3- or α5β1-integrins. Similar cellular adhesion was mediated by plasmin and thrombin-cleaved OPN showing that plasmin can be a potent regulator of OPN activity. These data show that OPN is highly susceptible to cleavage near its integrin-binding motifs, and the protein is a novel substrate for plasmin and cathepsin D.
Keywords: Cell Adhesion, Integrin, Plasmin, Proteolytic Enzymes, Thrombin, Cathepsin D, Osteopontin
Introduction
Osteopontin (OPN)2 is a highly acidic phosphorylated glycoprotein containing an integrin-binding Arg-Gly-Asp (RGD) sequence. OPN is implicated in a diversity of physiological processes such as bone mineralization, inhibition of ectopic calcification, wound healing, inflammation, regulation of immune cell functions, and tumor growth (1–3). OPN is synthesized by a variety of cells and is present in most tissues and body fluids, including blood, urine, and milk (1).
OPN is present in milk in very high concentrations (∼138 mg/liter) (4), but the function is not clear. However, several functions can be hypothesized, for instance OPN can inhibit the formation of renal stones by inhibiting growth and aggregation of calcium crystals (1). Similarly, OPN can be speculated to inhibit unintentional calcium crystallization and precipitation in milk. OPN has also been implicated in mammary gland development and differentiation (5). Furthermore, a significant proportion of the milk OPN is expected to pass through the gut and into the intestines largely intact upon milk consumption, as the protein is relatively resistant to proteolysis by neonatal gastric juice (6). This opens the possibility that OPN or OPN fragments could play a role in the infant immune response, as milk OPN can induce the expression of interleukin-12 from human intestinal lamina propria mononuclear cells (4).
Many of the cellular functions propagated by OPN are mediated through interactions with integrin receptors. The αvβ6, α5β1, α8β1, αvβ1, αvβ5, and αvβ3 integrins bind OPN via the RGD sequence (1, 7, 8), whereas the α4β1 and α9β1 integrins bind the cryptic non-RGD motif Ser-Val-Val-Tyr-Gly-Leu-Arg (SVVYGLR) (9, 10), and the monocyte αXβ2-integrin receptor interacts with the highly acidic parts of OPN (11).
OPN is extensively altered through post-translational modifications such as phosphorylation, sulfation, and glycosylation. These modifications have significant implications on the interaction with integrins (8). Another modification that can alter the functionality of OPN is proteolytic processing. Recently, intracellular cleavage of OPN by caspase-8 at Asp119–Phe120 and Asp141–Gly142 has been shown to be a regulatory switch in determining cell death of cancer cells (12). OPN is also a substrate for thrombin and matrix metalloproteinase (MMP)-2, -3, -7, and -9 (13–17). Thrombin hydrolyzes human OPN at Arg152–Ser153, whereas MMPs cleave nearby at Gly150–Leu151, which in all cases results in the generation of N-terminal OPN fragments containing an exposed RGD145 sequence. Thrombin and MMP cleavage of OPN can modulate its functionality, as e.g. adhesion of tumor cells to OPN via the αVβ3-integrin is significantly increased after thrombin and MMP cleavage (15). Proteolytic processing of OPN also affects cell adhesion to other integrins, as the α5β1-integrin receptor only binds OPN in an RGD-dependent manner after thrombin cleavage (18). Thrombin cleavage of OPN is also required for α9β1 interaction with the resulting N-terminal fragment through the cryptic motif 146SVVYGLR152 (7, 9). Small cleavage differences can have significant effects on the interaction between OPN and integrins. For instance, the α5β1 and α9β1 integrins can bind thrombin-cleaved but not MMP-cleaved OPN (7). Likewise, enhanced RGD-dependent cell binding of fibroblast-like synoviocytes to thrombin-cleaved OPN is abrogated upon removal of Arg152 by carboxypeptidase B (19). Hence, it is clearly demonstrated in a number of studies that proteolytic processing is an important regulator of OPN functionality.
Proteolytic fragments of OPN have been observed in both mineralized tissues and in body fluids. Based on antibody detection, the existence of thrombin-cleaved OPN has been suggested in human bone marrow (20), during blood coagulation, and in milk (13, 14). Furthermore, fragments with the C-terminal amino acids Leu151 and Arg152 have been observed in the synovial fluid of patients with rheumatoid arthritis (19). The exact in vivo proteolytic cleavage positions in OPN have been identified in only a few studies. Two C-terminal components isolated from porcine bone appeared to be generated by trypsin-like cleavage at Arg-Ser and Lys-Ala bonds (homologous to Arg160–Pro161 and Lys187–Ala188 in human OPN) (21). Two OPN forms, beginning at Val3 and Phe53, that possess crystal growth inhibitory activity have been purified from human urine (22, 23). In a later study, two additional urinary OPN fragments generated from hydrolysis of the Arg228–Leu229 and Tyr230–Lys231 bonds were characterized (24). In milk, large fragments resulting from hydrolysis of the Thr26–Trp27 and Leu38–Leu39 bonds and smaller C-terminal forms cleaved at Arg159–Arg160 and Lys187–Ala188 have been identified and isolated (25).
The characterized OPN fragments from bone, urine, and milk do not appear to have been generated by thrombin or MMP digestion, as the cleavage sites do not fit the specificity of these proteases. This indicates that some yet unidentified proteolytic enzymes cleave OPN in vivo. By a proteomic approach, several exo- and endopeptidases have been identified in human milk, including plasmin, prothrombin, cathepsins, and kallikrein (26). Plasmin is the principal protease in milk, and its activity therein has been thoroughly described (27–29). The lysosomal endopeptidase cathepsin D is also well known to be present in soluble forms in milk (26, 30).
In this study, N-terminal OPN fragments have been purified and characterized from human milk. All fragments had been generated by proteolytic cleavage in the near vicinity of the RGD sequence, where six novel cleavage sites were identified. In vitro experiments showed that plasmin and cathepsin D, which both are endogenous to milk, could hydrolyze OPN at sites corresponding to those identified in vivo. Cell binding assays showed that OPN cleaved by plasmin and thrombin, but not cathepsin D, exhibited similar increased ability to mediate cell adhesion compared with the full-length protein. This shows that the novel OPN proteases, plasmin and cathepsin D, can regulate the activity of the protein.
EXPERIMENTAL PROCEDURES
Materials
Percoll, phorbol 12-myristate 13-acetate, plasmin, thrombin, cathepsin D, bovine serum albumin, 97% 18O-water, endoproteinase Asp-N, fibronectin, toluidine blue, and mouse IgG1 isotype control monoclonal antibody (MOPC-21) were from Sigma. The narrow-bore reversed-phase chromatography C2/C18 PC 2.1/10 and the Superdex 75 PC 3.2/30 columns were from GE Healthcare. Reagents used for sequencing were purchased from Applied Biosystems (Warrington, UK). 2,5-Dihydroxybenzoic acid was from LaserBio Labs (Sophia-Antipolis Cedex, France). Monoclonal antibodies against αvβ3 (clone LM609) and α5β1 (clone JBS5) were from Chemicon (Temecula, CA). All other chemicals used were of analytical grade.
Cell Lines and Culture
The MDA-MB-435 human breast cancer cell line (a kind gift from Dr. David T. Denhardt, Rutgers University, NJ) was maintained in Dulbecco's modified Eagle's medium with Glutamax (Invitrogen) supplemented with 10% fetal bovine serum and 1% antibiotics (penicillin/streptomycin). K562 cells (a kind gift from Dr. Thomas Vorup-Jensen, Aarhus University, Denmark) were cultured in RPMI 1640 medium with 2 mm glutamine, 10 units/ml penicillin, 10 μg/ml streptomycin, and 10% fetal bovine serum.
Polyclonal OPN Antibodies
Polyclonal antiserum against human milk OPN was raised in rabbits at DAKO (Glostrup, Denmark). OPN-specific polyclonal antibodies were isolated from the rabbit serum on a protein A-Sepharose column. The specificity of the antibodies was checked and verified by Western blot analyses of milk samples and purified OPN.
Western Blotting
10 μl of purified OPN (10 ng/μl) and skim milk (diluted 150 times) were loaded onto 16% Tris-Tricine gels, fractionated by SDS-PAGE, and electrophoretically transferred to Hybond-P polyvinylidene difluoride membranes (GE Healthcare) for immunodetection. The membranes were blocked in 2% Tween in Tris-buffered saline before addition of polyclonal rabbit OPN antibodies (5 μg/ml). OPN was detected with alkaline phosphatase-conjugated secondary immunoglobulins.
N-terminal Sequencing
Amino acid sequence analyses were performed on an Applied Biosystems PROCISE HT protein sequencer with on-line identification of phenylthiohydantoin-derivatives.
Purification of Osteopontin
OPN was purified from human milk from a pool of donors essentially as described previously (31). The full-length protein and N-terminal fragments were separated by RP-HPLC on a Vydac C18 column connected to a GE Healthcare system. Separation was carried out at 40 °C in 0.1% trifluoroacetic acid, and the proteins were eluted by increasing the concentration of a buffer consisting of 75% (v/v) propan-2-ol in 0.1% trifluoroacetic acid stepwise from 10 to 30% at a flow rate of 0.85 ml/min. Proteins were monitored by measuring the effluent at 226 nm. The collected fractions were analyzed by N-terminal sequencing, mass spectrometry, and SDS-PAGE, and proteins were visualized by Coomassie staining and Western blotting. Urinary OPN was purified from human urine as described previously (24). The amount of purified OPN was determined by amino acid analysis.
Mass Spectrometric Analysis of OPN
Purified full-length OPN and N-terminal fragments were analyzed by MS using a Voyager DE-PRO matrix-assisted laser desorption ionization time-of-flight (MALDI-TOF) mass spectrometer (Applied Biosystems). Samples for MS analyses were prepared by mixing the sample with a saturated solution of 2,5-dihydroxybenzoic acid in a 1:1 ratio directly on the MS target probe. All spectra were obtained in positive linear ion mode using a nitrogen laser at 337 nm and an acceleration voltage of 20 kV. Typically, 50–100 laser shots were added per spectrum and calibrated with external standards. The masses were assigned using the half-height method.
Determination of the C Terminus of OPN Fragments
N-terminal OPN fragments were digested with endoproteinase Asp-N using an enzyme to substrate ratio of 1:75 (w/w) in 50 mm sodium phosphate buffer (pH 8.0) containing H216O/H218O (1:1) at 37 °C for 18 h. The generated peptides were separated by RP-HPLC on a narrow-bore reversed-phase chromatography C2/C18 PC 2.1/10 column connected to a SMART system (GE Healthcare). Separation was carried out in 0.1% trifluoroacetic acid (buffer A) and eluted with a gradient of 60% acetonitrile in 0.1% trifluoroacetic acid (buffer B) developed over 54 min (0–9 min, 0% buffer B; 9–49 min, 0–50% buffer B; 49–54 min, 50–100% buffer B) at a flow rate of 0.15 ml/min. The peptides were detected in the effluent by measuring the absorbance at 214 nm. Peptides were characterized by mass spectrometric analyses using positive reflector-ion mode. The theoretical peptide masses were calculated using the GPMAW program (Lighthouse Data, Odense, Denmark). Urinary OPN was digested with Asp-N as described for milk OPN. The resulting peptides were desalted and concentrated on a Zip-Tip column containing C18 reversed-phase material (Millipore, MA) before MS analysis.
In Vitro Digestion of OPN
Purified full-length milk OPN was digested with cathepsin D using an enzyme to substrate ratio of 1:75 (w/w) in 0.1 m sodium acetate at 37 °C for 6 h and with plasmin or thrombin (30 milliunits/μg OPN) in 0.1 m ammonium bicarbonate at 37 °C for 1 h. The generated fragments from the thrombin digest were separated by RP-HPLC as described above. The cathepsin D and plasmin digests were fractionated by gel filtration on a Superdex 75 PC 3.2/30 column connected to a SMART system (GE Healthcare). The gel filtration column was equilibrated with 0.1 m ammonium bicarbonate and operated at a flow rate of 0.05 ml/min. Proteins were monitored in the effluent at 214 nm. All fractions from the separations were analyzed by mass spectrometric and amino acid sequence analyses. All MS spectra were obtained in both the positive reflector ion and positive linear ion mode. The fractions containing N-terminal OPN fragments of interest were lyophilized and further analyzed by Western blotting, and their C termini were determined by cleavage with endoproteinase Asp-N as described above. The amount of purified N-terminal OPN fragments was determined by amino acid analysis.
Cell Adhesion Assays
Flat-bottom 96-well tissue culture-treated polystyrene microtiter plates (Corning Glass) were coated with 100 μl of full-length OPN or proteolytically generated N-terminal fragments (50 pmol/ml) in phosphate-buffered saline at 4 °C overnight and then blocked with 1% bovine serum albumin. MDA-MB-435 cells were trypsinized, washed twice, and then resuspended to 5 × 105 cells/ml in Dulbecco's modified Eagle's medium containing 1 mg/ml bovine serum albumin. K562 cells were pelleted and resuspended to 1 × 106 cells/ml in a HEPES buffer as described previously (11) and stimulated with 100 ng/ml phorbol 12-myristate 13-acetate. For blocking of integrin function, cells were incubated for 30 min at 37 °C in the presence of neutralizing antibodies at a concentration of 5 μg/ml or an IgG1 isotype control monoclonal antibody. Subsequently, cells (100 μl) were added to coated wells and incubated for 2 h (MDA-MB-435) or 30 min (K562) at 37 °C in a humidified atmosphere with 5% CO2. Nonadhered cells were removed by washing twice with 75 μl of Percoll (73% Percoll, 0.9% NaCl), and adherent cells were fixed with 50 μl of fixative (10% glutaraldehyde in Percoll). Fixed cells were stained with 100 μl of 0.5% toluidine blue and solubilized in 50 μl of 0.5% Triton X-100 before reading at 630 nm using a Microplate Autoreader EL 311 (Bio-Tek Instruments Inc., Winooski, VT).
RESULTS
Purification and Analysis of Human Milk OPN
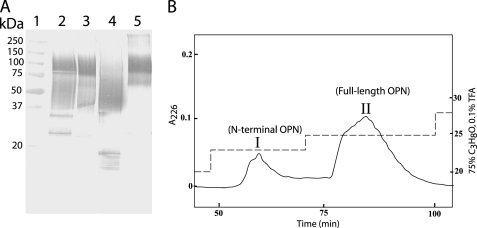
OPN in human milk was analyzed by Western blotting with highly specific antibodies. Several OPN species reacting with the antibody were observed (Fig. 1A, lane 2) as follows: two distinct bands migrating at ∼25 and ∼35 kDa, a broad indistinct band migrating at ∼40–75 kDa, and a band migrating at ∼100 kDa. To further analyze these different forms, OPN was purified from human milk. Western blotting showed that the species migrating at ∼40–75 and ∼100 kDa had been purified (Fig. 1A, lane 3). These forms were separated by RP-HPLC using stepwise increasing concentrations of propan-2-ol as elution buffer. The elution profile revealed two peaks (I and II in Fig. 1B), each of which contained one of the two purified OPN forms (Fig. 1A, lanes 4 and 5), as judged by Western blotting. Sequence analyses showed that both components had the N-terminal sequence 1IPVKQA, which corresponds to the N-terminal sequence of OPN. Combined with linear MALDI-TOF-MS analysis (data not shown), this indicates that peaks I and II in Fig. 1B represent an N-terminal fragment of OPN and the full-length protein, respectively.
FIGURE 1.
Analysis of human milk OPN. A, Western blot analysis using a polyclonal OPN antibody. Lane 1, molecular mass standards. Lane 2, skim milk. Lane 3, OPN purified from milk. Lane 4, fraction I from B. Lane 5, fraction II from B. B, RP-HPLC of purified human milk OPN. Separation was performed on a Vydac C18 column connected to a GE Healthcare system by stepwise increasing concentrations of 75% propan-2-ol in 0.1% trifluoroacetic acid (TFA) (dashed line) at a flow rate of 0.85 ml/min. Proteins were monitored at 226 nm (solid line).
Identification of in Vivo Cleavage Sites in OPN
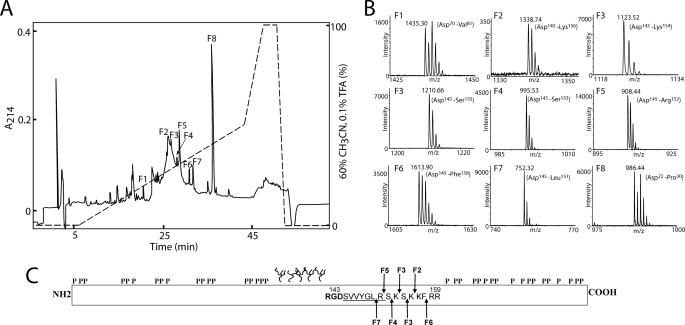
To identify the cleavage sites of OPN in milk, the C-terminal amino acid sequences of the purified N-terminal OPN fragments (peak I in Fig. 1B) had to be assigned. Identification of the C terminus of proteins and peptides is not as straightforward as N-terminal sequencing using Edman chemistry. Instead, the purified N-terminal fragments were digested with endoproteinase Asp-N in a buffer containing equal amounts of H216O and H218O. Asp-N-catalyzed hydrolysis leads to incorporation of one water oxygen atom in the carboxyl group of the cleaved peptides (32). This will result in 50% incorporation of an 16O and 18O atom, respectively, in all generated peptides except for the C-terminal peptide that is not affected by the hydrolysis. Hence, a double signal spaced by 2 Da will be observed for all internal peptides, whereas the peptide representing the original C-terminal peptide will not contain any 18O atom, and therefore it will retain its normal isotopic distribution when analyzed by MALDI-TOF-MS.
The digest of the N-terminal OPN fragments with Asp-N in 50% H218O was separated by RP-HPLC (Fig. 2A), and all fractions were analyzed by MALDI-TOF-MS in reflector-ion mode. As expected, most peptides were represented by double signals spaced by 2 Da identifying them as internal peptides (peaks F1 and F8 in Fig. 2B show the MS spectra of two representative internal peaks). However, peptides in six fractions (F2–F7 in Fig. 2) had a natural isotopic distribution, indicating that they represent C-terminal peptides. The masses of these peptides all corresponded to cleavage of OPN a few residues to the C-terminal side of the integrin binding RGD145 sequence (Fig. 2C). This shows that OPN in milk is very susceptible to proteolytic cleavage in the near vicinity of this important sequence motif.
FIGURE 2.
Reversed-phase HPLC separation and MS analysis of N-terminal OPN digested with Asp-N in a buffer containing 50% H218O. A, peptides were separated on a narrow-bore reversed-phase chromatography C2/C18 PC 2.1/10 column operated by a SMART system (GE Healthcare). Separation was carried out in 0.1% trifluoroacetic acid (TFA), and peptides were eluted with a gradient of 60% acetonitrile in 0.1% trifluoroacetic acid (dashed line) at a flow rate of 0.15 ml/min. The peptides were detected in the effluent by measuring the absorbance at 214 nm (solid line). B, all fractions were analyzed by MS, and representative data from fractions F1 to F8 are shown. Only the relevant m/z region is shown. In fractions F1 and F8, two internal peptides are identified based on the characteristic isotopic pattern caused by incorporation of 18O. Peptides present in fractions F2–F7 were identified as C-terminal peptides as these retain their natural isotopic distribution. C, schematic representation of OPN showing the identified cleavage sites in milk OPN. The location of phosphorylations and O-glycosylations is based on the post-translational modification pattern from human milk OPN (31). The RGD sequence is shown in boldface, and the cryptic integrin-binding site SVVYGLR is underlined. The cleavage sites identified in fraction F2–F7 are shown with arrows and are all located in the near vicinity of the integrin-binding motifs.
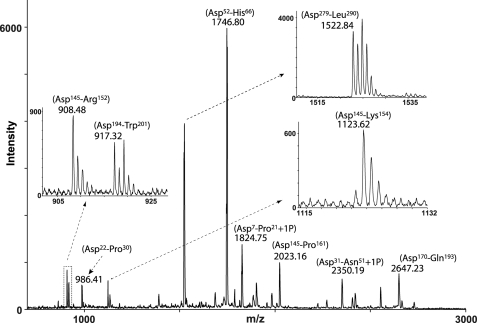
To investigate whether OPN fragments similar to those identified in milk also exist in other body fluids, a mixture of both cleaved and uncleaved OPN purified from urine was subjected to C-terminal analysis as described for milk OPN. The peptides generated by Asp-N were analyzed by MALDI-TOF-MS without prior separation (Fig. 3). The isotopic distribution indicated that two peptides, Asp145–Arg152 and Asp145–Lys154, were C-terminal peptides, whereas the other peptides appeared with the characteristic isotopic pattern of internal peptides.
FIGURE 3.
Identification of cleavage products of OPN purified from human urine. OPN was purified from urine, and the isolated protein batch consists of both OPN fragments as well as the full-length protein. The urinary OPN forms were digested with Asp-N in a buffer containing 50% H218O and analyzed by MS without prior separation of the resulting peptides. Two peptides corresponding to Asp145–Arg152 and Asp145–Lys154 were identified as C-terminal peptides, whereas other OPN peptides were observed with the monoisotopic pattern characteristic of internal peptides.
Proteolytic Enzymes in Milk That Cleave OPN
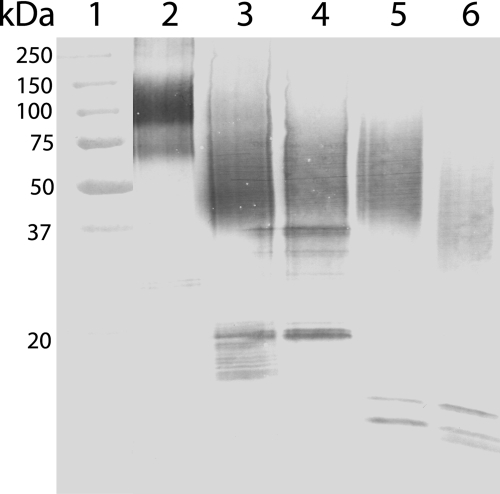
Plasmin and cathepsin D are the best characterized endopeptidases in milk, whereas thrombin is the most studied OPN protease in general. To test whether the N-terminal OPN fragments identified in milk (Fig. 2) could be generated by these proteases, the full-length protein (peak II in Fig. 1B) was incubated with these enzymes. The digests were analyzed by Western blotting and showed that thrombin, plasmin, and cathepsin D all generate OPN fragments appearing as a smear in SDS-PAGE at ∼40–75 kDa (Fig. 4). This is similar to the migration of the N-terminal fragment isolated from milk, suggesting that these proteases could be cleaving OPN in vivo. Next, the exact cleavage sites in OPN generated by thrombin, plasmin, and cathepsin D were investigated. The full-length protein was digested with the three proteases, and the resulting fragments and peptides were separated by RP-HPLC or gel filtration (data not shown). All fractions were analyzed by MALDI-TOF-MS (Table 1), and the N termini of the peptides were identified by sequencing, and in some cases the C termini were also determined by Asp-N cleavage in 50% H218O followed by MS as described above. These analyses showed that thrombin, as expected, generated the N-terminal fragment Ile1–Arg152. Moreover, minor thrombin cleavage was also observed at Arg228–Leu229 in the C-terminal part of the protein. This minor thrombin cleavage site corresponds to a similar cleavage site identified in bovine OPN (33). Two N-terminal fragments, Ile1–Arg152 and Ile1–Lys154, were purified from the plasmin digest. This shows that thrombin and plasmin have overlapping specificities with regard to cleavage of OPN. However, judged by sequencing and the signal intensity in MS, plasmin cleavage at Lys154–Ser155 seems to be preferred over Arg152–Ser153. Finally, cathepsin D hydrolyzed the Leu38–Leu39 and Leu151–Arg152 bonds resulting in an OPN fragment consisting of residues Leu39–Leu151. This corresponds well with an OPN fragment starting at Leu39 previously purified from human milk (25).
FIGURE 4.
OPN cleavage by thrombin, plasmin, and cathepsin D. Western blot analysis using a polyclonal OPN antibody. Lane 1, molecular weight standards. Lane 2, full-length OPN purified from milk (fraction II in Fig. 1B). Lane 3, N-terminal OPN fragments purified from human milk (fraction I in Fig. 1B). Lane 4–6, full-length OPN digested with thrombin (lane 4), plasmin (lane 5), and cathepsin D (lane 6).
TABLE 1.
Characterization of OPN fragments generated by thrombin, plasmin and cathepsin D
| Treatmentsa |
Peptideb | Observed massc | Calculated massd | Mass difference/commente | |
|---|---|---|---|---|---|
| 1st digest | 2nd digest | ||||
| Da | Da | ||||
| T1 | Ile1–Arg152 | 28,000† | 16,891 | 11109 (P and O-glycans) | |
| Asp-N | Asp145–Arg152 | 908.48 | 908.48 | Normal isotope pattern | |
| T2 | Ser153–Arg228 | 9043.05†/8961.76† | 8642.20 | 400.85 (5P)/319.56 (4P) | |
| T3 | Ser153–Asn298 | 17,888.52† | 16,843.08 | 1045.44 (13P) | |
| T4 | Leu229–Asn298 | 8864.33† | 8219.90 | 644.43 (8P) | |
| P1 | Ile1–Arg152 | 28,200† | 16,891 | 11,309 (P and O-glycans) | |
| Ile1–Lys154 | 17,106 | 11,094 (P and O-glycans) | |||
| Asp-N | Asp145–Arg152 | 908.49 | 908.48 | Normal isotope pattern | |
| Asp-N | Asp145–Lys154 | 1123.64 | 1123.60 | Normal isotope pattern | |
| P2 | Ser153–Lys187 | 4325.60 | 4085.94 | 239.66 (3P) | |
| P3 | Ser155–Lys187 | 4110.60/4030.59 | 3870.82 | 239.78 (3P)/159.77 (2P) | |
| P4 | Phe158–Lys187 | 3767.47/3687.40 | 3527.59 | 239.88 (3P)/159.81 (2P) | |
| P5 | Arg160–Lys187 | 3464.16/3384.20 | 3224.43 | 239.73 (3P)/159.77 (2P) | |
| P6 | Ala188–Lys225 | 4519.15†/4438.67 | 4198.91 | 317.78 (4P)/239.76 (3P) | |
| P7 | Ala188–Lys231 | 5294.22†/5214.29† | 4977.28 | 316.94 (4P)/237.01 (3P) | |
| P8 | Arg232–Lys283 | 6620.45† | 6142.62 | 477.83 (6P) | |
| P9 | Phe284–Asn298 | 1930.56 | 1690.80 | 239.76 (3P) | |
| CD1 | Ile1–Thr26 | 3078.35 | 2838.41 | 239.94 (3P) | |
| CD2 | Trp27–Leu38 | 1439.60 | 1439.72 | ||
| CD3 | Trp27–Phe53 | 3202.35 | 3042.44 | 159.91 (2P) | |
| CD4 | Leu39–Leu151 | 22,200† | 12,492 | 9708 (P and O-glycans) | |
| Lys54–Leu151 | 10,888 | 11,312 (P and O-glycans) | |||
| Asp-N | Asp145–Leu151 | 752.29 | 752.38 | Normal isotope pattern | |
| CD5 | Arg152–Leu195 | 5289.17 | 5049.49 | 239.68 (3P) | |
| CD6 | Asn196–Leu229 | 4115.70/4037.73† | 3875.74 | 239.96 (3P)/159.75 (2P) | |
| CD7 | Ser251–Leu269 | 2594.90 | 2339.05 | 255.85 (3P, Mox) | |
| CD8 | Val270–Asn298 | 3643.30 | 3323.68 | 319.62 (4P) | |
a Fractions were from the digestion of OPN with thrombin (T), plasmin (P), or cathepsin D (CD). In some cases, the fragments were further digested with endoproteinase Asp-N in buffer containing 50% H218O.
b Peptides were identified by N-terminal sequence and MALDI-TOF-MS analyses.
c Observed molecular masses (MH+) were determined by MALDI-TOF-MS. Unless mentioned otherwise, masses are monoisotopic. † means the molecular masses were determined in linear mode.
d Theoretical calculated masses are shown.
e Differences between observed and calculated masses are shown. The type of modification corresponding to the mass difference is given in parentheses as follows: phosphorylation (P) and oxidation of methionine (Mox). The C terminus of fragments secondarily digested with Asp-N in 50% H218O was identified by observation of peptides with a normal isotopic pattern.
The determined OPN cleavage sites of the three proteases are summarized in Fig. 5A. Thrombin cleaves OPN at Arg152–Ser153, as reported in the literature, and to a minor degree at Arg228–Leu229. Plasmin preferably cleaves to the C-terminal side of lysine or arginine residues in the C-terminal part of OPN, whereas cathepsin D predominantly cleaves between two hydrophobic amino acids, especially after leucine residues. However, the cleavages at Phe53–Lys54 and Leu151–Arg152 show that this protease can also hydrolyze bonds involving basic amino acids. As seen in Fig. 5B, the region close to the RGD motif containing the identified cleavage sites is very well conserved among all analyzed mammalian sequences. This could suggest that regulatory proteolytic cleavage in this region could be important for OPN functionality.
FIGURE 5.
Plasmin, thrombin, and cathepsin D cleavage sites in OPN. A, cleavage sites for plasmin (P), thrombin (T), and cathepsin D (CD) are indicated with black arrows. Previously identified cleavage sites for MMPs (15) are shown with gray arrows. The RGD motif is highlighted in boldface, and the SVVYGLR sequence is underlined. B, alignment of mammalian OPN sequences. The region containing the identified cleavage sites in human milk is boxed. The integrin-binding motifs RGD and SVVYGLR are highlighted in gray. Glycosylation sites identified in human milk OPN (31) are indicated with a diamond, and corresponding residues in other OPN species are highlighted in black.
Adhesion Assay
OPN is well recognized for its ability to communicate with cells through integrins. This event can be modulated by MMP and thrombin cleavage close to the RGD motif. As plasmin and cathepsin D also cleave OPN close to the RGD sequence, it could be expected that processing by these proteases would likewise influence the ability of OPN to interact with cells. To assess this, the OPN fragments generated by plasmin (Ile1–Arg152/Ile1–Lys154), thrombin (Ile1–Arg152), and cathepsin D (Leu39–Leu151) were purified and examined for their ability to mediate adhesion of different cell types. Likewise, full-length OPN and the mixture of N-terminal fragments were purified, and equimolar concentrations of all forms were used in all binding assays.
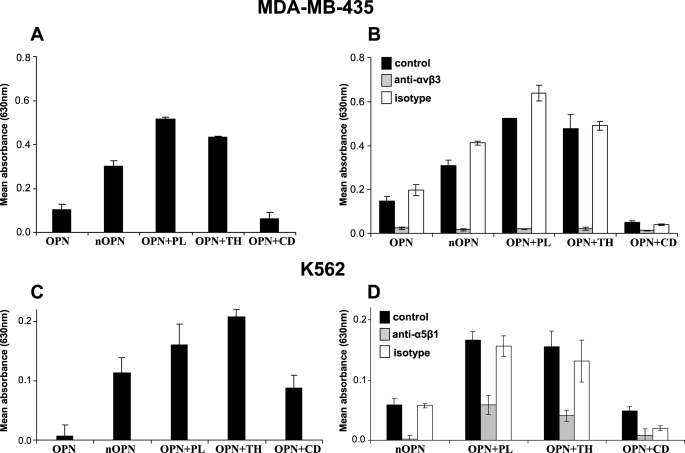
The human cancer cell line MDA-MB-435 has previously been shown to interact weakly with highly phosphorylated OPN in an RGD-dependent manner (34). In this study, weak adherence of these cells to highly phosphorylated full-length milk OPN was also observed (Fig. 6A). The adhesion was increased 3-fold when the mixture of N-terminal OPN fragments was used as binding substrate. Likewise, the binding to the fragments generated by thrombin and plasmin hydrolysis was more than four times higher compared with the intact protein. In contrast, the cathepsin D-cleaved OPN supported only minimal adhesion. To investigate which integrins were responsible for the binding, the adhesion assay was performed in the presence of a blocking antibody against αVβ3. As seen in Fig. 6B, the anti-αVβ3 antibody, but not the isotype control, completely inhibited the adhesion of the cells to all forms of OPN.
FIGURE 6.
Adhesion of cells to cleaved OPN. Adhesion assays were performed with two different cell types to surfaces coated with 5 pmol of full-length OPN or protease-cleaved human milk. OPN is referred to as OPN (full-length), nOPN (mixture of purified N-terminal fragments), OPN+PL (OPN cleaved with plasmin), OPN+TH (OPN cleaved with thrombin), and OPN+CD (OPN cleaved with cathepsin D). Bars show mean values for three wells per protein ± S.D. Data shown are representative of three independent experiments for each cell line. Adhesion to 1% bovine serum albumin was used as background and was subtracted from all values. A, adhesion of MDA-MB-435 cells to surfaces coated with the specified OPN forms. B, adhesion of MDA-MB-435 cells to wells coated with OPN after preincubation in the presence or absence of an antibody against the αVβ3-integrin or an isotype IgG control (5 μg/ml). C, adhesion of K562 cells to surfaces coated with the specified OPN forms. D, adhesion of K562 cells to wells coated with OPN after preincubation in the presence or absence of an antibody against the α5β1-integrin or an isotype IgG control (5 μg/ml).
It has been reported that K562 cells can adhere to thrombin-cleaved OPN in an RGD-dependent manner through the α5β1-integrin receptor but not to the full-length molecule (18). Thus, the binding specificity of this cell line to the different forms of OPN was investigated (Fig. 6C). The adhesion to full-length OPN was very weak and much lower than that mediated by the N-terminal mixture and the cathepsin D fragment, whereas the strongest adherence of the K562 cells was mediated by the plasmin and thrombin fragments. In all cases, the adhesion could be inhibited by an anti-α5β1 antibody (Fig. 6D).
DISCUSSION
Seven N-terminal OPN fragments, generated by proteolysis in the region Leu151–Phe158 right next to the integrin-binding RGD145 sequence, have been identified in human milk. OPN has previously been shown to be a substrate for thrombin (13, 14) and several MMPs (15–17). However, six of the observed cleavage sites identified in this study do not correspond to thrombin or MMP cleavage. This shows that other proteases can cleave OPN in vivo, and these may therefore be important in the regulation of OPN activity. The two endogenous milk proteases, plasmin and cathepsin D, were shown to be able to generate three of the identified fragments.
The characterized N-terminal OPN fragments only differ by single amino acid residues at their C terminus. The identified C-terminal amino acids were Leu151, Arg152, Ser153, Lys154, Ser155, Lys156, and Phe158 (Fig. 2). The proteolytic scenario giving rise to this population of fragments could either be the action of different individual proteases cleaving at specific sites, or alternatively, OPN could initially be cleaved by an endopeptidase and then subsequently be trimmed by carboxypeptidases. Both alternatives are possible, as both endo- and exopeptidases have been identified in human milk (26).
Plasmin and cathepsin D have been recognized as the major proteolytic enzymes in milk (27, 28, 30). Their activity toward OPN in milk is further supported by a previous characterization of four different milk OPN fragments resulting from cleavage at Thr26–Trp27, Leu38–Leu39, Arg159–Arg160, and Lys187–Ala188 (25). Cleavage at the first two positions was also observed in this study when OPN was digested with cathepsin D and at the latter two when it was digested with plasmin (Table 1 and Fig. 5). The identified OPN fragment cleaved at Arg152–Ser153 could be generated in vitro by both thrombin and plasmin, so both proteases could potentially be generating this fragment in milk.
Plasmin and thrombin are both alkaline serine proteinases that hydrolyze peptide bonds at the C-terminal side of the basic amino acids lysine and arginine. However, their specificity is somewhat different as thrombin preferentially cleaves after arginine residues, whereas plasmin has the highest specificity for lysine residues, although this is not as pronounced as the selectivity of thrombin (35). These specificities fit well with the cleavage pattern observed in this study (Fig. 5). Plasmin has long been regarded as the principal protease in milk (27, 28). Human milk contains both active plasmin and the inactive zymogen plasminogen, as well as the two activators of the zymogen, tissue-type plasminogen activator and urokinase-type plasminogen activator (27, 36), whereas the absence or only trace amounts of the plasmin inhibitors α2-antiplasmin and α2-macroglobulin have been demonstrated (37). In human milk, several casein fragments and peptides resulting from plasmin cleavage at lysine residues have been identified (29). The concentration of active plasmin in human milk has been estimated to ∼0.1 μg/ml, whereas the content of plasminogen is ∼0.2 μg/ml (36). The inactive zymogen prothrombin has been identified in human colostrum in a proteomics based study (26), and in a classic early study it has been estimated to be present in human milk at a concentration below 0.25% of the concentration in plasma (38). This roughly corresponds to a milk concentration of prothrombin of ∼0.275 μg/ml (39) and is close to the concentration of plasminogen. However, to our knowledge activated thrombin has never been reported in milk.
The identification of two urinary OPN fragments cleaved at Arg152–Ser153 and Lys154–Ser155 also suggests plasmin or thrombin cleavage of OPN in urine. However, neither prothrombin nor thrombin is present in urine under normal conditions (40, 41), whereas both plasminogen and plasmin activity have been described in urine (42, 43). Collectively, the presence of OPN fragments with exposed SVVYGLR152 sequences in milk and urine is therefore most likely to originate from plasmin activity, although thrombin in the literature has been regarded as the primary protease responsible for cleavage of OPN in vivo.
Likewise, it is highly likely that plasmin cleavage of OPN plays a significant role in other physiological tissues and fluids. In plasma, for instance, the concentrations of prothrombin and plasminogen are more or less the same (∼100–150 μg/ml) (39, 44) as is the case in milk. However, the normal concentrations of the plasmin-α2-antiplasmin (∼0.4 μg/ml) and thrombin-antithrombin III (∼2 ng/ml) complexes (45) indicate that the activation level of plasmin is significantly higher than that of thrombin in plasma. Furthermore, the presence of cleaved OPN assumed to originate from thrombin activity has been correlated with the inflammatory disease rheumatoid arthritis (19, 46). As elevated levels of both thrombin and plasmin have been associated with rheumatoid arthritis (47), plasmin as well as thrombin processing of OPN could potentially be involved in this pathophysiological process.
Cathepsin D is an aspartic endopeptidase located in the lysosomes of cells, but it is also found in a soluble form in milk (26, 30). Cathepsin D has optimum activity at acidic pH, and consequently, the conditions in milk are not optimal as the pH here tends to be neutral. However, proteolytic activity of cathepsin D released by lactating rat mammary epithelial cells has been observed under physiological conditions (48). One mechanism could involve intracellular proteolysis after internalization of the substrate, followed by cathepsin D cleavage within the lysosomes and then release of the degradation products in milk. Alternatively, endopeptidase activity of cathepsin D secreted from various tissues has recently been observed outside the cell under physiological conditions in a manner not involving internalization but dependent on a local acid secretion driven by cell proton extruders (49). The presence of both cathepsin D and OPN has been reported extracellularly in human serum, milk, and urine, as well as in different types of cancer (1, 50). It is therefore likely that OPN, as in milk, could be present in cathepsin D-modified forms in many other physiological compartments. For instance, a fragment suggested as being the result of thrombin and subsequently carboxypeptidase B activity has recently been observed in the synovial fluid of patients with rheumatoid arthritis (19). However, this species is also identical to the OPN fragment generated by cathepsin D, and as this protease has also been associated with the progression of rheumatoid arthritis (51), it could be modifying OPN in this disease.
The role of OPN in milk is not clear; however, it could be involved in cell signaling by engaging integrins in milk or in the gastrointestinal tract. Proteolysis of OPN mediated by plasmin and cathepsin D in milk, as well as in other fluids or tissues, could influence its functionalities as thrombin and MMP cleavage increases cell adhesion and migration facilitated by the protein (7, 8, 15). Consistent with this, a major increase in adhesion of the human cell lines MDA-MB-435 mediated by the αVβ3-integrin and K562 via the α5β1-integrin was observed when the mixture of N-terminal OPN from milk and plasmin- or thrombin-cleaved OPN was used as substrate compared with the full-length protein. Adhesion of both cell types to the mixture of N-terminal fragments was lower than the binding to the plasmin/thrombin-cleaved protein. This indicates that some of the N-terminal fragments are not recognized by the αVβ3- or α5β1-integrins. Indeed, the fragment generated by cathepsin D cleavage at Leu151–Arg152 mediated a lower degree of adhesion than the plasmin- and thrombin-generated fragments. This is in line with a recent study showing that the OPN fragment Ile1–Leu151 mediated significantly lower RGD-dependent cell adhesion than the thrombin fragment Ile1–Arg152 (19).
Plasmin- and thrombin-cleaved OPN mediated equally strong adhesion of the tested cell types through the αVβ3- and α5β1-integrins. Increased cell adhesion of MMP- and thrombin-cleaved OPN via the αVβ3-integrin has been reported (15). The interaction between the α5β1-integrin and OPN is also RGD-dependent but is more strictly controlled as it requires thrombin cleavage (18) and is diminished by MMP-3 (7) and cathepsin D cleavage. Collectively, this shows that plasmin and thrombin cleavage of OPN produces N-terminal fragments with similar affinity for the αVβ3- and α5β1-integrins.
All fragments identified in milk contained various versions of the cryptic SVVYGLR motif. For instance, cathepsin D-cleaved OPN contains a truncated version, SVVYGL, of this binding site, whereas plasmin generates an N-terminal form containing the exact motif, SVVYGLR (minor fragment), and a form with two additional amino acids, SVVYGLRSK (major fragment). All fragments could potentially interact with cells independent of the RGD sequence via the α4β1 and α9β1 integrins. In particular, plasmin-cleaved OPN is interesting, as the major cleavage site of this protease is positioned only two amino acids to the C-terminal side of the thrombin cleavage site and because plasmin- and thrombin-cleaved OPN were observed to be equal in mediating cellular adhesion.
In this study, several OPN fragments generated by cleavage in the Leu151–Phe158 region have been characterized in human milk. Here, it is shown that besides previously described fragments generated by MMPs and thrombin, several other biologically active OPN fragments exist in vivo. In addition, the protein was identified to be a novel substrate for the endopeptidases plasmin and cathepsin D. Plasmin- and thrombin-cleaved OPN showed similar functional properties in relation to cell adhesion, and consequently, plasmin could as such be as relevant for regulation of OPN activity as the well documented thrombin cleavage of OPN.
This work was supported by grants from The Carlsberg Foundation, The Novo Nordisk Foundation, The Danish Dairy Research Foundation, Arla Foods amba, and The Milk Protein Research Consortium.
- OPN
- osteopontin
- RGD
- Arg-Gly-Asp
- SVVYGLR
- Ser-Val-Val-Tyr-Gly-Leu-Arg
- MMP
- matrix metalloproteinase
- MALDI-TOF
- matrix-assisted laser desorption ionization time-of-flight
- RP-HPLC
- reverse phase-high pressure liquid chromatography
- Tricine
- N-[2-hydroxy-1,1-bis(hydroxymethyl)ethyl]glycine
- MS
- mass spectrometry.
REFERENCES
- 1.Sodek J., Ganss B., McKee M. D. (2000) Crit. Rev. Oral Biol. Med. 11, 279–303 [DOI] [PubMed] [Google Scholar]
- 2.Bellahcène A., Castronovo V., Ogbureke K. U., Fisher L. W., Fedarko N. S. (2008) Nat. Rev. Cancer 8, 212–226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wang K. X., Denhardt D. T. (2008) Cytokine Growth Factor Rev. 19, 333–345 [DOI] [PubMed] [Google Scholar]
- 4.Schack L., Lange A., Kelsen J., Agnholt J., Christensen B., Petersen T. E., Sørensen E. S. (2009) J. Dairy Sci. 92, 5378–5385 [DOI] [PubMed] [Google Scholar]
- 5.Nemir M., Bhattacharyya D., Li X., Singh K., Mukherjee A. B., Mukherjee B. B. (2000) J. Biol. Chem. 275, 969–976 [DOI] [PubMed] [Google Scholar]
- 6.Chatterton D. E. W., Rasmussen J. T., Heegaard C. W., Sørensen E. S., Petersen T. E. (2004) Trends Food Sci. Technol. 15, 373–383 [Google Scholar]
- 7.Yokosaki Y., Tanaka K., Higashikawa F., Yamashita K., Eboshida A. (2005) Matrix Biol. 24, 418–427 [DOI] [PubMed] [Google Scholar]
- 8.Kazanecki C. C., Uzwiak D. J., Denhardt D. T. (2007) J. Cell. Biochem. 102, 912–924 [DOI] [PubMed] [Google Scholar]
- 9.Yokosaki Y., Matsuura N., Sasaki T., Murakami I., Schneider H., Higashiyama S., Saitoh Y., Yamakido M., Taooka Y., Sheppard D. (1999) J. Biol. Chem. 274, 36328–36334 [DOI] [PubMed] [Google Scholar]
- 10.Bayless K. J., Davis G. E. (2001) J. Biol. Chem. 276, 13483–13489 [DOI] [PubMed] [Google Scholar]
- 11.Schack L., Stapulionis R., Christensen B., Kofod-Olsen E., Skov Sørensen U. B., Vorup-Jensen T., Sørensen E. S., Höllsberg P. (2009) J. Immunol. 182, 6943–6950 [DOI] [PubMed] [Google Scholar]
- 12.Kim H. J., Lee H. J., Jun J. I., Oh Y., Choi S. G., Kim H., Chung C. W., Kim I. K., Park I. S., Chae H. J., Kim H. R., Jung Y. K. (2009) Proc. Natl. Acad. Sci. U.S.A. 106, 15326–15331 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Senger D. R., Perruzzi C. A., Gracey C. F., Papadopoulos A., Tenen D. G. (1988) Cancer Res. 48, 5770–5774 [PubMed] [Google Scholar]
- 14.Senger D. R., Perruzzi C. A., Papadopoulos A., Tenen D. G. (1989) Biochim. Biophys. Acta 996, 43–48 [DOI] [PubMed] [Google Scholar]
- 15.Agnihotri R., Crawford H. C., Haro H., Matrisian L. M., Havrda M. C., Liaw L. (2001) J. Biol. Chem. 276, 28261–28267 [DOI] [PubMed] [Google Scholar]
- 16.Takafuji V., Forgues M., Unsworth E., Goldsmith P., Wang X. W. (2007) Oncogene 26, 6361–6371 [DOI] [PubMed] [Google Scholar]
- 17.Dean R. A., Overall C. M. (2007) Mol. Cell. Proteomics 6, 611–623 [DOI] [PubMed] [Google Scholar]
- 18.Barry S. T., Ludbrook S. B., Murrison E., Horgan C. M. (2000) Biochem. Biophys. Res. Commun. 267, 764–769 [DOI] [PubMed] [Google Scholar]
- 19.Sharif S. A., Du X., Myles T., Song J. J., Price E., Lee D. M., Goodman S. B., Nagashima M., Morser J., Robinson W. H., Leung L. L. (2009) Arthritis Rheum. 60, 2902–2912 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Grassinger J., Haylock D. N., Storan M. J., Haines G. O., Williams B., Whitty G. A., Vinson A. R., Be C. L., Li S., Sørensen E. S., Tam P. P., Denhardt D. T., Sheppard D., Choong P. F., Nilsson S. K. (2009) Blood 114, 49–59 [DOI] [PubMed] [Google Scholar]
- 21.Zhang Q., Domenicucci C., Goldberg H. A., Wrana J. L., Sodek J. (1990) J. Biol. Chem. 265, 7583–7589 [PubMed] [Google Scholar]
- 22.Shiraga H., Min W., VanDusen W. J., Clayman M. D., Miner D., Terrell C. H., Sherbotie J. R., Foreman J. W., Przysiecki C., Neilson E. G., Hoyer J. R. (1992) Proc. Natl. Acad. Sci. U.S.A. 89, 426–430 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sørensen S., Justesen S. J., Johnsen A. H. (1995) Urol. Res. 23, 327–334 [DOI] [PubMed] [Google Scholar]
- 24.Christensen B., Petersen T. E., Sørensen E. S. (2008) Biochem. J. 411, 53–61 [DOI] [PubMed] [Google Scholar]
- 25.Sørensen S., Justesen S. J., Johnsen A. H. (2003) Protein Expr. Purif. 30, 238–245 [DOI] [PubMed] [Google Scholar]
- 26.Palmer D. J., Kelly V. C., Smit A. M., Kuy S., Knight C. G., Cooper G. J. (2006) Proteomics 6, 2208–2216 [DOI] [PubMed] [Google Scholar]
- 27.Heegaard C. W., Larsen L. B., Rasmussen L. K., Højberg K. E., Petersen T. E., Andreasen P. A. (1997) J. Pediatr. Gastroenterol. Nutr. 25, 159–166 [DOI] [PubMed] [Google Scholar]
- 28.Kelly A. L., O'Flaherty F., Fox P. F. (2006) Int. Dairy J. 16, 563–572 [Google Scholar]
- 29.Ferranti P., Traisci M. V., Picariello G., Nasi A., Boschi V., Siervo M., Falconi C., Chianese L., Addeo F. (2004) J. Dairy Res. 71, 74–87 [DOI] [PubMed] [Google Scholar]
- 30.Vĕtvicka V., Vágner J., Baudys M., Tang J., Foundling S. I., Fusek M. (1993) Biochem. Mol. Biol. Int. 30, 921–928 [PubMed] [Google Scholar]
- 31.Christensen B., Nielsen M. S., Haselmann K. F., Petersen T. E., Sørensen E. S. (2005) Biochem. J. 390, 285–292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Schnölzer M., Jedrzejewski P., Lehmann W. D. (1996) Electrophoresis 17, 945–953 [DOI] [PubMed] [Google Scholar]
- 33.Sørensen E. S., Højrup P., Petersen T. E. (1995) Protein Sci. 4, 2040–2049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Christensen B., Kazanecki C. C., Petersen T. E., Rittling S. R., Denhardt D. T., Sørensen E. S. (2007) J. Biol. Chem. 282, 19463–19472 [DOI] [PubMed] [Google Scholar]
- 35.Weinstein M. J., Doolittle R. F. (1972) Biochim. Biophys. Acta 258, 577–590 [DOI] [PubMed] [Google Scholar]
- 36.Koryckadahl M., Dumas B. R., Chene N., Martal J. (1983) J. Dairy Sci. 66, 704–711 [Google Scholar]
- 37.Lindberg T., Ohlsson K., Weström B. (1982) Pediatr. Res. 16, 479–483 [DOI] [PubMed] [Google Scholar]
- 38.Schønheyder F., Thomsen S. B. (1942) Acta Physiol. Scand. 4, 309–316 [Google Scholar]
- 39.Koldas M., Uras F. (2001) Thromb. Res. 102, 221–227 [DOI] [PubMed] [Google Scholar]
- 40.Vaziri N. D., Toohey J., Paule P., Hung E., Darwish R., Barton C. H., Alikhani S. (1984) Am. J. Med. 77, 433–436 [DOI] [PubMed] [Google Scholar]
- 41.Kitamoto Y., Tomita K., Imamura T. (2004) Ann. Clin. Biochem. 41, 133–137 [DOI] [PubMed] [Google Scholar]
- 42.Oda T., Tamura K., Yoshizawa N., Sugisaki T., Matsumoto K., Hattori M., Sawai T., Namikoshi T., Yamada M., Kikuchi Y., Suzuki S., Miura S. (2008) Nephrol. Dial. Transplant. 23, 2254–2259 [DOI] [PubMed] [Google Scholar]
- 43.Cao Y., Veitonmaki N., Keough K., Cheng H., Lee L. S., Zurakowski D. (2000) Int. J. Mol. Med. 5, 547–551 [DOI] [PubMed] [Google Scholar]
- 44.Vinazzer H. (1988) Haemostasis 18, Suppl. 1, 41–45 [DOI] [PubMed] [Google Scholar]
- 45.Taguchi O., Gabazza E. C., Yoshida M., Yamakami T., Kobayashi H., Shima T. (1996) Clin. Chim. Acta 244, 69–81 [DOI] [PubMed] [Google Scholar]
- 46.Yamamoto N., Sakai F., Kon S., Morimoto J., Kimura C., Yamazaki H., Okazaki I., Seki N., Fujii T., Uede T. (2003) J. Clin. Invest. 112, 181–188 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ohba T., Takase Y., Ohhara M., Kasukawa R. (1996) J. Rheumatol. 23, 1505–1511 [PubMed] [Google Scholar]
- 48.Lkhider M., Castino R., Bouguyon E., Isidoro C., Ollivier-Bousquet M. (2004) J. Cell Sci. 117, 5155–5164 [DOI] [PubMed] [Google Scholar]
- 49.Piwnica D., Fernandez I., Binart N., Touraine P., Kelly P. A., Goffin V. (2006) Mol. Endocrinol. 20, 3263–3278 [DOI] [PubMed] [Google Scholar]
- 50.Benes P., Vetvicka V., Fusek M. (2008) Crit. Rev. Oncol. Hematol. 68, 12–28 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Sohar N., Hammer H., Sohar I. (2002) Biol. Chem. 383, 865–869 [DOI] [PubMed] [Google Scholar]