Abstract
Chemotactic cells must sense shallow extracellular gradients and produce localized intracellular responses. We previously showed that the temporal and spatial activation of two protein kinase B (PKB) homologues, PkbA and PkbR1, in Dictyostelium discoideum by phosphorylation of activation loops (ALs) and hydrophobic motifs had important roles in chemotaxis. We found that hydrophobic motif phosphorylation depended on regulation of TorC2 (target of rapamycin complex 2); however, the regulation of AL phosphorylation remains to be determined at a molecular level. Here, we show that two PDK (phosphoinositide-dependent protein kinase) homologues, PdkA and PdkB, function as the key AL kinases. Cells lacking both PdkA and PdkB are defective in PKB activation, chemotaxis, and fruiting body formation upon nutrient deprivation. The pleckstrin homology domain of PdkA is sufficient to localize it to the membrane, but transient activation of PdkA is independent of PIP3 as well as TorC2 and dispensable for full function. These results confirm the importance of the TorC2-PDK-PKB pathway in chemotaxis and point to a novel mechanism of regulation of PDKs by chemoattractant.
Keywords: Akt PKB, Chemotaxis, Dictyostelium, Phosphatidylinositol-dependent Kinase-1 (Pdk1), Tor Complex (Torc)
Introduction
Many cells can detect extracellular chemical gradients and move toward or away from higher concentrations in a process referred to as chemotaxis or directed cell migration. In embryogenesis, chemotaxis is used repeatedly to rearrange cells, for instance, during primordial germ cell migration, organ formation, and wiring of the nervous system. In the adult, chemotaxis mediates trafficking of immune cells and participates in wound healing and in maintenance of tissue architecture and allows stem cells to target to and persist in their niches. Chemotaxis also plays an important role in pathological states such as excessive inflammation and cancer metastasis.
Current studies indicate that chemotaxis involves a network of interconnected signaling pathways (1). In one important series of events, chemoattractants activate PI3Ks2 that produce the accumulation of phosphatidylinositol 3,4,5-trisphosphate (PIP3) and the protrusion of pseudopodia at the leading edge of the cell (2). It is thought that the effects of PIP3 are mediated by the local recruitment and activation of AKT/PKB. Unregulated overproduction of PIP3, as occurs in cells lacking the lipid phosphatases, PTEN or SHIP1, causes ectopic projections outside of the leading edge and impairs chemotaxis (3, 4). Similarly, direct activation of PI3K, bypassing the receptor, is sufficient to trigger cell extensions (5). Surprisingly, however, cells lacking PIP3 production can still carry out chemotaxis (6–8). The apparent paradox was explained in Dictyostelium discoideum by showing that chemoattractants still trigger the phosphorylation of PKB substrates in the absence of PIP3 (9). This is because the cAMP signaling pathways converge on two PKB homologues, a PIP3-dependent enzyme, PkbA, with a PIP3-specific pleckstrin homology (PH) domain at its N terminus and a PIP3-independent enzyme, PkbR1, which is tethered to the plasma membrane via N-terminal myristoylation (10, 11). The activation of the PKBs, including the PIP3-independent regulation of PkbR1, is restricted to the leading edge of chemotaxing cells, and the substrates of these kinases are believed to regulate the cytoskeleton (9). So far, six novel substrates have been found to be targets of the PKBs. These include the cytoskeletal protein, TalinB, the Ras guanine nucleotide exchange factors GefN and GefS, the PI(4)P-dependent PI5 kinase PI5K, the Rho GTPase-activating enzyme GacQ, and the p21-activated kinase PakA (9, 12). It is not yet understood how phosphorylation of these substrates links chemoattractant signaling to the cytoskeleton.
When cells are stimulated with the chemoattractant, PkbA and PkbR1 are rapidly and transiently phosphorylated within their hydrophobic motifs (HMs) and activation loops (ALs), as has been described for their mammalian counterparts (9, 13–15). Phosphorylation of the HMs of the two PKBs is mediated by TorC2 (target of rapamycin complex 2). One of the earliest genetic defects in chemotaxis to be described in D. discoideum was in the gene PianissimoA (piaA), later shown to be a subunit of TorC2 (16, 17). In PkbA, AL phosphorylation also requires recruitment of the enzyme to the membrane by transient accumulation of PIP3. Because AL phosphorylation in PkbR1 is independent of PIP3, the mechanism of its regulation is not known. It may rely, for example, on an observed absolute prerequisite for HM phosphorylation. However, a version of PkbR1 containing phosphomimetic substitution in the HM, expressed in cells lacking PiaA, is still phosphorylated on the AL with normal kinetics, suggesting an independent layer of regulation of AL phosphorylation (9).
To understand the spatial and temporal regulation of the PKBs by chemoattractant, we sought to determine the kinases that phosphorylate the ALs and study their regulation. In mammalian cells, AL phosphorylation in AKT/PKB is mediated by PDK1 (14, 18). There are two PDK1 homologues in the D. discoideum genome, PdkA and PdkB. In this report, we determine the roles of these enzymes in activation of PkbA and PkbR1 and in regulation of the chemotactic response.
EXPERIMENTAL PROCEDURES
Cell Growth and Differentiation
Cells were cultured axenically in HL5 medium at 22 °C. pdkA−, pdkB−, and pdkA−pdkB− cells were constructed in an AX3 background using a disruption cassette containing the blasticidin S-resistant (BSR) gene. The PDKA disruption plasmid was made by the following steps. First, the region between +744 and +1561 (where +1 is the first base of the open reading frame in the pdkA genomic DNA) was deleted, and a SmaI site was created by PCR using pCR4-PDKA as a template and primers YK176 and YK177. Next, the BSR cassette was inserted into the SmaI site, and the DNA was digested with the EcoRI before transformation into D. discoideum cells. PDKB disruption plasmid was also made by the same strategy as PDKA. The region between +849 and +1967 of the pdkB genomic DNA was replaced by the BSR cassette using pCR4-PDKB and primers YK178 and YK179. The DNA was digested with NotI and PmeI. The pdkA−pdkB− double mutant cells were made as follows. The BSR gene in pdkA− cells was removed by transformation with a Cre expression plasmid. Then, the pdkB gene was disrupted as above. All mutants were confirmed by Southern blot analysis. To assess developmental phenotypes, cells growing in HL5 medium were washed with development buffer (DB) and plated on 1.5% non-nutrient DB agar. For differentiation, cells grown in HL5 were washed with DB and starved in DB for 1 h at 2 × 107 cells/ml and then pulsed with 60 nm of cAMP every 6 min for 4 h (19).
Plasmid Construction
A pdkA genomic DNA fragment was amplified by PCR using AX3 genomic DNA as a template and primers YK151 and YK166 and cloned into pCR4 and finally denoted as pCR4-PDKA. A pdkB genomic DNA fragment was amplified by PCR using AX3 genomic DNA as a template and primers YK154 and YK155 and cloned into pCR4 and finally denoted as pCR4-PDKB. For GFP fusion constructs, both DNA were cloned into the BglII and XhoI sites of pEX-GFP. For PDKA deletion series, each region was amplified by PCR and cloned in pEX-GFP.
Immunoblotting
Differentiated cells were shaken at 200 rpm in DB with 5 mm caffeine for 30 min, washed with DB twice, resuspended at 2 × 107 cells/ml in DB, and kept on ice before assay. Cells were stimulated with 1 μm cAMP, lysed in SDS sample buffer at various time points, and boiled for 5 min. Samples were subjected to electrophoresis on 4–15% SDS-PAGE gels. After electrophoresis, proteins were transferred to a polyvinylidene difluoride membrane. The membrane was blocked with 5% skim milk in Tris-buffered saline with 0.1% Tween 20 and probed with the indicated antibodies. The GFP polyclonal antibody was homemade. All phospho-specific antibodies were purchased from Cell Signaling Technology including anti-phospho-PKB substrate antibody (110B7), anti-phospho-PDK docking motif antibody (18A2), anti-phospho-protein kinase C (pan) antibody (190D10).
Micropipette Assay for Chemotaxis
Differentiated cells were plated on a chambered coverglass (Lab-Tek, Nalgen Nunc) and allowed to adhere to the surface. A micropipette filled with 10 μm cAMP was placed into the field of view. The response of the cells was recorded by time-lapse video every 30 s for 30 min. Chemotactic parameters were analyzed by software packages provided by Y. Xiong and P.A. Iglesias.3 “Speed” was calculated as the total length of the track divided by the elapsed time. “Chemotaxis speed” was calculated as the distance (d1 − d2), where d1 and d2 are the start and the end point of the migration path to the micropipette, divided by the elapsed time. “Chemotactic index” was calculated by an instantaneous, weighted chemotaxis index. For each frame, the cosine of the angle between the direction of movement and the direction to the micropipette was determined. These values were weighted according to the length of the movement step and averaged. “Persistence” was calculated as the shortest linear length between the start and end point of the migration path divided by the total length traveled by the cell.
Protein Translocation to Membrane Assay
Differentiated cells were placed on a chambered coverglass and stimulated at 1 μm cAMP. The images were taken at time-lapse intervals of 2 s for 1 min. For the biochemical translocation assay, differentiated cells were stimulated with 1 μm cAMP and lysed through a 5-μm pore membrane with an equal amount of basal buffer. The membrane-enriched fraction was collected by microcentrifugation.
RESULTS
PdkA and PdkB Are the Activation Loop Kinases of PkbA and PkbR1
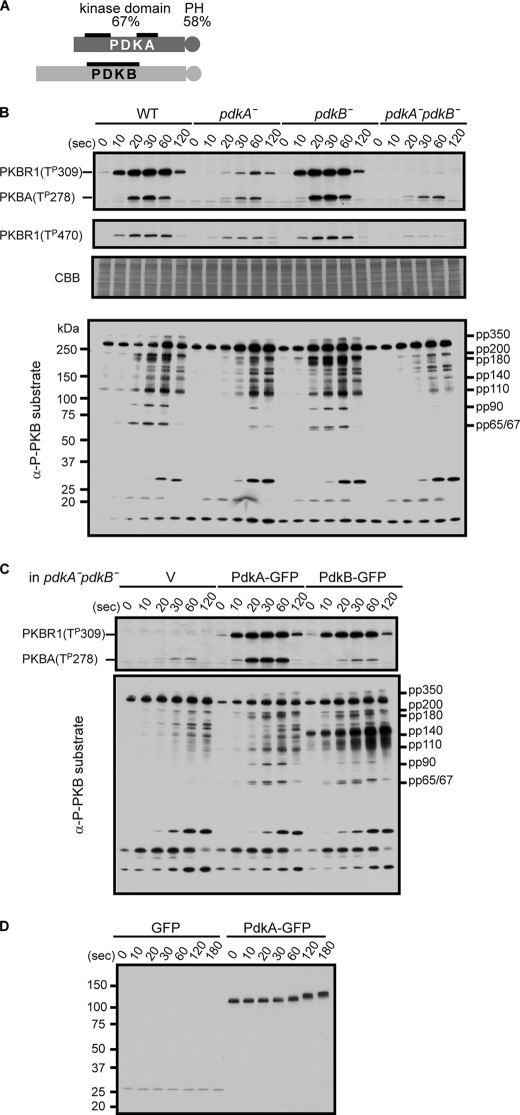
To examine whether PDK homologues in D. discoideum function as the AL kinases, we generated single and double-null mutants of the two PDK1 (phosphoinositide-dependent protein kinase 1) family kinases PdkA (DDB0216243) and PdkB (DDB0216246) in the D. discoideum genome by homologous recombination and examined AL and HM phosphorylations in PkbA and PkbR1. Within the kinase domain, PdkA and PdkB share 52 and 47% identity with mammalian (human) PDK1. Furthermore, like mammalian PDK1, both have a PH domain at the C terminus (Fig. 1A) (20). As previously shown, in wild-type cells, the phosphorylation levels of PkbR1 and PkbA at Thr309 and Thr278 in the ALs, respectively, increased transiently and subsided within 2 min (Fig. 1B) (9). In pdkA− cells, these AL phosphorylations of both PkbR1 and PkbA were reduced, whereas in pdkB− cells, there was no significant difference from wild-type cells. However, AL phosphorylation of PkbR1 was further reduced in pdkA−pdkB− cells to only 3% of that in wild-type cells. In contrast, the AL phosphorylation of PkbA was not further reduced in pdkA−pdkB− cells compared with the pdkA− cells. We also examined phosphorylation of the HM of PkbR1 at Thr470 in each of the cell lines. In wild-type cells, HM phosphorylation increased transiently with kinetics similar to the AL phosphorylation. Compared with wild-type, the integrated levels of phosphorylation were 41% in pdkA−, 100% in pdkB− cells, and 5% in pdkA−pdkB− cells. Previously, we found that alanine substitution of the AL phosphorylation site of PkbR1 caused an ∼50% reduction of HM phosphorylation (9).
FIGURE 1.
PdkA and PdkB are the activation loop kinases of PkbA and PkbR1. A, PdkA and PdkB are schematically represented and share 67 and 58% similarities in a kinase (thick black bars) and PH domain (circles), respectively. B, wild-type (WT), pdkA−, pdkB−, and pdkA−pdkB− cells were stimulated with 1 μm cAMP, and cell extracts were prepared at the indicated times. The phosphorylation levels of the AL, Thr309 for PkbR1 and Thr278 for PkbA, the HM, Thr470 for PkbR1, and the PKB substrates were detected by immunoblotting with α-phospho-protein kinase C (pan) antibody, α-phospho-PDK docking motif antibody, and α-phospho-PKB substrate antibody, respectively. pp350, pp200, pp180 (including GefN), pp140, pp110 (including GefS and PI5K), pp90, and pp65/67 (including GacQ) were already assigned as PKB substrates in α-phospho-PKB substrate antibody staining experiments. The protein-transferred membrane was stained with Coomassie Brilliant Blue (CBB) as a loading control. C, AL phosphorylations of PkbR1 and PkbA (upper panel) and PKB substrates phosphorylations (lower panel) were compared in pdkA−pdkB− cell extracts expressing vector (V), PdkA-GFP, or PdkB-GFP prepared as in B. D, wild-type cells expressing GFP or PdkA-GFP were treated as in B, and cell extracts taken at the indicated times were probed with an α-GFP antibody.
To examine the consequences of the reduced AL phosphorylations of PKB kinases, the cell extracts from each of the cell lines were immunoblotted by the anti-phosphorylation-specific PKB substrates antibodies. As previously shown, pp350, pp200, pp180, pp140, pp110, pp90, and pp65/67 comprise substrates of the PKBs (9). In wild-type cells, phosphorylation in these bands increases rapidly and then subsides within several minutes. The phosphorylation levels of these bands were decreased in accordance with the decreased phosphorylation levels of PkbR1 and PkbA at the AL site. These observations show the importance of AL phosphorylation of the PKBs.
To verify the specificity of these defects in AL phosphorylations, PdkA-GFP or PdkB-GFP were ectopically expressed in the pdkA−pdkB− cells. As shown in the upper panel of Fig. 1C, PdkA-GFP expression restored the AL phosphorylations of PkbR1 and PkbA to wild-type levels. However, PdkB-GFP only functioned for PkbR1 phosphorylation. This is consistent with the observation that in pdkA−pdkB− versus pdkA− cells, only AL phosphorylation of PkbR1 was further reduced (Fig. 1B). In both cell lines, expression of PdkA-GFP or PdkB-GFP restored the phosphorylations of PKB substrates (Fig. 1C, lower panel). Together, these results show that PdkA is a major AL kinase for PkbR1 and PkbA, and PdkB has a minor activity for PkbR1. The residual signals remaining in the pdkA−pdkB− cells suggest that unknown protein kinases also play a minor role for the AL phosphorylations of PkbR1 and PkbA.
Our data suggest that, although PdkA-GFP and PdkB-GFP were overexpressed, this did not cause excessive phosphorylation of the PKBs (compare Fig. 1, B and C and supplemental Fig. S1A). The two proteins were expressed at similar levels and migrated at 125 kDa and 150 kDa, as expected from the calculated molecular weights of PdkA and PdkB, respectively (supplemental Fig. S1A). PdkB-GFP itself is apparently a PKB substrate, as the overexpressed protein showed up as a heavily phosphorylated additional band at 150 kDa in the PKB substrate profile (Fig. 1C and see below). It should be noted that cells overexpressing PkbR1 showed a significant increase of AL phosphorylation of PkbR1 (data not shown). Taken together, these data indicate that PkbR1, not either PDK, is limiting for this phosphorylation event.
Both PdkA and PdkB appear to undergo cAMP-induced modifications. First, we noticed that within 1 min after cAMP stimulation, the mobility of PdkA-GFP shifted from an apparent molecular weight of 125 to 140 (Fig. 1D). GFP from cAMP-stimulated, GFP-expressing cells did not show any change in mobility (Fig. 1D). The retardation typically subsided by 5 min after stimulation, but it was maintained persistently in the presence of 10 mm dithiothreitol, which inhibits extracellular phosphodiesterase (supplemental Fig. S1B). Taken together, these results show that the modifications of PdkA are regulated by cAMP and do not display adaptation. Second, while PdkB-GFP did not show an apparent mobility shift, as noted above, strong staining with the anti-phosphorylation-specific PKB substrates antibodies was observed at ∼150 kDa, specifically in cells expressing PdkB-GFP, and the intensity increased with cAMP stimulation (Fig. 1C, lower panel). This band is phosphorylated PdkB-GFP, as the size corresponded to PdkB-GFP; the band disappeared in the cell extracts after immunodepletion by an anti-GFP antibody (data not shown); and PdkB has a putative PKB phosphorylation motif (RERSST at Thr735) recognized by anti-phosphorylation-specific PKB substrates antibodies.
PDK Activity Is Required for Proper Chemotaxis
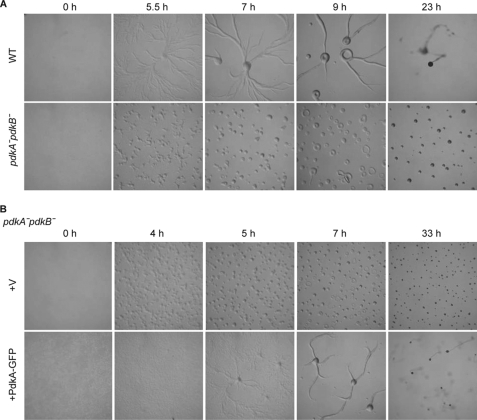
We wondered whether the PDKs were required for the developmental process whereby D. discoideum cells spontaneously aggregate into streams of cells that form a mound, which ultimately transforms into a fruiting body. Since pkbR1− and pkbA−pkbR1− cells arrest at the mound stage, we expected that, as PdkA and PdkB were the AL kinases for the PKBs, they should be also required near this stage of development (9, 10). When wild-type cells were placed on a non-nutrient agar, they made streams beginning ∼5 h and finally formed fruiting bodies (Fig. 2A, upper panel). While pdkA− or pdkB− single null cells formed fruiting bodies like wild-type (data not shown), pdkA−pdkB− cells arrested at the mound stage like pkbR1− and pkbA−pkbR1− cells (Fig. 2A, lower panel). Curiously, pdkA−pdkB− cells started to aggregate rather suddenly, without typical streams, at an early development stage (Fig. 2A, lower panel). We have shown that pkbA−pkbR1− cells are defective in the cell-to-cell signal process required for streaming, and these results suggest that pdkA−pdkB− cells are also important in signal relay.4 To confirm whether the phenotypes of pdkA−pdkB− cells depend on PDK activities, the pdkA−pdkB− cells expressing PdkA-GFP were examined. While cells expressing control vector still showed defects in streams and fruiting body formation, the PdkA-GFP-expressing cells reversed the both defects (Fig. 2B).
FIGURE 2.
Cells lacking PdkA and PdkB are defective in early development. A, wild-type and pdkA−pdkB− cells were starved on a non-nutrient agar plate and allowed to form fruiting bodies. The images were taken at the indicated times. B, pdkA−pdkB− cells expressing vector (V) or PdkA-GFP were treated as in A.
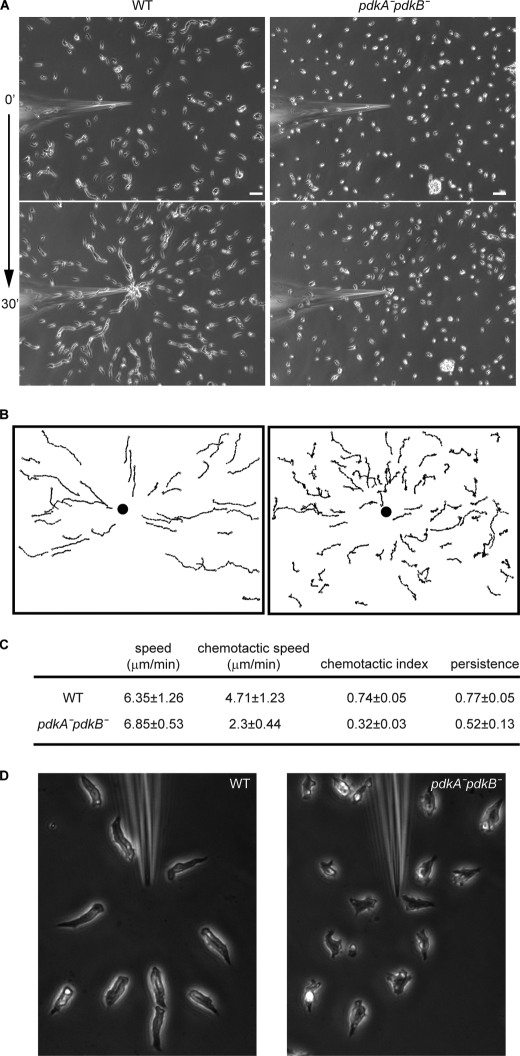
Because pdkA−pdkB− cells had reduced PKB activities and showed similar phenotypes to pkbR1− cells during nutrient deprivation, we expected there would be a defect in their chemotactic activity. Wild-type cells responded to gradients produced at the tip of a micropipette filled with 10 μm cAMP and moved directionally with the usual streams, whereas the chemotaxis of the pdkA−pdkB− cells was impaired (Figs. 3, A and B and supplemental Movies S1 and S2). Compared with wild-type, pdkA−pdkB− cells showed a reduced chemotactic index (0.74 versus 0.32) but a nearly identical speed (Fig. 3C). Also, the mutant cells did not display streams in this assay, again suggesting a defect in signal relay (Fig. 3A). In more magnified images shown in Fig. 3D, wild-type cells had a typical elongated polarized shape, whereas the pdkA−pdkB− cells were less polarized. These defects were not due to impaired development as wild-type and pdkA−pdkB− cells showed similar cAR1 expression profile (data not shown).
FIGURE 3.
Cells lacking PdkA and PdkB are defective in chemotaxis. A, wild-type (WT) and pdkA−pdkB− cells were spread on a coverslip and exposed to cAMP gradients produced by a micropipette containing 10 μm cAMP. The response of cells was recorded by time-lapse video at a 30 s for 30 min frame rate. The first and last images are shown. Scale bars, 50 μm. B, a tracing of the centroid of individual cells is shown. A filled circle indicates the position of needle. C, speed, chemotaxis speed, chemotaxis index, and persistence were quantified by the methods described under “Experimental Procedures” from the movies of three independent assays. D, cell morphology of wild-type and pdkA−pdkB− cells were compared at a 32× magnification.
To show that the chemotaxis defects were specific, we examined the pdkA−pdkB− cells expressing PdkA-GFP. Similar to the pdkA−pdkB− cells, the vector-transfected cells had a reduced chemotactic activity. On the other hand, the PdkA-GFP expressing cells showed significantly better chemotaxis with the appearance of streams (supplemental Fig. S3). Together, these results show that the PDKs are required for chemotaxis, streaming, and progression through the mound stage of development.
Temporal and Spatial Regulation of the Localization of PdkA and PdkB
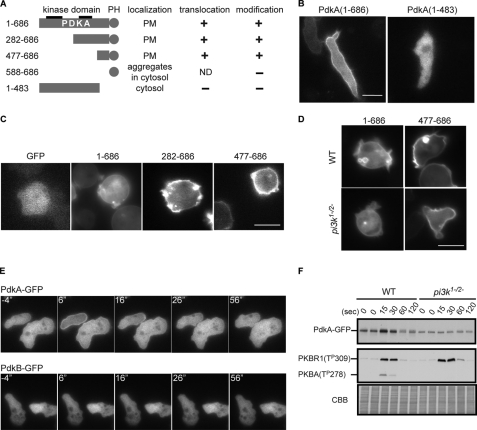
Next, we investigated PdkA and PdkB localizations using the GFP-fused proteins. In mammalian cells, the PH domain of PDK1 can bind to PIP3 and has an important function for AL phosphorylation, although the localization of PDK1 is controversial (21). PdkA-GFP was associated with the plasma membrane in chemotaxing cells and also bound to intracellular vesicles in growth stage cells (Figs. 4, B and C). When chemotaxing cells were treated with caffeine to inhibit de novo cAMP production, the basal membrane binding of PdkA-GFP was reduced. Under these conditions, PdkA-GFP transiently translocated to the plasma membrane, peaking within 10 s after cAMP stimulation (Fig. 4E). The translocation of PdkA-GFP was also observed by disrupting cells mechanically and enriching the membrane fraction. PdkA-GFP accumulated in membrane-enriched fraction within 15 s of after addition of chemoattractant and returned to the cytosol by 1 min (Fig. 4F). On the other hand, PdkB-GFP was localized in the cytosol in chemotaxing cells as well as in growth stage (Fig. 4E and data not shown). Moreover, PdkB-GFP did not change its cytosolic localization upon cAMP stimulation (Fig. 4E).
FIGURE 4.
Spatial and temporal regulation of the localization of PdkA and PdkB. A, the subcellular localization, translocation activities, and chemoattractant-induced modification of a series of deletions of PdkA are summarized. PM, plasma membrane; +, −, or ND means positive, no response, or not determined for chemoattractant-induced translocation or modification, respectively. B, localization of PdkA-(1–686)-GFP and PdkA-(1–483)-GFP were shown in wild-type (WT) cells during chemotaxis. C, fluorescent images of cells expressing GFP and a series of PdkA deletion proteins fused with GFP are shown. D, localizations of PdkA-(1–686)-GFP or PdkA-(477–686)-GFP are compared in wild-type and pi3k1−2− cells. E, to evaluate translocation activity, wild-type cells expressing PdkA-GFP or PdkB-GFP were stimulated with 1 μm cAMP, and fluorescent images were taken every 2 s. Representative responses are shown by a series of time-lapsed images. F, wild-type and pi3k1−2− cells were stimulated with 1 μm cAMP and lysed at the indicated times. The membrane-enriched fraction was collected by centrifugation and probed with α-GFP antibody (upper panel) and α-phospho-protein kinase C (pan) antibody (middle panel). Coomassie Brilliant Blue (CBB) staining of the membrane for the immunoblotting was used as a loading control. Scale bars represent 10 μm.
To delineate the region that is responsible for the localization and/or translocation of PdkA-GFP as well as the modification of PdkA described earlier, we constructed a series of deletions and assessed their activities. A C-terminal region between residues 477 and 686, including the PH domain, was sufficient for both the basal membrane localization as well as the chemoattractant-induced translocation (Fig. 4, A and C). As a construct carrying only a PH domain (amino acids 588–686) produced punctate fluorescence, probably due to artificial aggregations, we could not further delineate the region. Consistently, an N-terminal region containing the first 483 residues was localized to the cytosol and did not translocate in response to cAMP (Fig. 4B and data not shown). Interestingly, while the mobility of PdkA-GFP shifted during chemoattractant treatment, that of PdkA-(1–483)-GFP was constant (Fig. 4A). Further deletion analysis of PdkA-GFP confirmed that the sites of modification were mainly located between residues 477 and 547 (supplemental Fig. S4). These data suggest that the region containing the PH domain of PdkA is necessary and sufficient for the basal membrane localization and translocation to the membrane as well as the cAMP-induced mobility shift.
We used pi3k1−2− cells, previously shown to have greatly decreased PIP3 production, to evaluate the dependence of the localization and translocation of PdkA-GFP on PIP3 (22, 23). While the basal membrane localization still remained in pi3k1−2− cells, the localization on membrane ruffles and/or on the macropinosome-like structures was lost (Fig. 4D). We also found that PdkA-GFP was localized on small unidentified intracellular vesicles. The translocation of PdkA-GFP upon cAMP addition was lost in pi3k1−2− cells when assessed either microscopically or biochemically (data not shown and Fig. 4F). As a control, AL phosphorylations of PkbR1 and PkbA were assessed in the same fractions. Although the AL of PkbR1 was phosphorylated similarly in wild-type and pi3k1−2− cells, the AL phosphorylation of PkbA was disrupted in pi3k1−2− cells. These data are consistent with the previous evidence that ALs of PkbR1 and PkbA are phosphorylated in PIP3-independent and -dependent manners, respectively. These results suggest that PdkA binds to the membrane by two different mechanisms; the basal membrane binding is independent of PIP3, and the localization on membrane ruffles and the macropinosome-like structures and the translocation are dependent on PIP3.
PH Domain of PdkA Is Dispensable for Function
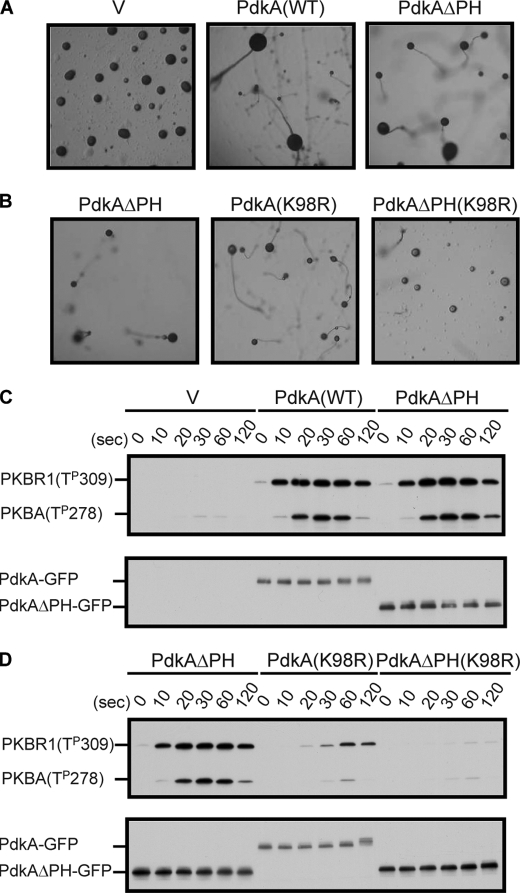
To evaluate importance of the PH domain-containing region of PdkA in vivo, the version of PdkA lacking the PH domain (amino acids 1–483), fused with GFP (PdkAΔPH-GFP), was expressed in pdkA−pdkB− cells and the ability to rescue the developmental and biochemical defects was investigated. As previously shown, pdkA−pdkB− cells arrested at the mound stage upon nutrient deprivation. Surprisingly, the expression of PdkAΔPH-GFP could restore fruiting body formation as effectively as wild-type PdkA-GFP (Fig. 5A). To further assess the role of the PH domain, we created a chimeric PdkA containing K98R mutation, which weakens kinase activity, along with the deletion of the PH domain-containing region. In pdkA−pdkB− cells expressing this chimeric PdkA, compared with a version containing K98R alone, fruiting body formation was partially impaired (Fig. 5B). The mutation K98M, predicted to have no ATP-binding activity, completely eliminated fruiting body formation as well as AL phosphorylations (supplemental Fig. S5). Next, we examined the ability of PdkAΔPH-GFP to phosphorylate the AL of PkbR1 and PkbA. Surprisingly, PdkAΔPH-GFP and PdkA-GFP expressing cells displayed the same intensities of bands (Fig. 5C, upper panel). However, in cells expressing the PdkA(K98R) or the chimeric protein, AL phosphorylation of PkbR1 was reduced by 28 or 1.3%, respectively (Fig. 5D). The levels of expression of PdkAΔPH-GFP were slightly higher than those of PdkA-GFP (Fig. 5C, lower panel). These results showed that the PH domain and modifications of PdkA are dispensable for the AL phosphorylations and the fruiting body formation. However, the PH domain-containing region may have a role for PdkA functions when the proteins are expressed endogenously or the kinase domain is impaired.
FIGURE 5.
The PH domain of PdkA is dispensable for function. A, pdkA−pdkB− cells expressing vector (V), PdkA-GFP, and PdkAΔPH-GFP were starved on a non-nutrient agar plate and allowed to form fruiting bodies. B, pdkA−pdkB− cells expressing PdkAΔPH-GFP, PdkA(K98R)-GFP, or PdkAΔPH(K98R)-GFP were treated as in A. C, cell extracts from pdkA−pdkB− cells expressing vector (V), PdkA-GFP, and PdkAΔPH-GFP were prepared at the indicated times after 1 μm cAMP stimulation and probed with α-phospho-protein kinase C (pan) antibody (upper panel) and α-GFP antibody (lower panel). D, the levels of AL phosphorylations of PkbR1 and PkbA (upper panel) and the expression of PdkA (lower panel) were compared in cells from B.
DISCUSSION
We have shown that PdkA and PdkB are chemoattractant-mediated kinases for PkbA and PkbR1 and that activation of the PKBs, through coordinated phosphorylation on ALs by the PDKs and on HMs by TorC2 have important roles in chemotaxis. We showed previously that PkbA and PkbR1 were heavily dependent on TorC2-catalyzed HM phosphorylation. In this report, we focused on the molecular mechanism of regulation of AL phosphorylation. Assessment of specific PDK-deficient cell lines demonstrated that PdkA is the major AL kinase of PkbR1 and PkbA, that PdkB contributes to phosphorylation of PkbR1, and that additional minor AL-directed activities are present in cells. The pdkA−pdkB− cells were defective in PKB activation and chemotaxis and failed to form proper multicellular structures upon nutrient deprivation. These defects could be largely reversed by expression of PdkA-GFP. This protein localized on the plasma membrane and was additionally recruited by chemotactic stimulation whereas PdkB-GFP was found in the cytosol. The PH domain of PdkA was sufficient for basal localization, which was independent of PI3K, as well as for stimulated recruitment to the membrane, which depended on PI3K. Surprisingly, the PH domain appears to be dispensable for the function of PdkA when the protein is overexpressed, but the PH domain enhances function when the kinase activity of PdkA is compromised. These findings provide further evidence of the significance of the TorC2-PDK-PKB pathway in chemotaxis.
The Role of AL Phosphorylation of PkbR1 and PkbA and the Specificity of PdkA and PdkB
The phosphorylation of the highly conserved site within the AL, thought to bring about a conformational change in the molecule, is essential for PKB activity. We have previously shown that the alanine substitution T309A of PkbR1 caused a nearly complete loss of function. However, because phosphomimetic substitutions T309D and T309E were also nonfunctional, we could not conclude that phosphorylation per se had an essential role (9). Here, we show that pdkA−pdkB− cells display greatly reduced phosphorylation at Thr309 of PkbR1 and Thr278 of PkbA and that kinase activities, assessed by endogenous PKB substrates, are coordinately decreased. Because, as noted above, minor phosphorylations of PkbR1 and PkbA are still present in pdkA−pdkB− cells and there are no PDK homologues other than PdkA and PdkB in the D. discoideum genome, another related kinase can also phosphorylate the ALs. Interestingly, pdkA−pdkB− cells also display decreased HM phosphorylation of PkbR1, consistent with our previously reported observation that T309A in PkbR1 reduced HM phosphorylation by 50% by an unknown mechanism. It is possible that a conformational change caused by AL phosphorylation makes PkbR1 a better substrate for TorC2 or renders the HM resistant to dephosphorylation. Our data suggest that the concomitant phosphorylations at the AL and HM contribute the stable activation of PKBs.
Although PdkA and PdkB are structurally very similar, there are key differences in their functions. First, the degree of a contribution to AL phosphorylation is different. Although pdkA− cells show greatly reduced AL phosphorylation of both PKBs, pdkB− cells maintain nearly wild-type levels. However, when PdkA-GFP or PdkB-GFP was ectopically expressed at similar levels in pdkA−pdkB− cells, the AL phosphorylation of PkbR1 was recovered in a similar way. These data suggest that normally PdkA is more highly expressed than PdkB, leading to the corresponding activities for AL phosphorylations of the PKBs in wild-type cells. Second, the specificity for PkbR1 and PkbA differs for the two PDKs. While PdkA can phosphorylate both PkbR1 and PkbA, PdkB only phosphorylates PkbR1. This preference could arise from the respective kinase domains, as the deletion of PH domain of PdkA does not change specificity. Third, while PdkA localizes on the plasma membrane on structures that resemble macropinosomes and on intracellular vesicles, PdkB localizes in the cytosol. These locations could be due differences in the PH domains, as the association of PdkA with the membranous structures depends on its PH domain. Preliminary examination of the binding of the PH domain of PdkA to lipids using phosphatidylinositol phosphate strips has shown only weak interaction with phosphatidylinositol 3,4-bisphosphate and phosphatidic acid.5 In pi3k1−2− cells, the macropinosome labeling and the recruitment to the plasma membrane were lost, but the basal association with the membrane and intracellular vesicles remained. The molecule responsible for the basal membrane binding remains to be determined. Mammalian PDK1 can physically interact with SGK1 and S6K, which are in the same family of AGC kinases as PkbR1, via the HM in a phosphorylation-dependent manner (24). However, PdkA-GFP or the PH domain of PdkA (477–686)-GFP still localized on the plasma membrane in pkbR1− cells.6
Regulation of PdkA by Chemoattractants
We have previously shown that phosphorylation of the AL of PkbR1 is strictly dependent on prior HM phosphorylation. This result is reminiscent of the phosphorylation of the ALs of SGK1 and S6K by PDK1 in mammalian cells (20). In these instances, PDK1 activity requires phosphorylation-dependent binding to the HM via a PDK1-interacting fragment pocket. Important residues in the PDK1-interacting fragment pocket of PDK1 are conserved in PdkA and PdkB. So far, we have not observed a co-immunoprecipitation of PdkA with PkbR1 in a manner dependent on HM phosphorylation.6 However, a weak interaction might be sufficient to promote the phosphorylation of the AL of PkbR1. Alternatively, phosphorylation of the HM may cause a conformational change in PkbR1 that is a prerequisite for phosphorylation by the PDKs.
We previously reported that a version of PkbR1, bearing the phosphomimetic mutation, T470E, at the key site in the HM, was still transiently phosphorylated at Thr309 in the AL in piaA− cells that lack TorC2 activity, implying that the time course of phosphorylation at the AL is not controlled simply by the profile of HM phosphorylation (9). Rather, it appears that PdkA is transiently activated by chemoattractant stimulation. That is, either the activity of PdkA is regulated or the chemoattractant somehow renders PkbR1 as a better substrate for PdkA. A likely upstream regulator of PdkA-mediated phosphorylation is RasC. When RasC is disrupted, phosphorylation of PkbR1 HM as well as AL is reduced. Furthermore, when constitutively active RasC, RasC(Q62L), is expressed in cells, the phosphorylation of both HM and AL are significantly prolonged.4 The exact molecular mechanism of this regulation of PdkA by RasC awaits for further investigation. Since we showed here that PdkA lacking a PH domain still functions, another portion of the molecule including a kinase domain is regulated upon stimulation.
We have shown that the PH domain of PdkA is dispensable but may enhance the function of the enzyme under certain conditions. The lack of requirement for the PH domain is unexpected but might be consistent with the results from a knock-in mouse of a PH domain mutant of PDK1 that cannot bind to PIP3. Cells from these mice showed reduced AL phosphorylation of AKT, but 35% of the activity was still remaining, suggesting that the PH domain of PDK1 is important but not essential for function (25).
Involvement of PdkA and PdkB in Chemotaxis
The pdkA−pdkB− cells displayed physiological defects that closely resembled those of cells lacking PKB activity. The pdkA−pdkB− cells arrested at the multicellular “mound stage” of development, like the pkbR1− cells and the pkbA−pkbR1− cells (9, 10). They were also defective in chemotaxis like the pkbR1− cells (9). These data suggest that PdkA and PdkB function mainly through PkbR1 and PkbA in the early differentiation phase even though other AGC family kinases are present in D. discoideum cells. These similarities are consistent with the levels of AL phosphorylation observed in each of the strains. However, the defective phenotypes were not exactly the same in pdkA−pdkB− cells and pkbR1− or pkbA− pkbR1− cells. First, the chemotaxis defect, based on the chemotaxis index, in pkbR1− cells appeared stronger than that in pdkA−pdkB− cells. Second, although pkbR1− cells maintained a polarized shape during chemotaxis, pdkA−pdkB− cells were less polarized. Third, the pkbA− pkbR1− cells did not grow in liquid media without addition of killed bacteria, but pdkA−pdkB− cells did. These differences might be due to slightly different remaining levels of PKB activities and different thresholds of each biological event for PKB activity. It would be interesting to compare the phenotypes of pkbA− pkbR1− cells with the cells that completely lack AL phosphorylations of these PKBs.
These findings firmly establish that the TorC2-PDK-PKB pathway plays an important role in regulating directed cell migration. This model implies that regulated phosphorylation of a series of specific PKB substrates would regulate directed migration. We have previously identified six of the proteins that are targets of the TorC2-PDK-PKB pathway. This hypothesis is quite distinct from many other theories about how chemoattractants link to the cytoskeleton. The multiple points of regulation in the TorC2-PDK-PKB pathway make this pathway well suited to confine signaling to the leading edge of cells as we have previously reported. It is interesting that the TorC2-PDK-PKB module, known widely for its role in regulating cell growth transitions, also plays a prominent role in directed migration. It is possible that early phosphorylation events triggered by chemoattractants regulate migration and that subsequent phosphorylation of the other substrates regulates cell growth.
Supplementary Material
Acknowledgments
We acknowledge Yuan Xiong and Pablo A. Iglesias for providing analysis software packages for chemotactic parameters and members of the Devreotes laboratory. We appreciate the dictyBase database for annotation of the pdkA and pdkB gene.
This work was supported, in whole or in part, by National Institutes of Health Grants GM 28007 and GM 34933 (to P. N. D.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Table 1, Figs. S1 and S3–S5, and Movies S1 and S2.
Y. Xiong and P. A. Iglesias, manuscript in submission.
H. Cai, S. Das, Y. Kamimura, C. A. Parent, and P. N. Devreotes, manuscript in preparation.
Y. Kamimura and P. N. Devreotes, unpublished results.
Y. Kamimura and P. N. Devreotes, unpublished data.
- PI3K
- phosphatidylinositol 3-kinase
- PDK
- phosphoinositide-dependent protein kinase
- TorC2
- target of rapamycin complex 2
- PKB
- protein kinase B
- PIP3
- phosphatidylinositol 3,4,5-trisphosphate
- AL
- activation loop
- HM
- hydrophobic motif
- PiaA
- PianissimoA
- BSR
- blasticidin S-resistant
- PH
- pleckstrin homology
- GFP
- green fluorescent protein
- DB
- development buffer.
REFERENCES
- 1.King J. S., Insall R. H. (2009) Trends Cell Biol. 19, 523–530 [DOI] [PubMed] [Google Scholar]
- 2.Franca-Koh J., Kamimura Y., Devreotes P. (2006) Curr. Opin. Genet. Dev. 16, 333–338 [DOI] [PubMed] [Google Scholar]
- 3.Iijima M., Devreotes P. (2002) Cell 109, 599–610 [DOI] [PubMed] [Google Scholar]
- 4.Nishio M., Watanabe K., Sasaki J., Taya C., Takasuga S., Iizuka R., Balla T., Yamazaki M., Watanabe H., Itoh R., Kuroda S., Horie Y., Förster I., Mak T. W., Yonekawa H., Penninger J. M., Kanaho Y., Suzuki A., Sasaki T. (2007) Nat. Cell Biol. 9, 36–44 [DOI] [PubMed] [Google Scholar]
- 5.Inoue T., Meyer T. (2008) PLoS One 3, e3068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chen L., Iijima M., Tang M., Landree M. A., Huang Y. E., Xiong Y., Iglesias P. A., Devreotes P. N. (2007) Dev. Cell 12, 603–614 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.van Haastert P. J., Keizer-Gunnink I., Kortholt A. (2007) J. Cell Biol. 177, 809–816 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ferguson G. J., Milne L., Kulkarni S., Sasaki T., Walker S., Andrews S., Crabbe T., Finan P., Jones G., Jackson S., Camps M., Rommel C., Wymann M., Hirsch E., Hawkins P., Stephens L. (2007) Nat. Cell Biol. 9, 86–91 [DOI] [PubMed] [Google Scholar]
- 9.Kamimura Y., Xiong Y., Iglesias P. A., Hoeller O., Bolourani P., Devreotes P. N. (2008) Curr. Biol. 18, 1034–1043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Meili R., Ellsworth C., Firtel R. A. (2000) Curr. Biol. 10, 708–717 [DOI] [PubMed] [Google Scholar]
- 11.Meili R., Ellsworth C., Lee S., Reddy T. B., Ma H., Firtel R. A. (1999) EMBO J. 18, 2092–2105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chung C. Y., Potikyan G., Firtel R. A. (2001) Mol. Cell 7, 937–947 [DOI] [PubMed] [Google Scholar]
- 13.Alessi D. R., Andjelkovic M., Caudwell B., Cron P., Morrice N., Cohen P., Hemmings B. A. (1996) EMBO J. 15, 6541–6551 [PMC free article] [PubMed] [Google Scholar]
- 14.Alessi D. R., James S. R., Downes C. P., Holmes A. B., Gaffney P. R., Reese C. B., Cohen P. (1997) Curr. Biol. 7, 261–269 [DOI] [PubMed] [Google Scholar]
- 15.Sarbassov D. D., Guertin D. A., Ali S. M., Sabatini D. M. (2005) Science 307, 1098–1101 [DOI] [PubMed] [Google Scholar]
- 16.Chen M. Y., Long Y., Devreotes P. N. (1997) Genes Dev. 11, 3218–3231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lee S., Comer F. I., Sasaki A., McLeod I. X., Duong Y., Okumura K., Yates J. R., 3rd, Parent C. A., Firtel R. A. (2005) Mol. Biol. Cell 16, 4572–4583 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Alessi D. R., Deak M., Casamayor A., Caudwell F. B., Morrice N., Norman D. G., Gaffney P., Reese C. B., MacDougall C. N., Harbison D., Ashworth A., Bownes M. (1997) Curr. Biol. 7, 776–789 [DOI] [PubMed] [Google Scholar]
- 19.Kamimura Y., Tang M., Devreotes P. (2009) Methods Mol. Biol. 571, 255–270 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mora A., Komander D., van Aalten D. M., Alessi D. R. (2004) Semin. Cell Dev. Biol. 15, 161–170 [DOI] [PubMed] [Google Scholar]
- 21.Bayascas J. R. (2008) Cell. Cycle 7, 2978–2982 [DOI] [PubMed] [Google Scholar]
- 22.Funamoto S., Meili R., Lee S., Parry L., Firtel R. A. (2002) Cell 109, 611–623 [DOI] [PubMed] [Google Scholar]
- 23.Huang Y. E., Iijima M., Parent C. A., Funamoto S., Firtel R. A., Devreotes P. (2003) Mol. Biol. Cell 14, 1913–1922 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Biondi R. M., Kieloch A., Currie R. A., Deak M., Alessi D. R. (2001) EMBO J. 20, 4380–4390 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bayascas J. R., Wullschleger S., Sakamoto K., García-Martínez J. M., Clacher C., Komander D., van Aalten D. M., Boini K. M., Lang F., Lipina C., Logie L., Sutherland C., Chudek J. A., van Diepen J. A., Voshol P. J., Lucocq J. M., Alessi D. R. (2008) Mol. Cell Biol. 28, 3258–3272 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.