Abstract
Recently, microRNAs have emerged as regulators of cancer metastasis through acting on multiple signaling pathways involved in metastasis. In this study, we have analyzed the level of miR-10b and cell motility and invasiveness in several human esophageal squamous cell carcinoma cell lines. Our results reveal a significant correlation of miR-10b level with cell motility and invasiveness. Overexpression of miR-10b in KYSE140 cells increased cell motility and invasiveness, whereas inhibition of miR-10b in EC9706 cells reduced cell invasiveness, although it did not alter cell motility. Additionally, we identified KLF4, a known tumor suppressor gene that has been reported to suppress esophageal cancer cell migration and invasion, as a direct target of miR-10b. Furthermore, overexpression of miR-10b in KYSE140 and KYSE450 cells led to a reduction of endogenous KLF4 protein, whereas silencing of miR-10b in EC9706 cells caused up-regulation of KLF4 protein. Coexpression of miR-10b and KLF4 in KYSE140 cells and coexpression of small interfering RNA for KLF4 mRNA and miR-10b-AS in EC9706 cells partially abrogated the effect of miR-10b on cell migration and invasion. Finally, analyses of the miR-10b level in 40 human esophageal cancer samples and their paired normal adjacent tissues revealed an elevated expression of miR-10b in 95% (38 of 40) of cancer tissues, although no significant correlation of the miR-10b level with clinical metastasis status was observed in these samples.
Keywords: Cell/Migration, RNA/MicroRNA, Transcription/Zinc Finger, Tumor/Metastases, Tumor/Suppressor, KLF4, miR-10b
Introduction
MicroRNAs, a class of small noncoding RNAs, have been identified as a new kind of gene expression regulators through targeting mRNAs for translational repression or cleavage (1–3). Lately, emerging evidence suggests important roles for miRNAs2 in apoptosis (4), hematopoietic development (5), cell proliferation (6), skin morphogenesis (7), and neural development (8). Deregulation of miRNAs has also been reported in a variety of tumors, including breast cancer, leukemia, lung cancer, and colon cancer (9–11), which indicated a significant correlation between miRNAs and human malignancy. miRNA expression profiling in esophageal cancers revealed a distinct miRNA signature (12, 13), and some miRNAs showed correlation with several clinicopathologic parameters (13). Furthermore, miR-21 is reported to regulate the proliferation and invasion in ESCC (14), and the miR-106b-25 polycistron is activated by genomic amplification and is potentially involved in esophageal neoplastic progression (15), providing evidence of a causal role for miRNAs in esophageal cancer development.
Recent studies show that miRNAs may act as activators or inhibitors of tumor metastasis by acting on multiple signaling pathways involved in metastasis (16–18). Ma et al. (16) found that miR-10b initiates invasion and metastasis in breast cancer. miR-10b, induced by the prometastatic transcription factor TWIST1, proceeds to inhibit translation of mRNA of HOXD10, a transcription factor already known for its roles in cell motility (19), resulting in increased expression of a pro-metastatic gene, RHOC. This study has provided the first evidence for a role of miRNA in tumor metastasis. Subsequently several additional miRNAs have been reported to act on various steps of metastasis (17, 18).
Krüppel-like factor 4 (KLF4), a zinc finger protein of the Krüppel-like factor family, plays a role in cell cycle regulation, differentiation, and rises in response to DNA damage, serum starvation, and contact inhibition (20, 21). In line with these studies, the loss of KLF4 expression has been reported in several human tumors, including colorectal, stomach, esophageal, and bladder cancers (22–25), which indicates its tumor suppressor role. However, KLF4 also exhibits oncogenic properties. Overexpression of KLF4 could be detected in oropharynx (26) and mammary carcinomas (27). Moreover, it has been reported to inhibit metastasis of several cancers, including esophageal (28), pancreatic (23), and colorectal cancer cells (29). In addition, KLF4 mRNA is targeted by miR-145 (30), implicating the post-transcriptional control of KLF4.
In this study, we have identified a significant correlation between the level of miR-10b and human ESCC cell motility and invasiveness. Furthermore, we have verified a functional role for miR-10b in ESCC cell migration and invasion. Additionally we have identified KLF4 mRNA as a direct target of miR-10b and shown that KLF4 can partly inhibit ESCC cell migration and invasion initiated by miR-10b. Finally, we found elevated expression of miR-10b in 95% (38 of 40) of human esophageal cancer tissues compared with the normal counterparts, although no significant correlation of miR-10b expression with clinical metastasis status was observed.
EXPERIMENTAL PROCEDURES
Cell Line, Cell Culture, and Transfection
Human ESCC cell lines KYSE30, KYSE70, KYSE140, KYSE150, KYSE410, KYSE450, KYSE510, and EC9706 were all established from human ESCC patients (31–33). Among them, the KYSE series cells were generous gifts from Dr. Y. Shimada at Hyogo College of Medicine (Hyogo, Japan), and EC9706 was established and maintained in our laboratory.
Lipofection 2000 (Invitrogen) was used for DNA plasmid transfection, and Hiperfect (Qiagen) was used for oligonucleotide transfection according to the manufacturer's protocols. miRNA inhibitor (miR-10b-AS) was chemically enhanced by 2′O-4′C-methylene (2′-OME) modification. siRNA sequence for KLF4 knockdown was GGACGGCTGTGGATGGAAA (34). Single strand 2′-OME-antisense enhanced green fluorescent protein was used as a negative control for miR-10b-AS and double strand nonsense oligonucleotide for siRNA.
Tissue Specimens
Forty paired tissue specimens (tumor and adjacent normal mucosa) of primary human ESCC were provided by the First Affiliated Hospital of Anhui Medical University (Anhui Province, China). All of the tissues were obtained at the time of surgery and immediately stored in liquid nitrogen until use. None of the patients had received radiotherapy or chemotherapy before surgery. Patients diagnosed with metastasis had lymph node metastasis verified by pathological analysis. For all the samples, clinicopathologic characteristics (age, gender, differentiation, tumor depth, and tumor node metastasis) are shown in Table 1. This study was approved by the ethical committees of the Chinese Academy of Medical Sciences Cancer Institute and the First Affiliated Hospital of Anhui Medical University, and informed consent was obtained from each patient.
TABLE 1.
The clinicopathologic characteristics of 40 ESCC patients
| Clinicopathologic characteristics | Case distribution |
|---|---|
| Age (median) | 38–72 (57) |
| Sex (male/female) | 30/10 |
| Differentiation (well/moderate/poor) | 14/17/9 |
| Tumor depth (submucosa/muscularis propria/adventitia) | 4/9/27 |
| Lymph node metastasis (negative/positive) | 20/20 |
Real Time Quantitative PCR
For real time RT-PCR of miR-10b and KLF4 mRNA, total RNA was isolated using TRIzol solution (Invitrogen). Reverse transcription was performed using Moloney murine leukemia virus reverse transcriptase (Epicenter, Paris, France). miR-10b was reverse transcripted by the looped primer, which binds to six nucleotides at the 3′ portion of miR-10 molecules. Reverse transcription of KLF4 mRNA was performed according to the manufacturer's protocol. Real time PCR was performed using RT Real-TimeTM SYBR Green (SuperArray, Frederick, MD) according to the manufacturer's protocol. The U6 small nuclear RNA and glyceraldehyde-3-phosphate dehydrogenase mRNA were used as internal controls for miR-10b and KLF4 mRNA, respectively. All of the reactions were run in triplicate. The ΔΔCt method for relative quantification of gene expression was used to determine miRNAs and KLF4 mRNA expression levels. The looped RT primer for miR-10 was GTCGTATCCAGTGCGTGTCGTGGAGTCGGCAATTGCACTGGATACGACCACAAA. The PCR primers for miR-10b were GGATACCCTGTAGAACCGAA and CAGTGCGTGTCGTGGAGT. The PCR primers for miR-10a were GGATACCCTGTAGATCCGAA and CAGTGCGTGTCGTGGAGT. The primers for U6 were TGGGGTTATACATTGTGAGAGGA and GTGTGCTACGGAGTTCAGAGGTT. The primers for KLF4 mRNA were CGAACCCACACAGGTGAGAA and TACGGTAGTGCCTGGTCAGTTC. The primers for glyceraldehyde-3-phosphate dehydrogenase mRNA were GGTCGGAGTCAACGGATTTG and ATGAGCCCCAGCCTTCTCCAT.
Plasmid Construction
The genomic sequence of the human miR-10b gene was cloned from mixed DNA of normal esophageal tissues, inserted into pcDNA3.0 (Invitrogen) and named pcDNA3.0-miR-10b. The full-length 3′-UTR of KLF4 was subcloned into the pIS0 luciferase plasmid (35) to generate pIS0-KLF4-3′-UTR. Mutant construct of KLF4 3′-UTR, named pIS0-KLF4-3′-UTR-mut, which carried a substitution of four nucleotides within the core binding sites of KLF4 3′-UTR, was carried out using overlapping extension PCR. pcDNA3.1- KLF4 was previously constructed in our lab (36). The primers for miR-10b were GGCGGATCCCAAGCCCATTAGGCTACCTG and CCGGAATTCTGAGGAGCTTCTGGAGAGGA. The primers for KLF4 3′-UTR were CCGGAGCTCTTTTAAATCCCAGACAGTGGATATG and CGGTCTAGAGGTTTATTTAAAACTTAATTCTCACCTTG. The primers for mutation were TTTATTGGGTGATATCCACAACTTCCA and GATATCACCCAATAAATTATATCCGTGA.
Migration and Invasion Assay
For migration assay, 1 × 105 KYSE140 cells and 0.5 × 105 EC9706 cells were plated onto 24-well Boyden chambers (Corning, Lowell, MA) with an 8-μm pore polycarbonate membrane. For invasion assay, 1.5 × 105 KYSE140 cells and 1 × 105 EC9706 cells were plated on chambers precoated with 20 μg of Matrigel. In both assays, the cells were plated in medium without serum, and medium containing 10% fetal bovine serum in the lower chamber served as chemoattractant. After 18 or 24 h, the cells that did not migrate or invade through the pores were removed by cotton swabs. The inserts were fixed and stained, and three random fields for each insert were counted. The results were averaged among three independent experiments.
Luciferase Reporter Assay
The cells of 90% confluence in 96-well plates were transiently transfected with firefly luciferase reporter gene constructs and miR-10b-expressing plasmid. After 48 h, luciferase activity was measured using a dual luciferase reporter assay system according to the manufacturer's protocol (Promega, Madison, WI), normalized for transfection efficiency by cotransfecting with 0.5 ng of pRL-SV40 Renilla. For each plasmid construct, three independent transfection experiments were performed in triplicate.
Western Blot
Western blots were performed as previously described (37). Protein bands were detected by chemiluminescence using the ECL system (Vigorous Biotech, Beijing, China) according to the manufacture's protocol. The intensity of the bands was quantified by densitometry.
Half-life Analysis
KYSE140 cells of 90% confluence were transfected with pcDNA3.0-miR-10b and control plasmid. 48 h later, the cells were treated with 100 μg/ml cyclohexamide. At the indicated time points, the cell extracts were prepared and analyzed by Western blot with KLF4 and control β-actin antibodies. The band intensities were quantified by densitometry, and the resulting KLF4/β-actin ratios were plotted in a nonlinear regression plot.
Statistical Analysis
Statistical analysis was performed on SPSS13.0. Pearson's correlation analysis was used to examine the correlation between miR-10b and migration and invasion potential and the correlation between miR-10b and KLF4 protein levels of human ESCC cell lines. Differences between two groups were explored by Student's t test. For comparison of paired tissues, paired Student's t test was used to determine the statistical significance. The values were presented as the means ± S.D. A p value of <0.01 was considered to be statistically significant, which is labeled with an asterisk in the figures.
RESULTS
miR-10b Expression Correlates with Cell Motility and Invasiveness in Human ESCC Cell Lines
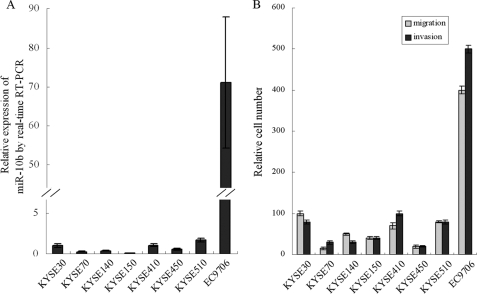
Real time RT-PCR (quantitative PCR) analysis of eight human ESCC cell lines revealed that miR-10b levels varied considerably among the cell lines (Fig. 1A). The highest expression level of miR-10b was observed in EC9706 cells, which was ∼70-fold higher than that in KYSE70, KYSE140, and KYSE150 cells.
FIGURE 1.
Correlation of miR-10b with migration and invasion potential in human ESCC cell lines. A, miR-10b expression in eight ESCC cell lines was measured by real time quantitative RT-PCR, and the U6 small nuclear RNA was used as an internal control. B, migration and invasion potential was assessed by Transwell assays and quantified by relative cell numbers. Pearson's correlation analysis was used to examine the correlation between miR-10b and the migration and invasion potential of human ESCC cell lines.
To test the correlation between the miR-10b level and migration and invasion potential in human ESCC cell lines, we performed Transwell assays on the ESCC cell lines (Fig. 1B). In the KYSE series, statistically significant correlations of the miR-10b level with both cell migration and invasion potential were observed (r = 0.74 and 0.76, respectively, with a p value of <0.05). Taking into account the EC9706 cell line, highly significant correlations were found between miR-10b and cell migration and invasion potential (r = 0.98 and 0.97, respectively, with a p value of <0.001). These results suggested a causal role for miR-10b in the migration and invasion of human ESCC cell lines.
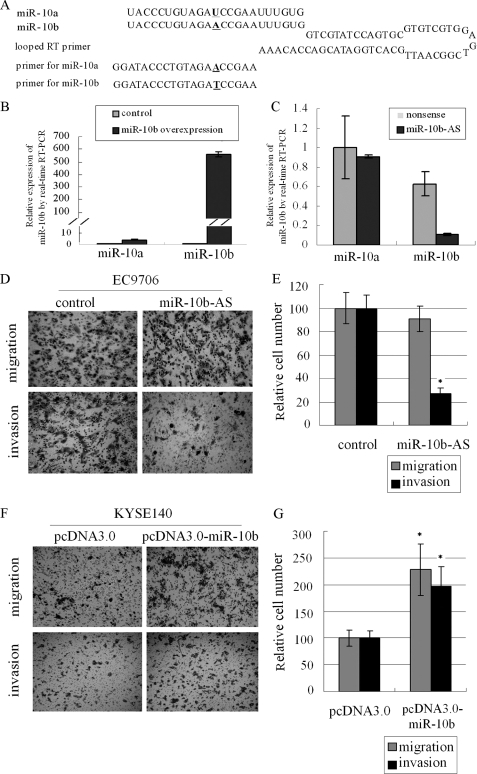
miR-10b is closely related to miR-10a, which differs from miR-10b by a single nucleotide (Fig. 2A). To rule out potential cross-reactivity of miR-10a, we overexpressed miR-10b in KYSE140 cells. Total RNA was converted to cDNA using a looped RT primer (Fig. 2A). Mature miR-10b and miR-10a were measured by real time PCR using miRNA-specific primers (Fig. 2A). The results showed that the miR-10b level was dramatically elevated (>1000-fold), whereas the increase of miR-10a was elevated much less (Fig. 2B). Transfection of the antisense inhibitor miR-10b-AS into EC9706 significantly reduced the level of miR-10b but not that of miR-10a (Fig. 2C), indicating that miR-10b PCR primers were relatively specific, exhibiting weak cross-reactivity with cDNA reverted from mature miR-10a.
FIGURE 2.
miR-10b promotes migration and invasion of human ESCC cell lines. A, sequences of miR-10a and miR-10b, looped primer used for RT and miRNA specific primers for real time PCR. B and C, in B KYSE140 cells transfected by pcDNA3.0-miR-10b and control vector and in C EC9706 cells transfected by miR-10b-AS, miR-10a, and miR-10b expression were measured by real time quantitative RT-PCR. D–G, migration and invasion potential in both cell lines (D and F) treated as above were measured by Transwell assays and were expressed as relative cell numbers (E and G). The results were the means of three independent experiments ± S.D.
miR-10b Promotes Human ESCC Cell Migration and Invasion
To determine whether miR-10b regulates human ESCC cell migration and invasion, we selected EC9706 cells, which show strong migration and invasion potential, and KYSE140 cells, which show weak migration and invasion potential, for further study. We first performed loss-of-function analysis by silencing miR-10b with miR-10b-AS in EC9706 cells. Inhibition of miR-10b in EC9706 cells led to a 73% reduction in invasion assay compared with control cells, whereas no change was observed in the migration potential (Fig. 2, D and E). We then transfected pcDNA-3.0-miR-10b into KYSE140 cells, and a 2-fold increase in cell migration as well as invasion potential was observed (Fig. 2, F and G). In addition, overexpression of miR-10b had no effect on cell proliferation in KYSE140 cells (data not shown). These observations indicated a positive role for miR-10b in migration and invasion of human ESCC cell lines.
KLF4 mRNA Is a Direct Target of miR-10b
To explore the molecular mechanism of miR-10b function, we used TargetScan4.0 to predict targets of miR-10b in human. Several genes, which have been reported to suppress cell migration or invasion, were selected for further analysis. Among them, KLF4 was of particular interest, because it was found to be down-regulated in human esophageal cancer (22, 25, 38, 39) and correlated with cancer metastasis or migration of cancer cells including human esophageal cancer (28).
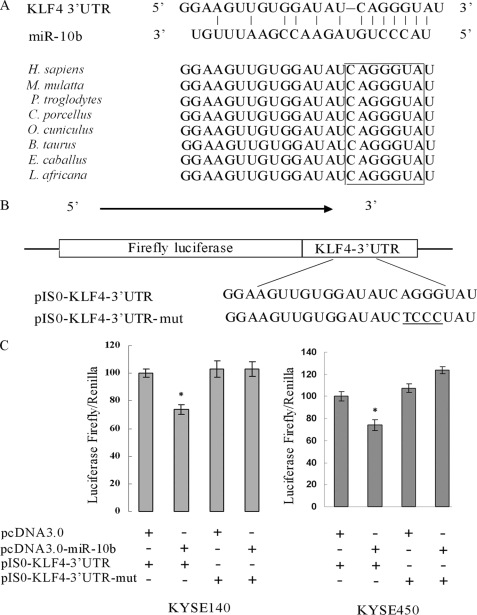
As shown in Fig. 3A, there was a miR-10b binding site at nucleotides 295–301 of the KLF4 3′-UTR. Comparing the human sequence for interspecies homology, we found that the miR-10b target sequence at nucleotides 295–301 of the KLF4 3′-UTR is highly conserved among different species (Fig. 3A). To determine whether KLF4 is a direct target of miR-10b, we constructed pIS0-KLF4-3′-UTR and pIS0-KLF4-3′-UTR-mut (Fig. 3B). Cotransfection of KYSE140 cells with pIS0-KLF4–3′-UTR and pcDNA3.0-miR-10b caused a 30% decrease in the luciferase activity compared with the negative control. This suppression was rescued by the four-nucleotide substitution in the core binding sites (Fig. 3C). The similar effect was also found in KYSE450 cells (Fig. 3C).
FIGURE 3.
The KLF4 3′-UTR is a target of miR-10b. A, a miR-10b target site resides at nucleotides 295–301 of the KLF4 3′-UTR and is highly conserved in different species. Upper panel, sequence alignment of miR-10b with binding sites on the KLF4 3′-UTR. Lower panel, sequence of the miR-10b binding site within the KLF4 3′-UTR of eight species. B, diagram of the luciferase reporter plasmids: plasmid with the full-length KLF4-3′-UTR insert (pIS0-KLF4-3′-UTR) and plasmid with a mutant KLF4-3′-UTR (pIS0-KLF4-3′-UTR-mut) which carried a substitution of four nucleotides within the miR-10b binding site. C, luciferase activity assay demonstrates a direct targeting of the 3′-UTR of KLF4 by miR-10b. KYSE140 and KYSE450 cells were transfected with pcDNA3.0-miR-10b and pIS0-KLF4–3′-UTR/pIS0-KLF4-3′-UTR-mut. pRL-SV40 Renilla was used for normalization of transfection efficiency. After 48 h, the luciferase activities were measured. H. sapiens, Homo sapiens; M. mulatta, Macaca mulatta; P. troglodytes, Pan troglodytes; C. porcellus, Cavia porcellus; O. cuniculus, Oryctolagus cuniculus; B. taurus, Bos taurus; E. caballus, Equus caballus; L. africana, Loxodonta africana.
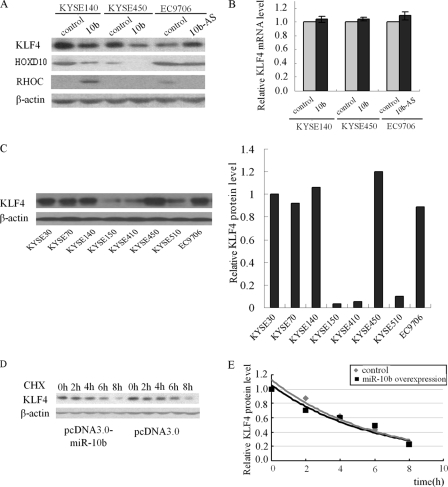
miR-10b Regulates Endogenous KLF4 Protein Expression
To test whether miR-10b expression affected endogenous KLF4 expression, we transfected pcDNA3.0- miR-10b and control plasmid into KYSE140 and KYSE450 cells; a decrease of KLF4 protein level in KYSE140 and KYSE450 cells was observed. Consistent with these results, silencing of miR-10b in EC9706 cells showed an increase in the KLF4 protein level (Fig. 4A). Meanwhile we found that HOXD10 was also regulated by miR-10b in these three cell lines, with a corresponding change of RHOC level (Fig. 4A). Real time quantitative PCR of KLF4 mRNA showed that miR-10b overexpression or inhibition had no effect on KLF4 mRNA level (Fig. 4B), which indicated the post-transcriptional regulation of miR-10b on KLF4 mRNA. We then examined the KLF4 protein levels in eight ESCC cell lines (Fig. 4C) and compared them with miR-10b levels, but no significant correlation was found between the KLF4 protein levels and the miR-10b levels.
FIGURE 4.
miR-10b regulates KLF4 expression at the post-transcriptional level. A, KYSE140 and KYSE450 cells were transfected with pcDNA3.0-miR-10b and control vector. EC9706 were transfected with miR-10b-AS and nonsense sequence. After 48 h, KLF4, HOXD10, and RHOC protein levels were analyzed by Western blot. B, in parallel, KLF4 mRNA in the three cell lines treated as above was measured by real time RT-PCR. Glyceraldehyde-3-phosphate dehydrogenase was used as internal control. C, KLF4 protein level was tested by Western blot, and the band intensities of KLF4 protein were quantified by densitometry and normalized against β-actin. Pearson's correlation analysis was used to examine the correlation between miR-10b and KLF4 protein levels of human ESCC cell lines. D and E, KYSE140 cells, at 48h after transfection of pcDNA3.0 and pcDNA-3.0-miR-10b, were treated with 100 μg/ml cyclohexamide (CHX) and harvested at the indicated time points. Whole cell extracts were analyzed by Western blot in D. Band intensities of KLF4 protein were quantified by densitometry and analyzed in a nonlinear regression plot in E. The results are the means of three independent experiments.
KLF4 is an unstable protein, and ubiquitin-mediated proteasome is involved in its degradation. When new protein synthesis was blocked, KLF4 underwent rapid turnover and exhibited a relatively short half-life of 2 h (40). To rule out any effect of miR-10b on KLF4 protein stability, the half-life of KLF4 was measured. KYSE140 cells transfected with miR-10b-expressing plasmid were treated with cyclohexamide to halt new protein synthesis. At the indicated time points, the cell extracts were prepared and analyzed by Western blot (Fig. 4D). Analysis of KLF4/β-actin ratios by regression plot showed that the half-life of KLF4 in miR-10b overexpressing cells was comparable with control cells (Fig. 4E). Therefore, miR-10b does not change the rate of KLF4 protein degradation. The decreased KLF4 expression in miR-10b overexpressing cells was due to the post-transcriptional regulation of miR-10b.
KLF4 Partly Suppresses Migration and Invasion Initiated by miR-10b
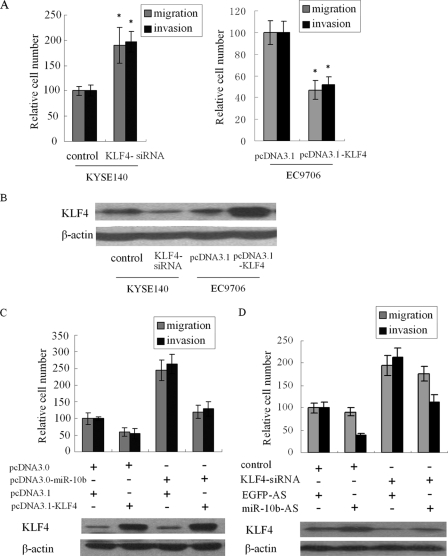
Next, we investigated whether KLF4 suppressed cell migration and invasion in KYSE140 and EC9706 cells. KLF4 has been reported to suppress metastasis of several cancers including esophageal cancer cells (28). As shown in Fig. 5 (A and B), silencing of KLF4 by siRNA in KYSE140 cells led to increased cell migration and invasiveness, whereas overexpression of KLF4 in EC9706 cells resulted in a decrease in cell migration and invasion, demonstrating a negative role for KLF4 protein in the migration and invasion in human ESCC cells.
FIGURE 5.
KLF4 partly suppresses migration and invasion initiated by miR-10b. A, KYSE140 cells were transfected with siRNA targeting KLF4 mRNA and nonsense sequence, and EC9706 cells were transfected with pcDNA3.1-KLF4 and pcDNA3.1. After 48 h, Transwell assays were performed to evaluate the migration and invasion potential of these cells. B, KLF4 protein levels in these cells were analyzed by Western blot. C and D, pcDNA3.1-KLF4, which carried the whole KLF4 coding sequence without 3′-UTR, and pcDNA3.0-miR-10b were cotransfected into KYSE140 cells (C). EC9706 cells were transfected with siRNA for KLF4 mRNA and miR-10b-AS (D). Then migration and invasion assays were performed to evaluate the effect of KLF4 on the function of miR-10b. In parallel, KLF4 protein levels were analyzed by Western blot.
To further establish a functional connection between miR-10b and KLF4, we tested whether KLF4 deregulation was required for regulation of miR-10b on cell migration and invasion. pcDNA3.1-KLF4, which carried the whole KLF4 coding sequence without 3′-UTR, and pcDNA3.0-miR-10b were cotransfected into KYSE140 cells. Interestingly, we found that expression of KLF4 partly abrogated migration and invasion initiated by miR-10b in KYSE140 cells (Fig. 5C). To further test this, we cotransfected EC9706 cells with siRNA for KLF4 mRNA and miR-10b-AS and found that the effect of miR-10b-AS was partially attenuated by siRNA for KLF4 mRNA (Fig. 5D). These results indicate that KLF4 serves as a target of miR-10b, contributing to the effect of miR-10b on cell migration and invasion. In parallel, the KLF4 protein level of both experiments was analyzed by Western blot.
miR-10b Is Significantly Elevated in Human ESCC Tissues
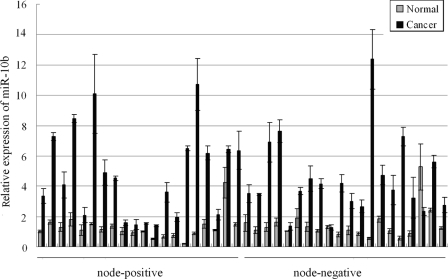
Finally, we studied whether the expression level of miR-10b in vivo was associated with the metastasis outcome in human ESCC patients. We measured the miR-10b level in 40 tumor specimens and their paired normal adjacent tissues. Among these tissues, 20 pairs were node-positive, and the others were node-negative, which was verified by pathological analysis. As shown in Fig. 6, an elevated miR-10b level was detected in 95% (38 of 40) of the tumor tissues compared with their corresponding normal tissues, with a mean fold of 3.39. The elevation of miR-10b levels in cancer tissues was highly significant with a p value of <0.001, indicating the important role of miR-10b in human ESCC. However, Student's t test, used to examine the difference of miR-10b level between node-positive and node-negative tissues, yielded no significant correlation of miR-10b levels with clinical metastasis status, with a p value of 0.242.
FIGURE 6.
miR-10b is significantly up-regulated in human ESCC tissue samples. miR-10b expression in 40 paired tissue specimens (cancer and adjacent normal mucosa) of primary human ESCC was measured by relative quantitative RT-PCR. The U6 small nuclear RNA was used as an internal control. Student's t test was used to examine the difference of miR-10b levels between node-positive and node-negative tissues.
DISCUSSION
miRNAs, a class of small noncoding RNAs, have emerged as important regulators of protein post-transcriptional regulation. Increased numbers of reports reveal the aberrant expression of several miRNAs in tumors with a correlation with certain oncogenes or tumor suppressors or in response to chemotherapy or radiation (9, 41, 42). In the past 2 years, accumulating data have pointed to a regulatory role for miRNAs in cancer metastasis, which was first discovered by Ma et al. (16), who found that miR-10b initiates breast cancer invasion and metastasis. In this study, we investigated the role and the functional target of miR-10b in human ESCC.
We first analyzed the miR-10b levels and migration and invasion potential in eight ESCC cell lines. The results showed that miR-10b level and migration and invasion potential varied significantly among different cell lines. The migration potential of the cell lines that we observed was consistent with a previous study in which they evaluated migration potential of 12 ESCC cell lines (43). Statistically significant correlations were observed between miR-10b expression and the migration and invasion potential in ESCC cells, which suggested a causal role for miR-10b in human ESCC cell migration and invasion in culture. We subsequently confirmed that miR-10b promoted migration and invasion in human ESCC cells, which was similar to that in breast cancer cells (16).
Next we explored the molecular mechanism underlying its function and found that KLF4 was a direct target of miR-10b. KLF4 is a transcription factor that is involved in cell cycle regulation, apoptosis, and differentiation; its expression can be increased by DNA damage, serum deprivation, and contact inhibition (20, 21). Recently, KLF4 has been shown to inhibit cell migration and invasion in TE2 of human ESCC cell line (28). In addition, KLF4 can inhibit pancreatic cancer metastasis in vivo (23). Consistent with these results, KLF4 overexpression reduces cell migration and invasion in RKO colon cancer cells (29). These observations, taken together, indicate that KLF4 has a negative role in migration and invasion of cancer cells, which is opposite to the positive role of miR-10b. To explore the role of KLF4 in the promotion of migration and invasion by miR-10b, we first confirmed the suppression role of KLF4 protein on cell migration and invasion in KYSE140 and EC9706 cells. To further establish the functional connection between miR-10b and KLF4, we overexpressed KLF4 and miR-10b in KYSE140 cells and silenced KLF4 and miR-10b in EC9706 cells and found that KLF4 protein could partly abrogate the function of miR-10b. Therefore, KLF4 protein might have a role in miR-10b regulation of cell migration and invasion. KLF4 overexpression suppressed both cell migration and invasion in EC9706 cells, whereas silencing of miR-10b did not lead to changes of EC9706 cell migration. Additionally, KLF4 overexpression did not completely counteract the effect of miR-10b, which indicated the existence of other targets of miR-10b. In fact, it is already known that HOXD10 is a direct and functional target of miR-10b, which resulted in an increased expression of a well characterized pro-metastatic gene RHOC (16). We confirmed that HOXD10 is regulated by miR-10b with a corresponding change of RHOC in the three ESCC cell lines.
We further investigated the KLF4 protein levels in eight ESCC cell lines and compared them with miR-10b levels, but no significant correlation was found between KLF4 protein levels and miR-10b levels. Because KLF4 has an important role in many biological processes, its expression may be regulated by many different regulators (21, 44). Therefore, miR-10b, as one of its post-transcriptional regulators, could partly influence its protein level but may not have a correlation with its expression level.
miR-10b was initially found to be down-regulated in breast cancer specimens compared with normal breast tissues (45); the down-regulation in breast cancer was further confirmed by others (16, 46). Conversely, miR-10b was subsequently found to be significantly up-regulated in human glioblastoma multiforme (47) and pancreatic adenocarcinomas (48). In the present study, we detected miR-10b level in 40 primary human ESCC tissues and their corresponding normal counterparts and found that 95% (38 of 40) of the cancer tissues had miR-10b expression levels higher than those of the paired normal tissues, with a p value of <0.001, which was in line with the discoveries in human glioblastoma multiforme (47) and pancreatic adenocarcinomas (48). However, no significant correlation was found between the miR-10b level and the metastasis outcome of the patients, which gave rise to the controversy of two groups (16, 46). Ma et al. (16) showed that miR-10b was overexpressed in about 50% (9 of 18) of metastasis-positive tissues compared with metastasis-free tissues or normal breast tissues. However, Gee et al. (46) studied 219 patients and found that miR-10b did not have any significant correlation with factors such as the presence or number of tumor-involved lymph nodes. Our data demonstrated no significant correlation of miR-10b levels with clinical metastasis status. However, this is not necessarily in conflict with the finding that miR-10b promoted the migration and invasion of human ESCC cells. First, the primary cancer was a heterogeneous mass of cells, such as mesenchymal cells, blood cells, and cancer cells. Metastatic or invasive cells might be only a small fraction in the whole cancer samples. Thus it is unlikely to correlate miR-10b with the clinical metastasis status in whole tissue samples. Second, the few metastatic or invasive cells may be at the growing edge of cancer, which may not be contained in the dissected samples. In addition, although miR-10b has been shown to initiate cancer metastasis, considering the multi-steps in cancer metastasis, we are not sure in which steps miR-10b may be involved. Moreover, 40 pairs of cancer samples may not be enough to test the correlation of miR-10b with the clinical metastasis status. Therefore, it is not surprising that no correlation was found between the miR-10b level and the clinical metastasis status in whole cancer samples.
In animal genomes, multiple precursors are found to produce similar mature products (49, 50), which form miRNA families. miR-10a, a close relative of miR-10b, is located at chromosome 17q21 within the cluster of the HOXB genes. The two miRNAs differ only in one nucleotide in their central nucleotides. They are identical at nucleotides 2–8, which seems to be the most important segment for target recognition (51). miR-10a has been studied in several tumors (52–54). According to TargetScan4.0 analysis, the KLF4 mRNA target site for miR-10b overlaps with miR-10a. Moreover, comparing with miR-10b, miR-10a could form a perfectly complementary miRNA-target pairs with KLF4 mRNA. Thus miR-10a might have a similar role in human esophageal cancer, which needs to be further investigated.
In conclusion, we have shown for the first time that the miR-10b level was significantly correlated with the migration and invasion potential of human ESCC cells. Furthermore, miR-10b overexpression promoted cell migration and invasion in human ESCC cell lines. Additionally, we identified KLF4 mRNA as a direct and functional target of miR-10b. Finally, we revealed elevated expression of miR-10b in ESCC tissue specimens, although there was no significant correlation of miR-10b levels with clinical metastasis status. A future challenge will be to identify additional targets of miR-10b to elucidate its role in esophageal cancer.
Acknowledgments
We thank Dr. Y. Shimada for providing KYSE human esophageal carcinoma cell lines and Dr. D. P. Bartel for pIS0 plasmid.
This work was supported by National Basic Research Program of China Grant 2006CB910407 and National Natural Science Foundation of China Grant 30721001.
- miRNA
- microRNA
- ESCC
- esophageal squamous cell carcinoma
- siRNA
- small interfering RNA
- KLF
- Krüppel-like factor
- RT
- reverse transcription
- UTR
- untranslated region
- RHOC
- ras homolog gene family, member C.
REFERENCES
- 1.Pillai R. S. (2005) RNA 11, 1753–1761 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zamore P. D., Haley B. (2005) Science 309, 1519–1524 [DOI] [PubMed] [Google Scholar]
- 3.Bartel D. P. (2004) Cell 116, 281–297 [DOI] [PubMed] [Google Scholar]
- 4.Brennecke J., Hipfner D. R., Stark A., Russell R. B., Cohen S. M. (2003) Cell 113, 25–36 [DOI] [PubMed] [Google Scholar]
- 5.Chen C. Z., Li L., Lodish H. F., Bartel D. P. (2004) Science 303, 83–86 [DOI] [PubMed] [Google Scholar]
- 6.Yi R., Poy M. N., Stoffel M., Fuchs E. (2008) Nature 452, 225–229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yi R., O'Carroll D., Pasolli H. A., Zhang Z., Dietrich F. S., Tarakhovsky A., Fuchs E. (2006) Nat. Genet. 38, 356–362 [DOI] [PubMed] [Google Scholar]
- 8.Schratt G. M., Tuebing F., Nigh E. A., Kane C. G., Sabatini M. E., Kiebler M., Greenberg M. E. (2006) Nature 439, 283–289 [DOI] [PubMed] [Google Scholar]
- 9.Calin G. A., Dumitru C. D., Shimizu M., Bichi R., Zupo S., Noch E., Aldler H., Rattan S., Keating M., Rai K., Rassenti L., Kipps T., Negrini M., Bullrich F., Croce C. M. (2002) Proc. Natl. Acad. Sci. U.S.A. 99, 15524–15529 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Calin G. A., Sevignani C., Dumitru C. D., Hyslop T., Noch E., Yendamuri S., Shimizu M., Rattan S., Bullrich F., Negrini M., Croce C. M. (2004) Proc. Natl. Acad. Sci. U.S.A. 101, 2999–3004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Calin G. A., Liu C. G., Sevignani C., Ferracin M., Felli N., Dumitru C. D., Shimizu M., Cimmino A., Zupo S., Dono M., Dell'Aquila M. L., Alder H., Rassenti L., Kipps T. J., Bullrich F., Negrini M., Croce C. M. (2004) Proc. Natl. Acad. Sci. U.S.A. 101, 11755–11760 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Feber A., Xi L., Luketich J. D., Pennathur A., Landreneau R. J., Wu M., Swanson S. J., Godfrey T. E., Litle V. R. (2008) J. Thorac. Cardiovasc. Surg. 135, 255–260 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Guo Y., Chen Z., Zhang L., Zhou F., Shi S., Feng X., Li B., Meng X., Ma X., Luo M., Shao K., Li N., Qiu B., Mitchelson K., Cheng J., He J. (2008) Cancer Res. 68, 26–33 [DOI] [PubMed] [Google Scholar]
- 14.Hiyoshi Y., Kamohara H., Karashima R., Sato N., Imamura Y., Nagai Y., Yoshida N., Toyama E., Hayashi N., Watanabe M., Baba H. (2009) Clin. Cancer Res. 15, 1915–1922 [DOI] [PubMed] [Google Scholar]
- 15.Kan T., Sato F., Ito T., Matsumura N., David S., Cheng Y., Agarwal R., Paun B. C., Jin Z., Olaru A. V., Selaru F. M., Hamilton J. P., Yang J., Abraham J. M., Mori Y., Meltzer S. J. (2009) Gastroenterology 136, 1689–1700 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ma L., Teruya-Feldstein J., Weinberg R. A. (2007) Nature 449, 682–688 [DOI] [PubMed] [Google Scholar]
- 17.Huang Q., Gumireddy K., Schrier M., le Sage C., Nagel R., Nair S., Egan D. A., Li A., Huang G., Klein-Szanto A. J., Gimotty P. A., Katsaros D., Coukos G., Zhang L., Puré E., Agami R. (2008) Nature Cell Biol. 10, 202–210 [DOI] [PubMed] [Google Scholar]
- 18.Asangani I. A., Rasheed S. A., Nikolova D. A., Leupold J. H., Colburn N. H., Post S., Allgayer H. (2008) Oncogene 27, 2128–2136 [DOI] [PubMed] [Google Scholar]
- 19.Myers C., Charboneau A., Cheung I., Hanks D., Boudreau N. (2002) Am. J. Pathol. 161, 2099–2109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Shields J. M., Christy R. J., Yang V. W. (1996) J. Biol. Chem. 271, 20009–20017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhang W., Geiman D. E., Shields J. M., Dang D. T., Mahatan C. S., Kaestner K. H., Biggs J. R., Kraft A. S., Yang V. W. (2000) J. Biol. Chem. 275, 18391–18398 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ohnishi S., Ohnami S., Laub F., Aoki K., Suzuki K., Kanai Y., Haga K., Asaka M., Ramirez F., Yoshida T. (2003) Biochem. Biophys. Res. Commun. 308, 251–256 [DOI] [PubMed] [Google Scholar]
- 23.Wei D., Kanai M., Jia Z., Le X., Xie K. (2008) Cancer Res. 68, 4631–4639 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhao W., Hisamuddin I. M., Nandan M. O., Babbin B. A., Lamb N. E., Yang V. W. (2004) Oncogene 23, 395–402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Luo A., Kong J., Hu G., Liew C. C., Xiong M., Wang X., Ji J., Wang T., Zhi H., Wu M., Liu Z. (2004) Oncogene 23, 1291–1299 [DOI] [PubMed] [Google Scholar]
- 26.Foster K. W., Ren S., Louro I. D., Lobo-Ruppert S. M., McKie-Bell P., Grizzle W., Hayes M. R., Broker T. R., Chow L. T., Ruppert J. M. (1999) Cell Growth Differ. 10, 423–434 [PubMed] [Google Scholar]
- 27.Foster K. W., Frost A. R., McKie-Bell P., Lin C. Y., Engler J. A., Grizzle W. E., Ruppert J. M. (2000) Cancer Res. 60, 6488–6495 [PubMed] [Google Scholar]
- 28.Yang Y., Goldstein B. G., Chao H. H., Katz J. P. (2005) Cancer Biol. Ther. 4, 1216–1621 [DOI] [PubMed] [Google Scholar]
- 29.Dang D. T., Chen X., Feng J., Torbenson M., Dang L. H., Yang V. W. (2003) Oncogene 22, 3424–3430 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Xu N., Papagiannakopoulos T., Pan G., Thomson J. A., Kosik K. S. (2009) Cell 137, 647–658 [DOI] [PubMed] [Google Scholar]
- 31.Shimada Y., Imamura M., Wagata T., Yamaguchi N., Tobe T. (1992) Cancer 69, 277–284 [DOI] [PubMed] [Google Scholar]
- 32.Kanda Y., Nishiyama Y., Shimada Y., Imamura M., Nomura H., Hiai H., Fukumoto M. (1994) Int. J. Cancer 58, 291–297 [DOI] [PubMed] [Google Scholar]
- 33.Han Y., Wei F., Xu X., Cai Y., Chen B., Wang J., Xia S., Hu H., Huang X., Han Y., Wu M., Wang M. (2002) ZhonghuaYi. XueYi. Chuan Xue Za Zhi. 19, 455–457 [PubMed] [Google Scholar]
- 34.Rowland B. D., Bernards R., Peeper D. S. (2005) Nat. Cell Biol. 7, 1074–1082 [DOI] [PubMed] [Google Scholar]
- 35.Yekta S., Shih I. H., Bartel D. P. (2004) Science 304, 594–596 [DOI] [PubMed] [Google Scholar]
- 36.Zhou Q., Hong Y., Zhan Q., Shen Y., Liu Z. (2009) Cancer Res. 69, 8284–8292 [DOI] [PubMed] [Google Scholar]
- 37.Zhang L., Ding F., Cao W., Liu Z., Liu W., Yu Z., Wu Y., Li W., Li Y., Liu Z. (2006) Clin. Cancer Res. 12, 1639–1646 [DOI] [PubMed] [Google Scholar]
- 38.Bianchi F., Hu J., Pelosi G., Cirincione R., Ferguson M., Ratcliffe C., Di Fiore P. P., Gatter K., Pezzella F., Pastorino U. (2004) Clin. Cancer Res. 10, 6023–6028 [DOI] [PubMed] [Google Scholar]
- 39.Yasunaga J., Taniguchi Y., Nosaka K., Yoshida M., Satou Y., Sakai T., Mitsuya H., Matsuoka M. (2004) Cancer Res. 64, 6002–6009 [DOI] [PubMed] [Google Scholar]
- 40.Chen Z. Y., Wang X., Zhou Y., Offner G., Tseng C. C. (2005) Cancer Res. 65, 10394–10400 [DOI] [PubMed] [Google Scholar]
- 41.He L., He X., Lim L. P., de Stanchina E., Xuan Z., Liang Y., Xue W., Zender L., Magnus J., Ridzon D., Jackson A. L., Linsley P. S., Chen C., Lowe S. W., Cleary M. A., Hannon G. J. (2007) Nature 447, 1130–1134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Meng F., Henson R., Lang M., Wehbe H., Maheshwari S., Mendell J. T., Jiang J., Schmittgen T. D., Patel T. (2006) Gastroenterology 130, 2113–2129 [DOI] [PubMed] [Google Scholar]
- 43.Tong T., Zhong Y., Kong J., Dong L., Song Y., Fu M., Liu Z., Wang M., Guo L., Lu S., Wu M., Zhan Q. (2004) Clin. Cancer Res. 10, 7304–7310 [DOI] [PubMed] [Google Scholar]
- 44.Ghaleb A. M., Aggarwal G., Bialkowska A. B., Nandan M. O., Yang V. W. (2008) Mol. Cancer Res. 6, 1920–1927 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Iorio M. V., Ferracin M., Liu C. G., Veronese A., Spizzo R., Sabbioni S., Magri E., Pedriali M., Fabbri M., Campiglio M., Ménard S., Palazzo J. P., Rosenberg A., Musiani P., Volinia S., Nenci I., Calin G. A., Querzoli P., Negrini M., Croce C. M. (2005) Cancer Res. 65, 7065–7070 [DOI] [PubMed] [Google Scholar]
- 46.Gee H. E., Camps C., Buffa F. M., Colella S., Sheldon H., Gleadle J. M., Ragoussis J., Harris A. L. (2008) Nature 455, E8–E9 [DOI] [PubMed] [Google Scholar]
- 47.Ciafrè S. A., Galardi S., Mangiola A., Ferracin M., Liu C. G., Sabatino G., Negrini M., Maira G., Croce C. M., Farace M. G. (2005) Biochem. Biophys. Res. Commun. 334, 1351–1358 [DOI] [PubMed] [Google Scholar]
- 48.Bloomston M., Frankel W. L., Petrocca F., Volinia S., Alder H., Hagan J. P., Liu C. G., Bhatt D., Taccioli C., Croce C. M. (2007) J. Am. Med. Assoc. 297, 1901–1908 [DOI] [PubMed] [Google Scholar]
- 49.Tanzer A., Stadler P. F. (2004) J. Mol. Biol. 339, 327–335 [DOI] [PubMed] [Google Scholar]
- 50.Tanzer A., Amemiya C. T., Kim C. B., Stadler P. F. (2005) J. Exp. Zool. Mol. Dev. Evol. 304, 75–85 [DOI] [PubMed] [Google Scholar]
- 51.Lewis B. P., Burge C. B., Bartel D. P. (2005) Cell 120, 15–20 [DOI] [PubMed] [Google Scholar]
- 52.Tan Y., Zhang B., Wu T., Skogerbø G., Zhu X., Guo X., He S., Chen R. (2009) BMC Mol. Biol. 10, 12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Veerla S., Lindgren D., Kvist A., Frigyesi A., Staaf J., Persson H., Liedberg F., Chebil G., Gudjonsson S., Borg A., Månsson W., Rovira C., Höglund M. (2009) Int. J. Cancer 124, 2236–2242 [DOI] [PubMed] [Google Scholar]
- 54.Agirre X., Jiménez-Velasco A., San José-Enériz E., Garate L., Bandrés E., Cordeu L., Aparicio O., Saez B., Navarro G., Vilas-Zornoza A., Pérez-Roger I., García-Foncillas J., Torres A., Heiniger A., Calasanz M. J., Fortes P., Román-Gómez J., Prósper F. (2008) Mol. Cancer Res. 6, 1830–1840 [DOI] [PubMed] [Google Scholar]