Abstract
The PARP-3 protein is closely related to the PARP-1 and PARP-2 proteins, which are involved in DNA repair and genome maintenance. Here, we characterized the biochemical properties of human PARP-3. PARP-3 is able to ADP-ribosylate itself as well as histone H1, a previously unknown substrate for PARP-3. PARP-3 is not activated upon binding to DNA and is a mono-ADP-ribosylase, in contrast to PARP-1 and PARP-2. PARP-3 interacts with PARP-1 and activates PARP-1 in the absence of DNA, resulting in synthesis of polymers of ADP-ribose. The N-terminal WGR domain of PARP-3 is involved in this activation. The functional interaction between PARP-3 and PARP-1 suggests that it may have a role in DNA repair. However, here we report that PARP-3 small interfering RNA-depleted cells are not sensitive to the topoisomerase I poison camptothecin, inducing DNA single-strand breaks, and repair these lesions as efficiently as wild-type cells. Altogether, these results suggest that the interaction between PARP-1 and PARP-3 is unrelated to DNA single-strand break repair.
Keywords: ADP-ribosylation, DNA/Repair, Enzymes, Enzymes/Mechanisms, Histones/Modification, Protein/DNA Interactions
Introduction
Poly(ADP-ribosylation) is a ubiquitous protein modification that is important in the regulation of transcription, cell proliferation, differentiation, and apoptosis. Poly(ADP-ribosylation) occurs in almost all nucleated cells of mammals, plants, and lower eukaryotes but is absent in yeast. It represents an immediate cellular response to DNA damage induced by ionizing radiation, alkylating agents, and oxidants (1, 2). PARP-1 (poly(ADP-ribose) polymerase 1) is an abundant nuclear protein that is activated by DNA strand breaks to modify acceptor proteins with poly(ADP-ribose) (PAR)4 (3). PARP-1 protects DNA breaks and chromatin structure and recruits DNA repair and checkpoint proteins to sites of damage (4–6). ADP-ribose is produced from NAD+ by cleavage of the glycosidic bond between nicotinamide and ribose, a reaction catalyzed by PARPs. Hydrolysis of the high energy bond between the nicotinamide and ribose produces a free energy of −34.3 kJ/mol (−8.2 kcal/mol). This energy is used by PARPs for post-translational modification of proteins by synthesizing ADP-ribose polymers attached to the proteins. PARPs can auto-modify themselves or heteromodify other proteins (1, 2).
Human PARPs constitute a large family of 17 proteins encoded by different genes and displaying a conserved catalytic domain. In addition to a catalytic domain, PARP family members typically contain one or more additional motifs or domains, including zinc fingers, BRCT (BRCA1 C terminus-like) motifs, ankyrin repeats, macrodomains and different types of protein/protein interaction sites (7, 8). PARP-1 has a highly conserved structure, including an N-terminal DNA-binding domain, a central auto-modification domain, and a C-terminal catalytic domain.
Of the human PARP enzymes, at least PARP-1, PARP-2, and tankyrase-1 are required for the maintenance of genome stability. Inhibition of PARP-1 is synthetic lethal with defects in homologous recombination and is currently tested as a monotherapy for heritable breast and ovarian cancers deficient in the BRCA1 or BRCA2 gene (9–11). Several PARP inhibitors are currently being tested in clinical trials for cancer treatment. However, it is unclear whether the efficiency of the PARP inhibitors is due to inhibition of PARP-1 alone or if they also inhibit other PARP family members. Therefore, to better understand the effects of PARP inhibition in cancer treatment, information on other PARP family members is needed. In particular, PARP-3 is highly related to PARP-1 and PARP-2, but there is little information on the biochemical properties of this enzyme. PARP-3 is a 60-kDa protein containing an N-terminal WGR (tryptophan-, glycine-, and arginine-rich) domain and a C-terminal catalytic domain. The crystal structure of the human PARP-3 catalytic domain was determined recently (Protein Data Bank code 3C4H) (12) and was found to be highly similar to mammalian PARP-1 and PARP-2 catalytic domains (13, 14). The WGR domain contains conserved tryptophan, glycine, and arginine residues, but the function of this domain remains unknown. It has been speculated that it could have a nucleic acid-binding function (15).
There are contradicting data about the nuclear localization of PARP-3. It was reported that PARP-3 localizes to the centrosome, particularly to the daughter centriole (16), but another group showed that the protein mostly associates with polycomb group bodies (17). Functional analysis revealed that overexpression of PARP-3 interferes with the G1/S cell cycle but does not influence centrosomal duplication (16). It was shown that PARP-3 interacts with PARP-1 and co-immunoprecipitates with the catalytic subunit of DNA-dependent protein kinase, DNA ligases III and IV, Ku80, and Ku70, proteins involved in DNA repair (16, 17). Later, it was shown that PARP-3 also binds to histones H3C and H2BE (18). Whether PARP-3 has a function in the DNA damage response or not is still unknown.
The aim of our study was to investigate the biochemical properties of PARP-3 to better understand its cellular functions. Despite the high degree of similarity to the catalytic domains of PARP-1 and PARP-2, PARP-3 possesses certain functional differences. We show that PARP-3 is an active enzyme with auto- and trans-ADP-ribosylation activity. PARP-3 is a mono-ADP-ribosylase in contrast to PARP-1 and PARP-2. We also show that PARP-3 can activate PARP-1 in the absence of DNA.
EXPERIMENTAL PROCEDURES
ADP-ribosylation Assay
Expression and purification of recombinant full-length PARP-3 and truncated PARP-3 (Lys178–His532; tPARP-3) were as described previously (12). PARP-1 was purchased from Alexis Corp. The ability of PARP-3 to catalyze the ADP-ribosylation of itself and histone H1 was assayed using 6-biotin-17-NAD (Bio-NAD+; Trevigen, Inc.). 0.8 μm PARP-3 or tPARP-3 was incubated at 22 °C in PARP reaction buffer (50 mm Tris-HCl buffer (pH 8.0) and 2 mm MgCl2) with 25 μm Bio-NAD+, 75 μm NAD+, and 2.4 μm histone H1 (New England Biolabs) in the absence or presence of 5 μg/ml activated DNA (Sigma). The reaction was stopped by the addition of lithium dodecyl sulfate (LDS) sample buffer (Invitrogen) and freezing on dry ice at the following time points: 0, 0.5, 1, 1.5, 2, 5, 10, 15, and 20 min. The samples were separated on NuPAGE Novex 4–12% BisTris gels (Invitrogen) and blotted onto polyvinylidene difluoride membranes (GE Healthcare). The blots were probed with anti-biotin antibody (Roche Diagnostics), followed by incubation with horseradish peroxidase-conjugated secondary antibody (Thermo Scientific), and protein bands were visualized using SuperSignal West Femto chemiluminescence substrate (Thermo Scientific).
Inhibition of Auto-ADP-ribosylation of PARP-3 with Different Inhibitors
250 nm purified PARP-3 was preincubated for 5 min at room temperature in PARP reaction buffer with varying concentrations of PARP-1 inhibitors: 3-aminobenzamide, 1,5-dihydroisoquinoline, 1,8-naphthalimide, 4-amino-1,8-naphthalimide, DR2313(2-methyl-3,5,7,8-tetrahydrothiopyrano[4,3-d]pyrimidine-4-one), PJ34 (N-(6-oxo-5,6-dihydrophenanthridin-2-yl)-(N,N-dimethylamino)acetamide), and KU0058948. The reaction was started by the addition of 25 μm Bio-NAD+. After 5 min of incubation at room temperature, the reaction was stopped by dilution in LDS sample buffer. Samples were analyzed as described above. For comparison, the activity of 10 nm PARP-1 was assayed in the presence of varying concentrations of the same inhibitors and under the same reaction conditions. PARP-1 was preincubated with the inhibitors for 5 min, and reactions were started by the addition of Bio-NAD+ and activated DNA. IC50 values for the PARP inhibitors against PARP-3 and PARP-1 were estimated from the intensities of the ADP-ribose signal after quantification using Adobe Photoshop CS2.
Interaction between PARP-3 and PARP-1
To confirm the reported interaction between PARP-3 and PARP-1 and to check how strong the interaction is, a binding experiment was performed. U2OS cells overexpressing Myc-PARP-1 were lysed in 50 mm HEPES (pH 7.5), 150 mm NaCl, 1 mm EDTA, 1 mm EGTA, 10% glycerol, 1% Triton X-100, and Complete protease inhibitors (Roche Applied Science) and spun in a microcentrifuge to clear the lysate. Anti-Myc antibody conjugated to agarose beads (sc-40, Santa Cruz Biotechnology) was added to the cellular extract and incubated overnight on a rotating unit at 4 °C. Beads were washed with lysis buffer containing 300 mm NaCl and incubated with purified PARP-3 for 4 h. Beads were then washed four times with lysis buffer containing 1 m NaCl or 0.1% SDS, and co-immunoprecipitated proteins were eluted by boiling the beads in LDS sample buffer. After electrophoresis, samples were blotted onto polyvinylidene difluoride membrane and probed with mouse anti-PARP-1 monoclonal antibody (sc-8007) and goat anti-PARP-3 polyclonal antibody (sc-30625) (Santa Cruz Biotechnology). Additional steps were performed as described above.
To investigate if the interaction between PARP-3 and PARP-1 has any effect on the activity of these proteins, 500 nm PARP-3 or tPARP-3 was incubated in PARP reaction buffer with 25 μm Bio-NAD+ and 75 μm NAD+ in the absence or presence of 20 nm PARP-1 without the addition of activated DNA. After 2.5, 5, and 10 min of incubation at room temperature, the reactions were stopped by dilution in LDS sample buffer. In additional experiments prior to incubation with PARP-3, PARP-1 was inhibited with KU0058948 at 50 nm, a concentration that inhibits PARP-1 but not PARP-3 to any large extent. The samples were analyzed as described above. To analyze the effect of Ca2+ on the activity of PARP-1 in the presence of PARP-3, a time course experiment was conducted as described above in the presence of 2 mm CaCl2.
To check the dependence of PARP-1 activity on the amount of PARP-3 present, 20 nm PARP-1 was incubated with different concentrations of PARP-3 in PARP reaction buffer in the presence of 25 μm Bio-NAD+ and 75 μm NAD+ and in the absence of DNA for 5 min. The reaction was stopped by dilution in LDS sample buffer, and the samples were analyzed as described above.
Inhibition with meta-Iodobenzylguanidine (MIBG)
250 nm purified PARP-3 was incubated with 12.5 μm Bio-NAD+ in PARP reaction buffer and with different concentrations of MIBG for 30 min at room temperature. The reaction was stopped by dilution in LDS sample buffer.
Hydrolysis with HgCl2
125 nm purified PARP-3 was incubated with 12.5 μm Bio-NAD+ in PARP reaction buffer for 30 min at room temperature. HgCl2 was added to the reaction mixture to 1 mm and incubated for 1, 2, or 4 h. Control reactions were supplemented with 1 mm NaCl and incubated as described above. The reactions were stopped by dilution in LDS sample buffer.
Hydrolysis with Hydroxylamine
1 μm or 100 nm purified PARP-3 or 10 nm purified PARP-1 was incubated with 25 μm Bio-NAD+ in PARP reaction buffer for 30 min at room temperature. Activated DNA (5 μg/ml) was added to the PARP-1 reactions. The reactions were thereafter mixed with neutral NH2OH to 1 m and incubated for 1, 2, or 4 h at room temperature. The control reactions were mixed with NaCl to 1 m and treated as described above. The reactions were stopped by dilution in LDS sample buffer.
Synthesis and Separation of ADP-ribose Polymers
10 μg of PARP-3 was incubated with 1 mm NAD+ in PARP reaction buffer. 1 μg of PARP-1 was incubated with 1 mm NAD+ and 5 μg/ml activated DNA. After 30 and 60 min of incubation at 37 °C, aliquots of the reaction products were taken for analysis by Western blotting with the antibody to PARP-3 (sc-30625) and the antibody to PARP-1 (sc-8007). The reaction was stopped by the addition of ice-cold trichloroacetic acid to a final concentration of 20% (w/v). PARs were isolated from trichloroacetic acid precipitates by the method of Panzeter and Althaus (19). Polymers were loaded on a 20% polyacrylamide gel (19:1 acrylamide/bisacrylamide) in 100 mm Tris borate buffer (pH 8.3) and 2 mm EDTA. Following electrophoretic separation, the gel was stained using a Pierce color silver stain kit following the manufacturer's instructions.
Small Interfering RNA (siRNA) and Camptothecin Treatment
siRNA against PARP-1, PARP-2, or PARP-3 was designed in-house as described (9). Scrambled siRNA was purchased from Dharmacon and Qiagen. For clonogenic survival assay, 1 × 104 SW480 cells grown overnight in 6-well plates were transfected with 100 nm siRNA using Oligofectamine reagent (Invitrogen) according to the manufacturer's instructions. Cells were then cultured in normal growth medium for 48 h prior to trypsinization and replating in the presence or absence of increasing doses of camptothecin for 24 h. Depletion was confirmed by reverse transcription-PCR as described previously (9).
RESULTS
Auto- and Trans-ADP-ribosylation Activity of PARP-3
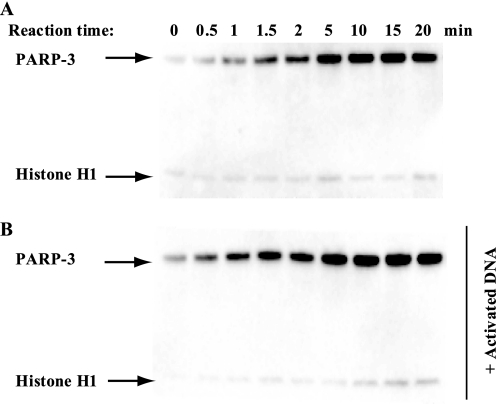
We investigated the ADP-ribosylation activity of both truncated PARP-3 (amino acids 178–532, lacking the WGR domain) and full-length PARP-3 (amino acids 1–532) using Bio-NAD+ and histone H1 as a substrate. PARP-3 was able to ADP-ribosylate itself (auto-ADP-ribosylation) as well as histone H1 (trans-ADP-ribosylation), a previously unknown substrate of PARP-3 (Fig. 1A). We investigated if PARP-3 activity is increased by the addition of activated DNA, as is the case for PARP-1. We found that the addition of activated DNA did not influence PARP-3 activity (Fig. 1B). The ratio between the substrate specificities of PARP-3 for PARP-3 and histone H1 did not reveal any preference for either of the two substrates. However, in the presence of DNA, PARP-3 was the preferred substrate. This is likely to be an effect of DNA binding to histones and preventing them from interacting with the enzyme. tPARP-3 possesses both auto-ADP-ribosylase activity and trans-ADP-ribosylase activity using histone H1 as a substrate (supplemental Fig. S1), showing that the N-terminal part with the WGR domain is not crucial for the ADP-ribosylase activity. Further characterization was performed using full-length PARP-3.
FIGURE 1.
Enzymatic activity of PARP-3. A, PARP-3 is able to ADP-ribosylate itself and histone H1 as measured using the substrate Bio-NAD+ and performing a time course experiment. 0.8 μm PARP-3 was incubated with 25 μm Bio-NAD+, 75 μm NAD+, and 2.4 μm histone H1. B, effect of DNA on PARP-3 enzymatic activity. 0.8 μm PARP-3 was incubated with 25 μm Bio-NAD+, 75 μm NAD+, and 2.4 μm histone H1 in the presence of 5 μg/ml activated DNA.
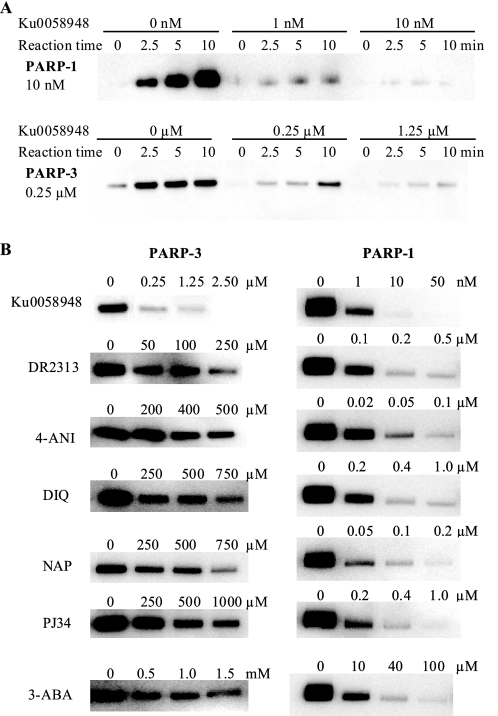
The inhibitory potency of a panel of PARP-1 inhibitors and binders was analyzed. We showed previously that at equimolar concentrations of inhibitor and the substrate Bio-NAD+, only the highly potent PARP-1-specific inhibitor KU0058948 shows PARP-3 inhibition (12). To estimate the inhibitory potency of KU0058948 for PARP-3 compared with PARP-1, time course experiments in the presence of increasing concentrations of KU0058948 were performed (Fig. 2A). Initial rates were determined at each KU0058948 concentration, and the activity was plotted against inhibitor concentration (supplemental Fig. S2). 0.25 μm KU0058948 reduced the activity of 0.25 μm PARP-3 to 18%. The apparent Ki at 25 μm Bio-NAD+ was estimated to be ∼10 nm using the quadratic binding equation. The Km for Bio-NAD+ was determined to 130 μm, which means that the apparent Ki is close to the true Ki because most of the enzyme molecules are free to bind inhibitor molecules at the concentration of Bio-NAD+ used. Comparison of the inhibition of PARP-1 and PARP-3 with KU0058948 (Fig. 2, A and B) shows that the compound was a far less potent inhibitor of PARP-3 than of PARP-1. However, KU0058948 was the most potent PARP-3 inhibitor of those tested. Despite the strong structural similarity between the PARP-1 and PARP-3 catalytic domains (12), PARP-3 also displayed considerably lower sensitivity compared with PARP-1 to other known PARP-1 inhibitors (Fig. 2B). IC50 values are presented in supplemental Table 1.
FIGURE 2.
Inhibition of PARP-3 and PARP-1 activity by various inhibitors. A, shown is the dose-dependent inactivation of auto-ADP-ribosylation of PARP-3 and PARP-1 by KU0058948. B, 250 nm PARP-3 or 10 nm PARP-1 was incubated with 25 μm Bio-NAD+ and different concentrations of various inhibitors for 5 min. PARP-1 samples were supplemented with activated DNA. 4-ANI, 4-amino-1,8-naphthalimide; DIQ, 1,5-dihydroisoquinoline; NAP, 1,8-naphthalimide; 3-ABA, 3-aminobenzamide.
PARP-3 Binds to and Activates PARP-1 in the Absence of DNA
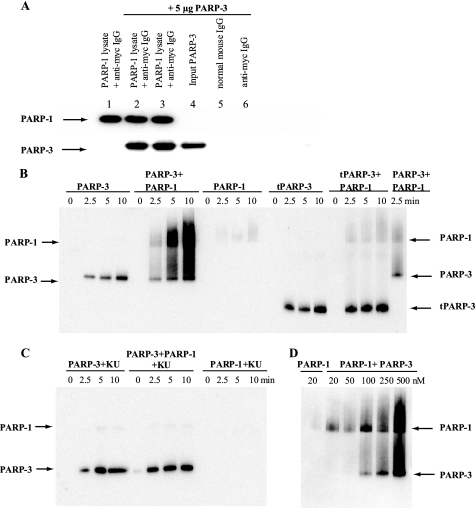
The biological role of PARP-3 is not known, but it may be linked with the role of PARP-1 in maintenance of genome stability, as an interaction between the proteins has been reported (16, 17). Here, we confirmed an interaction between full-length PARP-3 and PARP-1 in immunoprecipitation experiments (Fig. 3A). The interaction was able to withstand 1 m NaCl or 0.1% SDS, suggesting that the interaction is strong.
FIGURE 3.
Interaction between PARP-3 and PARP-1. A, shown is the immunoprecipitation of PARP-3 with PARP-1. Myc-PARP-1 was pulled out from U2OS cell lysate by anti-Myc antibody-agarose beads, and purified PARP-3 was added. The protein interaction was analyzed by Western blotting. Lane 1, no PARP-3 added; lanes 2 and 3, PARP-3 incubated with Myc-PARP-1 and anti-Myc antibody beads and washed with 1 m NaCl (lane 2) or 0.1% SDS (lane 3), respectively; lane 4, PARP-3 only; lanes 5 and 6, PARP-3 incubated with normal IgG-agarose beads or anti-Myc antibody-agarose beads, respectively. B, shown are the results from the activity assay using PARP-3, tPARP-3, and PARP-1 in the absence of activated DNA. PARP-3 or tPARP-3 was incubated with Bio-NAD+ and NAD+ in the absence or presence of PARP-1. At time 0 and after 2.5, 5, and 10 min of incubation at room temperature, the reaction was stopped by dilution in LDS sample buffer. C, to selectively inhibit PARP-1 activity, the protein was preincubated with 50 nm KU0058948 (KU) before the activity assay was performed. D, the activity of 20 nm PARP-1 was assayed for 5 min in the presence of different concentrations of PARP-3.
To test a possible role of this interaction, we analyzed the activity of PARP-3 in the presence of PARP-1 and in the absence of activated DNA. Interestingly, we found that PARP-3 activated PARP-1, probably by forming catalytically active heterodimers with PARP-1 possessing auto- and trans-poly(ADP-ribosylation) activity (Fig. 3B). Incubation of tPARP-3 with PARP-1 produced a weak PARP-1 auto-ADP-ribosylation signal but did not show any trans-ADP-ribosylation activity of PARP-1 (Fig. 3B). At the same time, the auto-ribosylation activity of tPARP-3 was not affected. These results suggest that the N-terminal part of PARP-3 containing the WGR domain is needed for activating PARP-1 and is probably involved in the formation of catalytically active heterodimers between PARP-1 and PARP-3. Also, the site(s) of auto-modification are not exclusively located in the N-terminal part of PARP-3 that is absent in tPARP-3 because tPARP-3 is ADP-ribosylated.
The PAR chains formed by the PARP-3/PARP-1 heterodimer are catalyzed by PARP-1 because incubation of PARP-1 with KU0058948 (50 nm) abrogated the synthesis of poly-ADP-ribose, without affecting the PARP-3 activity (Fig. 3C). Incubation of PARP-1 with PARP-3 at different ratios revealed that PARP-1 auto-ADP-ribosylation is preferred compared with trans-ADP-ribosylation (Fig. 3D). The addition of Ca2+ to the reaction mixture produced only a minor increase in the PARP-1 activation by PARP-3 (supplemental Fig. S3).
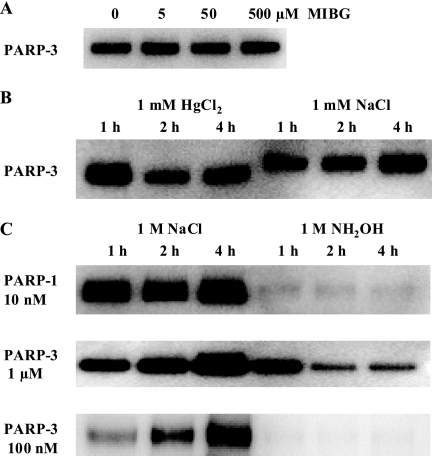
Chemical Stability of PARP-3 Auto-modification
The covalent attachment of ADP-ribose units from NAD+ to various substrates can occur on different amino acids, including arginine, asparagine, serine, cysteine, and glutamic acid (2). To identify the amino acid residues of PARP-3 that are modified, we investigated the chemical stability of the modification. The inhibitor of arginine-specific mono(ADP-ribose) transferases, MIBG (20), had no effect on the enzymatic activity of PARP-3 (Fig. 4A), indicating that arginine residues most probably are not modified in PARP-3. The chemical bonds between ADP-ribose and various amino acids have different sensitivities to acidic, alkaline, or other types of treatment. Auto-modified PARP-3 was insensitive to HgCl2, which cleaved the linkage between cysteine and ADP-ribose (Fig. 4B). Ester bonds between ADP-ribose and acidic amino acids can be hydrolyzed by hydroxylamine (2, 21). Auto-modified PARP-3 and PARP-1 were sensitive to hydroxylamine treatment (Fig. 4C), indicating that ADP-ribosylation occurs on glutamic and/or aspartic acid residues at least under the conditions used in our experiments.
FIGURE 4.
Sensitivity of auto-ADP-ribosylation of PARP-3 to different treatments. A, PARP-3 was auto-modified with Bio-NAD+ in the presence of different concentrations of MIBG at room temperature for 30 min. B, PARP-3 was auto-modified with Bio-NAD+ at room temperature for 30 min. HgCl2 was added to the reaction mixture to 1 mm and incubated for 1, 2, or 4 h. Control reactions were supplemented with 1 mm NaCl and incubated as described above. C, PARP-3 was auto-modified with Bio-NAD+ at room temperature for 30 min. The modified protein was then treated with 1 m neutral NH2OH for 1, 2, or 4 h at room temperature. PARP-1 was auto-modified with Bio-NAD+ in the presence of activated DNA for 30 min at room temperature. The control reactions were mixed with NaCl to 1 m and were treated as described above.
PARP-3 Is a Mono-ADP-ribosylase
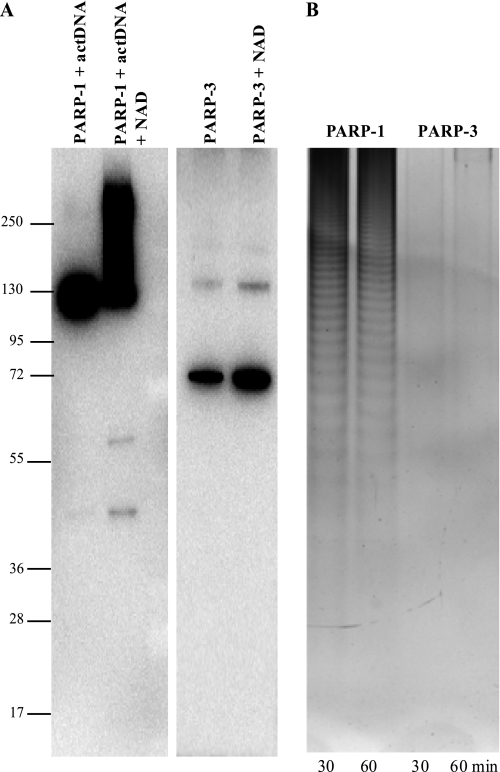
PARP-3 is structurally very similar to the catalytic domains of PARP-1 and PARP-2, which are both poly(ADP-ribose) polymerases. Therefore, it is reasonable to believe that also PARP-3 would be a poly(ADP-ribose) polymerase. Here, we compared the ADP-ribosylase activity of PARP-1 and PARP-3. To determine whether the multiple ADP-ribose modifications were linked together as a polymer or monomers attached at one or multiple sites, the ADP-ribose moieties were cleaved from the proteins and resolved by electrophoresis using a sequencing gel format (Fig. 5B). The gel demonstrated that PARP-3 could not synthesize long polymers as PARP-1. Analysis of PARP-3 auto-ADP-ribosylation by Western blotting showed only one band at the size of PARP-3, suggesting that PARP-3 is a mono-ADP-ribosylase (Fig. 5A).
FIGURE 5.
Characterization of ADP-ribosylation. PARP-3 was incubated with NAD+, and PARP-1 was incubated with NAD+ in the presence of activated DNA (actDNA) at 37 °C for 30 and 60 min. A, aliquots of the reaction products were analyzed by Western blotting using mouse monoclonal antibody to PARP-1 (sc-8007) and goat polyclonal antibody to PARP-3 (sc-30625). B, ADP-ribose polymers were detached from the proteins and separated on 20% polyacrylamide gel. The gel was stained with silver.
PARP-3 Is Not Involved in the Response to Camptothecin and DNA Single-strand Break (SSB) Repair
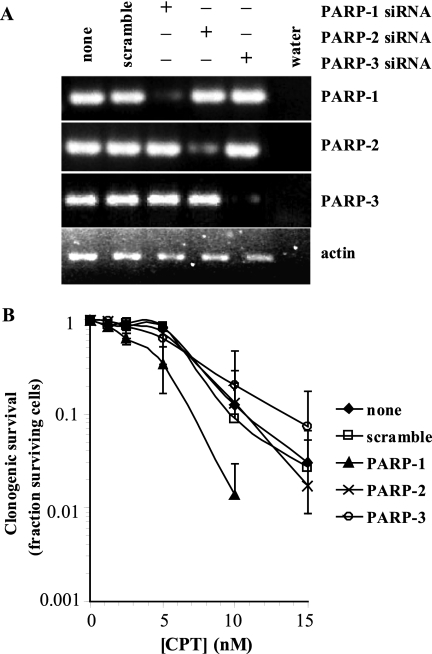
To test a possible role of PARP-3 in the DNA damage response, we determined the sensitivity of PARP-1 and PARP-3 siRNA-depleted human SW480 cells to the topoisomerase I inhibitor camptothecin (CPT), which kills PARP-1-defective or PARP-1-inhibited cells by increasing the persistence of SSBs (22). The CPT analog irinotecan (CPT-11) is currently used in combination therapies and is presently in clinical trials. We found that PARP-1 and not PARP-3 siRNA depletion sensitized cells to CPT (Fig. 6), suggesting that PARP-3 is not involved in the SSB response and that unwelcomed inhibition of PARP-3 by PARP-1 inhibitors is not likely to influence the PARP-1 inhibitor and irinotecan combination treatment used in patients.
FIGURE 6.
PARP-3 is not required for survival against camptothecin-induced damage. A, reverse transcription-PCR analysis of RNA lysates after 48 h of transfection with siRNA. B, clonogenic survival of siRNA-treated cells in the presence of CPT. PARP enzymes were depleted for 48 h prior to 24 h of treatment with CPT.
DISCUSSION
In this study, we characterized the biochemical properties of human PARP-3. PARP-3 was described recently as a non-active protein compared with PARP-1 and PARP-2 (23). This conclusion was made because neither polymer formation nor auto-modification of PARP-3 was observed after a 10-s reaction. Along with two previous reports from independent laboratories (16, 17), we have shown that PARP-3 is an active enzyme. In the presence of biotinylated NAD+, the protein was found to be able to ADP-ribosylate itself as well as histone H1, a previously unknown substrate for PARP-3. However, despite the strong structural similarity between the catalytic domains of PARP-1 and PARP-3, these proteins possess certain functional differences. For example, PARP-3 was considerably less sensitive to known PARP-1 inhibitors (Fig. 2), consistent with what has been reported previously (10, 24). The assessment of PARP-3 activity in the presence of various PARP-1 inhibitors showed that KU0058948 was the most active PARP-3 inhibitor; however, it inhibited PARP-1 much more potently than PARP-3. Therefore, the use of PARP-1 inhibitors in the clinic is unlikely to give any side effects by inhibiting PARP-3.
PARP-3 is not activated upon binding to DNA and is a mono-ADP-ribosylase in contrast to PARP-1 and PARP-2. PARP-3 has auto-ADP-ribosylation activity but, in contrast to PARP-1, did not show the size shift on polyacrylamide gels as a result of PARP-bound PAR (Figs. 1 and 5A). Moreover, when ADP-ribose moieties were cleaved from the proteins after the auto-ADP-ribosylation reaction and resolved by electrophoresis, it was demonstrated that PARP-3 could not synthesize long polymers as PARP-1 (Fig. 5B). It has been suggested that the presence or absence of a catalytic glutamate defines the enzymatic activity of PARPs as a polymerase or a mono-transferase (21). However, PARP-3 has a glutamate (Glu514) in the catalytic domain (12) but still lacks polymerase activity, showing that the active-site glutamate is not the sole feature that determines mono- versus poly-ADP-ribosylase activity.
The covalent attachment of ADP-ribose units to various protein substrates can occur on different amino acids, including arginine, serine, cysteine, asparagine, and glutamic acid (2). Early experiments showed that the glutamic acid residues in histone H1 (Glu3, Glu16, and Glu115) and histone H2B (Glu3) and the carboxyl group of the C-terminal lysine residue in histone H1 (Lys219) are the sites of ADP-ribosylation (25, 26). Analysis of the chemical stability of the ADP-ribose-PARP linkage against different treatments can also give information about which amino acid residues are the sites of modification. We showed that auto-modified PARP-3, like PARP-1, was sensitive to hydroxylamine treatment at pH 7, indicating that ADP-ribosylation under the reaction conditions used in our assays occurred on glutamic and/or aspartic acid residues. Mutational analysis in the work of Altmeyer et al. (23) led to the identification of three lysine residues in the PARP-1 auto-modification domain (Lys498, Lys521, and Lys524) as the sites of ADP-ribosylation. Recently, the first successful identification of the ADP-ribosylation sites in PARP-1 was done by mass spectrometry (27). In this study, the glutamic and aspartic acid residues in the PARP-1 auto-modification domain (Glu488, Glu491, and Asp387) were identified as the acceptor amino acid residues. It should be mentioned that both studies showed that auto-modification of PARP-1 was not limited to the auto-modification domain but could occur beyond this region.
Whether PARP-3 has any function in the DNA damage response is still unknown. We have shown that PARP-3 siRNA-depleted cells are not sensitive to camptothecin and that they repair DNA SSBs as efficiently as wild-type cells, suggesting that the interaction between PARP-1 and PARP-3 is unrelated to DNA SSB repair. However, we found that in the absence of activated DNA, PARP-3 can activate PARP-1. It was shown previously that an alternative 55-kDa short product of the PARP-1 gene consisting of the catalytic domain can synthesize PAR independently of DNA strand breaks (28). The 40-kDa C-terminal catalytic domain of avian PARP-1 also possesses auto-poly(ADP-ribosylation) activity independent of DNA (29). It can also form catalytically competent heterodimers with full-length PARP-1. PARP-1 and PARP-2 have been shown to heterodimerize and heteromodify each other but in the presence of activated DNA (30).
The interaction between PARP-1 and PARP-2 or PARP-3 has led to speculations that the two structurally similar PARPs may regulate PARP-1 activity under different circumstances. PARP-2 was shown recently to be critical for PARP-1-mediated activation of homologous recombination at stalled replication forks (6). The protein levels of PARP-3 appear to be very low in all mammalian tissues (31). However, its expression is restricted to a subset of cell types like the neuroglial cells in the brain and spinal cord and epithelial cells forming the ducts of the prostate, salivary glands, liver, and pancreas. This expression pattern indicates that the expression of PARP-3 is tightly regulated. Thus, there is a possibility that the expression and localization of PARP-3 are ways to control PARP-1 activity in cells and cell signaling through PARP-1-mediated formation of PAR chains via PARP-1/PARP-3 interactions.
Supplementary Material
This work was supported by the Swedish Cancer Society, the Swedish Children's Cancer Foundation, the Swedish Research Council, the Swedish Pain Relief Foundation, and the Medical Research Council (to T. H.) and by the Lars Hierta Memorial Foundation (to A.-S. J.). The Structural Genomics Consortium is a registered charity (1097737) that receives funds from the Canadian Institutes for Health Research, the Canadian Foundation for Innovation, Genome Canada through the Ontario Genomics Institute, GlaxoSmithKline, the Karolinska Institutet, the Knut and Alice Wallenberg Foundation, the Ontario Innovation Trust, the Ontario Ministry for Research and Innovation, Merck & Co., Inc., the Novartis Research Foundation, the Swedish Agency for Innovation Systems, the Swedish Foundation for Strategic Research, and the Wellcome Trust.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1–S3 and Table 1.
- PAR
- poly(ADP-ribose)
- PARP
- poly(ADP-ribose) polymerase
- tPARP-3
- truncated PARP-3
- Bio-NAD+
- 6-biotin-17-NAD+
- LDS
- lithium dodecyl sulfate
- BisTris
- 2-[bis(2-hydroxyethyl)amino]-2-(hydroxymethyl)propane-1,3-diol
- MIBG
- meta-iodobenzylguanidine
- siRNA
- small interfering RNA
- SSB
- single-strand break
- CPT
- camptothecin.
REFERENCES
- 1.Schreiber V., Dantzer F., Ame J. C., de Murcia G. (2006) Nat. Rev. Mol. Cell Biol. 7, 517–528 [DOI] [PubMed] [Google Scholar]
- 2.Hassa P. O., Haenni S. S., Elser M., Hottiger M. O. (2006) Microbiol. Mol. Biol. Rev. 70, 789–829 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Satoh M. S., Lindahl T. (1992) Nature 356, 356–358 [DOI] [PubMed] [Google Scholar]
- 4.Allinson S. L., Dianova I. I., Dianov G. L. (2003) Acta Biochim. Pol. 50, 169–179 [PubMed] [Google Scholar]
- 5.Ahel I., Ahel D., Matsusaka T., Clark A. J., Pines J., Boulton S. J., West S. C. (2008) Nature 451, 81–85 [DOI] [PubMed] [Google Scholar]
- 6.Bryant H. E., Petermann E., Schultz N., Jemth A. S., Loseva O., Issaeva N., Johansson F., Fernandez S., McGlynn P., Helleday T. (2009) EMBO J. 28, 2601–2615 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hakmé A., Wong H. K., Dantzer F., Schreiber V. (2008) EMBO Rep. 9, 1094–1100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hassa P. O., Hottiger M. O. (2008) Front. Biosci. 13, 3046–3482 [DOI] [PubMed] [Google Scholar]
- 9.Bryant H. E., Schultz N., Thomas H. D., Parker K. M., Flower D., Lopez E., Kyle S., Meuth M., Curtin N. J., Helleday T. (2005) Nature 434, 913–917 [DOI] [PubMed] [Google Scholar]
- 10.Farmer H., McCabe N., Lord C. J., Tutt A. N., Johnson D. A., Richardson T. B., Santarosa M., Dillon K. J., Hickson I., Knights C., Martin N. M., Jackson S. P., Smith G. C., Ashworth A. (2005) Nature 434, 917–921 [DOI] [PubMed] [Google Scholar]
- 11.Helleday T., Petermann E., Lundin C., Hodgson B., Sharma R. A. (2008) Nat. Rev. Cancer 8, 193–204 [DOI] [PubMed] [Google Scholar]
- 12.Lehtiö L., Jemth A. S., Collins R., Loseva O., Johansson A., Markova N., Hammarström M., Flores A., Holmberg-Schiavone L., Weigelt J., Helleday T., Schüler H., Karlberg T. (2009) J. Med. Chem. 52, 3108–3111 [DOI] [PubMed] [Google Scholar]
- 13.Ruf A., Mennissier de Murcia J., de Murcia G., Schulz G. E. (1996) Proc. Natl. Acad. Sci. U.S.A. 93, 7481–7485 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Oliver A. W., Amé J. C., Roe S. M., Good V., de Murcia G., Pearl L. H. (2004) Nucleic Acids Res. 32, 456–464 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tao Z., Gao P., Hoffman D. W., Liu H. W. (2008) Biochemistry 47, 5804–5813 [DOI] [PubMed] [Google Scholar]
- 16.Augustin A., Spenlehauer C., Dumond H., Ménissier-De Murcia J., Piel M., Schmit A. C., Apiou F., Vonesch J. L., Kock M., Bornens M., De Murcia G. (2003) J. Cell Sci. 116, 1551–1562 [DOI] [PubMed] [Google Scholar]
- 17.Rouleau M., McDonald D., Gagné P., Ouellet M. E., Droit A., Hunter J. M., Dutertre S., Prigent C., Hendzel M. J., Poirier G. G. (2007) J. Cell. Biochem. 100, 385–401 [DOI] [PubMed] [Google Scholar]
- 18.Ewing R. M., Chu P., Elisma F., Li H., Taylor P., Climie S., McBroom-Cerajewski L., Robinson M. D., O'Connor L., Li M., Taylor R., Dharsee M., Ho Y., Heilbut A., Moore L., Zhang S., Ornatsky O., Bukhman Y. V., Ethier M., Sheng Y., Vasilescu J., Abu-Farha M., Lambert J. P., Duewel H. S., Stewart I. I., Kuehl B., Hogue K., Colwill K., Gladwish K., Muskat B., Kinach R., Adams S. L., Moran M. F., Morin G. B., Topaloglou T., Figeys D. (2007) Mol. Syst. Biol. 3, 1–17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Panzeter P. L., Althaus F. R. (1990) Nucleic Acids Res. 18, 2194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Loesberg C., van Rooij H., Smets L. A. (1990) Biochim. Biophys. Acta 1037, 92–99 [DOI] [PubMed] [Google Scholar]
- 21.Kleine H., Poreba E., Lesniewicz K., Hassa P. O., Hottiger M. O., Litchfield D. W., Shilton B. H., Lüscher B. (2008) Mol. Cell 32, 57–69 [DOI] [PubMed] [Google Scholar]
- 22.Smith L. M., Willmore E., Austin C. A., Curtin N. J. (2005) Clin. Cancer Res. 11, 8449–8457 [DOI] [PubMed] [Google Scholar]
- 23.Altmeyer M., Messner S., Hassa P. O., Fey M., Hottiger M. O. (2009) Nucleic Acids Res. 37, 3723–3738 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jones P., Altamura S., Boueres J., Ferrigno F., Fonsi M., Giomini C., Lamartina S., Monteagudo E., Ontoria J. M., Orsale M. V., Palumbi M. C., Pesci S., Roscilli G., Scarpelli R., Schultz-Fademrecht C., Toniatti C., Rowley M. (2009) J. Med. Chem. 52, 7170–7185 [DOI] [PubMed] [Google Scholar]
- 25.Ogata N., Ueda K., Kagamiyama H., Hayaishi O. (1980) J. Biol. Chem. 255, 7616–7620 [PubMed] [Google Scholar]
- 26.Ogata N., Ueda K., Hayaishi O. (1980) J. Biol. Chem. 255, 7610–7615 [PubMed] [Google Scholar]
- 27.Tao Z., Gao P., Liu H. W. (2009) J. Am. Chem. Soc. 131, 14258–14260 [DOI] [PubMed] [Google Scholar]
- 28.Sallmann F. R., Vodenicharov M. D., Wang Z. Q., Poirier G. G. (2000) J. Biol. Chem. 275, 15504–15511 [DOI] [PubMed] [Google Scholar]
- 29.Mendoza-Alvarez H., Alvarez-Gonzalez R. (2004) J. Mol. Biol. 336, 105–114 [DOI] [PubMed] [Google Scholar]
- 30.Schreiber V., Amé J. C., Dollé P., Schultz I., Rinaldi B., Fraulob V., Ménissier-de Murcia J., de Murcia G. (2002) J. Biol. Chem. 277, 23028–23036 [DOI] [PubMed] [Google Scholar]
- 31.Rouleau M., El-Alfy M., Lévesque M. H., Poirier G. G. (2009) J. Histochem. Cytochem. 57, 675–685 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.