Abstract
Compound A (CpdA), a dissociated glucocorticoid receptor modulator, decreases corticosteroid-binding globulin (CBG), adrenocorticotropic hormone (ACTH), and luteneinizing hormone levels in rats. Whether this is due to transcriptional regulation by CpdA is not known. Using promoter reporter assays we show that CpdA, like dexamethasone (Dex), directly transrepresses these genes. Results using a rat Cbg proximal-promoter reporter construct in BWTG3 and HepG2 cell lines support a glucocorticoid receptor (GR)-dependent transrepression mechanism for CpdA. However, CpdA, unlike Dex, does not result in transactivation via glucocorticoid-responsive elements within a promoter reporter construct even when GR is co-transfected. The inability of CpdA to result in transactivation via glucocorticoid-responsive elements is confirmed on the endogenous tyrosine aminotransferase gene, whereas transrepression ability is confirmed on the endogenous CBG gene. Consistent with a role for CpdA in modulating GR activity, whole cell binding assays revealed that CpdA binds reversibly to the GR, but with lower affinity than Dex, and influences association of [3H]Dex, but has no effect on dissociation. In addition, like Dex, CpdA causes nuclear translocation of the GR, albeit to a lesser degree. Several lines of evidence, including fluorescence resonance energy transfer, co-immunoprecipitation, and nuclear immunofluorescence studies of nuclear localization-deficient GR show that CpdA, unlike Dex, does not elicit ligand-induced GR dimerization. Comparison of the behavior of CpdA in the presence of wild type GR to that of Dex with a dimerization-deficient GR mutant (GRdim) strongly supports the conclusion that loss of dimerization is responsible for the dissociated behavior of CpdA.
Keywords: Gene Regulation, Radioreceptor Assays, Receptor Regulation, Steroid Hormone Receptor, Transcription Target Genes, Compound A, Dissociated Glucocorticoid
Introduction
CpdA4 is a more stable analogue of the labile compound found in the Namibian plant, Salsola tuberculatiformis Botschantzev, which causes prolonged gestation in sheep and contraception in rats (1). CpdA, like the shrub, causes contraception in rats (2). Further investigation at a molecular level has shown that CpdA competitively inhibits sheep adrenal cytochrome P450-dependent steroid 11β-hydroxylase (P450c11), the enzyme responsible for the final step in the synthesis of glucocorticoids (GCs) (2, 3). In addition, CpdA binds to and displaces endogenous GCs from rat and sheep CBG, the plasma globulin that binds GCs with high affinity (4). Studies in Wistar rats suggest that the latter mechanism predominates in vivo with significantly increased free corticosterone levels due to displacement from CBG by CpdA and concomitant decreases in CBG, ACTH, and LH levels (5). Although the significant decreases in CBG, ACTH, and LH levels may be ascribed solely to feedback regulation mediated by the increase in a free, biologically active GC concentration, it was postulated that direct interaction of CpdA with the GR should not be discounted (5). The fact that CpdA interacts with the two GC-binding proteins, P450c11 and CBG, implies that interaction and signaling through the GR may be a distinct possibility. Indeed, our recent article (6) describes such an interaction within the context of anti-inflammatory action.
The GR is a ligand-dependent transcription factor mediating the effects of GCs (7). In the absence of ligand, the GR is predominantly cytoplasmic. Upon ligand binding the GR translocates to the nucleus, where the activated receptor can transactivate or transrepress specific genes (8). Several models for transcriptional modulation by the GR have been presented (9–11). Broadly speaking, transactivation is mediated by binding of a GR dimer to glucocorticoid response elements (GREs) in the promoter region of GC-responsive genes, followed by recruitment of coactivators, chromatin remodeling, and increased gene transcription (12). Although transrepression may also be mediated via direct binding to DNA, via negative GREs (9), it mostly proceeds, without direct DNA binding by the GR, via protein-protein interactions that require binding of the GR monomer to other transcription factors, such as NFκB, AP-1, and C/EBP (10, 13–16). This last mechanism is often called tethering.
The current study establishes that CpdA, like Dex, directly transrepresses the three genes, Cbg, proopiomelanocortin (Pomc), and gonadotropin-releasing hormone (Gnrh), shown to be involved in the in vivo response to CpdA and that these results are thus not only due to the increase in free corticosterone (5). Further investigation indicates that CpdA transrepression of Cbg is GR-mediated. Despite its ability to transrepress GC-sensitive genes, CpdA is unable to transactivate GRE-containing promoters. Analysis of the molecular mechanism of action of CpdA via the GR indicates that CpdA binds reversibly to endogenous rat GR, influences association but not dissociation of [3H]Dex, and causes nuclear translocation of liganded GR, although to a lesser extent than Dex. However, CpdA, unlike Dex, does not result in dimerization of the GR. The implications of loss of dimerization are explored further by comparing the activity of CpdA via the wtGR with that of Dex via a dimerization-deficient GR mutant (GRdim) (16). Nuclear translocation behavior and transrepression mediated by CpdA via the wtGR does not differ significantly from that observed with Dex via the GRdim strongly supporting a mechanism whereby the inability of CpdA to elicit ligand-induced dimerization of the GR is responsible for its dissociated behavior.
EXPERIMENTAL PROCEDURES
Test Compounds
Dex, phorbol 12-myristate 13-acetate (PMA), aldosterone, 4,5α-dihydrotestosterone (DHT), 17β-estradiol (E2), spironolactone, and mifepristone (RU486) were obtained from Sigma, ICI 182,780 from Tocris, R5020 from PerkinElmer Life Sciences, and hydroxyflutamide was a kind gift from Dr. C. Tendler (Schering Plough Research Institute). Compound A (CpdA; 2-(4-acetoxyphenyl)-2-chloro-N-methylethylammonium chloride) was synthesized as described previously (2).
Plasmids
G. L. Hammond kindly provided the rat Cbg proximal promoter reporter construct (rCBG295Luc) (17). The Gnrh promoter reporter construct (3446mGnRHluc) was donated by D. DeFranco (18). The rat Pomc promoter reporter construct (JA300) and the expression vector for NUR77 (pCMX-Nur77) were gifts from J. Drouin (19). The β-galactosidase reporter plasmid (pPGKβGeopbA) was a gift from P. Soriano (Fred Hutchinson Cancer Research Centre, Seattle, WA) and the pGL2-basic empty vector was obtained from Promega. The rat GRα (pSVGR1) expression vector was a gift from R. Miesfield (20), whereas the human GR (pRS-hGRα) and human MR (pRS-hMR) were gifts from R. M. Evans (Howard Hughes Medical Institute, La Jolla, CA). CFP-tagged GR (pECFP-hGRα) and YFP-tagged GR (pEYFP-hGRα) were gifts from J. Cidlowski (21). Mouse wtGR (pcDNA3.1-GRWT) and the mouse GRdim (pcDNA3.1-GRdim) mutant were gifts from H. Reichardt (22). Rat GR (pTC2-wtGRrat) and c-myc-tagged nuclear translocation mutant GR (myGRNL1−) were gifts from R. Haché (23). Human ERα (pcDNA3-ERα) was a gift from D. Harnish (24). Human PRB (pSG5hPRB) was obtained from S. Simons Jr. (25) and human AR (pSVARo) was obtained from A. Brinkmann (26). (GRE)250hIL6PLuc and the FLAG-tagged GR pEFFlaghGRα (molecular mass, 96 kDa) were constructed as previously described (27, 28), whereas the (GRE)2tkLuc construct, and GFP-tagged GR (pEGFP-C2-GR, molecular mass, 128.5 kDa) were provided by S. Okret (Dept. of Medical Nutrition, Karolinska Institute, Sweden). The GRE-containing promoter reporter construct (pTAT-GRE2-Elb-luc) was a gift from G. Jenster (29), the ERE-containing promoter reporter construct (pGL2–3x-ERE-TATA-luc) was from D. McDonnell (30), whereas the IL6-luc promoter reporter construct (p(IL6κB)350hu.IL6Pluc+) has been described previously (31).
Cell Culture
BWTG3 (LEGEST, University of Gent, Belgium), HepG2 (Highveld Biological Association, South Africa), and COS-1 cells (ATCC) were cultured in high glucose (4.5 g/ml) Dulbecco's modified Eagle's medium (DMEM) (Sigma) supplemented with 10% FCS (Highveld Biologicals, South Africa), 100 IU/ml of penicillin, 100 μg/ml of streptomycin (Invitrogen), 2 mm glutamine (Merck), 44 mm sodium bicarbonate (Invitrogen), and 1 mm sodium pyruvate (Invitrogen). In addition, BWTG3 and HepG2 cells had 0.1 mm nonessential amino acids (Invitrogen) added to their medium. GT1-7 neuronal cells (a kind gift from P. Mellon, University of California, La Jolla, CA) were maintained in DMEM with 25 mm HEPES, 4500 mg/liter of glucose and pyroxidine, supplemented with 10% fetal bovine serum. AtT-20 (ATCC) pituitary tumor cells in suspension were cultured in Kaighn's modification of Ham's F-12K medium (Invitrogen), supplemented with 15% horse serum and 2.5% fetal bovine serum.
Competitive Whole Cell Binding Assays
Assays were performed essentially as described in Ref. 32 with the following modifications. Forty-eight h after plating BWTG3 cells (2 × 105 cells/well in 24-well tissue culture plates) were washed three times with pre-warmed phosphate-buffered saline (PBS) and then incubated in medium (minus FCS or penicillin-streptomycin) with 20 nm [3H]Dex (specific activity of 89 Ci/mmol; AEC-Amersham Biosciences) and varying concentrations of unlabeled test compounds for 1 h at 37 °C. Cells were then placed on ice and after 1 h washed three times, for 15 min each, with ice-cold PBS containing 0.2% (w/v) bovine serum albumin (BSA). Cells were lysed with 100 μl of lysis buffer (PE Biosystems) and binding was determined by scintillation counting. Total binding was normalized to protein concentration (Bradford assay) and expressed as percentage binding (top plateau for Dex binding designated as 100% binding and bottom plateau as 0% binding).
Kinetic Whole Cell Binding Assays
Binding assays were done in the presence of 1 nm [3H]Dex in the absence or presence of CpdA. For association binding experiments, binding was measured for different times (0–240 min) after addition of radioligand in the absence (total binding) or presence of 10 μm unlabeled Dex (nonspecific binding). The binding experiment then proceeded as described for competitive binding. Specific binding (total binding − nonspecific binding) was plotted. For dissociation binding experiments, the cells were first incubated in the presence of 1 nm [3H]Dex with or without CpdA for 2 h at 37 °C after which the medium was replaced with fresh medium without radioligand and CpdA. Binding at different time points (0–240 min) proceeded as described for competitive binding and plotted.
Reversibility of Whole Cell Binding Assay
The assay was done by preincubating cells with 10 μm CpdA or 10 μm Dex for 1 h at 37 °C. Cells were then washed three times with pre-warmed PBS containing 0.2% (w/v) BSA and incubated for 1 h at 37 °C with 1 nm [3H]Dex in the presence or absence of 10 μm unlabeled Dex. The binding experiment then proceeded as described for competitive binding. Specific binding (total binding − nonspecific binding) was plotted.
Whole Cell Binding Assays to Determine Steroid Receptor Content of BWTG3 Cells
This binding assay was done using 20 nm [3H]Dex (specific activity of 85 Ci/mmol; AEC-Amersham Biosciences) for GR, 20 nm [3H]E2 (specific activity of 84 Ci/mmol; AEC-Amersham Biosciences) for ER, 20 nm [3H]R5020 (specific activity of 84.6 Ci/mmol; AEC-Amersham Biosciences) for PR, 20 nm [3H]mibolerone (specific activity of 76.8 Ci/mmol; AEC-Amersham Biosciences) for AR, and 9 nm [3H]aldosterone (specific activity of 87.9 Ci/mmol; AEC-Amersham Biosciences) for MR. Cognate unlabeled ligands were added in 1000-fold excess. The binding experiment proceeded essentially as described for competitive binding except that the incubation at 37 °C was done for 4 h. Specific binding (total binding − nonspecific binding) was calculated in femtomole/mg of protein using a counting efficiency of 40%.
Promoter Reporter Construct Studies
Cells (BWTG3, HepG2, COS-1, GT1-7, and AtT-20 cells) were plated in relevant complete medium at the densities indicated in the figure legends. Cells were transfected 24 h later with constructs (as indicated in figure legends) in medium without FCS or, for GT1-7 and AtT-20 cells, medium with 10% dextran-coated charcoal-stripped fetal bovine serum (Highveld Biologicals, South Africa) using FuGENETM 6 transfection reagent (Roche Applied Science) or for GT1-7 cells, with the Lipofectamine method (Invitrogen), as described by the manufacturer. Cells plated into 10-cm tissue culture dishes were replated 24 h after transfection at the densities indicated in the figure legends. Test compounds were added to cells 24 or 4 h (GT1-7 cells) after transfection or replating and incubated for 24 or 20 h (GT1-7 cells). Induction in BWTG3, HepG2, and COS-1 cells occurred in medium without FCS and antibiotics except for assays investigating ER activity, where phenol red-free medium was used, whereas induction in GT1-7 and AtT-20 cells occurred in medium with 10% dextran-coated charcoal-stripped fetal bovine serum. After induction cells were lysed with 100 μl of lysis buffer (PE Biosystems), and frozen at −20 °C. Luciferase activity was determined using the luciferase assay kit (Promega) and β-galactosidase activity was measured using the Galacto-star assay kit (PE Biosystems) according to the manufacturer's instructions. Light emission was measured in a luminoskan plate reader (Labsystems). Luciferase relative light units were normalized with β-galactosidase values to correct for transfection efficiency. For replated cells, protein concentration was measured using the Bradford method and luciferase relative light units normalized with protein concentrations to correct for plating efficiency.
RNA Isolation
Twenty-four h after plating BWTG3 or HepG2 cells (3 × 105 cells/plate in 10-cm tissue culture dishes) medium was changed to Opti-MEM (Invitrogen) and cells were incubated for a further 24 h. Cells were then treated with test compounds for 72 (HepG2 cells) or 24 h (BWTG3 cells), and total RNA was extracted according to the TRIzol method, as described by the manufacturer (Sigma). After extraction, the final RNA pellet was dissolved in 50 μl of formazol (Molecular Research Center, Inc.) for Northern blotting, or 50 ml of diethyl pyrocarbonate water for reverse transcription-PCR, and kept at −70 °C until used.
Northern Blotting
Northern blotting was essentially preformed as previously described (33). Briefly, 20 μg of total RNA was loaded and run on a 1% formaldehyde-agarose gel and transferred to a nylon membrane (Hybond-N+, Amersham Biosciences). RNA was fixed on the membrane using a UV cross-linker for 12 s. The membrane was prehybridized in a hybridization oven at 50 °C for 1 h with pre-warmed “Dig easy Hyb” solution (Roche Applied Science). Plasmids carrying human CBG cDNA (cDNA kindly provided by G. L. Hammond) were amplified in a DH5α competent Escherichia coli strain and digested with EcoRI to obtain the CBG cDNA insert of 1.2 kb. Hybridization was performed overnight at 50 °C with [32P]CBG cDNA probes labeled with [α-32P]deoxycytidine triphosphate using the random priming technique (Amersham Biosciences megaprime labeling kit). Membranes were washed twice for 5 min in 2× SSC, 0.1% SDS at room temperature, followed by two washes for 15 min in 0.1× SSC, 0.1% SDS at 50 °C. Membranes were exposed between 24 and 48 h at −70 °C followed by densitometric scanning of the autoradiograms using the UN-SCAN-IT program. Membranes were stripped using a hot 0.5% SDS solution and reprobed with [32P]β-actin cDNA, provided by I. Parker, UCT, South Africa. Autoradiography was for less than 24 h, followed by densitometric scanning and normalizing the CBG values with the corresponding β-actin values.
Reverse Transcription-PCR
Total RNA (5 μg) was reverse transcribed with oligo(dT)15 primers (Promega) using the avian myeloblastosis virus reverse transcriptase enzyme (Promega) and followed by a PCR on the obtained cDNA with Taq DNA polymerase (Promega) and primers, specific for mouse glyceraldehyde-3-phosphate dehydrogenase, rCbg, and rTat (primer sequences available on request).
Tyrosine Aminotransferase Assay
Twenty-four h after plating BWTG3 cells (2.5 × 105 cells/well/6-well tissue culture plate) medium was changed to medium with stripped FCS for a further 24 h. Cells were then treated with test compounds for 4 h after which cells were washed twice with PBS and lysed with 250 μl of lysis buffer (10 mm Tris-HCl, pH 7.5, 0.25 m sucrose, 0.1 mm phenylmethylsulfonyl fluoride, and 10 μg/ml of aprotinin) and frozen at −20 °C. On thawing, lysates were briefly sonicated and centrifuged (10,000 × g for 10 min at 4 °C). TAT activity was determined according to the method of Diamondstone (34). TAT activity was expressed as absorbance at 330 nm/min and graphed as percentage of control.
Immunofluorescence Analysis
COS-1 cells were plated into 10-cm tissue culture dishes and transiently transfected 24 h later with constructs as indicated in the figure legends using FuGENE 6. Cells were replated 24 h later onto coverslips in 6-well plates in medium and at the densities indicated in the figure legends. BWTG3 cells were grown on coverslips in 6-well plates in complete medium at densities indicated in the figure legends. Twenty-four h later cells were serum-starved for 24 h in medium as indicated in the figure legends. Cells were treated with test compounds as indicated in the legends. After induction, cells were placed on ice, rinsed with 1 ml of methanol, and incubated at −20 °C for 15 min with another 1 ml of methanol. Cells were then washed three times with ice-cold PBS plus 0.2% BSA and transferred to new 6-well plates containing 2 ml of blocking buffer (PBS with 3% (v/v) newborn calf serum and 1% (v/v) BSA). Cells were incubated for 1 h at room temperature and then washed twice with ice-cold PBS plus 0.2% BSA. To visualize GR, cells were incubated with the primary rabbit anti-GR antibody, P-20 (Santa Cruz biotechnology) diluted 1:100 in blocking buffer for BWTG3 cells, or H-300 (Santa Cruz biotechnology) diluted 1:1000 in blocking buffer for COS-1 cells. To visualize c-myc-tagged GR cells were incubated with a mouse anti-c-myc antibody, 9E10 (Sigma), diluted 1:500. Cells were then washed three times with ice-cold PBS plus 0.2% BSA and incubated for 1 h at room temperature with secondary antibody, Alexa Fluor 488-tagged or 594-tagged anti-rabbit antibody (Molecular Probes) as indicated in the figure legends, diluted 1:500 in blocking buffer. Nuclei were visualized using either propidium iodide staining including RNase (30 min at 37 °C), Hoechst 33258 stain (Sigma, according to the manufacturer's instructions), or 4′,6-diamidino-2-phenylindole staining for 5 min at room temperature as indicated in the figure legends. Cells were then washed three times with ice-cold PBS, mounted on glass slides, and analyzed using the microscopes indicated in the figure legends. Cells were assessed for intracellular localization of protein signal in a double-blind fashion with 45–50 cells counted in each sample. Cells were either allocated to one of three groups, predominantly nuclear, predominately cytoplasmic, or evenly distributed between the cytoplasm and nucleus or allocated as nuclear or not.
Co-immunoprecipitation
COS-1 cells (2 × 106 cells per plate in 10-cm tissue culture dishes) were transiently transfected 24 h after plating with 38.6 ng of hGR (19.3 ng of pEFFlaghGRα and 19.3 ng of pEGFP-C2-GR) and 11.5 μg of pGL2-basic using the DEAE-dextran method. Cells were re-plated 24 h later into 6-well plates (6 × 105 cells/well) in medium with stripped FCS and after 24 h treated with ethanol, Dex (1 μm), or CpdA (10 μm) for 1 h. After induction cells were washed twice with PBS before extraction on ice in Buffer A (10 mm Hepes, pH 7.5 (Invitrogen), 1.5 mm MgCl2, 10 mm KCl, 0.1% Nonidet P-40 (Roche Applied Science), and Complete Mini protease inhibitor mixture (Roche Applied Science)). After two cycles of freeze-thaw cells were collected, centrifuged at 14,000 × g for 15 min, and the supernatant collected. Protein concentrations were determined using the Bradford method and 20 μg of protein/sample set aside for Western blots (input). Lysates (200 μg of protein/sample) were precipitated with 30 μl of EZview Red ANTI-FLAG M2 Affinity Gel beads (Sigma), pre-washed 4 times with Buffer A in the presence of 0.5% BSA, in a total volume of 250 μl for 16 h at 4 °C. Beads were washed four times with 200 μl of Buffer A supplemented with 0.5% Triton X-100 (BDH) and 150 mm NaCl. 20 μl of Laemmli buffer (62.5 mm Tris-HCl, pH 6.8, 10% (v/v) glycerol, 1.25% (m/v) SDS, 0.00125% (m/v) bromphenol blue, and 2.5% β-mercaptoethanol) was then added to beads, which were boiled for 7.5 min at 95 °C. For Western blotting immune precipitates (20 μl) were separated on a 10% SDS-PAGE gel, together with the inputs of total cell lysate (20 μg). Following electrophoresis, proteins were electroblotted and transferred to Hybond-ECL nitrocellulose membrane (Amersham Biosciences), which were probed for GR (H-300 antibody from Santa Cruz Biotechnology diluted 1:3000) and visualized using ECL peroxidase-labeled anti-rabbit antibody (AEC-Amersham Biosciences) and ECL Western blotting detection reagents (GE Healthcare) on Hyperfilm (Amersham Biosciences).
Fluorescence Resonance Energy Transfer
COS-1 cells (2 × 106 cells/plate in 10-cm tissue culture dishes) were transiently transfected with 5.8 μg of pECFP-hGRα and 5.8 μg of pEYFP-hGRα for the FRET experiment and 5.8 μg of pECFP-hGRα, or 5.8 μg of pEYFP-hGRα, plus 5.8 μg of pGL2-basic for controls using the DEAE-dextran method. Twenty-four h later cells were replated (2 × 106 cells/well) into 8-well Lab-Tek chambered coverglass plates (Nunc) in medium with stripped FCS. Twenty-four h after replating cells were analyzed in the temperature-controlled chamber (37 °C) of an Olympus Cells system attached to an IX 81 inverted fluorescence microscope (Olympus Corp.) equipped with an F-view-II-cooled CCD camera (Soft Imaging Systems). The light source was a 150-watt xenon lamp, part of the MT20 excitation source. Cells were observed with a ×60 objective lens and the Cell® imaging software used for image acquisition and analysis. The YFP filter set excites at S500/20x (Chroma) and emission is detected at S535/30m, whereas the CFP filter set excites at S430/25x and emission is detected at S470/30m. Cells that express similar CFP-GR and YFP-GR levels were selected and treated with solvent, 10 μm CpdA or 10 μm Dex in DMEM containing no supplements. CFP, YFP, and FRET images were taken every minute over a 30-min period. FRET fluorescence was detected using a filter set with S430/25x excitation and S535/30m emission. An exposure time of 1500 ms at 100% light intensity was used. The signals measured in the FRET channel were corrected for cross-talk from the cyan and yellow channels using the following equation: nFRET = FRET signal − (a × YFP signal) − (b × CFP signal). n is normalized FRET and a and b were determined by measuring the crossover into the FRET channel of the YFP and CFP signals, respectively, in cells expressing each fusion protein on its own (35). In our system, ∼59% of the CFP signal and 2.6% of the YFP signal were detected in the FRET channel. Background subtraction was carried out using an area where no cells were present.
RESULTS
CpdA Causes Transrepression of Promoter Reporter Constructs of Three Genes Involved in the in Vivo Response to CpdA Administration in Rats
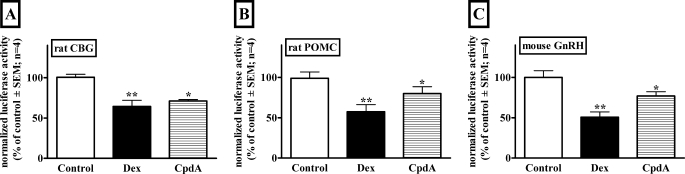
CpdA administration was previously shown to significantly repress CBG, ACTH, and LH levels in Wistar rats, while concomitantly increasing free corticosterone levels (5). Whereas this repression by CpdA might be attributed to the increase in biologically active corticosterone, the possibility of direct action of CpdA via the GR was not precluded. To test this assumption, we investigated the direct transcriptional effect of CpdA in cell lines on the promoter reporter constructs of Cbg, Pomc, and Gnrh. The latter two promoters were chosen as the Pomc gene encodes the precursor to ACTH (36), whereas GnRH levels reflect circulating LH levels (37). It is clear (Fig. 1) that CpdA, like Dex, significantly (p < 0.05) represses all three promoter reporter constructs.
FIGURE 1.
Transrepression of promoter reporter constructs of three genes involved in the in vivo response to CpdA administration in rats. A, rat CBG. BWTG3 cells (5 × 104 cells/well in 24-well tissue culture plates) were transiently transfected with 360 ng of rat Cbg promoter reporter construct (ratCBG295Luc), 200 ng of pGL2-basic, and 40 ng of β-galactosidase reporter plasmid (pPGKβGeopbA). B, rat Pomc. AtT-20 cells (2.5 × 105 cells/well in 24-well tissue culture plates) were transiently transfected with 240 ng of rat POMC promoter reporter construct (JA300), 60 ng of rat GR expression vector (pSVGR1), and 60 ng of rat expression vector for Nur77 (CMX-Nur77). C, mouse Gnrh. GT1-7 cells (1.25 × 105 cells/well in 24-well tissue culture plates) were transiently transfected with 600 ng of mouse GnRH promoter reporter construct (3446mGnRHluc). Twenty-four (A and B) or 4 (C) h after transfection test compounds were added at a concentration of 1 μm. Control wells received an equal amount of ethanol. Cells were treated for 24 (A and B) or 20 (C) h, respectively. Luciferase values were normalized for β-galactosidase (A) or protein (B and C) values and plotted as a percentage of the average control ± S.E. (error bars). Statistical analysis was done to compare values in the presence of test compounds relative to the corresponding control using one-way analysis of variance followed by Dunnett's multiple comparisons post test (*, p < 0.05; **, p < 0.01).
CpdA Transrepresses Rat CBG via a GR-dependent Mechanism in Hepatic Cell Lines but Does Not Transactivate GRE-containing Promoters or Genes
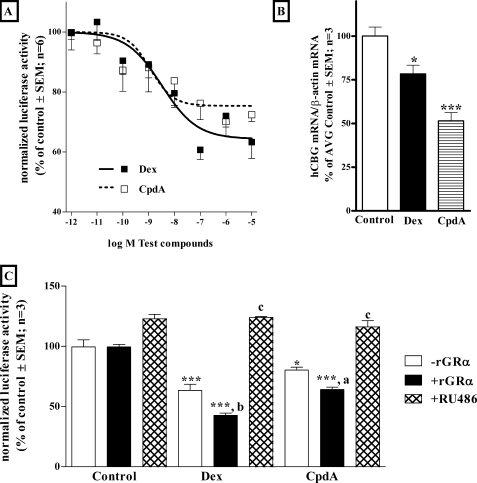
Having shown that CpdA transrepresses, we chose to focus on the CBG promoter, as little work has been done on GC responsiveness of this promoter in contrast to the POMC and GnRH promoters. Dose-response curves of rat CBG promoter reporter transrepression in BWTG3 cells (Fig. 2A) revealed no significant (p > 0.05) difference between the potency (log EC50) of Dex (−8.43) and CpdA (−8.93). However, analysis of the efficacy (maximal repression) indicates that Dex represses 34.5 ± 2.7%, which is significantly (p < 0.05) more than CpdA (24.7 ± 3.8%). Both CpdA and Dex also significantly repress CBG mRNA levels in HepG2 (Fig. 2B) and BWTG3 cells (supplemental Fig. S1), which establishes that transcriptional regulation of the Cbg promoter correlates with regulation at the mRNA level.
FIGURE 2.
Transrepression of the rat Cbg proximal promoter reporter construct and endogenous gene in liver cell lines. A, dose-response curves of transrepression of the rat CBG295Luc promoter reporter construct by CpdA and Dex in the presence of endogenous GR. The rat CBG295Luc promoter reporter construct (360 ng) plus 200 ng of pGL2-basic and 40 ng of pPGKβGeopbA were transiently transfected into BWTG3 cells (5 × 104 cells/well in 24-well tissue culture plates). Twenty-four h after transfection cells were treated with increasing concentrations of test compounds, as indicated, and lysed after 24 h. Control wells received an equal amount of ethanol. Luciferase values were normalized for β-galactosidase and plotted as a percentage of the average control ± S.E. (error bars). Log EC50 and percentage repression values were determined by fitting a sigmoidal dose-response curve with the variable slope. B, transrepression of human CBG mRNA. HepG2 cells were treated with Dex (1 μm) or CpdA (10 μm) for 72 h. Control wells received an equal amount of ethanol. Total RNA was analyzed with Northern blot analysis, using hCBG cDNA, stripped, and reprobed with β-actin to control for loading. CBG results are presented as normalized relative to β-actin and as % of average control ± S.E. (error bars). Statistical analysis was done to compare values in the presence of test compounds relative to the corresponding control using one-way analysis of variance followed by Dunnett's multiple comparisons post test (*, p < 0.05; ***, p < 0.001). C, transrepression of the rat CBG295Luc promoter reporter construct by CpdA and Dex in the absence or presence of co-transfected rGRα and RU486. BWTG3 cells (5 × 104 cells/well in 24-well tissue culture plates) were transiently transfected with rat CBG295Luc (360 ng), 200 ng of rGRα (pSVGR1) or pGL2-basic as indicated, and 40 ng of pPGKβGeopbA. Twenty-four h after transfection, test compounds were added (CpdA at 10 μm; Dex at 1 μm; RU486 at 20 μm) and cells were lysed after 24 h. Control wells received an equal amount of ethanol. Luciferase values were normalized for β-galactosidase and plotted as a percentage of the average control ± S.E. (error bars). Statistical analysis was done to (i) compare values in the presence of test compounds relative to the corresponding control using one-way analysis of variance followed by Dunnett's multiple comparison's post test (*, p < 0.05; **, p < 0.01), and to (ii) compare values without GR (−rGRα) to values with co-transfected GR (+rGRα) and RU486 (+RU486) for each compound tested using one-way analysis of variance followed by Dunnett's multiple comparisons post test (a, p < 0.05; b, p < 0.01; c, p < 0.001).
To test whether transrepression potential is dependent on the GR, BWTG3 cells were co-transfected with the rat GRα expression vector (rGRα) in the presence of the rat CBG295Luc reporter construct (Fig. 2C, closed bars). Co-transfection with rGRα in BWTG3 cells significantly increased the transrepression response of Dex (p < 0.01) and CpdA (p < 0.05). To strengthen the case that the observed repression of the rat CBG295Luc promoter reporter construct by CpdA was mediated by the GR, the effect of the glucocorticoid antagonist, RU486, on transrepression in BWTG3 cells was also investigated (Fig. 2C, hatched bars). RU486 relieved the transrepression observed in the presence of both CpdA and Dex suggesting that repression of the rat CBG295Luc promoter reporter construct by Dex, as well as CpdA, is mediated by the GR.
Having shown that CpdA can result in transrepression of GC-responsive genes, such as Cbg, we were interested in whether it could also result in transactivation of GC-responsive genes and thus evaluated the ability of CpdA to transactivate two GRE-containing promoters. BWTG3 cells were transiently transfected with a (GRE)2tkLuc (Fig. 3A) or a (GRE)250hIL6PLuc promoter reporter construct (supplemental Fig. S2A). The result with both constructs shows that in the presence of endogenous GR (Figs. 3A and supplemental S2A, open bars), Dex significantly (p < 0.01) transactivates the GRE-dependent promoter constructs while co-transfection of rat GRα (Figs. 3A and supplemental S2A, closed bars) almost doubled the transactivation response of Dex. However, even at 10 times the concentration of Dex, CpdA did not result in transactivation, even when GR was co-transfected. The glucocorticoid antagonist, RU486, abolishes Dex (Figs. 3B and supplemental S2B, hatched bars) transactivation but CpdA does not. Dex, but not CpdA, up-regulates TAT mRNA (Fig. 3C) and protein activity levels (Fig. 3D) in BWTG3 cells, which confirms the inability of CpdA to transactivate an endogenous gene containing a GRE motif.
FIGURE 3.
Transactivation of a GRE-containing promoter reporter construct and endogenous gene in a liver cell line. A, transactivation of a GRE-containing promoter by CpdA and Dex in BWTG3 cells, in the absence or presence of co-transfected rGRα. BWTG3 cells (5 × 104 cells/well in 24-well tissue culture plates) were transiently transfected with 360 ng of GRE-driven promoter reporter construct ((GRE)2tkLuc), 200 ng of rGRα (pSVGR1) or pGL2-basic as indicated, and 40 ng of pPGKβGeopbA. Twenty-four h after transfection, test compounds were added (CpdA at 10 μm; Dex at 1 μm) and cells were lysed after 24 h. Control wells received an equal amount of ethanol. Luciferase values were normalized for β-galactosidase and values plotted as a percentage of the average control ± S.E. (error bars). Statistical analysis was done to (i) compare values in the presence of test compounds relative to the corresponding control using one-way analysis of variance followed by Dunnett's multiple comparison's post test (*, p < 0.05; **, p < 0.01) and to (ii) compare values without GR (−rGRα) to values with co-transfected GR (+rGRα) for each compound tested using a two-tailed unpaired t test (a, p < 0.05; b, p < 0.01). B, transactivation of GRE-containing promoter by CpdA and Dex in BWTG3 cells, in the absence or presence of RU486. The (GRE)2tkLuc promoter reporter construct (360 ng) was transiently transfected into BWTG3 cells (5 × 104 cells/well in 24-well tissue culture plates), together with 200 ng of rGRα (pSVGR1) and 40 ng of pPGKβGeopbA. Cells were treated with 10 μm test compounds as indicated. Control wells received an equal amount of ethanol. Luciferase values were normalized for β-galactosidase and plotted as a percentage of the average ± S.E. transactivation by Dex alone. Statistical analysis was done to compare values in the presence of test compounds relative to the corresponding controls using one-way analysis of variance followed by Dunnett's multiple comparison's post test (*, p < 0.05; **, p < 0.01; ***, p < 0.001). C, transactivation of TAT mRNA. BWTG3 cells were treated with Dex (1 μm) or CpdA (10 μm) for 72 h. Control wells received an equal amount of ethanol. Total RNA was reverse transcribed and the cDNA obtained subjected to PCR analysis with primers to detect mTAT and glyceraldehyde-3-phosphate dehydrogenase (GAPDH) (housekeeping gene used as an internal control) in separate reactions. PCR products were separated on agarose gel and visualized under UV light after EtBr staining. The figure is representative of three independent experiments. D, transactivation of TAT activity. BWTG3 cells were treated with Dex (1 μm) or CpdA (10 μm) for 4 h. Control wells received an equal amount of ethanol. Cell lysate was prepared and assayed for TAT activity ± S.E. (error bars). Statistical analysis was done to compare values in the presence of test compounds relative to control using one-way analysis of variance followed by Dunnett's multiple comparisons post test (*, p < 0.05).
CpdA Binds Reversibly to the Mouse GR, Influences Association of [3H]Dex but Has No Effect on Dissociation, and Causes Nuclear Translocation of GR
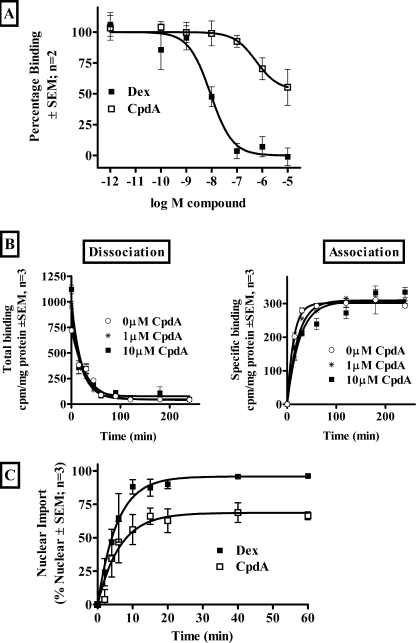
Binding of CpdA to mGR was investigated, using whole cell competitive binding, in BWTG3 cells that express endogenous GR (Fig. 4A). CpdA binds GR with a significantly (p < 0.05) lower affinity (Kd = 81.8 nm) than Dex (Kd = 1.29 nm). In addition, CpdA displayed an atypical binding curve in displacing only 47% of the [3H]Dex. CpdA can cyclize to an aziridine with alkylating potential (2) and thus to eliminate the possibility that CpdA is covalently modifying the GR and thus changing its affinity for Dex, BWTG3 cells were preincubated with CpdA or Dex, thoroughly washed, and then tested for binding of [3H]Dex (supplemental Fig. S3). Kinetic GR binding studies (Fig. 4B) show that CpdA slows the association of Dex resulting in a significant (p < 0.01) increase in the half-life (t½) of association (from 9.01 min in the absence of CpdA to 17.45 min in the presence of 10 μm CpdA), but that it does not significantly (p > 0.05) affect the dissociation of Dex from GR.
FIGURE 4.
Binding of CpdA to rodent GR and translocation to nucleus. A, competitive whole cell binding in BWTG3 cells of 20 nm [3H]Dex in the presence of increasing concentrations of unlabeled Dex or CpdA. The results shown are from two independent experiments performed in quadruplicate. Points indicate average ± S.E. Curve fitting was performed using non-linear regression and one-site competition to obtain log EC50 and maximal displacement. Ki values were obtained using the method of Cheng and Prusoff (68). B, kinetics of binding in BWTG3 cells of 1 nm [3H]Dex in the absence (0 μm CpdA) and presence of CpdA (1 and 10 μm). The results shown are from three independent experiments performed in triplicate ± S.E. (error bars). To obtain t½ curve fitting was performed using non-linear regression and one phase exponential association and one phase exponential decay, respectively. C, nuclear translocation of transiently transfected rGR in COS-1 cells. pTC2-wtGRrat (11.6 μg) was transiently transfected into COS-1 cells (2 × 106 cells/plate in 10-cm tissue culture dishes) and replated 24 h later (3 × 105 cells/well in a 6-well plate) into DMEM supplemented with 10% stripped FCS for 24 h before being treated with Dex (1 μm) or CpdA (10 μm) for 0, 2, 4, 6, 10, 15, 20, 40, or 60 min. After fixation, cells were subjected to immunostaining with rabbit anti-GR, followed by anti-rabbit Alexa 488 as a secondary antibody. Hoechst 33258 stain was used to visualize nuclei. Cells were analyzed using an IX 81 Olympus microscope. The percentage of cells showing nuclear localization of GR was quantified for three independent experiments ± S.E. (error bars). One-phase exponential association curve fitting was conducted to determine t½ and maximal localization.
Kinetic studies of the nuclear translocation of GR in COS-1 cells transiently transfected with rat GR (Fig. 4C) show that the nuclear import rate (t½) induced by CpdA is not significantly (p > 0.05) slower than that induced by Dex. However, the nuclear localization plateaus at a significantly (p < 0.01) lower level than seen with Dex (68.6 ± 3.4% nuclear for CpdA versus 95.8 ± 2.6% nuclear for Dex). These results are comparable with those found in BWTG3 cells where CpdA (1 μm), like Dex (1 μm), induces nuclear translocation of endogenous GR, albeit to a slightly lesser degree (72% nuclear) than Dex (100% nuclear) at 30 min (supplemental Fig. S4).
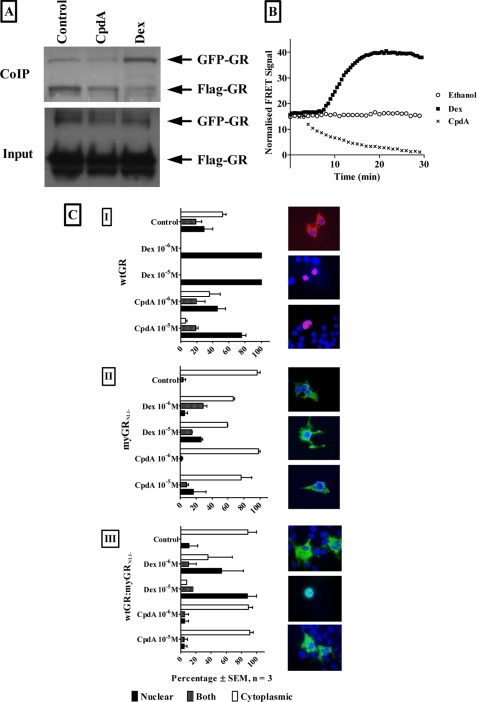
CpdA, Unlike Dex, Does Not Induce Dimerization of the GR
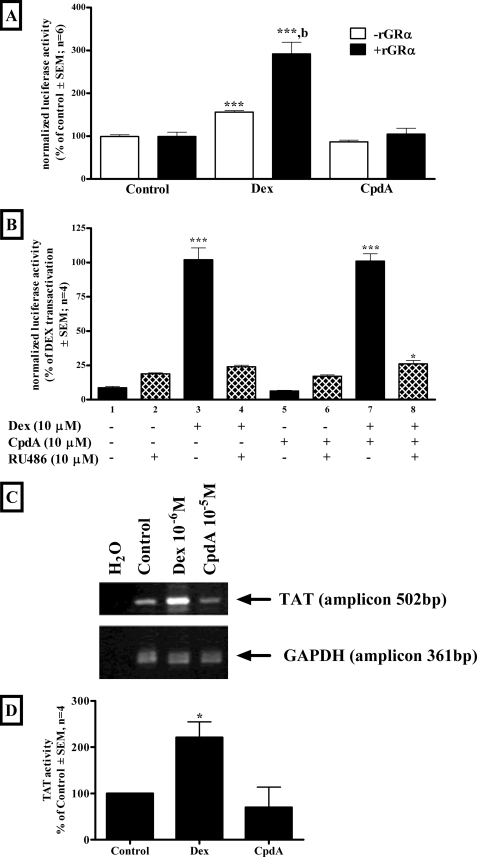
It has been postulated that the dissociated GC behavior could result from differential interaction of the liganded GR with co-activators and co-repressors (38) or from loss of GR dimerization (16, 39). We investigated the latter mechanism using a co-immunoprecipitation assay of transiently transfected FLAG- and GFP-tagged GR, immunoprecipitated with FLAG antibody, which shows that CpdA, unlike Dex, does not result in enrichment of GFP-tagged GR and thus GR dimerization (Fig. 5A). In fact CpdA appears to decrease baseline GR dimerization as confirmed with FRET (Fig. 5B), where addition of CpdA results in a decrease in FRET indicating a decrease in GR dimerization, whereas addition of Dex results in an increase in FRET indicating an increase in GR dimerization. The inability of CpdA to elicit GR dimerization is further confirmed with an elegant approach first used by Savory et al. (40). Briefly, a c-myc-tagged, nuclear localization signal 1 (NL1)-deficient mutant GR (myGRNL1−) is co-transfected with wtGR in COS-1 cells (Fig. 5C). Co-transfection of the wtGR, with an intact NL1, enables the defective mutant GR (myGRNL1−) to translocate to the nucleus within the context of a wtGR:myGRNL1− dimer where the wtGR NL1 suffices for translocation of the dimer. Transfection of the wtGR alone was visualized by anti-GR antibody and confirms that Dex and CpdA can cause nuclear translocation of the wtGR (Fig. 5C, I, closed bars). Transfection with the nuclear translocation mutant, myGRNL1−, alone (Fig. 5C, II) was visualized using an anti-myc antibody and shows that neither Dex nor CpdA can cause substantial nuclear translocation of this mutant GR and thus the GR remains mainly cytoplasmic (Fig. 5C, II, open bars). However, if the wtGR is cotransfected with the nuclear translocation mutant GR (Fig. 5C, III) and just the mutant receptor is visualized with an anti-myc antibody, nuclear translocation (Fig. 5C, III, closed bars) of the mutant GR is only observed with Dex (89% nuclear at 10 μm), but not with CpdA (5% nuclear at 10 μm).
FIGURE 5.
Dex, but not CpdA, results in ligand-induced dimerization of the GR in COS-1 cells. A, co-immunoprecipitation of differentially tagged GR. COS-1 cells were transiently transfected with FLAG-tagged GR (pEFFlaghGRα, mass 96 kDa), GFP-tagged GR (pEGFP-C2-GR, mass 128.5 kDa), and pGL2-basic and treated with ethanol (control), 10−6 m Dex or 10−5 m CpdA for 1 h. Cellular extracts were immunoprecipitated with anti-FLAG beads. Western blots were probed with anti-GR antibody. The figure is representative of three independent experiments. B, FRET analysis of GR dimerization. COS-1 cells were transiently transfected with CFP-tagged GR (pECFP-hGRα) and YFP-tagged GR (pEYFP-hGRα) and treated with ethanol (control), 10−5 m Dex or CpdA for 30 min while FRET intensity was monitored on the Olympus IX 81 motorized inverted microscope at 37 °C. Corrected FRET is plotted against time. The figure is representative of three independent experiments. C, Dex, but not CpdA, induces nuclear localization of the GR mutant when co-transfected with wtGR in COS-1 cells. COS-1 cells (3 × 106 per plate in 10-cm tissue culture dishes) were transfected (I) with 12 μg of WT rat GRα (pSVGR1) alone (wtGR), (II) with 12 μg of c-myc-tagged nuclear translocation mutant GR alone (myGRNL1−), or (III) with 12 μg DNA in an 8:1 ratio of wtGR:myGRNL1−(wtGR:myGRNL1−) and replated 24 h later (5 × 105 cells/well in a 6-well plate) into Opti-MEM for 24 h before induction with Dex (1 or 10 μm) or CpdA (1 or 10 μm) for 60 min. Control wells received an equal amount of ethanol. Localization of constructs was visualized as follows: (I) wtGR alone by indirect immunofluorescence using a rabbit anti-GR antibody followed by an Alexa Fluor 594-tagged anti-rabbit antibody (anti-GR; red); (II and III) myGRNL1− alone or wtGR:myGRNL1− by indirect immunofluorescence using a mouse anti-c-myc antibody followed by an Alexa Fluor 488-tagged anti-mouse antibody (anti-myc; green). Nuclei were visualized with 4′,6-diamidino-2-phenylindole staining (blue). Cells were analyzed using a Zeiss confocal LSM410 microscope. The percentage of cells showing nuclear (closed bars), both (hatched bars), or cytoplasmic (open bars) localization of GR was quantified for three independent experiments and is presented graphically ± S.E. (error bars). Representative micrographs for control, Dex (10 μm), and CpdA (10 μm) are presented to the right of each graph.
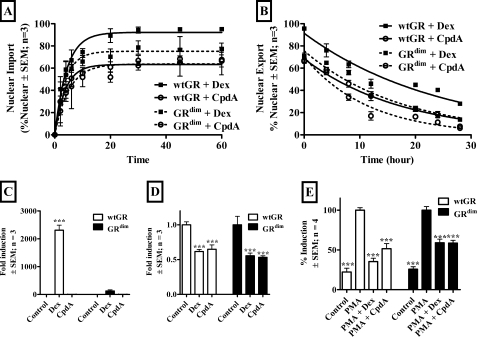
Implications of Loss of GR Dimerization Induced by CpdA
To test the implications of the loss of GR dimerization observed with CpdA we investigated the behavior of Dex and CpdA via both wild type GR (wtGR) and a GR dimerization mutant (GRdim) (16). Nuclear import (Fig. 6A) shows that, although there is no significant (p > 0.05) difference in nuclear import rate (t½), the maximal import is significantly (p < 0.01) higher for Dex via wtGR (93.28 ± 1.40%) than for Dex via GRdim (75.76 ± 3.18%), which is not significantly different from that attained with CpdA via wtGR (65.83 ± 1.19%) or CpdA via GRdim (64.60 ± 3.91%). Nuclear export (Fig. 6B) in contrast does show a significant (p < 0.05) difference in nuclear export rate (t½) between Dex via wtGR (18.74 ± 1.56 h) and Dex via GRdim (12.15 ± 0.32 h), CpdA via wtGR (12.56 ± 0.62 h), and CpdA via GRdim (7.89 ± 0.95 h). Transactivation of a GRE-containing promoter construct (Fig. 6C) indicates that Dex, unlike CpdA, significantly (p < 0.001) transactivates through wtGR, but not via GRdim. This is in contrast to the ability of Dex, like CpdA, to repress the Cbg (Fig. 6D) and Il-6 (Fig. 6E) promoters via both wtGR and GRdim.
FIGURE 6.
Implications of loss of GR dimerization induced by CpdA. A, nuclear import of transiently transfected mouse GR and mouse GRdim mutant in COS-1 cells. COS-1 cells (2 × 106 cells/plate in 10-cm tissue culture dishes) were transiently transfected with 7.5 μg of pcDNA3.1-GRWT or pcDNA3.1-GRdim and replated 24 h later (3 × 105 cells/well in a 6-well plate) in DMEM supplemented with 10% stripped FCS. After 24 h cells were treated with Dex (1 μm) or CpdA (10 μm) for 0, 2, 4, 6, 10, 20, 30, 45, or 60 min. After fixation, cells were subjected to immunostaining with anti-GR, followed by anti-rabbit Alexa 488 as a secondary antibody. Hoechst 33258 stain was used to visualize nuclei. Cells were analyzed using an IX 81 Olympus microscope. The percentage of cells showing nuclear localization of GR was quantified for three independent experiments ± S.E. (error bars). One-phase exponential association curve fitting was conducted to determine t½ and maximal localization. B, nuclear export of transiently transfected mouse GR and mouse GRdim mutant in COS-1 cells. COS-1 cells (2 × 106 cells/plate in 10-cm tissue culture dishes) were transiently transfected with 7.5 μg of pcDNA3.1-GRWT or pcDNA3.1-GRdim and replated 24 h later (3 × 105 cells/well in a 6-well plate) in DMEM supplemented with 10% stripped FCS for 24 h before being treated with Dex (1 μm) or CpdA (10 μm) for 45 min, washed, and monitored over 0, 4, 8, 12, 20, 24, and 28 h. After fixation, cells were subjected to immunostaining with rabbit anti-GR, followed by anti-rabbit Alexa 488 as a secondary antibody. Hoechst 33258 stain was used to visualize nuclei. Cells were analyzed using an IX 81 Olympus microscope. The percentage of cells showing nuclear localization of GR was quantified for three independent experiments ± S.E. (error bars). One phase exponential decay curve fitting was conducted to determine t½. C, transactivation of the transiently transfected GRE-containing promoter reporter construct via mouse GR or mouse GRdim mutant. COS-1 cells (1 × 104 cells/well in 96-well tissue culture plates) were transiently transfected with 100 ng of pTAT-GRE2-Elb-luc, 10 ng of pcDNA3.1-GRWT or pcDNA3.1-GRdim as indicated, and 10 ng of pPGKβGeopbA. Twenty-four h after transfection cells were induced with Dex (1 μm) or CpdA (10 μm) for 24 h. Control wells received an equal amount of ethanol. Luciferase values were normalized for β-galactosidase and values plotted as fold-induction relative to average control ± S.E. (error bars). D, transrepression of the transiently transfected rat CBG295Luc promoter reporter construct via mouse GR or mouse GRdim mutant. COS-1 cells (2 × 106 cells/plate in 10-cm tissue culture dishes) were transiently transfected with 9 μg of CBG295Luc and 3 ng of pcDNA3.1-GRWT or pcDNA3.1-GRdim as indicated. Twenty-four h after transfection cells were replated (5 × 104 cells/well in 24-well tissue culture plates). Cells were induced for 24 h with Dex (1 μm) or CpdA (10 μm) 24 h after replating. Control wells received an equal amount of ethanol. Luciferase values were normalized with protein concentration and values plotted as fold-induction relative to average control ± S.E. (error bars). E, transrepression of the transiently transfected IL6-luc promoter reporter construct via mouse GR or mouse GRdim mutant. COS-1 cells (2 × 106 cells/plate in 10-cm tissue culture dishes) were transiently transfected with 9 μg of p(IL6κB)350hu.IL6Pluc+ and 0.9 μg of pcDNA3.1-GRWT or pcDNA3.1-GRdim as indicated. Twenty-four h after transfection cells were replated (5 × 104 cells/well in 24-well tissue culture plates). Cells were induced for 24 h with PMA (10 ng/ml) alone or with Dex (1 μm) or CpdA (10 μm) 24 h after replating. Control wells received an equal amount of ethanol. Luciferase values were normalized with protein concentration and values plotted as percentage induction with PMA induction alone as 100% ± S.E. (error bars). Statistical analysis was done to compare values in the presence of test compounds relative to the corresponding control (C and D) or PMA induction alone (E) using one-way analysis of variance followed by Dunnett's multiple comparisons post test (*, p < 0.05; **, p < 0.01; ***, p < 0,001).
Steroid Receptor Specificity of CpdA
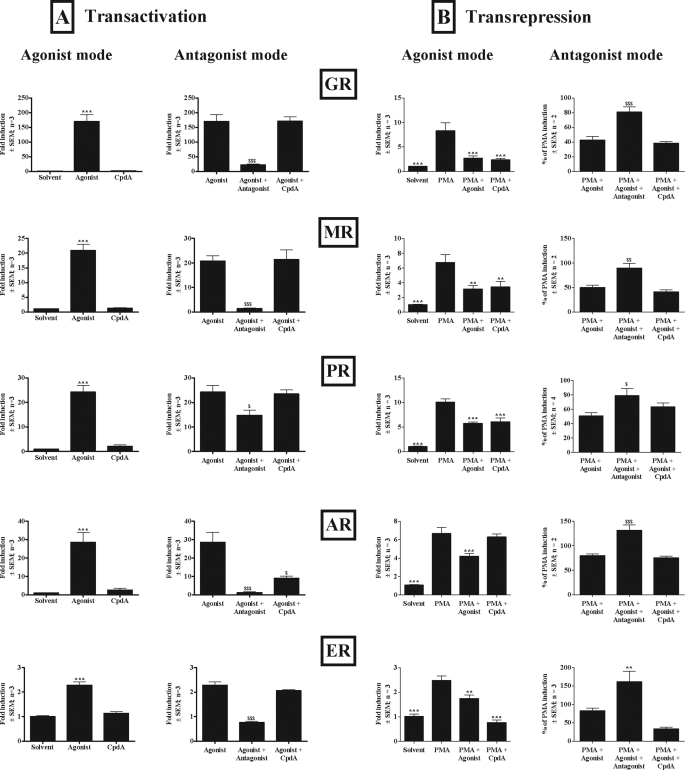
Having established that CpdA repression of Cbg is via the GR (Fig. 2), we set out to test the steroid receptor specificity of CpdA in COS-1 cells transiently transfected with GR, MR, PR, AR, or ER (Fig. 7). To test for transactivation via GR, MR, PR, and AR a GRE-containing promoter reporter was co-transfected as these receptors share a consensus response element (41, 42), whereas for transactivation via ER an ERE-containing promoter reporter was co-transfected. An NFκB-binding site-containing promoter reporter was cotransfected to test for transrepression. The agonists used were Dex for GR, aldosterone for MR, R5020 for PR, DHT for AR, and E2 for ER, whereas the antagonists used were RU486 for GR, spironolactone for MR, RU486 for PR, hydroxyflutamide for AR, and ICI 182,780 for ER. Both agonist and antagonist modes were tested and the results show that under these conditions CpdA acts as an AR antagonist in transactivation and as a GR, MR, PR, and ER agonist in transrepression.
FIGURE 7.
Steroid receptor specificity of CpdA. A, transactivation of transiently transfected GRE-containing promoter reporter construct via GR, MR, PR, or AR, or ERE-containing promoter reporter construct via ER. COS-1 cells (1 × 105 cells/well in 24-well tissue culture plates) were transiently transfected with 300 ng of pTAT-GRE2-Elb-luc, 30 ng of pPGKβGeopbA, and 30 ng of pRS-hGRα, pRS-hMR, pSG5hPRB, or pSVARo as indicated, or 1200 ng of pGL2–3x-ERE-TATA-luc, 30 ng of pPGKβGeopbA, and 30 ng of pcDNA3-ERα. Twenty-four h after transfection cells were induced for 24 h with solvent (ethanol), agonist (10−6 m), or CpdA (10−5 m) (agonist mode), or with agonist (10−6 m), agonist (10−6 m) plus antagonist (10−6 m), or agonist (10−6 m) plus CpdA (10−5 m) (antagonist mode). Luciferase values were normalized with β-galactosidase and values plotted as fold-induction ± S.E. (error bars) relative to average solvent. Statistical analysis was done to compare values in the presence of test compounds relative to solvent (*, p < 0.05; **, p < 0.01; ***, p < 0.00) for agonist mode or corresponding agonist ($, p < 0.05; $$, p < 0.01; $$$, p < 0.001) for antagonist mode using one-way analysis of variance with Dunnett's multiple comparisons test as post test. B, transrepression of transiently transfected IL-6 promoter reporter construct via GR, MR, PR, AR, or ER. COS-1 cells (2 × 106 cells/plate in 10-cm tissue culture dishes) were transiently transfected with 9 μg of p(IL6κB)350hu.IL6Pluc+ and 0.9 μg of pRS-hGRα, pRS-hMR, pSG5hPRB, pSVARo, or pcDNA3-ERα as indicated. Twenty-four h after transfection cells were replated (5 × 104 cells/well in 24-well tissue culture plates). Cells were induced for 24 h with solvent (ethanol), PMA (10 ng/ml), PMA plus agonist (10−6 m), or PMA plus CpdA (10−5 m) (agonist mode) or with PMA, PMA plus agonist (10−6 m), PMA plus agonist (10−6 m) and antagonist (10−6 m), or PMA plus agonist (10−6 m) and CpdA (10−5 m) (antagonist mode) 24 h after replating. Luciferase values were normalized with protein concentration and values plotted as fold-induction ± S.E. (error bars) relative to average solvent (agonist mode) or as percentage of PMA induction (antagonist mode). Statistical analysis was done to compare values in the presence of test compounds relative to PMA induction (*, p < 0.05; **, p < 0.01; ***, p < 0.00) for agonist mode or PMA plus corresponding agonist induction ($, p < 0.05; $$, p < 0.01; $$$, p < 0.001) for antagonist mode using one-way analysis of variance with Dunnett's multiple comparisons test as post test. Agonists used: Dex for GR; aldosterone for MR; R5020 for PR; DHT for AR; E2 for ER. Antagonists used: RU486 for GR; spironolactone for MR; RU486 for PR; hydroxyflutamide for AR; ICI 182,780 for ER.
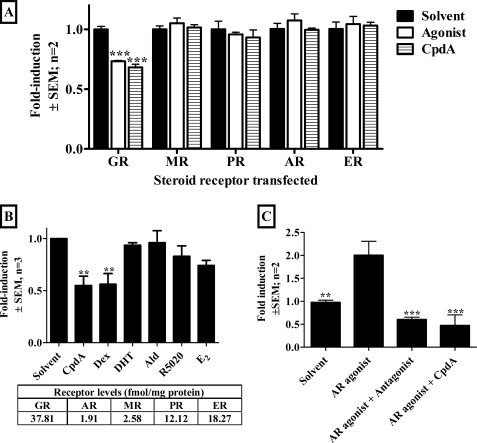
The promiscuous steroid receptor specificity of CpdA in transrepression via the NFκB-binding site, using COS-1 cells and PMA as a stimulus, was of concern, especially as previous data had shown transrepression of a NF-κB-driven promoter construct only via GR when using tumor necrosis factor as a proinflammatory stimulus in HEK293T cells (6). Therefore, further investigation focused on transrepression in the context of the Cbg proximal promoter and at the CBG protein level. The Cbg promoter is only significantly (p < 0.001) transrepressed by CpdA and the cognate agonist via the GR and not via the other steroid receptors (Fig. 8A). As steroid receptor levels were quite low in this experiment we investigated transactivation of a GRE- or ERE-containing promoter reporter at the same levels of transfected steroid receptors and found that the receptors were active at these concentrations (supplemental Fig. S5). In addition, at the CBG protein level only Dex and CpdA, but not the other steroid receptor ligands, show significant (p < 0.01) transrepression via endogenous steroid receptors in BWTG3 cells (Fig. 8B). Whole cell binding assays indicate that BWGT3 cells contain mainly GR, ER, and PR and little AR and MR (Fig. 8B, table inset). Thus taken in isolation these results (Fig. 8B) only support the findings that repression of Cbg by CpdA in BWTG3 cells occurs through GR. However, if the results are considered together with that in Fig. 8A it suggests increased steroid receptor specificity of CpdA in the context of the Cbg promoter and at CBG protein levels. Interestingly, the antagonism of the AR by CpdA observed with a GRE-containing promoter reporter (Fig. 7A) is confirmed at the protein activity level, with CpdA, like hydroxyflutamide, significantly (p < 0.001) antagonizing DHT transactivation of the TAT protein activity levels in BWTG3 cells transfected with AR (Fig. 8C).
FIGURE 8.
CpdA shows greater steroid receptor specificity in context of the Cbg promoter construct and at the CBG, but not TAT, protein level. A, steroid receptor specificity of transrepression of the transiently transfected rat CBG295Luc promoter reporter construct via GR, MR, PR, AR, or ER. COS-1 cells (2 × 106 cells/plate in 10-cm tissue culture dishes) were transiently transfected with 9 μg of CBG295Luc and 3 ng of pRS-hGRα, pRS-hMR, pSG5hPRB, pSVARo, or pcDNA3-ERα as indicated. Twenty-four h after transfection cells were replated (5 × 104 cells/well in 24-well tissue culture plates) in medium with 10% dextran-coated charcoal-stripped FCS and 1% antibiotics, except for assays investigating ER activity, where phenol red-free medium with 10% dextran-coated charcoal-stripped FCS and 1% antibiotics was used. Cells were induced, in DMEM without supplements except in assays investigating ER activity, where phenol red-free medium with 10% dextran-coated charcoal-stripped FCS and 1% antibiotics was used, for 24 h with solvent (ethanol), agonist (10−6 m), or CpdA (10−5 m) 24 h after replating. Luciferase values were normalized with protein concentration and values plotted as fold-induction ± S.E. (error bars) relative to average control. Statistical analysis was done for each steroid receptor to compare values in the presence of test compounds relative to the corresponding control using two-way analysis of variance followed by Bonferroni post tests (*, p < 0.05; **, p < 0.01; ***, p < 0.001). B, transrepression of CBG protein levels by steroid receptor ligands via endogenous steroid receptors in BWTG3 cells. BWTG3 cells (2.5 × 105 cells/well in 6-well tissue culture plates) were induced 24 h after plating with solvent (ethanol), steroid receptor agonist (10−6 m) or CpdA (10−5 m) in medium with 10% dextran-coated charcoal-stripped FCS and antibiotics except for assays investigating ER activity, where phenol red-free medium with 10% dextran-coated charcoal-stripped FCS and antibiotics was used. After 24 h cells were lysed, and lysates were separated on a SDS-PAGE gel and transferred to a Hybond-ECL nitrocellulose membrane, which was probed for CBG (S1000–76Z) and actin (sc-1616). Proteins were visualized using ECL peroxidase-labeled anti-goat antibody and ECL Western blotting detection reagents on Hyperfilm. For determination of CBG levels Hyperfilm bands of CBG and actin were quantified and results expressed as intensity of the CBG band relative to actin band ± S.E. (error bars). Statistical analysis was done to compare values in the presence of test compounds relative to the corresponding control using one-way analysis of variance followed by Dunnett's multiple comparisons post test (*, p < 0.05; **, p < 0.01; ***, p < 0,001). Table inset: whole cell binding was done in BWTG3 cells to determine endogenous steroid receptor levels by incubating BWTG3 cells with [3H]Dex for GR, [3H]E2 for ER, [3H]R5020 for PR, [3H]mibolerone for AR, or [3H]aldosterone for MR. Cognate unlabeled ligands were added in 1000-fold excess. Specific binding is presented in femtomole/mg of protein. C, transactivation of TAT activity in BWTG3 cells transfected with AR. BWTG3 cells (2.5 × 105 cells/well in 6-well tissue culture plates) were transiently transfected with 150 ng of pSVARo and induced 24 h after transfection with solvent (ethanol), DHT, AR agonist (10−6 m), DHT, AR agonist (10−6 m) plus hydroxyflutamide, AR antagonist (10−6 m), or DHT, AR agonist (10−6 m) plus CpdA (10−5 m) in medium with 10% dextran-coated charcoal-stripped FCS and antibiotics. Cell lysate was prepared and assayed for TAT activity ± S.E. (error bars) 4 h after induction. Statistical analysis was done to compare values in the presence of test compounds relative to control using one-way analysis of variance followed by Dunnett's multiple comparisons post test (*, p < 0.05). Agonists used: Dex for GR; aldosterone (Ald) for MR; R5020 for PR; DHT for AR; E2 for ER. Antagonists used: hydroxyflutamide for AR.
DISCUSSION
Glucocorticoids remain the most effective treatment for a variety of inflammatory diseases but prolonged treatment, especially at high doses, results in severe side effects (39, 43). Selective GR agonists that retain the beneficial anti-inflammatory action of GCs but display fewer side effects are actively sought. Specifically, it has been suggested that compounds that dissociate between transactivation involved in many of the side effects of GCs, and transrepression, the major mechanism whereby GCs mediate their anti-inflammatory action, may be useful (12, 44, 45). Recently, several compounds displaying such dissociation both in vitro and in vivo have been reported: AL-438 (38), ZK 209614 (46), and CpdA (6, 28).
CpdA was initially described within the context of contraception in rats (1), where in vivo studies in rats showed decreases in CBG, ACTH, and LH levels (5). Our results suggest that repression of these proteins is due to a direct transcriptional effect of CpdA.
CBG is a central player in GC disposition and of interest in inflammation as a negative acute-phase protein (47) and modulator of the acute-phase response (48), (49). Our results strongly support a mechanism whereby CpdA results in GR-mediated repression of the Cbg gene, as additional co-transfected GR increased transrepression, whereas addition of the GR antagonist, RU486, abrogated CpdA-induced transrepression.
CpdA, however, was unable to induce transactivation of GRE-containing promoter-reporter constructs or to induce an increase in TAT mRNA or activity. TAT, a liver enzyme, involved in gluconeogenesis, is up-regulated by GCs (50) and is involved in the metabolic side effects of GC therapy (39). Thus CpdA does not act as either a GR agonist or antagonist in transactivation of GRE-containing promoter reporters or endogenous genes despite acting as an agonist in repressing CBG. The ability of CpdA to dissociate transrepression from transactivation shown previously in the context of inflammation (6, 28), and prostate cancer (51) is thus confirmed and strengthened by our biochemical results.
Our results also show that CpdA binds reversibly to the endogenous GR in BWTG3 cells with an affinity that is about 63-fold less than that for Dex, which is in contrast to previous results in L929sA cells (6) where CpdA was shown to have a 4-fold higher affinity for the GR than Dex. The results do, however, agree with those in DU145 cells (51) where CpdA was shown to bind the GR to a lesser extent than Dex. In addition, we also confirmed previous work showing that CpdA causes nuclear translocation of the GR (6, 28, 51). Interestingly, however, we show that although the nuclear import rate is similar, GR treated with CpdA does not result in full nuclear localization.
It has been suggested that GR ligands that dissociate transrepression from transactivation may be identified by selecting for compounds that prevent dimerization of the GR, as transgenic mice expressing a dimerization-deficient GR (GRdim/dim) retain transrepression, but not transactivation, capacity (16). Our results, in agreement with a previous study (28), indeed show that CpdA prevents ligand-induced dimerization, whereas FRET studies show that CpdA in fact decreases basal GR dimerization.
Using a GRdim mutant (22) we show that the behavior of Dex via this mutant is comparable with that of CpdA via the wtGR. Dex-induced nuclear localization of the GRdim mutant is similar to that induced by CpdA via the wtGR in that full nuclear localization is not attained in import studies, whereas in export studies the export rate (t½) is similar for CpdA via the wtGR and Dex via the GRdim mutant. In addition, we show that Dex via the GRdim mutant, like CpdA via the wtGR, cannot transactivate a GRE-containing promoter. Transactivation of GRE-containing genes is generally mediated through direct binding of the GR to DNA (9, 10) and is suggested to require GR dimerization (16, 52), which neither CpdA acting through wtGR nor Dex acting via the GRdim mutant would elicit. Thus these results strongly support the idea that the lack of CpdA-induced dimerization of the GR may contribute to its inability to transactivate. Transrepression by the GR, in contrast, may be via negative GREs, or via a tethering mechanism that does not require DNA binding by the GR, but rather protein-protein interactions with other transcription factors (10, 53, 54). The first mechanism is reported to require dimerization of the GR, whereas the latter mechanism may be mediated by monomeric GR (9, 55). We show that CpdA acting via the wtGR, like Dex via the GRdim mutant, can repress an Il-6 promoter construct. This construct contains three NFκB elements binding NFκB proteins that have been shown to interact with GR via a tethering mechanism (10, 14), which does not require GR dimerization (16). In addition, we show that the Cbg promoter is repressed as effectively by Dex acting via the GRdim mutant as through the wtGR illustrating that dimerization of the GR is not required for transrepression. Although plasma levels of CBG are suppressed during prolonged administration of GCs, and physiological and physical stressors (56) and GCs decrease rat Cbg hepatic transcription (57), promoter studies that identify possible cis-acting sequence elements involved in GC regulation are lacking, nor have negative GREs been identified in the Cbg promoter (17). Our results thus suggest that CpdA and Dex repression of Cbg gene expression does not require GR dimerization and therefore probably proceeds via a tethering mechanism. The fact that the Pomc (58, 59) and Gnrh (18) promoters, which we show to be repressed by CpdA, have also been reported to be repressed by the GR via a tethering mechanism strengthens this hypothesis.
Although our results suggest that CpdA acts via the GR in transrepression of Cbg gene expression, investigation of CpdA action via other receptors yielded some unexpected results when promoters containing transcription binding sites, specifically for GRE and NFκB, were investigated in COS-1 cells. CpdA acts as an AR antagonist in transactivation of a GRE-binding site containing the promoter reporter in our cell model. This agrees with previous work (51, 60), however, we did not find the antagonism of transactivation via the GR shown by Yemelyanov et al. (51) nor did we observe PR antagonism as shown by Tanner et al. (60). Whether the discrepancies related to GR and PR antagonism reflect differences in the ligand used or depend on clonal variation in post-translational receptor modifications and/or receptor protein stability of cell lines used is not clear yet. Nevertheless, both our work and previous results (51, 60), indicate that CpdA does not act as an agonist in transactivation.
With respect to transrepression our results show that CpdA is not only a GR agonist but may also act via MR, PR, and ER in transrepressing a NFκB-binding site containing promoter reporter in COS-1 cells induced with PMA. This is in contrast to previous work investigating transrepression, via the same construct, but in HEK293T cells, with tumor necrosis factor rather than PMA as the pro-inflammatory stimulus, which showed that CpdA acted only via GR (6).
Further examination of the steroid receptor specificity of CpdA in transrepression, however, indicates that within the context of the Cbg minimal promoter and at the CBG protein level, CpdA displays a much greater steroid receptor specificity in only transrepressing CBG via the GR, and not via MR, PR, AR, and ER. In contrast to this, we found that the AR antagonist activity displayed by CpdA on a GRE-containing synthetic reporter in COS-1 cells was mimicked in BWTG3 cells for expressed AR-mediated regulation of endogenous TAT activity. It has been suggested that the flanking sequences around GREs may play an important role in receptor specificity (61) and that individual GREs retain specific “architectural signatures” that includes distinct GR-binding sites as well as binding motifs for other transcription factors (62). Thus by investigating only isolated transcription binding sites the role of these flanking sequences that may contribute to the composite elements (63) enriched for motives to GR as well as other transcription factors may be overlooked. We suggest then that the Cbg promoter may contain such additional motifs that contribute to the steroid receptor specificity of CpdA. In addition, our results suggest that the steroid receptor specificity of the anti-inflammatory action of CpdA in transrepressing pro-inflammatory cytokines may be co-determined by cell type, pro-inflammatory stimulus, promoter context, as well as cell-specific regulation of hormone receptor expression/activities.
In summary, our results suggest that the ability of CpdA to elicit a conformational change in the GR, which abrogates dimerization, is sufficient to explain the observed dissociation between transactivation and transrepression. However, our results do not exclude the possibility that additional mechanisms, such as an impaired ability of CpdA-liganded GR to bind to DNA, also play a role. Transrepression models shown to require DNA binding but not dimerization, such as shown for the keratin genes (64), would be useful to investigate this possibility. Investigation of genes shown to transactivate via tethering mechanisms, where no DNA binding or dimerization is required, such as for the β-casein gene where GR was shown to transactivate through tethering to signal transducers and activators of transcription (STAT) 5 (65), could also be useful to shed light on this aspect. It may be that CpdA elicits a conformational change in the GR that results in differential recruitment of cofactors as have been shown for the dissociated GC, AL-438 (38). In support of this hypothesis, it has previously been shown that, unlike Dex, CpdA bound to the GR does not recruit GRIP-1, SRC-1A, NcoR, or SMRT, suggesting that CpdA, acting via the GR, may recruit different cofactors as compared with Dex (32).
Altogether, CpdA represents a novel, non-steroidal, dissociated GR modulator, which abrogates GR dimerization. This makes CpdA a very attractive candidate to investigate for future therapeutic applications (28, 51, 66). However, its use as a GR ligand that can shed light on the fundamental mechanisms underlying the regulation of gene expression by the GR, specifically the role of GR dimerization, should not be underestimated. Within this context the decrease in maximal nuclear import and increase in nuclear export rate for CpdA-bound wtGR, as for Dex-bound GRdim, as compared with Dex-bound wtGR, shown in the current paper, suggests that loss of GR dimerization results in a GR species that exits the nucleus at a significantly faster rate and that GR dimerization contributes to the amount of GR present in the nucleus upon ligand activation. Previous work has suggested that the hinge region of the GR contains a solution dimerization domain (23) involved in cytosolic dimerization of the liganded GR, a nuclear localization signal, NL1 (40), that mediates liganded and unliganded GR nuclear location, and a nuclear retention signal that actively retains the GR in the nucleus (67). Loss of NL1 and nuclear retention signal results in increased nuclear export and reduced nuclear occupancy, which closely resembles the effect found with CpdA and GRdim in the current study. However, the regions involved do not overlap (hinge region: amino acids 505–547 and GRdim mutation at amino acid 458 in the ligand binding domain) suggesting a potentially novel insight regarding additional factors that influence nuclear retention of the GR.
Supplementary Material
Acknowlegment
We thank Carmen Langeveldt for excellent technical assistance.
This work was supported in part by Flemish-South African bilateral agreement BIL99/39 and grants from the Medical Research Council, National Research Foundation, and Stellenbosch University (South Africa) (to A. L. and J. H.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1–S5.
- CpdA
- Compound A (2-(4acetoxyphenyl)-2-chloro-N-methylethylammonium chloride)
- ACTH
- adrenocorticotropic hormone
- CBG
- corticosteroid-binding globulin
- Dex
- dexamethasone
- DHT
- 4,5α-dihydrotestosterone
- GRdim
- dimerization-deficient GR mutant
- E2
- 17β-estradiol
- FRET
- fluorescence resonance energy transfer
- GnRH
- gonadotropin-releasing hormone
- GC
- glucocorticoid
- GR
- glucocorticoid receptor
- GRE
- glucocorticoid response elements
- LH
- luteneinizing hormone
- RU486
- mifepristone
- NL1
- nuclear localization signal 1
- PMA
- 12-myristate 13-acetate
- P450c11
- P450-dependent steroid 11β-hydroxylase
- POMC
- proopiomelanocortin
- TAT
- tyrosine aminotransferase
- wt
- wild type
- CFP
- cyan fluorescent protein
- YFP
- yellow fluorescent protein
- ER
- estrogen receptor
- ERE
- estrogen response element
- IL
- interleukin
- DMEM
- Dulbecco's modified Eagle's medium
- FCS
- fetal calf serum
- PBS
- phosphate-buffered saline
- BSA
- bovine serum albumin.
REFERENCES
- 1.Swart P., Swart A. C., Louw A., van der Merwe K. J. (2003) Bioessays 25, 612–619 [DOI] [PubMed] [Google Scholar]
- 2.Louw A., Swart P., de Kock S. S., van der Merwe K. J. (1997) Biochem. Pharmacol. 53, 189–197 [DOI] [PubMed] [Google Scholar]
- 3.Louw A., Allie F., Swart A. C., Swart P. (2000) Endocr. Res. 26, 729–736 [DOI] [PubMed] [Google Scholar]
- 4.Louw A., Swart P., Allie F. (2000) Biochem. Pharmacol. 59, 167–175 [DOI] [PubMed] [Google Scholar]
- 5.Louw A., Swart P. (1999) Endocrinology 140, 2044–2053 [DOI] [PubMed] [Google Scholar]
- 6.De Bosscher K., Vanden Berghe W., Beck I. M., Van Molle W., Hennuyer N., Hapgood J., Libert C., Staels B., Louw A., Haegeman G. (2005) Proc. Natl. Acad. Sci. U.S.A. 102, 15827–15832 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Beato M., Truss M., Chávez S. (1996) Ann. N.Y. Acad. Sci. 784, 93–123 [DOI] [PubMed] [Google Scholar]
- 8.Beato M., Klug J. (2000) Hum. Reprod. Update 6, 225–236 [DOI] [PubMed] [Google Scholar]
- 9.Newton R. (2000) Thorax 55, 603–613 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.De Bosscher K., Vanden Berghe W., Haegeman G. (2003) Endocr. Rev. 24, 488–522 [DOI] [PubMed] [Google Scholar]
- 11.Zhou J., Cidlowski J. A. (2005) Steroids 70, 407–417 [DOI] [PubMed] [Google Scholar]
- 12.Adcock I. M. (2000) Pulm. Pharmacol. Ther. 13, 115–126 [DOI] [PubMed] [Google Scholar]
- 13.McEwan I. J., Wright A. P., Gustafsson J. A. (1997) Bioessays 19, 153–160 [DOI] [PubMed] [Google Scholar]
- 14.Webster J. C., Cidlowski J. A. (1999) Trends Endocrinol. Metab. 10, 396–402 [DOI] [PubMed] [Google Scholar]
- 15.Hayashi R., Wada H., Ito K., Adcock I. M. (2004) Eur. J. Pharmacol. 500, 51–62 [DOI] [PubMed] [Google Scholar]
- 16.Reichardt H. M., Tuckermann J. P., Göttlicher M., Vujic M., Weih F., Angel P., Herrlich P., Schütz G. (2001) EMBO J. 20, 7168–7173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Underhill D. A., Hammond G. L. (1995) Gene 162, 205–211 [DOI] [PubMed] [Google Scholar]
- 18.Chandran U. R., Warren B. S., Baumann C. T., Hager G. L., DeFranco D. B. (1999) J. Biol. Chem. 274, 2372–2378 [DOI] [PubMed] [Google Scholar]
- 19.Therrien M., Drouin J. (1993) Mol. Cell. Biol. 13, 2342–2353 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Miesfeld R., Rusconi S., Godowski P. J., Maler B. A., Okret S., Wikström A. C., Gustafsson J. A., Yamamoto K. R. (1986) Cell 46, 389–399 [DOI] [PubMed] [Google Scholar]
- 21.Schaaf M. J., Cidlowski J. A. (2003) Mol. Cell. Biol. 23, 1922–1934 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Waddell D. S., Baehr L. M., van den Brandt J., Johnsen S. A., Reichardt H. M., Furlow J. D., Bodine S. C. (2008) Am. J. Physiol. Endocrinol. Metab. 295, E785–E797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Savory J. G., Préfontaine G. G., Lamprecht C., Liao M., Walther R. F., Lefebvre Y. A., Haché R. J. (2001) Mol. Cell. Biol. 21, 781–793 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Harnish D. C., Scicchitano M. S., Adelman S. J., Lyttle C. R., Karathanasis S. K. (2000) Endocrinology 141, 3403–3411 [DOI] [PubMed] [Google Scholar]
- 25.Kastner P., Bocquel M. T., Turcotte B., Garnier J. M., Horwitz K. B., Chambon P., Gronemeyer H. (1990) J. Biol. Chem. 265, 12163–12167 [PubMed] [Google Scholar]
- 26.Brinkmann A. O., Faber P. W., van Rooij H. C., Kuiper G. G., Ris C., Klaassen P., van der Korput J. A., Voorhorst M. M., van Laar J. H., Mulder E. (1989) J. Steroid Biochem. 34, 307–310 [DOI] [PubMed] [Google Scholar]
- 27.De Bosscher K., Vanden Berghe W., Vermeulen L., Plaisance S., Boone E., Haegeman G. (2000) Proc. Natl. Acad. Sci. U.S.A. 97, 3919–3924 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Dewint P., Gossye V., De Bosscher K., Vanden Berghe W., Van Beneden K., Deforce D., Van Calenbergh S., Müller-Ladner U., Vander Cruyssen B., Verbruggen G., Haegeman G., Elewaut D. (2008) J. Immunol. 180, 2608–2615 [DOI] [PubMed] [Google Scholar]
- 29.Sui X., Bramlett K. S., Jorge M. C., Swanson D. A., von Eschenbach A. C., Jenster G. (1999) J. Biol. Chem. 274, 9449–9454 [DOI] [PubMed] [Google Scholar]
- 30.Weatherman R. V., Chang C. Y., Clegg N. J., Carroll D. C., Day R. N., Baxter J. D., McDonnell D. P., Scanlan T. S., Schaufele F. (2002) Mol. Endocrinol. 16, 487–496 [DOI] [PubMed] [Google Scholar]
- 31.Vanden Berghe W., Plaisance S., Boone E., De Bosscher K., Schmitz M. L., Fiers W., Haegeman G. (1998) J. Biol. Chem. 273, 3285–3290 [DOI] [PubMed] [Google Scholar]
- 32.Ronacher K., Hadley K., Avenant C., Stubsrud E., Simons S. S., Jr., Louw A., Hapgood J. P. (2009) Mol. Cell. Endocrinol. 299, 219–231 [DOI] [PubMed] [Google Scholar]
- 33.Sadie H., Styger G., Hapgood J. (2003) Endocrinology 144, 1958–1971 [DOI] [PubMed] [Google Scholar]
- 34.Diamondstone T. I. (1966) Anal. Biochem. 16, 395–401 [Google Scholar]
- 35.Trón L., Szöllósi J., Damjanovich S., Helliwell S. H., Arndt-Jovin D. J., Jovin T. M. (1984) Biophys. J. 45, 939–946 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Drouin J., Trifiro M. A., Plante R. K., Nemer M., Eriksson P., Wrange O. (1989) Mol. Cell. Biol. 9, 5305–5314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Adelman J. P., Mason A. J., Hayflick J. S., Seeburg P. H. (1986) Proc. Natl. Acad. Sci. U.S.A. 83, 179–183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Coghlan M. J., Jacobson P. B., Lane B., Nakane M., Lin C. W., Elmore S. W., Kym P. R., Luly J. R., Carter G. W., Turner R., Tyree C. M., Hu J., Elgort M., Rosen J., Miner J. N. (2003) Mol. Endocrinol. 17, 860–869 [DOI] [PubMed] [Google Scholar]
- 39.Schäcke H., Döcke W. D., Asadullah K. (2002) Pharmacol. Ther. 96, 23–43 [DOI] [PubMed] [Google Scholar]
- 40.Savory J. G., Hsu B., Laquian I. R., Giffin W., Reich T., Haché R. J., Lefebvre Y. A. (1999) Mol. Cell. Biol. 19, 1025–1037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Neubig R. R., Spedding M., Kenakin T., Christopoulos A. (2003) Pharmacol. Rev. 55, 597–606 [DOI] [PubMed] [Google Scholar]
- 42.Verrijdt G., Haelens A., Schoenmakers E., Rombauts W., Claessens F. (2002) Biochem. J. 361, 97–103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Barnes P. J. (2006) Eur. J. Pharmacol. 533, 2–14 [DOI] [PubMed] [Google Scholar]
- 44.Schäcke H., Rehwinkel H. (2004) Curr. Opin. Investig. Drugs 5, 524–528 [PubMed] [Google Scholar]
- 45.Barnes P. J. (2006) Eur. Respir. J. 27, 413–426 [DOI] [PubMed] [Google Scholar]
- 46.Schäcke H., Schottelius A., Döcke W. D., Strehlke P., Jaroch S., Schmees N., Rehwinkel H., Hennekes H., Asadullah K. (2004) Proc. Natl. Acad. Sci. U.S.A. 101, 227–232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Black P. H., Garbutt L. D. (2002) J. Psychosom. Res. 52, 1–23 [DOI] [PubMed] [Google Scholar]
- 48.Beishuizen A., Thijs L. G., Vermes I. (2001) Intensive Care Med. 27, 1584–1591 [DOI] [PubMed] [Google Scholar]
- 49.Ingenbleek Y., Bernstein L. (1999) Nutrition 15, 305–320 [DOI] [PubMed] [Google Scholar]
- 50.Rigaud G., Roux J., Pictet R., Grange T. (1991) Cell 67, 977–986 [DOI] [PubMed] [Google Scholar]
- 51.Yemelyanov A., Czwornog J., Gera L., Joshi S., Chatterton R. T., Jr., Budunova I. (2008) Cancer Res. 68, 4763–4773 [DOI] [PubMed] [Google Scholar]
- 52.Reichardt H. M., Tuckermann J. P., Bauer A., Schütz G. (2000) Z. Rheumatol. 59, Suppl. 2, II/1–II/5 [DOI] [PubMed] [Google Scholar]
- 53.Schoneveld O. J., Gaemers I. C., Lamers W. H. (2004) Biochim. Biophys. Acta 1680, 114–128 [DOI] [PubMed] [Google Scholar]
- 54.De Bosscher K., Haegeman G. (2009) Mol. Endocrinol. 23, 281–291 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Schäcke H., Hennekes H., Schottelius A., Jaroch S., Lehmann M., Schmees N., Rehwinkel H., Asadullah K. (2002) Ernst. Schering. Res. Found. Workshop 357–371 [DOI] [PubMed] [Google Scholar]
- 56.Deak T., Meriwether J. L., Fleshner M., Spencer R. L., Abouhamze A., Moldawer L. L., Grahn R. E., Watkins L. R., Maier S. F. (1997) Am. J. Physiol. Regul. Integr. Comp. Physiol. 273, R1998–R2004 [DOI] [PubMed] [Google Scholar]
- 57.Smith C. L., Hammond G. L. (1992) Endocrinology 130, 2245–2251 [DOI] [PubMed] [Google Scholar]
- 58.Martens C., Bilodeau S., Maira M., Gauthier Y., Drouin J. (2005) Mol. Endocrinol. 19, 885–897 [DOI] [PubMed] [Google Scholar]
- 59.Bilodeau S., Vallette-Kasic S., Gauthier Y., Figarella-Branger D., Brue T., Berthelet F., Lacroix A., Batista D., Stratakis C., Hanson J., Meij B., Drouin J. (2006) Genes Dev. 20, 2871–2886 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Tanner T. M., Verrijdt G., Rombauts W., Louw A., Hapgood J. P., Claessens F. (2003) Mol. Cell. Endocrinol. 201, 155–164 [DOI] [PubMed] [Google Scholar]
- 61.Jacobsen B. M., Jambal P., Schittone S. A., Horwitz K. B. (2009) Mol. Endocrinol. 23, 989–1000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.So A. Y., Cooper S. B., Feldman B. J., Manuchehri M., Yamamoto K. R. (2008) Proc. Natl. Acad. Sci. U.S.A. 105, 5745–5749 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.So A. Y., Chaivorapol C., Bolton E. C., Li H., Yamamoto K. R. (2007) PLoS Genet. 3, e94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Radoja N., Komine M., Jho S. H., Blumenberg M., Tomic-Canic M. (2000) Mol. Cell. Biol. 20, 4328–4339 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Stöcklin E., Wissler M., Gouilleux F., Groner B. (1996) Nature 383, 726–728 [DOI] [PubMed] [Google Scholar]
- 66.Zhang Z., Zhang Z. Y., Schluesener H. J. (2009) J. Immunol. 183, 3081–3091 [DOI] [PubMed] [Google Scholar]
- 67.Carrigan A., Walther R. F., Salem H. A., Wu D., Atlas E., Lefebvre Y. A., Haché R. J. G. (2007) J. Biol. Chem. 282, 10963–10971 [DOI] [PubMed] [Google Scholar]
- 68.Cheng Y., Prusoff W. H. (1973) Biochem. Pharmacol. 22, 3099–3108 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.