Abstract
Na+/H+ exchanger 3 (NHE3) plays an important role in neutral Na+ transport in mammalian epithelial cells. The Rho family of small GTPases and the PDZ (PSD-95/discs large/ZO-1) domain-based adaptor Shank2 are known to regulate the membrane expression and activity of NHE3. In this study we examined the role of βPix, a guanine nucleotide exchange factor for the Rho GTPase and a strong binding partner to Shank2, in NHE3 regulation using integrated molecular and physiological approaches. Immunoprecipitation and pulldown assays revealed that NHE3, Shank2, and βPix form a macromolecular complex when expressed heterologously in mammalian cells as well as endogenously in rat colon, kidney, and pancreas. In addition, these proteins co-segregated at the apical surface of rat colonic epithelial cells, as detected by immunofluorescence staining. When expressed in PS120/NHE3 cells, βPix increased membrane expression and basal activity of NHE3. Interestingly, the effects of βPix on NHE3 were abolished by cotransfection with dominant-negative Shank2 mutants and by treatment with Clostridium difficile toxin B, a Rho GTPase inhibitor, indicating that Shank2 and Rho GTPases are involved in βPix-mediated NHE3 regulation. Knockdown of endogenous βPix by RNA interference decreased Shank2-induced increase of NHE3 membrane expression in HEK 293T cells. These results indicate that βPix up-regulates NHE3 membrane expression and activity by Shank2-mediated protein-protein interaction and by activating Rho GTPases in the apical regions of epithelial cells.
Keywords: Cell/Epithelial, Cell/pH, G Proteins/Low Molecular Weight, Membrane/Proteins, Protein/Protein-Protein Interactions, Transport/Sodium
Introduction
Members of the Na+/H+ exchanger (NHE)2 family are integral membrane proteins that catalyze the extrusion of intracellular proton (H+) ions in exchange for extracellular sodium (Na+) ions and play vital roles in the regulation of cellular pH as well as transepithelial ion and water transport (1, 2). To date, eleven mammalian NHE proteins, including nine NHE isoforms belonged to the SLC9A family and two Na+/H+ antiporter proteins (3, 4), have been identified with unique tissue distribution and functional properties. As one of the better characterized isoforms, Na+/H+ exchanger 3 (NHE3, or SLC9A3) is known to be expressed in the apical membrane of epithelial cells of the renal proximal tubules and gastrointestinal tract where it plays a major role in acid-base and systemic fluid volume homeostasis (5). NHE3 knock-out mice have chronic diarrhea and altered salt and water homeostasis (6). NHE3 is known to be regulated by many hormones, neurotransmitters, and associated signaling systems such as cAMP, cGMP, and elevated intracellular calcium, but the underlying mechanisms are still only partially understood (5).
It has been shown that adaptor proteins with PDZ (PSD-95/discs large/ZO-1) domains play an important role in the membrane expression and acute regulation of NHE3 activity in polarized epithelia. For example, the NHERF family of adaptor proteins, which have two or four PDZ domains, are linked to cAMP-dependent inhibition of NHE3 in colon and kidney epithelia (7, 8). In addition, recent studies have indicated that another PDZ-based adaptor, Shank2, participates in the regulation of transepithelial salt and water transport by affecting NHE3 expression and activity (5, 9).
The Shank family of proteins was initially known for making molecular scaffolds in neuronal cells, where they serve as central coordinators of membrane and cytoplasmic protein complexes in the postsynaptic density (PSD) (10, 11). Shank polypeptides contain multiple sites for specific protein-protein interactions, including ankyrin repeats, an SH3 domain, a PDZ domain, a long proline-rich region, and a sterile α motif (SAM) (10). Currently there are three known members of the Shank family: Shank1, Shank2, and Shank3. Among them, Shank2 has been shown to be localized to the apical poles of pancreatic, colonic, hepatic, and renal epithelia and to modulate the activity of specific membrane transport proteins, such as the cystic fibrosis transmembrane conductance regulator (CFTR) and the type IIa sodium phosphate cotransporter (9, 12–14). In addition, Shank2 associates with NHE3 and up-regulates the membrane expression and basal activity of NHE3 in epithelial cells (5). However, the underlying mechanisms of these functions are still unclear.
Shank proteins interact with βPix and promote synaptic accumulation of βPix-associated signaling molecules at the PSD of excitatory synapses (15). It is believed that these associations may contribute to Shank-dependent organization of the PSD and to the regulation of dendritic spine dynamics (15). Pix (PAK-interacting exchange factor)/Cool (cloned out of library) is a family of guanine nucleotide exchange factors (GEFs) for the Rho family of small GTPases (16, 17). A very interesting feature is that Rho GTPases are critical for the retention and targeting of NHE3 in the apical membrane of epithelial cells (18, 19). Therefore, it is conceivable that βPix also associates with the Shank2-NHE3 complex and participates in NHE3 regulation by activating Rho GTPases near the apical pole in epithelial cells. In this study, we investigated the role of βPix in NHE3 regulation using integrated molecular and physiological approaches.
EXPERIMENTAL PROCEDURES
Materials and Cell Culture
Polyclonal antibodies against NHE3 (#1568 and #1314) (20, 21), Shank2 (#1136 and #3856) (11), and βPix (#1254 and #1257) (15) have been described previously. The anti-HA epitope monoclonal antibody (Cell Signaling Technology, Danvers, MA) and anti-GFP and β-actin polyclonal antibodies (Santa Cruz Biotechnology, Santa Cruz, CA) were purchased from commercial sources. The acetoxymethyl ester of 2′,7′-bis(carboxyethyl)-5(6)-carboxyfluorescein (BCECF) and Clostridium difficile toxin B (TxB) were purchased from Molecular Probes (Eugene, OR) and TechLab (Blacksburg, VA), respectively. Dithiobis(succinimidyl propionate) (DSP, cross-linking reagent), sulfo-NHS-SS-biotin, and NeutrAvidin were obtained from Pierce. All other chemicals, including nigericin, were purchased from Sigma. The pcDNA3-HA-rShank2 and pcDNA3.1-rShank2/CortBP1 plasmids (5, 9) containing wild-type Shank2/CortBP1 cDNA and the pcDNA3.1-rShank2(H109A) plasmid harboring the H109A mutation in the PDZ domain of Shank2 (14) (Fig. 1A) have been described previously. To generate pcDNA3.1-rShank2(ΔSAM), the SAM domain-deleted rShank2 (amino acids 1–1163 and amino acids Δ1164–1253) was PCR-amplified and subcloned into pcDNA3.1 using BamHI and NotI restriction sites. The pCMV-rNHE3 (22), pCMV-rNHE3′38HA3 (18, 23), and pEGFP-βPix (15) constructs have been described previously.
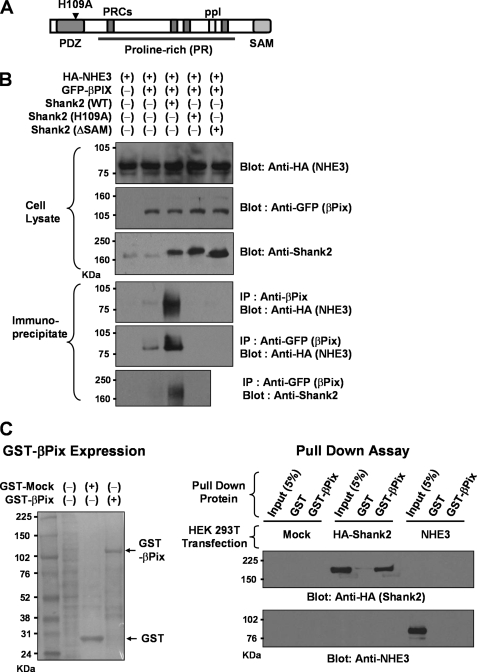
FIGURE 1.
Interaction of βPix, Shank2, and NHE3 in PS120/NHE3′38HA3 cells. A, a diagram depicting the domain structure of Shank2. B, immunoprecipitation (IP) was performed in PS120/NHE3′38HA3 cells that stably express HA epitope-tagged NHE3 (pCMV-rNHE3′38HA3). PS120/NHE3′38HA3 cells were cotransfected with the plasmids expressing GFP-βPix (pEGFP-βPix) and Shank2 (pcDNA3.1-rShank2). In some experiments, the Shank2 PDZ domain mutant (pcDNA3.1-rShank2/H109A) and SAM domain-deleted mutant (pcDNA3.1-rShank2/ΔSAM) were transfected to identify the role of these domains in protein complex formation. The PS120 cells were treated with the cross-linking agent DSP (2 mm) for 30 min at room temperature before harvesting. Protein samples were precipitated with anti-βPix (#1254) and anti-GFP antibodies and immunoblotting was carried out using monoclonal anti-HA, polyclonal anti-GFP, and polyclonal anti-Shank2 (#1136) antibodies. In immunoblotting of cell lysates, 50 μg of protein was loaded into each lane. Immunoprecipitation was performed using a total of 500 μg of cell lysate. C, pulldown assay. The GST-βPix fusion protein was expressed in E. coli and purified with glutathione-Sepharose 4B. HEK 293T cells were transfected with mock, pcDNA3-HA-Shank2, or pCMV-rNHE3 plasmids, and protein samples were incubated with 50 μg of GST alone or GST-βPix fusion protein. The pelleted protein by glutathione-Sepharose resin was immunoblotted with anti-HA or anti-NHE3 (#1568) antibodies. Left-hand image shows a Ponceau S stain of immunoblot (mock transfected), and right-hand images represent Shank2 and NHE3 immunoblots. GST-βPix showed a direct interaction with Shank2, but not with NHE3. PRCs, proline-rich clusters; ppI, proline-rich SH3 binding motif; and SAM, sterile α motif.
PS120 (NHE-deficient hamster fibroblast) and HEK 293T (human embryonic kidney) cells were maintained in Dulbecco's modified Eagle's medium-high glucose (Invitrogen) supplemented with 10% fetal bovine serum and penicillin (50 IU/ml)/streptomycin (50 μg/ml). Plasmids expressing NHE3 were stably transfected into PS120 cells using Lipofectamine Plus Reagent (Invitrogen). NHE3 stable transfectants were selected by resistance to the antibiotic Geneticin (G418, Invitrogen) and by an H+-killing method (24). Madin-Darby canine kidney (MDCK)-NHE3′38HA3-stable cells, described previously (18), were maintained in the 1:1 Dulbecco's modified Eagle's medium/nutrient mixture F-12 with 5% fetal bovine serum and with G418 selection (500 μg/ml). To knock down endogenous βPix expression in HEK 293T cells, 25-bp double-stranded RNA oligonucleotides specific for βPix were synthesized (Invitrogen) and transfected into cells using Lipofectamine 2000 (Invitrogen). The target small interfering RNA (siRNA) sequence was 5′-GGAGGATTATCATACAGATAGACAA-3′. A negative control RNA (StealthTM RNAi Negative Control Duplexes, catalog no. 12935-300, Invitrogen) was used in appropriate control experiments. Two days after transfection, cells were harvested in lysis buffer for immunoblotting.
Immunohistochemistry
Immunostaining of frozen sections was performed as reported previously (14). Briefly, colon tissue from Sprague-Dawley rats was embedded in OCT (Miles, Elkhart, IN), frozen in liquid N2, and cut into 4-μm sections. The sections were fixed and permeabilized by incubation in cold methanol for 10 min. Nonspecific binding sites were blocked by incubation for 1 h at room temperature with 0.1 ml of phosphate-buffered saline containing 5% goat serum, 1% bovine serum albumin, and 0.1% gelatin (blocking medium). After blocking, the sections were stained by incubating them with anti-Shank2 (#3856), anti-NHE3 (#1314), and/or anti-βPix (#1257) antibodies and then treated with fluorophore-tagged secondary antibodies. To cover the surface of the first primary antibody for double labeling using primary antibodies from the same host species, sections were incubated with unconjugated AffiniPure Fab Fragment Goat Anti-Rabbit IgG (H+L) (Jackson ImmunoResearch, West Grove, PA) overnight at 4 °C. Images were obtained with a Zeiss LSM 510 confocal microscope.
Immunoprecipitation and Immunoblotting
For immunoprecipitation, precleared colon, kidney, pancreas, or PS120 lysates (500 μg of protein) were mixed with the appropriate antibodies and incubated overnight at 4 °C in lysis buffer. The PS120 cells were treated with the cross-linking agent DSP (2 mm) for 30 min at room temperature before extraction. Immune complexes were collected by binding to protein G beads, which were subsequently washed four times with lysis buffer prior to electrophoresis. The immunoprecipitates or lysates (50 μg of protein) were suspended in SDS sample buffer and separated by SDS-PAGE. The separated proteins were transferred to nitrocellulose membranes, and the membranes were blocked by 1-h incubation at room temperature in blocking solution containing 5% nonfat dry milk. The membranes were then incubated with the appropriate primary and secondary antibodies, and protein bands were detected with enhanced chemiluminescence solutions (Amersham Biosciences).
Pulldown Assay
cDNA containing full-length βPix was generated by PCR amplification and subcloned into the glutathione S-transferase (GST) fusion vector pGEX4T-1 using SalI and NotI restriction sites. The GST-βPix fusion protein was expressed in Escherichia coli (strain B/BL21-DE3) and purified with glutathione-Sepharose 4B (Amersham Biosciences). For pulldown experiments, HEK 293T cells were transfected with pcDNA3-HA-Shank2 and pCMV-rNHE3. Two days after transfection, HEK 293T cells were lysed on ice in a 1% Triton X-100 buffer containing 200 mm NaCl, 2 mm MgCl2, 2 mm CaCl2, 10 mm HEPES (pH 7.4), and proteinase inhibitors (CompleteMini, Roche Applied Science). After centrifugation, the supernatant was incubated with 50 μg of GST fusion protein for overnight at 4 °C, followed by precipitation with glutathione-Sepharose 4B resin. The glutathione-Sepharose resin was pelleted and washed with wash buffer (3 × 5 min, 4 °C) prior to resuspension in SDS sample buffer and immunoblotting.
Cell-surface Biotinylation Assay
Cell-surface biotinylation of NHE3 was performed as described previously (5). Briefly, PS120/NHE3, MDCK/NHE3′38HA3, and HEK 293T cells were washed with ice-cold phosphate-buffered saline containing 0.1 mm CaCl2 and 1 mm MgCl2, and the plasma membrane proteins were then biotinylated by gently shaking the cells in phosphate-buffered saline containing sulfo-NHS-SS-biotin (Pierce) for 30 min at 4 °C. After biotinylation, the cells were washed extensively with quenching buffer and phosphate-buffered saline to remove excess biotin. The cells were then lysed, and NeutrAvidin solution (UltraLink Immobilized NeutrAvidin Beads 10%, Pierce) was added to the supernatant, and the mixture was incubated at 4 °C overnight. Avidin-bound complexes were pelleted (13,000 rpm) and washed three times. Biotinylated proteins were eluted in SDS sample buffer, resolved by SDS-PAGE, electrotransferred, and immunoblotted with the anti-NHE3 (#1568) antibody.
Measurement of Na+/H+ Exchange Activity
Na+/H+ exchange activity was measured using a standard protocol with some modifications (25). Briefly, cells grown on glass coverslips were loaded with a pH-sensitive fluorescent dye, BCECF, and intracellular pH (pHi) changes were measured. When Shank2 and βPix constructs were transiently expressed, a GFP-expressing plasmid was cotransfected, and pHi measurements were performed with cells expressing high levels of GFP as previously reported (26). As shown in supplemental Fig. 1, BCECF fluorescence showed at least 10-fold higher intensity than GFP fluorescence, and the background GFP fluorescence did not affect pHi measurements. The cells were acidified by an NH4+ (20 mm) pulse and subsequent perfusion with a Na+-free solution. The maximal Na+-dependent pHi recovery was measured in cells acidified to a pH of 6.4–6.5. The standard perfusion solution contained (mm): 140 NaCl, 5 KCl, 1 MgCl2, 1 CaCl2, 10 glucose, and 10 HEPES (pH 7.4 adjusted with NaOH). Na+-free solutions were prepared by replacing Na+ with N-methyl-d-glucamine+. The osmolarity of all solutions was adjusted to 310 mm with the major salt. The 490/440 nm ratios were calibrated intracellularly by perfusing the cells with solutions containing 145 mm KCl, 10 mm Tris, 5 μm nigericin with pH adjusted to 6.2–7.8, as described previously (5, 27). In each experiment, the intrinsic buffer capacity (βi) was calculated by measuring pHi in response to 5–20 mm NH4Cl pulses (5). However, any gene modulation did not significantly change βi. Therefore, all of the NHE activity values are expressed as ΔpH/min, and this value was directly analyzed without compensating for βi.
Statistical Analysis
The results of multiple experiments are presented as the means ± S.E. Statistical analysis was performed with analysis of variance followed by a Tukey multiple comparison test. p < 0.05 was considered statistically significant.
RESULTS
Shank2 Mediates Associations between NHE3 and βPix
To investigate a putative relationship between NHE3, βPix, and Shank2, PS120 cells that stably express a triple HA epitope-tagged form of NHE3 (NHE3′38HA3) were cotransfected with expression plasmids containing GFP-βPix singly or in combination with wild-type and dominant-negative forms of Shank2. Following 48 h of transfection, direct and indirect interactions among these proteins were examined by immunoprecipitation using the cross-linking agent DSP. As shown in Fig. 1B, an association between NHE3 and βPix was evident when wild-type Shank2 was coexpressed in the PS120/NHE3′38HA3 cells, whereas very little NHE3 immunoprecipitate was detected in cells transfected with βPix alone. This latter weak signal presumably reflects a complex of exogenous NHE3′38HA3 and βPIX with low levels of endogenous Shank2 that are present in PS120 cells (Fig. 1B). Although a weak association between βPix and NHE3 can be detected without using the cross-linking agent DSP, treatment with DSP evoked a much stronger interaction in immunoprecipitation (supplemental Fig. 2), implying that the association between βPix and NHE3 is mediated by an indirect interaction. The direct interaction of βPix with Shank2, but not with NHE3, was further confirmed by the pulldown assay using GST-βPix fusion protein (Fig. 1C).
The PDZ domain of Shank2 has been shown to mediate Shank2-NHE3 and Shank2-βPix interactions (5, 15). In general, the first histidine residue of the second α-helix of the PDZ domain (position αB1, His-109 in rShank2) plays an important role in class I PDZ interaction by forming a strong hydrogen bond between its N-3 nitrogen and the hydroxyl group of the −2 serine/threonine residue of the ligand (14). Interestingly, the dominant negative Shank2 PDZ (H109A) mutant completely abolished the association between NHE3 and βPix (Fig. 1B), indicating that the PDZ domain of Shank2 is critically involved in the NHE3-βPix association. Because Shank2 contains only one PDZ domain, it is unlikely that a Shank2 molecule can bind simultaneously to both NHE3 and βPix. Instead, Shank proteins can multimerize via the SAM domain, a domain known to mediate oligomerization (28). Thus, we examined the role of Shank2 multimerization in the NHE3-βPix association by using the SAM domain-deleted Shank2 (ΔSAM) construct. Notably, deletion of Shank2 SAM domain also completely abolished the NHE3-βPix interaction (Fig. 1B). Collectively, these results imply that oligomerization of Shank2-NHE3 and Shank2-βPix creates a large protein complex, resulting in association between NHE3 and βPix.
βPix Increases Surface Expression and Basal Activity of NHE3 in PS120 Cells in a Shank2-dependent Manner
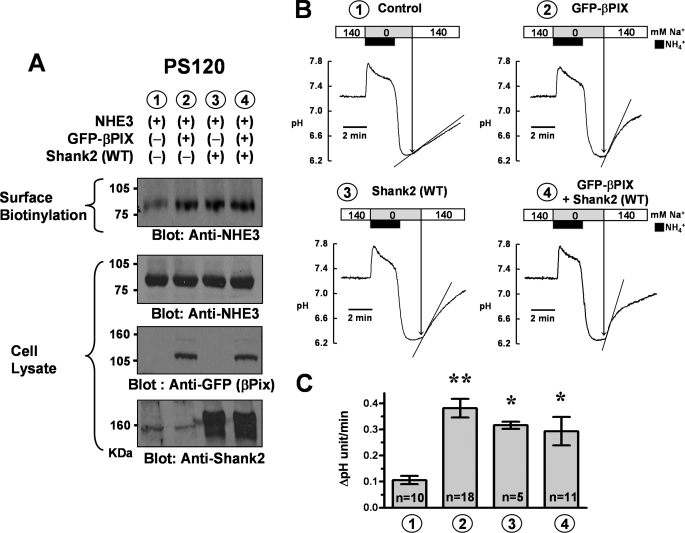
To investigate the functional role of βPix, the surface distribution and activity of NHE3 were investigated in PS120/NHE3 cells. In these experiments, we used non-HA-tagged NHE3 to better approximate its native structure. Plasma membrane expression of NHE3 was examined using a surface biotinylation assay. NHE activity was measured as the Na+-dependent increase in pHi after intracellular acidification induced by an NH4+ pulse as detailed under “Experimental Procedures.” Some studies measuring NHE3 kinetics in PS120 cells have been done under serum-deprived conditions, because serum deprivation for 18 h increases the surface expression of NHE3 (25). However, we used serum-supplemented conditions to preserve the innate regulation of NHE3 in PS120 cells. In addition, it has been shown that molecular scaffold-induced effects were better observed in the serum-supplemented condition (5). Notably, βPix increased the surface expression and activity of NHE3 (Fig. 2). The basal NHE activity of PS120/NHE3 cells was 0.106 ± 0.016 ΔpH/min, and this value was increased to 0.382 ± 0.036 ΔpH/min by βPix expression (Fig. 2C). As reported previously (5), Shank2 also up-regulated the surface expression and basal activity of NHE3. However, neither the surface expression nor the basal activity of NHE3 was further increased by coexpression of βPix and Shank2 (Fig. 2), suggesting that βPix and Shank2 share a common pathway in up-regulating NHE3.
FIGURE 2.
Effects of βPix expression on NHE3 surface expression and activity in PS120/NHE3 cells. PS120/NHE3 cells that stably express non-HA epitope-tagged NHE3 (pCMV-rNHE3) were cotransfected with pEGFP-βPix, pcDNA3.1-rShank2, and each mock plasmid. A, surface-biotinylated proteins and whole cell lysates were immunoblotted with anti-NHE3 (#1568), anti-GFP, and anti-Shank2 (#1136) antibodies. A low level of endogenous Shank2 expression is observed in lanes 1 and 2. Three separate experiments showed similar results. B, PS120/NHE3 cells were transfected with each plasmid, and NHE activities were measured as detailed under “Experimental Procedures.” The cells were kept in serum-supplemented conditions. C, a summary of multiple NHE activity measurements. *, p < 0.05; **, p < 0.01; difference from lane 1.
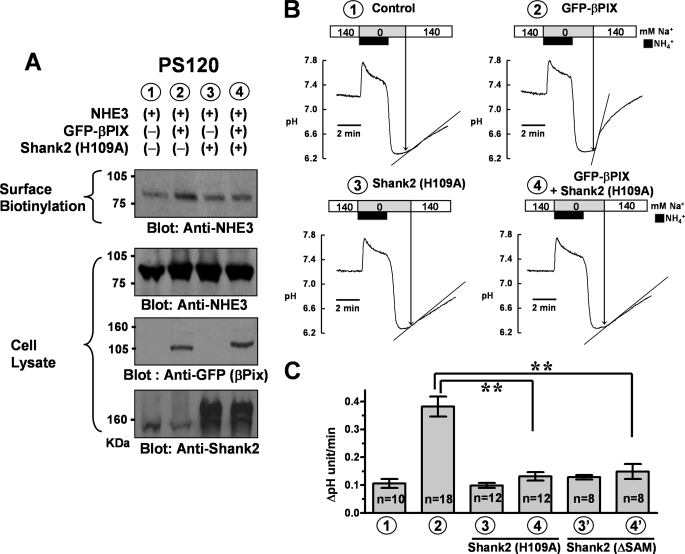
Next, we explored the role of Shank2 in the βPix-induced up-regulation of NHE3. In agreement with the results of our immunoprecipitation experiment (Fig. 1B), the Shank2 PDZ (H109A) mutant completely nullified the effects of βPix on NHE3 surface expression and activity (Fig. 3, A–C). Similar effects were observed with the Shank2 SAM domain-deleted (ΔSAM) mutant (Fig. 3C). These results indicate that the PDZ domain-mediated protein-protein interaction and multimerization of Shank2 molecules are required for the βPix-induced up-regulation of NHE3.
FIGURE 3.
Role of Shank2 in the βPix-induced up-regulation of NHE3 in PS120/NHE3 cells. PS120/NHE3 cells were cotransfected with pEGFP-βPix and/or pcDNA3.1-rShank2/H109A, and surface biotinylation and NHE activity measurements were performed. A, surface-biotinylated proteins and whole cell lysates were immunoblotted with anti-NHE3 (#1568), anti-GFP, and anti-Shank2 (#1136) antibodies. Four separate experiments showed similar results. B, PS120/NHE3 cells were transfected with each plasmid, and NHE activities were measured. C, a summary of multiple NHE activity measurements. Summarized results from cells transfected with pcDNA3.1-rShank2/ΔSAM (Shank2/ΔSAM) instead of pcDNA3.1-rShank2/H109A (Shank2/H109A) are also illustrated in the last two columns. Note that the dominant negative Shank2 PDZ domain mutant (H109A) and SAM domain-deleted mutant (ΔSAM) completely blocked the βPix-induced up-regulation of NHE3. **, p < 0.01; difference from lane 2.
βPix Is Required for Shank2-induced Up-regulation of NHE3
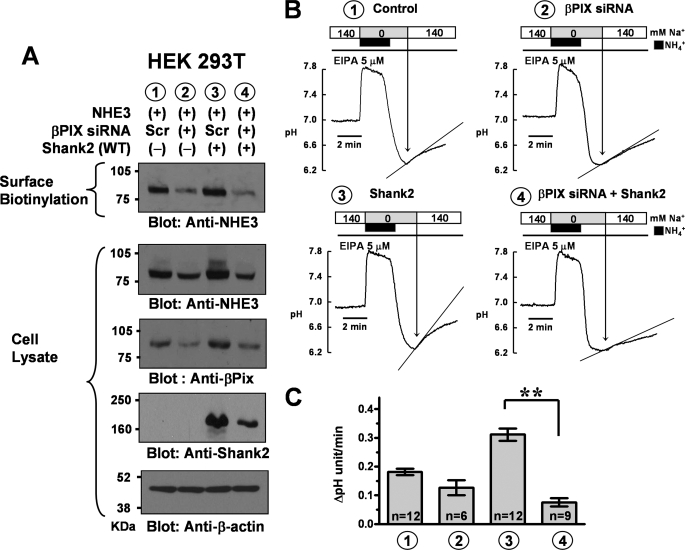
To determine whether βPix is involved in Shank2-induced up-regulation of NHE3, we used RNA interference to knock down βPix expression in HEK 293T cells that endogenously express human βPix. Compared with scrambled siRNA control, treatment with βPix siRNA induced a pronounced reduction in βPix protein expression (−82.9 ± 6.9%, p < 0.01, n = 4 as determined by densitometry) and a partial decrease in Shank2 expression (−44.9 ± 4.6%, p < 0.01, n = 4) and cytosolic expression of NHE3 (−28.0 ± 6.5%, p < 0.05, n = 4) without affecting β-actin expression (0.6 ± 4.4%, p = 0.90, n = 4). Interestingly, knock down of βPix was paralleled by the loss of NHE3 surface expression (−76.6 ± 6.7%, p < 0.01, n = 4, Fig. 4A). Although partial decrease in Shank2 (−45%) might contribute, the profound loss of βPix (−83%) would give a better explanation for the comparable decrease in NHE3 surface expression (−77%). Importantly, knock down of βPix completely abolished the Shank2-induced up-regulation of NHE3 surface expression. Comparable results were also observed in the NHE3 activity measurements (Fig. 4, B and C). These results suggest that association of βPix with the Shank2-NHE3 complex is an important underlying mechanism for the Shank2-induced NHE3 up-regulation reported previously (5).
FIGURE 4.
Effect of βPix knockdown on Shank2-induced up-regulation of NHE3 in HEK 293T cells. HEK 293T cells expressing NHE3 were cotransfected with pcDNA3.1-rShank2 or mock plasmids, and were treated with scrambled RNA or siRNAs against human βPix 1 day after plasmid transfection as detailed under “Experimental Procedures.” Forty-eight hours after siRNA treatment, cell-surface biotinylation and NHE measurements were carried out. A, surface-biotinylated proteins and whole cell lysates were immunoblotted with anti-NHE3 (#1568), anti-βPix (#1254), anti-Shank2 (#1136), and anti-β-actin antibodies. Three separate experiments showed similar results. B, NHE activities were measured in HEK 293T cells transfected with each plasmid and treated with siRNAs. Ethyl-isopropyl-amiloride (EIPA, 5 μm) was administered during NHE activity measurements to block endogenous NHE1 activity in HEK 293T cells. C, a summary of multiple NHE activity measurements. Note that βPix siRNA completely blocked the Shank2-induced up-regulation of NHE3. Scr: a negative control (scrambled) RNA (StealthTM RNAi Negative Control Duplexes, Invitrogen); **, p < 0.01; difference from lane 3.
Rho GTPases Are Involved in βPix-induced Up-regulation of NHE3
NHE3 requires an intact cytoskeleton for its optimal function. In most cells, small GTP-binding proteins of the Rho family are major regulators of the actin cytoskeleton (29, 30). In fact, Rho GTPases have been shown to play a critical role in the surface retention of NHE3 in MDCK epithelial cells (18). Because βPix works as an activating factor for Rho GTPases (16, 17), we considered the possibility that Rho GTPases are involved in the βPix-induced up-regulation of NHE3. Because the role of Rho proteins in the surface expression of NHE3 was best studied in MDCK cells (18), we analyzed surface expression of NHE3 in MDCK-NHE3′38HA3 cells after treatment with the Rho GTPase inhibitor C. difficile TxB. As depicted in Fig. 5, treatment with TxB resulted in a 66.2 ± 9.2% reduction in the surface expression of NHE3. More importantly, TxB completely eliminated the βPix-induced increase in NHE3 surface expression. These findings are consistent with the notion that Rho GTPases are involved in βPix-Shank2 complex-mediated regulation of NHE3.
FIGURE 5.
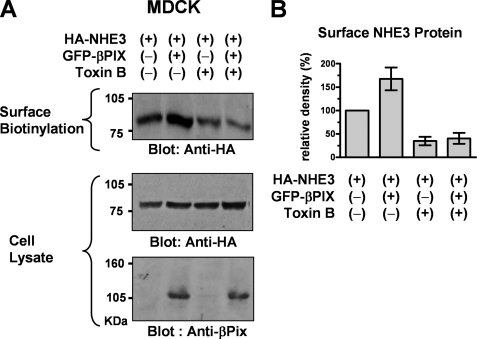
Effect of Rho GTPase inhibition on βPix-induced up-regulation of NHE3 in MDCK cells. MDCK cells stably expressing NHE3′38HA3 were transfected with pEGFP-βPix or mock plasmids. Forty-eight hours after transfection, cells were kept with or without the Rho GTPase inhibitor C. difficile TxB (4 μg/ml) for 4 h at 37 °C. A, surface NHE3 was visualized using surface biotinylation assay as described under “Experimental Procedures.” B, a summary of densitometric analysis from four separate experiments (relative density to NHE3 alone).
NHE3, βPix, and Shank2 Associate in Vivo
The relationship of NHE3, βPix, and Shank2 was examined next in epithelial tissues to explore its physiological relevance. Expression of Shank2 protein was initially confirmed in rat pancreas, ileum, colon, and kidney by immunoblotting (supplemental Fig. 3). As reported previously, kidney tissues express only the long isoform of Shank2 (Shank2E) (13), whereas colon and pancreas express both the short (CortBP1) and the long (Shank2E) forms of Shank2 (supplemental Fig. 3 and Fig. 6A). Importantly, coimmunoprecipitation results showed that NHE3 associates with both βPix and Shank2 in rat tissues, indicating that a protein complex of NHE3-βPix-Shank2 exists in rat epithelial tissues (Fig. 6A). Lastly, the localization of NHE3, βPix, and Shank2 was determined immunohistochemically in rat colon (Fig. 6B). NHE3 was principally expressed in the apical membrane of colonic epithelial cells. Although small fractions of βPix and Shank2 were observed in the basolateral area, these two proteins were highly concentrated in the apical pole. Consequently, NHE3, βPix, and Shank2 were all located near the apical membrane in colonic epithelial cells.
FIGURE 6.
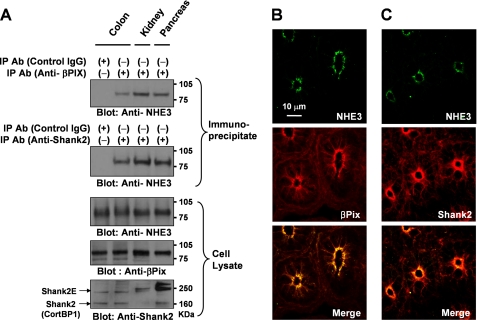
Formation of an NHE3, βPix, and Shank2 complex in vivo. A, coimmunoprecipitation of NHE3 with βPix and Shank2 in rat colon, kidney, and pancreas. Detergent extracts of each rat tissue fraction were immunoprecipitated with control (nonimmune IgG), anti-Shank2 (#3856), or βPix(#1257) antibodies and characterized by immunoblotting with anti-NHE3 (#1568), anti-βPix (#1254), and anti-Shank2 (#1136) antibodies. B and C, rat colon slices were immunofluorescently stained with anti-NHE3 (#1314), βPix (#1257), and Shank2 (#3856) antibodies. To perform a double labeling experiment using primary antibodies from the same host species, sections were incubated with unconjugated AffiniPure Fab Fragment Goat Anti-Rabbit IgG (H+L) after the first staining of NHE3 as described under “Experimental Procedures.” Then, the second labeling for βPix or Shank2 was conducted. Note that NHE3, βPix, and Shank2 are colocalized in the apical regions of colonic epithelial cells.
DISCUSSION
In the present study, we describe a novel mechanism of NHE3 regulation by βPix. NHE3 is expressed on the plasma membranes of many gastrointestinal organs and contributes to the maintenance of intracellular pH and volume, transcellular absorption of NaCl and NaHCO3, and fluid balance as well as regulation of systemic pH (31). NHE3 is both rapidly stimulated and inhibited as part of normal digestive physiology, and it contributes to multiple pathophysiological states when it is down-regulated for a prolonged period (7, 31–33). PDZ-based adapter molecules are important mediators of NHE3 regulation, participating in apical targeting, surface retention, and the acute control of NHE3 activity in epithelial cells (7). Shank2 is a PDZ-based adaptor enriched in the apical region of gastrointestinal and kidney epithelia and has been shown to directly bind and regulate NHE3 (5, 12). This association increases the membrane expression and basal activity of NHE3, but prevents the cAMP-dependent acute inhibition of NHE3 (5). It remains unknown how Shank2 up-regulates the membrane expression and activity of NHE3. Here, we show that βPix forms a protein complex with Shank2 and increases the membrane expression and activity of NHE3 in a Rho GTPase-dependent manner. Prevention of the cAMP-dependent inhibition of NHE3 might be mediated via the binding between the proline-rich domain of Shank2 and the N-terminal regions of phosphodiesterases that cleave cAMP, as has been demonstrated in CFTR regulation at the apical membrane of epithelial cells (9). βPix does not seem to be involved in the cAMP-dependent regulation of NHE3, because expression of βPix did not alter the cAMP effects on NHE3 activity (supplemental Fig. 4).
Pix proteins constitute a family of GEF proteins for the Rho GTPases (16, 17, 34). Pix was first cloned as p85SPR (SH3 domain-containing proline-rich protein), and the Pix family contains two members, αPix and βPix (22). These GEFs activate Rho small G-proteins by facilitating a switch from an inactive GDP-bound to an active GTP-bound state. The activation of Rho proteins influences vesicle movement, impacting endocytosis and exocytosis of integral plasma membrane proteins (35, 36). These encompass a variety of membrane transporters, including Na+ channels, K+ channels, nonselective cation channels, and CFTR (36–38). In addition, Rho GTPases play a pivotal role in apical retention and targeting of NHE3 in epithelial cells (18). As a major activator of Rho GTPases, GEF proteins also play important roles in cytoskeleton rearrangement, membrane trafficking, and transporter regulation (39, 40). However, the molecular nature of the GEF responsible for NHE3 regulation remains obscure. Our data indicate that βPix forms a protein complex with NHE3 in rat epithelial tissues such as colon, kidney, and pancreas (Fig. 6). In addition, βPix up-regulates NHE3 surface expression and activity (Fig. 2), an effect that is blocked by the Rho GTPase inhibitor TxB (Fig. 5). These findings strongly suggest that βPix is one of the major GEFs responsible for NHE3 regulation at the apical membrane of epithelial cells.
Expression of βPix protein increased the membrane expression and basal activity of NHE3, resembling the effects of Shank2 expression. Evidence presented in this study indicates that Shank2 is involved in the βPix-induced up-regulation of NHE3 and vice versa. For example, βPix-induced up-regulation of NHE3 was abolished by the Shank2 PDZ- and SAM-domain mutants (Fig. 3). Protein complex formation between βPix and NHE3 was also abolished by these dominant negative Shank2 mutants (Fig. 1). The PDZ domain of Shank2 mediates binding to NHE3 (5). It is also known that PDZ domain of Shank binds to the C-terminal PDZ-binding motif of βPix. Interestingly, Shank2 has only one PDZ domain. This raises the possibility that βPix and NHE3 may compete with each other to bind to Shank2, rather than form a protein complex. However, coimmunoprecipitation results showed that Shank2 actually mediates the association between βPix and NHE3 (Fig. 1). The C terminus of Shank2 contains a SAM domain, which is known to mediate oligomerization (28). SAM domains are small protein modules that are present in many different proteins in diverse cellular compartments and are involved in wide-ranging functions, including scaffolding, signal transduction, and transcriptional regulation (41). Unlike other common protein modules in Shank2, such as proline-rich and SH3 domains, they can bind to other SAM domains and self-associate, which suggests that Shank proteins can multimerize in a tail-to-tail manner. The finding that SAM domain-deleted Shank2 mutants abolished the association between NHE3 and βPix suggests that Shank2 exists as an oligomer, cross-linking multiple sets of protein complexes with NHE3 and βPix.
An interesting finding in this study is that association between βPix and NHE3 was readily detectable in rat epithelial tissues (Fig. 6), whereas the interaction was faint in the heterologous expression system without using the cross-linking agent DSP (supplemental Fig. 2). This raises a possibility that a new protein may be involved in the stabilization of βPix-Shank2-NHE3 complex in epithelial tissues. A further study that identifies the complete binding partners of βPix and Shank2 in epithelial cells will elucidate this question.
The regulatory mechanisms resulting from the association of NHE3 with Shank2 and βPix are summarized in Fig. 7. Oligomerization of Shank2 in the apical cytoskeleton forms a large protein complex. This recruits many regulatory and structural proteins, including NHE3 and βPix. In addition, Shank2 oligomerization may facilitate dimerization of βPix, which is required for activation of Rho GTPases (42). Consequently, activation of Rho GTPases up-regulates membrane expression and activity of NHE3 at the apical membrane of epithelial cells. Identification of βPix as a regulator of NHE3 will not only shed new light on electroneutral sodium and hydrogen transport in the gastrointestinal and renal epithelia, but also provide another avenue for the correction of disease states caused by fluid electrolyte imbalance.
FIGURE 7.
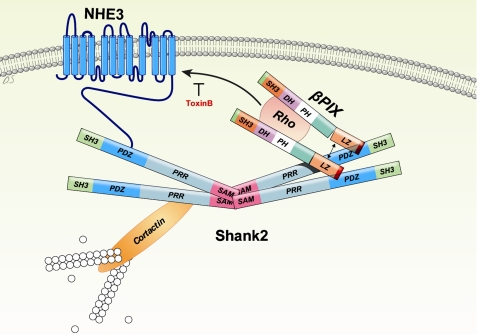
A model for the regulation of NHE3 through interaction with βPix and Shank2. The molecular machinery implicated in the regulation of NHE3 at the apical membrane of epithelial cells is illustrated. Both NHE3 and βPix form protein complexes through the PDZ domain of Shank2. The multimerization of Shank2 via its SAM domain recruits many regulatory and structural proteins, including NHE3 and βPix to the complex. Also, the dimerization of βPix facilitates the activation of Rho GTPases. Consequently, NHE3 is directed to the cell surface by its interaction with βPix and Shank2. SH3, Src homology3; SAM, sterile alpha motif; PRR, proline-rich region; DH, Dbl homology; PH, pleckstrin homology; and LZ, leucine zipper.
Supplementary Material
Acknowledgments
We thank Dr. Orson Moe (University of Texas Southwestern Medical Center, Dallas, TX) for kindly providing NHE3 antibodies. We also thank Dong-Su Jang for editorial assistance.
This work was supported in part by Grants R11-2007-040-01001-0 and R01-2007-000-20710-0 from the Korea Science and Engineering Foundation, Ministry of Education, Science and Technology, Korea, and by Grant A070001 from the Korea Healthcare Technology R&D Project, Ministry for Health, Welfare and Family Affairs, Republic of Korea.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. 1–4.
- NHE
- Na+/H+ exchanger
- GEF
- guanine nucleotide exchange factor
- PDZ
- PSD-95/Dlg/ZO-1
- PSD
- postsynaptic density
- PAK
- p21-activated kinase
- Pix
- PAK-interacting exchange factor
- PKA
- cAMP-dependent protein kinase
- SAM
- sterile α motif
- SH3
- Src homology-3
- HA
- hemagglutinin
- GFP
- green fluorescent protein
- BCECF
- acetoxymethyl ester of 2′,7′-bis(carboxyethyl)-5(6)-carboxyfluorescein
- TxB
- toxin B
- DSP
- dithiobis(succinimidyl propionate)
- CMV
- cytomegalovirus
- MDCK
- Madin-Darby canine kidney cell
- siRNA
- small interference RNA
- GST
- glutathione S-transferase
- HEK
- human embryonic kidney
- pHi
- intracellular pH
- CFTR
- cystic fibrosis transmembrane conductance regulator.
REFERENCES
- 1.Brett C. L., Donowitz M., Rao R. (2005) Am. J. Physiol. Cell Physiol. 288, C223–C239 [DOI] [PubMed] [Google Scholar]
- 2.Orlowski J., Grinstein S. (2004) Pflugers Arch. 447, 549–565 [DOI] [PubMed] [Google Scholar]
- 3.Fuster D. G., Zhang J., Shi M., Bobulescu I. A., Andersson S., Moe O. W. (2008) J. Am. Soc. Nephrol. 19, 1547–1556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Xiang M., Feng M., Muend S., Rao R. (2007) Proc. Natl. Acad. Sci. U.S.A. 104, 18677–18681 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Han W., Kim K. H., Jo M. J., Lee J. H., Yang J., Doctor R. B., Moe O. W., Lee J., Kim E., Lee M. G. (2006) J. Biol. Chem. 281, 1461–1469 [DOI] [PubMed] [Google Scholar]
- 6.Gawenis L. R., Stien X., Shull G. E., Schultheis P. J., Woo A. L., Walker N. M., Clarke L. L. (2002) Am. J. Physiol. Gastrointest. Liver Physiol. 282, G776–G784 [DOI] [PubMed] [Google Scholar]
- 7.Donowitz M., Li X. (2007) Physiol. Rev. 87, 825–872 [DOI] [PubMed] [Google Scholar]
- 8.Weinman E. J., Cunningham R., Shenolikar S. (2005) Pflügers Arch. 450, 137–144 [DOI] [PubMed] [Google Scholar]
- 9.Lee J. H., Richter W., Namkung W., Kim K. H., Kim E., Conti M., Lee M. G. (2007) J. Biol. Chem. 282, 10414–10422 [DOI] [PubMed] [Google Scholar]
- 10.Sheng M., Kim E. (2000) J. Cell Sci. 113, 1851–1856 [DOI] [PubMed] [Google Scholar]
- 11.Lim S., Naisbitt S., Yoon J., Hwang J. I., Suh P. G., Sheng M., Kim E. (1999) J. Biol. Chem. 274, 29510–29518 [DOI] [PubMed] [Google Scholar]
- 12.McWilliams R. R., Breusegem S. Y., Brodsky K. F., Kim E., Levi M., Doctor R. B. (2005) Am. J. Physiol. Cell Physiol. 289, C1042–C1051 [DOI] [PubMed] [Google Scholar]
- 13.McWilliams R. R., Gidey E., Fouassier L., Weed S. A., Doctor R. B. (2004) Biochem. J. 380, 181–191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kim J. Y., Han W., Namkung W., Lee J. H., Kim K. H., Shin H., Kim E., Lee M. G. (2004) J. Biol. Chem. 279, 10389–10396 [DOI] [PubMed] [Google Scholar]
- 15.Park E., Na M., Choi J., Kim S., Lee J. R., Yoon J., Park D., Sheng M., Kim E. (2003) J. Biol. Chem. 278, 19220–19229 [DOI] [PubMed] [Google Scholar]
- 16.Bagrodia S., Taylor S. J., Jordon K. A., Van Aelst L., Cerione R. A. (1998) J. Biol. Chem. 273, 23633–23636 [DOI] [PubMed] [Google Scholar]
- 17.Manser E., Loo T. H., Koh C. G., Zhao Z. S., Chen X. Q., Tan L., Tan I., Leung T., Lim L. (1998) Mol. Cell 1, 183–192 [DOI] [PubMed] [Google Scholar]
- 18.Alexander R. T., Furuya W., Szászi K., Orlowski J., Grinstein S. (2005) Proc. Natl. Acad. Sci. U.S.A. 102, 12253–12258 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hayashi H., Szászi K., Coady-Osberg N., Furuya W., Bretscher A. P., Orlowski J., Grinstein S. (2004) J. Gen. Physiol. 123, 491–504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Amemiya M., Loffing J., Lötscher M., Kaissling B., Alpern R. J., Moe O. W. (1995) Kidney Int. 48, 1206–1215 [DOI] [PubMed] [Google Scholar]
- 21.Bookstein C., Xie Y., Rabenau K., Musch M. W., McSwine R. L., Rao M. C., Chang E. B. (1997) Am. J. Physiol. 273, C1496–C1505 [DOI] [PubMed] [Google Scholar]
- 22.Lee J., Jung I. D., Chang W. K., Park C. G., Cho D. Y., Shin E. Y., Seo D. W., Kim Y. K., Lee H. W., Han J. W., Lee H. Y. (2005) Exp. Cell Res. 307, 315–328 [DOI] [PubMed] [Google Scholar]
- 23.Kurashima K., Szabó E. Z., Lukacs G., Orlowski J., Grinstein S. (1998) J. Biol. Chem. 273, 20828–20836 [DOI] [PubMed] [Google Scholar]
- 24.Bagrodia S., Cerione R. A. (1999) Trends Cell Biol. 9, 350–355 [DOI] [PubMed] [Google Scholar]
- 25.Ahn W., Kim K. H., Lee J. A., Kim J. Y., Choi J. Y., Moe O. W., Milgram S. L., Muallem S., Lee M. G. (2001) J. Biol. Chem. 276, 17236–17243 [DOI] [PubMed] [Google Scholar]
- 26.Lee M. G., Wigley W. C., Zeng W., Noel L. E., Marino C. R., Thomas P. J., Muallem S. (1999) J. Biol. Chem. 274, 3414–3421 [DOI] [PubMed] [Google Scholar]
- 27.Zhao H., Star R. A., Muallem S. (1994) J. Gen. Physiol. 104, 57–85 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Thanos C. D., Goodwill K. E., Bowie J. U. (1999) Science 283, 833–836 [DOI] [PubMed] [Google Scholar]
- 29.Hall A. (1998) Science 279, 509–514 [DOI] [PubMed] [Google Scholar]
- 30.Szászi K., Grinstein S., Orlowski J., Kapus A. (2000) Cell. Physiol. Biochem. 10, 265–272 [DOI] [PubMed] [Google Scholar]
- 31.Zachos N. C., Tse M., Donowitz M. (2005) Annu. Rev. Physiol. 67, 411–443 [DOI] [PubMed] [Google Scholar]
- 32.Moe O. W., Amemiya M., Yamaji Y. (1995) J. Clin. Invest. 96, 2187–2194 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Donowitz M., Janecki A., Akhter S., Cavet M. E., Sanchez F., Lamprecht G., Zizak M., Kwon W. L., Khurana S., Yun C. H., Tse C. M. (2000) Ann. N.Y. Acad. Sci. 915, 30–42 [DOI] [PubMed] [Google Scholar]
- 34.Oh W. K., Yoo J. C., Jo D., Song Y. H., Kim M. G., Park D. (1997) Biochem. Biophys. Res. Commun. 235, 794–798 [DOI] [PubMed] [Google Scholar]
- 35.Runnels L. W., Yue L., Clapham D. E. (2002) Nat. Cell Biol. 4, 329–336 [DOI] [PubMed] [Google Scholar]
- 36.Staruschenko A., Nichols A., Medina J. L., Camacho P., Zheleznova N. N., Stockand J. D. (2004) J. Biol. Chem. 279, 49989–49994 [DOI] [PubMed] [Google Scholar]
- 37.Cheng J., Wang H., Guggino W. B. (2005) J. Biol. Chem. 280, 3731–3739 [DOI] [PubMed] [Google Scholar]
- 38.Storey N. M., O'Bryan J. P., Armstrong D. L. (2002) Curr. Biol. 12, 27–33 [DOI] [PubMed] [Google Scholar]
- 39.Schiller M. R., Chakrabarti K., King G. F., Schiller N. I., Eipper B. A., Maciejewski M. W. (2006) J. Biol. Chem. 281, 18774–18786 [DOI] [PubMed] [Google Scholar]
- 40.Zheng Y. (2001) Trends Biochem. Sci. 26, 724–732 [DOI] [PubMed] [Google Scholar]
- 41.Qiao F., Bowie J. U. (2005) Sci. STKE 2005, re7. [DOI] [PubMed] [Google Scholar]
- 42.Koh C. G., Manser E., Zhao Z. S., Ng C. P., Lim L. (2001) J. Cell Sci. 114, 4239–4251 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.