Abstract
The FOXO family of forkhead transcription factors has a variety of important functions in stress response, metabolism, cell cycle, apoptosis, longevity, etc. The transcriptional activity and subcellular localization of FOXO are tightly regulated by post-translational modifications, including phosphorylation by various kinases. Here, we report that the transforming growth factor-β-activated kinase (TAK1)-Nemo-like kinase (NLK) pathway negatively regulates FOXO1. We show that NLK binds and phosphorylates FOXO1 at Pro-directed Ser/Thr residues in the transactivation domain. The phosphorylation by TAK1-NLK pathway inhibits the transcriptional activity of FOXO1 and excludes FOXO1 from the nucleus, which is independent of phosphatidylinositol 3-kinase/Akt pathway. Consistently, knockdown of TAK1-NLK pathway dephosphorylates FOXO1 and decreases phospho-Ser-329 FOXO1 level. It also induces translocation of FOXO1 into the nucleus and leads to an increase in mRNA levels of FOXO target genes and poly(ADP-ribose) polymerase cleavage. In addition, we show the interaction between NLK and FOXO1 is evolutionarily conserved in Drosophila. Collectively, these findings provide the first evidence that TAK1-NLK pathway is a novel regulator of FOXO1.
Keywords: Akt PKB, Apoptosis, Protein Translocation, Signal Transduction, Transforming Growth Factor Beta (TGFbeta), FOXO, NLK, TAK1
Introduction
FOXO proteins belong to the O class of forkhead box transcription factor superfamily, all of which have a forkhead box domain for DNA binding (1). FOXO family proteins are regulated by various signaling networks through post-translational modifications such as phosphorylation, acetylation, and ubiquitination. Complex regulation of FOXO modulates the expression of target genes involved in apoptosis, cell cycle, stress response, longevity, DNA repair, as well as glucose metabolism (1).
The best known example of the upstream signaling pathways of FOXO is the phosphatidylinositol 3-kinase (PI3K)3/Akt pathway that is activated by growth factors such as insulin and insulin-like growth factor-1 (2). DAF-16 of Caenorhabditis elegans was first identified as a target of the insulin pathway and has three Akt phosphorylation sites, which are conserved from worms to humans (3). The phosphorylation by Akt sequesters FOXO in the cytoplasm and inhibits its functions, leading to decreased expression of FOXO target genes (2). Recently, other inhibitory phosphorylation sites on FOXO transcription factors have been identified. CDK2 can phosphorylate FOXO1 at Ser-249 and Ser-298, resulting in not only inhibition of its transcriptional activity but also its exclusion from the nucleus (4). In addition, Ser-329 of human FOXO1 was shown to be phosphorylated in vitro by the dual specificity tyrosine-phosphorylated and -regulated kinase 1A (DYRK1A), and this phosphorylation inhibits FOXO1 activity and decreases FOXO1 levels in the nucleus (5). It is of note that both of these inhibitory phosphorylation sites are Pro-directed Ser/Thr residues, raising the possibility that other Pro-directed kinases such as MAPK or cyclin-dependent kinase family could phosphorylate these sites.
Drosophila nemo encodes a founding member of the NLK family, which was identified as a gene required for photoreceptor cell rotation during eye morphogenesis (6), and its orthologues were subsequently identified in worms and vertebrates (lit-1 and NLK, respectively) (7, 8). NLK was proposed to be classified into an atypical MAPK family along with ERK3/4, because it has a high homology to ERK1 kinase domain but lacks the tyrosine residue in the activation loop (9). In the past decade, a number of transcription factors and co-factors have been demonstrated to be phosphorylated and regulated by NLK. The first and best characterized substrate of NLK is the T-cell factor/lymphoid enhancer factor family that mediates the Wnt-dependent signaling pathway (10). Genetic and biochemical analyses have revealed that NLK associates and phosphorylates T-cell factor/lymphoid enhancer factor proteins (10–15), which result in a blockade of binding to its target promoters and a decrease in the nuclear level of T-cell factor/lymphoid enhancer factor molecules (10). In addition, NLK has been demonstrated to phosphorylate STAT3 (16), c-Myb (17), A-Myb (18), Drosophila Mad (19), cAMP-response element-binding protein-binding protein (18), and SET Domain Bifurcated 1 (SETDB1) (20). The results bring diverse consequences such as mesoderm induction by STAT3 (16), degradation of c-Myb (17), deacetylation of A-Myb (18), nuclear export of Mad (19), inhibition of cAMP-response element-binding protein-binding protein activity (18), and histone methylation by SETDB1 (20). These findings suggest that NLK regulates important gene expressions via its ability to phosphorylate diverse transcription factors and co-factors.
In this study, we show a functional interaction between NLK and FOXO1. NLK specifically associates with FOXO1 and directly phosphorylates at least eight Pro-directed Ser/Thr residues. This regulation affects the activity and localization of FOXO1. Additionally, we show that TAK1, an upstream kinase of NLK, also inhibits FOXO1. Our results reveal that the TAK1-NLK pathway is a novel upstream regulator of FOXO1.
EXPERIMENTAL PROCEDURES
cDNA Constructs and Antibodies
FOXO1-HA and 8×FK1tkLuc cDNA were generous gifts from Dr. W. H. Biggs III (The Ludwig Institute for Cancer Research). 3F10 anti-HA rat monoclonal antibody was purchased from Roche Applied Science; anti-HA mouse monoclonal antibody anti-PARP antibody was from Cell Signaling Technology; anti-FLAG M2 antibody was from Sigma; anti-GST antibody was from Upstate Biotechnology, Inc.; and 9E10 anti-Myc antibody was from Developmental Studies Hybridoma Bank.
Quantitative Real Time PCR
The expression of FOXO1 target mRNA was measured by quantitative real time PCR using an iCycler iQTM multicolor real time detection system (Bio-Rad). Total RNA was isolated from HEK293T cells with the Easy-BlueTM system (Intron Biotechnology, Korea), which were transfected with control (catalogue no. D-001210-01-20, Dharmacon), human NLK siRNA (catalogue no. MU-004763-01, Dharmacon), or human TAK1 siRNA (catalogue no. D-003790-06, Dharmacon) by X-tremeGENE siRNA transfection reagent (Roche Applied Science). After cDNA was synthesized from 2 μg of RNA using the Moloney murine leukemia virus reverse transcriptase (Promega), quantitative real time PCR was performed using specific primer sets. The primer sets were designed to span intron-exon borders to distinguish amplified cDNA from genomic DNA. The primer sequences used for PCR were as follows: human NLK-specific primers (5′-CCAGTGACTTTGAGCCTGTC-3′ for 5′ and 5′-GATGGCTGAGCAACAGTGG-3′ for 3′), human Bim-specific primers (5′-TCCCTACAGACAGAGCCACAAGAC-3′ for 5′ and 5′- AATACCCTCCTTGCATAGTAAGCG-3′ for 3′), and human p27Kip1 specific primers (5′-GCAATGCGCAGGAATAAGGA-3′ for 5′ and 5′-TCCACAGAACCGGCATTTG-3′ for 3′). The expression level from each sample was normalized to the mRNA expression level of a housekeeping gene β-actin.
Cell Culture and Transfection
HEK293T and COS1 cells were grown in Dulbecco's modified Eagle's medium supplemented with 10% fetal bovine serum (Invitrogen) at 37 °C in a humidified atmosphere with 5% CO2 and were transiently transfected using the standard calcium phosphate protocol or Lipofectamine Plus reagent (Invitrogen).
Preparation of Cell Lysates, Immunoprecipitation, and Immunoblots
Cell stimulation was terminated by washing cells with ice-cold PBS. Cell lysates were prepared in Lysis buffer A (20 mm Tris, pH 7.5, 100 mm NaCl, 1 mm EDTA, 2 mm EGTA, 50 mm β-glycerophosphate, 50 mm NaF, 1 mm sodium vanadate, 2 mm dithiothreitol, 1 mm phenylmethylsulfonyl fluoride, 10 μg/ml leupeptin, 1 μg/ml pepstatin A, and 1% Triton X-100) and subjected to immunoprecipitation and immunoblot according to the standard procedures (21).
Protein Kinase Assay
FLAG-NLK and FOXO1-HA were immunoprecipitated by anti-FLAG monoclonal antibody and 12CA5 anti-HA antibody coupled to protein G-Sepharose (Amersham Biosciences), respectively. The samples were washed twice with Lysis buffer A and then twice with the buffer containing 500 mm NaCl. Finally, the immune complexes were washed with NLK kinase assay buffer containing 10 mm HEPES, pH 7.0, 5 mm MgCl2, and 1 mm dithiothreitol. The phosphotransferase activities were assayed in a 30-μl reaction mixture consisting of NLK kinase assay buffer, 1 μg of bacterially purified GST-FOXO1 C1 protein, 10 μm ATP, and 10 μCi of [γ-32P]ATP at 30 °C for 20 min. To use a full-length FOXO1-HA protein as a substrate, the immune complexes containing FOXO1-HA isolated from HEK293T cell lysates were combined with the FLAG-NLK-containing immune complexes after kinase assay buffer wash, and the resulting immune complex mixtures were subjected to phosphotransferase assays. The kinase assay samples were subjected to SDS-PAGE and subsequent autoradiography.
Luciferase Assay
Luciferase assays were performed using a Dual-LuciferaseTM reporter assay kit (Promega) according to the manufacturer's instructions.
Immunocytochemistry
COS1 and HEK293T cells were grown on poly-l-lysine (Sigma)-coated 8-well μ-slide (ibidi) and transiently transfected with GFP-NLK WT, KN, FOXO1-HA, or GFP-FOXO1. Mouse anti-HA antibody and Alexa 568-conjugated anti-rabbit antibody (Invitrogen) were used for cell staining according to the manufacturer's instructions. The samples were observed by a laser scanning confocal microscope (Carl Zeiss).
RESULTS
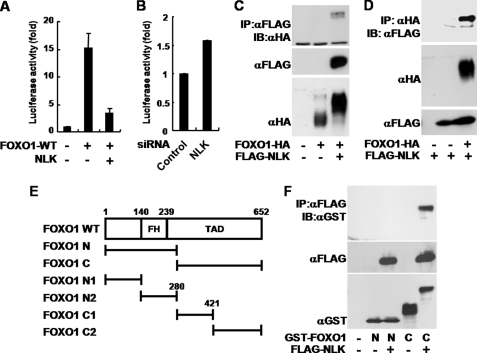
To investigate the relationship between NLK and FOXO1, we tested whether NLK regulates the transcriptional activity of FOXO1, using a luciferase assay system based on the 8×FK1Tk reporter containing multiple FOXO-binding sequences (22). Surprisingly, expression of NLK WT reproducibly suppressed the FOXO1-induced luciferase activity to a near control level (Fig. 1A). To examine the FOXO1 regulation in a more physiological condition, we observed the FOXO1-induced luciferase activity in an NLK knockdown condition. NLK knockdown by siRNA increased the transcriptional activity of FOXO1 (Fig. 1B).
FIGURE 1.
NLK interacts with FOXO1 and inhibits its transcriptional activity. A and B, COS1 cells were transiently transfected with p8×FK1tkLuc and pRL-TK Renilla reporter plasmids. FOXO1-HA WT and FLAG-NLK WT constructs (A) or siRNA of control and NLK (B) were co-transfected as indicated. Dual-Luciferase assays were performed. The values in the graph represent the mean of three independent cell preparations ± S.D. C, D, and F, HEK293T cells were transiently transfected as indicated. After 36 h of transfection, the cells were lysed for anti-FLAG (C), anti-HA (D), and anti-FLAG (F) immunoprecipitation (IP) assays. The precipitated proteins were subjected to immunoblot (IB) analyses (top and middle panels). Anti-HA (C), anti-FLAG (D), and anti-GST (F) immunoblots were completed for the whole cell lysates (bottom panels). The results shown are representative of three independent experiments. E, diagram of FOXO1 and its deletion mutants used in experiments. Numbers refer to the amino acid positions of mouse FOXO1. TAD, transactivation domain. FH, forkhead; N, N-terminal part of NLK; C, C-terminal part of NLK.
To understand the functional interaction between NLK and FOXO1, we tested whether FOXO1 and NLK physically interact with each other. We co-transfected FLAG-NLK and FOXO1-HA, and the cell lysates were immunoprecipitated with either anti-FLAG or anti-HA antibody. As a result, NLK was co-immunoprecipitated with FOXO1 and vice versa (Fig. 1, C and D). To determine the region of FOXO1 for NLK binding, we divided FOXO1 into two fragments (FOXO1 N and C), each of which represents almost half of the protein (Fig. 1E). After co-expression of NLK WT and the FOXO1 fragments, only FOXO1 C was co-immunoprecipitated with NLK WT (Fig. 1F).
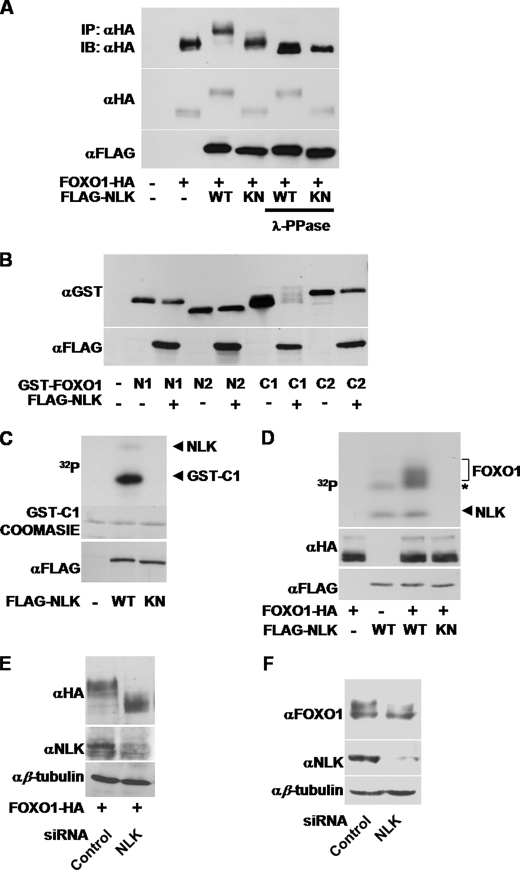
Notably, when FOXO1 was co-expressed with NLK WT, FOXO1 exhibited slow electrophoretic mobility on SDS-PAGE (Fig. 1, C, D, and F, and Fig. 2A) but no change in mobility when co-expressed with the NLK kinase-inactive mutant (KN, supplemental Fig. 1 and Fig. 2A). Similar results were obtained in other cell lines such as HEK293T, MCF7, and PC12 (supplemental Fig. 2). The slowly migrating forms of FOXO1 were almost completely eliminated by λ-phosphatase treatment after immunoprecipitation of FOXO1 (Fig. 2A). These data strongly suggest that NLK induces phosphorylation of FOXO1.
FIGURE 2.
NLK phosphorylates FOXO1 in vivo and in vitro. A, COS1 cells were transfected with denoted plasmids. After 36 h of transfection, the cells were lysed for anti-HA immunoprecipitation (IP) assays. The immune complexes containing FOXO1 were incubated with λ-phosphatase or buffer alone and then subjected to immunoblot (IB) analyses using anti-HA antibody (top panel). Anti-HA and anti-FLAG blots were also completed for the same whole cell lysates (middle and bottom panels). B, COS1 cells were transfected with FLAG-NLK, GST-FOXO1 N1, N2, C1, and C2 constructs. After 36 h of transfection, the cells were lysed for GST pulldown assays. The precipitated proteins were subjected to immunoblot analyses using anti-GST antibody (top panel). Anti-FLAG blots were also completed for the same whole cell lysates (bottom panel). C, HEK293T cells were transfected with FLAG-NLK WT or KN constructs. Expressed proteins were immunopurified with anti-FLAG antibody, and in vitro kinase assays were performed using GST-FOXO1 C1 as a substrate. The phosphorylated FOXO1 C1 proteins were visualized by autoradiography (top panel), and the C1 protein levels were compared by Coomassie Blue staining (middle panel). FLAG-NLK WT and KN protein levels were visualized by anti-FLAG immunoblot (bottom panel). D, FLAG NLK WT, KN, and full-length FOXO1-HA constructs were separately transfected in HEK293T cells. After 36 h of transfection, the cells were lysed for anti-HA or anti-FLAG immunoprecipitation. The immune complexes for NLK and FOXO1 were mixed together as indicated to conduct in vitro kinase assays. The phosphorylated FOXO1-HA proteins were visualized by autoradiography (top panel). FOXO1-HA and FLAG-NLK protein levels were visualized by anti-HA (middle panel) and anti-FLAG (bottom panel) immunoblots, respectively. The asterisk indicates NLK-associated phosphorylation activity, which does not seem to be related to FOXO1 phosphorylation. E and F, control or NLK siRNA was transfected with (E) or without (F) FOXO1-HA construct in COS1 cells. After 72 h of transfection, the cells were lysed for immunoblot analyses using anti-HA, anti-FOXO1, anti-NLK, and anti-β-tubulin antibodies.
To further map the phosphorylation site(s) of FOXO1, we divided FOXO1 into four fragments (Fig. 1E). Interestingly, only the C1 region of FOXO1 slowly migrated in SDS-PAGE when NLK was co-expressed (Fig. 2B). To determine whether NLK kinase directly phosphorylates FOXO1, we performed in vitro NLK assays using bacterially purified GST-FOXO1 C1 as a substrate. Consistent with the co-expression results (Fig. 2B), NLK WT strongly phosphorylated the C1 protein, whereas NLK KN did not (Fig. 2C). To verify that the FOXO1 full-length protein is also a substrate of NLK, we isolated the immune complexes from the cell lysates containing either NLK or FOXO1 and then mixed them together to conduct kinase assays. NLK WT, but not NLK KN, phosphorylated the FOXO1 full-length protein (Fig. 2D). Furthermore, NLK knockdown by siRNA increased the gel mobility of overexpressed and endogenous FOXO1 protein in SDS-PAGE (Fig. 2, E and F, respectively). Collectively, these results suggest that NLK can bind to FOXO1 and specifically phosphorylate it.
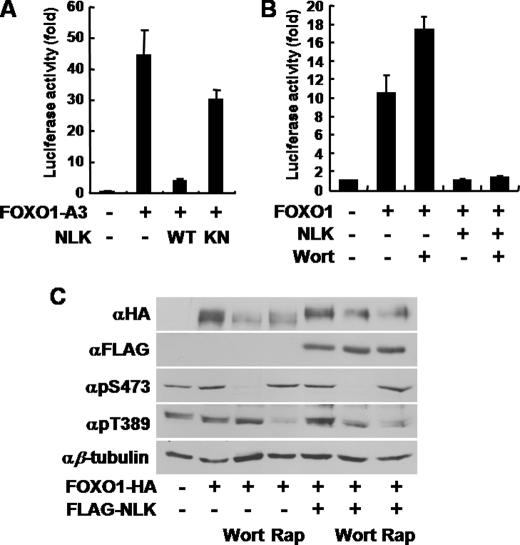
It has been well established that PI3K/Akt signaling pathway inhibits FOXO1 via phosphorylation and subsequent nuclear exclusion (2). To test whether the phosphorylation of FOXO1 by NLK is affected by PI3K/Akt pathway, we used an unphosphorylatable FOXO1 mutant by Akt (FOXO1-A3 (T24A/S256A/S319A) (2). Although FOXO1-A3 was more active than wild type, its transcriptional activity was almost completely abrogated by NLK WT (Fig. 3A) similar to the inhibition of FOXO1 WT by NLK WT (Fig. 1A). NLK KN, however, did not significantly inhibit FOXO1-A3 (Fig. 3A). In addition, NLK WT was able to block FOXO1 transcriptional activity induced by wortmannin, a potent inhibitor of PI3K (Fig. 3B), leading to the conclusion that NLK modulates FOXO1 regardless of PI3K/Akt activity. Moreover, consistent with Fig. 3, A and B, FOXO1 phosphorylation by NLK was not affected by wortmannin or rapamycin, a specific inhibitor of target of rapamycin (Fig. 3C). These results suggest that FOXO1 phosphorylation induced by NLK is different from that by Akt.
FIGURE 3.
NLK inhibits FOXO1 independent of PI3K/Akt pathway. A and B, COS1 cells were transiently transfected with p8×FK1tkLuc and pRL-TK Renilla reporter plasmids. A, FOXO1-A3 (unphosphorylatable FOXO1 by Akt) construct with NLK WT or KN construct was co-transfected as indicated. B, FOXO1-HA WT and FLAG-NLK constructs were co-transfected, with or without wortmannin (Wort), as indicated. Dual-Luciferase assays were performed. The values in the graph represent the mean of three independent cell preparations ± S.D. C, COS1 cells were transfected as indicated. Wortmannin and rapamycin (Rap) were treated for 30 min before cell lysates preparation. Cells were lysed and then subjected to immunoblot analyses.
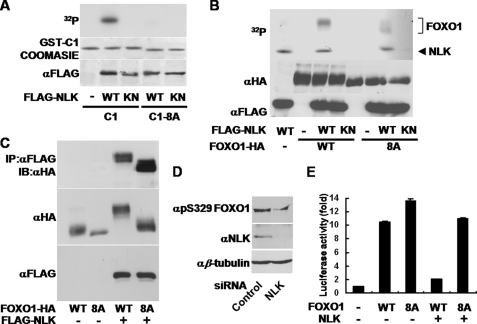
Given that NLK belongs to the MAPK family (8), we searched Pro-directed Ser/Thr residues in the C1 region of FOXO1. We found eight Ser/Thr residues (Ser-284, Ser-295, Ser-326, Ser-380, Ser-391, Thr-399, Ser-413, and Ser-415 in mouse FOXO1) as putative phosphorylation sites, which are all conserved among human, mouse, and rat (supplemental Fig. 3). To test whether these sites are phosphorylated by NLK, we replaced those Ser/Thr residues with Ala, one at a time, but the point mutants co-expressed with NLK WT did not produce a significant increase in mobility on SDS-PAGE (data not shown). Therefore, we decided to mutagenize all eight candidate residues to Ala. Subsequently, we performed in vitro NLK kinase assays using the C1 protein or the C1-8A protein (the eight residues were mutagenized to Ala) as a substrate. Surprisingly, the C1-8A protein was not phosphorylated by NLK (Fig. 4A). In addition, NLK WT barely phosphorylated the full-length FOXO1-8A protein (Fig. 4B). Moreover, co-expression of FOXO1-8A with NLK WT did not show slow-migrating forms (Fig. 4C, middle and bottom panels). Because FOXO1-8A still strongly associated with NLK WT (Fig. 4C, top panel), we strongly believed that the failure of the phosphorylation of FOXO1-8A by NLK is not due to its inability to bind to NLK. To further confirm the phosphorylation of FOXO1 by NLK at these amino acid residues, we used a phosphospecific antibody against Ser-329 of FOXO1, one of the putative NLK phosphorylation sites in FOXO1, and we found decreased phospho-FOXO1 signal when the cells were transfected with NLK siRNA (Fig. 4D). These results strongly suggest that some or all of these eight Ser/Thr residues in the transactivation domain of FOXO1 are major phosphorylation sites for NLK. However, we did not exclude the possibility that other Ser/Thr residues in regions other than C1 were phosphorylated by NLK.
FIGURE 4.
NLK phosphorylates multiple Ser/Thr residues of FOXO1. A, HEK293T cells were transfected with FLAG-NLK WT or KN construct. The cells were lysed for anti-FLAG immunoprecipitation assays. The precipitated proteins were subjected to in vitro kinase assay using GST-FOXO1 C1 or C1-8A as a substrate. The phosphorylated FOXO1 C1 and C1-8A proteins were visualized by autoradiography (top panel). The C1 and the C1-8A protein levels were compared by Coomassie Blue staining (middle panel). FLAG-NLK WT and KN protein levels were visualized by anti-FLAG immunoblot (bottom panel). B, FLAG-NLK WT, KN, FOXO1-HA WT, and 8A constructs were separately transfected in HEK293T cells. After 36 h of transfection, the cells were lysed for anti-FLAG or anti-HA immunoprecipitation. The various immune complexes for NLK and FOXO1 were mixed together as indicated and then subjected to in vitro kinase assays. The phosphorylated FOXO1-HA WT and 8A proteins were visualized by autoradiography (top panel). FOXO1-HA WT, 8A, FLAG-NLK WT, and KN protein levels were visualized by anti-HA (middle panel) and anti-FLAG (bottom panel) immunoblots, respectively. C, HEK293T cells were transfected as indicated. The cells were lysed for anti-FLAG immunoprecipitation (IP) assays. The precipitated proteins were subjected to immunoblot (IB) analyses using anti-HA antibody (top panel). Anti-HA and anti-FLAG blots were also completed for the same whole cell lysates (middle and bottom panels). D, control or NLK siRNA was transfected in COS1 cells. After 72 h of transfection, the cells were lysed for immunoblot analyses using anti-Ser(P)-329 (αpS329) (FOXO1), anti-NLK, and anti-β-tubulin antibodies. E, COS1 cells were transiently transfected with p8×FK1tkLuc and pRL-TK Renilla reporter plasmids. FOXO1-HA WT, 8A, and FLAG-NLK constructs were co-transfected as indicated. Dual-Luciferase assays were performed. The values in the graph represent the mean of three independent cell preparations ± S.D.
To investigate whether the FOXO1-8A mutation affects the transcriptional activity of FOXO1, we performed Dual-Luciferase reporter assays and found that the activity of FOXO1-8A was higher than that of FOXO1 WT (Fig. 4E), implicating that these phosphorylation sites are physiologically relevant. Moreover, NLK did not substantially inhibit the transcriptional activity of FOXO1-8A, whereas FOXO1 WT activity was almost completely abrogated by NLK (Fig. 4E), leading us to the conclusion that these phosphorylation sites mainly mediate the regulation of FOXO1 by NLK.
Next, to gain insight into the underlying mechanism of the regulation of FOXO1 by NLK, we introduced a GAL4-based luciferase assay system using pFR-luc, which contains five GAL4-binding sites (23). We generated GAL4 DNA binding domain-FOXO1 fusion constructs, of which activities are only coupled to the activity of the transactivation domain of FOXO1, but not the DNA binding domain of FOXO1. The luciferase activity of GAL4 DNA binding domain-FOXO1 WT was strongly inhibited by NLK (Fig. 5A) but that of FOXO1-8A was not significantly hindered (Fig. 5A). These results demonstrate that the FOXO1 phosphorylation by NLK impedes the activity of FOXO1 from activating transcriptional machinery.
FIGURE 5.
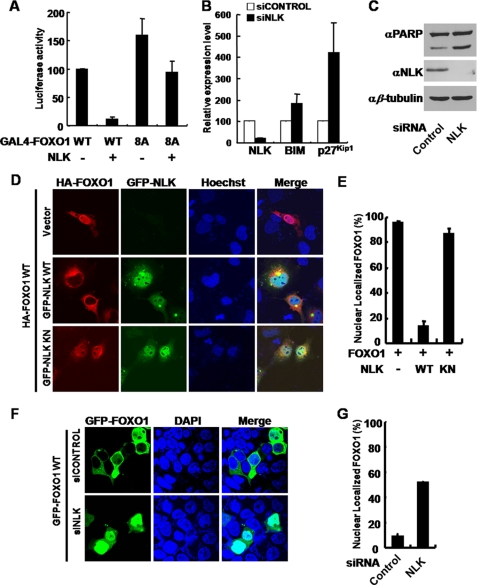
NLK regulates the transactivation activity and the localization of FOXO1. A, COS1 cells were transiently transfected with pFR-luc, which contains five GAL4-binding sites, and pRL-TK Renilla reporter plasmids. GAL4-FOXO1 WT, 8A, and FLAG-NLK constructs were co-transfected as indicated. Dual-Luciferase assays were performed. The values in the graph represent the mean of three independent cell preparations ± S.D. B, expression levels of NLK, Bim, and p27Kip1 transcripts in specific siRNA-transfected HEK293T cells were analyzed by reverse transcription-quantitative PCR. β-Actin was used as an internal control, n = 3. Bars indicate mean ± S.D. C, control or NLK siRNA was transfected in COS1 cells. After 72 h of transfection, the cells were lysed for immunoblot analyses using anti-PARP, anti-NLK, and anti-β-tubulin antibodies. D, COS1 cells were transiently transfected with FOXO1-HA alone, FOXO1-HA, and GFP-NLK WT, FOXO1-HA, and GFP-NLK KN constructs (top to bottom, respectively). After 24 h of transfection, the cells were fixed and subjected to immunocytochemistry. E, quantification of COS1 cells with nuclearly localized FOXO1 shown in D. FOXO1-HA alone, n = 54; FOXO1-HA and GFP-NLK WT, n = 54; FOXO1-HA and GFP-NLK KN, n = 40. Bars indicate mean ± S.D. F, HEK293T cells were transfected with siRNA of NLK and GFP-FOXO1 WT construct. After 72 h of transfection, cells were stained with 4′,6-diamidino-2-phenylindole (DAPI) (nucleus, blue). FOXO1 was depicted in green (GFP). The images shown are representative of three independent experiments. G, quantification of HEK293T cells with nuclearly localized FOXO1 shown in F. Control siRNA, n = 233; NLK siRNA, n = 153. Bars indicate mean ± S.D.
It has been already known that FOXO1 regulates the transcriptional level of Bim and p27Kip1, each of which modulates apoptosis and cell cycle arrest, respectively (24). We examined the transcript levels of both genes using reverse transcription-quantitative PCR. Consistent with the above data, the transcript levels of Bim and p27Kip1 were substantially increased in HEK293T cells transfected with NLK siRNA (Fig. 5B). In addition, we found that the level of PARP cleavage was increased in NLK knockdown conditions (Fig. 5C), suggesting that NLK modulates apoptosis through FOXO1. Consistently, co-expression of NLK with FOXO1 WT inhibited FOXO1-induced PARP cleavage but co-expression of NLK with FOXO1 8A mutant could not (supplemental Fig. 4). To further validate the regulation of FOXO1 by NLK, we examined the transcription levels of dInR and Thor/4E-BP, each of which is known to be regulated by FOXO1 in Drosophila (25). We found that the transcript levels of dInR and Thor/4E-BP were significantly increased in nemo null mutant fly (nmoP1/nmoj147-1, see supplemental Fig. 5). These results strongly support that NLK regulates FOXO1 transcriptional activity in vivo.
The phosphorylation of FOXO1 on the region C-terminal to the forkhead domain has been linked to its nuclear export (26). Notably, one of the NLK target sites, Ser-326, has been already known to be phosphorylated by DYRK, and this phosphorylation keeps FOXO1 molecules in the cytoplasm (5). Therefore, we speculated that NLK not only decreases the transcriptional activity of FOXO1 but also changes its localization. To test this hypothesis, COS1 cells were transfected with FOXO1 WT in the presence of GFP-NLK WT or KN, and its subcellular localization was determined. FOXO1 was mainly concentrated in the nucleus when expressed alone (Fig. 5, D and E). However, when FOXO1 was co-transfected with NLK WT, it dramatically translocated to the cytosol, especially in the perinuclear region, although NLK WT was mainly expressed in the nucleus (Fig. 5D). On the contrary, NLK KN did not translocate FOXO1 to the cytosol (Fig. 5, D and E) and FOXO1 8A mutant does not change its localization in the presence of NLK (supplemental Fig. 6), indicating that the phosphorylation by NLK is important for the nuclear export of FOXO1. To further confirm the effect of NLK on FOXO1 localization, HEK293T cells were transfected with NLK siRNA (supplemental Fig. 7A) with GFP-FOXO1 or FOXO1-HA. In this cell line, FOXO1 was mainly localized in the cytoplasm (Fig. 5F and supplemental Fig. 8) (27). Remarkably, however, down-regulation of NLK strongly induced translocation of FOXO1 to the nucleus (Fig. 5, F and G, and supplemental Fig. 8). Collectively, these results strongly suggest that the FOXO1 phosphorylation by NLK inhibits FOXO1 transcriptional activity and promotes its nuclear export.
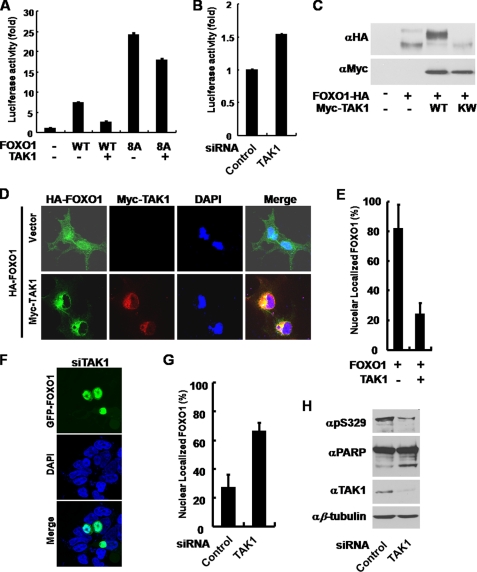
Previously, several studies showed that TAK1 is an upstream regulator of NLK (10, 13, 14, 16–18). Indeed, we found that TAK1 suppressed the FOXO1-induced luciferase activity, but it did not significantly suppress the FOXO1 8A mutant (Fig. 6A). In accordance with the results using NLK, knockdown of TAK1 increased the FOXO1 activity (supplemental Fig. 7B and Fig. 6B), and TAK1 induced slow electrophoretic mobility of FOXO1 on SDS-PAGE in a kinase activity-dependent manner (Fig. 6C). Furthermore, FOXO1 was expelled from the nucleus by TAK1 (Fig. 6, D and E). Consistent with the above results, the knockdown of TAK1 induced translocation of GFP-FOXO1 to the nucleus (Fig. 6, F and G). Finally, TAK1 down-regulation by siRNA decreased the phosphorylation of FOXO1 (Fig. 6H, top panel) and increased apoptosis (Fig. 6H, upper middle panel). These results led us to the conclusion that FOXO1 is regulated by the TAK1-NLK pathway in vivo.
FIGURE 6.
TAK1 regulates the transcriptional activity and the localization of FOXO1. A, COS1 cells were transiently transfected with p8×FK1tkLuc and pRL-TK Renilla reporter plasmids. FOXO1-HA WT, 8A, and Myc-TAK1 constructs were co-transfected as indicated. Dual-Luciferase assays were performed. The values in the graph represent the mean of three independent cell preparations ± S.D. B, COS1 cells were transfected with siRNA of control or TAK1 and p8×FK1tkLuc and pRL-TK Renilla reporter plasmids as indicated. Dual-Luciferase assays were performed. The values in the graph represent the mean of three independent cell preparations ± S.D. C, COS1 cells were transiently transfected with FOXO1-HA and Myc-TAK1 WT or KW constructs as indicated. After 36 h of transfection, the cells were lysed for anti-HA and anti-Myc immunoblot analyses. D, COS1 cells were transiently transfected with FOXO1-HA alone, FOXO1-HA, and Myc-TAK1 WT constructs. After 24 h of transfection, the cells were fixed and subjected to immunocytochemistry. E, quantification of COS1 cells with nuclearly localized FOXO1 shown in D. FOXO1-HA alone, n = 62; FOXO1-HA and Myc-TAK1 WT, n = 59. Bars indicate mean ± S.D. F, HEK293T cells were transfected with siRNA of TAK1 and GFP-FOXO1 WT construct. After 72 h of transfection, cells were stained with 4′,6-diamidino-2-phenylindole (DAPI) (nucleus, blue). FOXO1 was depicted in green (GFP). The images shown are representative of three independent experiments. G, quantification of HEK293T cells with nuclearly localized FOXO1 shown in F. Control siRNA, n = 74; NLK siRNA, n = 99. Bars indicate mean ± S.D. H, COS1 cells were transfected with siRNA of control or TAK1. After 72 h of transfection, the cells were lysed for immunoblot analyses using anti-Ser(P)-329 (αpS329) (FOXO1), αPARP, αTAK1, and αβ-tubulin antibodies.
DISCUSSION
FOXO1 regulation is achieved by post-translational modifications, such as phosphorylation, acetylation, and ubiquitination (1). These post-translational modifications alter FOXO1 transcriptional activity, subcellular localization, protein levels, and DNA-binding affinity (1). Among several upstream regulators of FOXO1, the PI3K/Akt pathway, specifically Akt, is the best known regulator of FOXO1 by phosphorylation (2). In this study, we showed that the TAK1-NLK pathway is a novel regulator of FOXO1, phosphorylating on the residues distinct from Akt phosphorylation sites (Figs. 3 and 4). FOXO1 regulation by TAK1-NLK pathway inhibits its transcriptional activity and exports it to the cytosol. In addition, we showed that down-regulation of NLK clearly up-regulated the transcription of Bim and p27Kip1 (Fig. 5B), well known FOXO1 target genes (22). These results strongly suggest that the TAK1-NLK pathway has an important role in regulating FOXO1 in a manner independent of the PI3K/Akt pathway.
NLK is involved in diverse signaling pathways by phosphorylating several transcription factors (16–20). Consistently, NLK null mice show complex phenotypes, such as growth retardation, neurological abnormalities, and hematopoietic defects (28). So far, it is not known which signaling pathway is responsible for producing these phenotypes. Therefore, it is necessary to further investigate the relationship between the phenotypes and the interaction of NLK and FOXO1.
As we described earlier, the phosphorylation of various transcription factors by NLK results in diverse biochemical consequences, for example translocation to the cytoplasm (19), degradation by ubiquitination (17), or decreased acetylation (18). Although we elucidated that phosphorylation of FOXO1 by NLK induced its nuclear export in this study, we also looked at the degradation and acetylation status of FOXO1. FOXO1 immune complexes were probed with anti-ubiquitin and -acetylation antibodies, but we did not see any difference in these post-translational modifications in the presence or absence of NLK (data not shown). Therefore, we strongly suggest that FOXO1 phosphorylation by NLK does not further induce ubiquitination and acetylation of FOXO1. The remaining question is as follows. What are the underlying mechanisms of the transcriptional inhibition and nuclear exclusion of FOXO1? It was revealed that 14-3-3 proteins bind to the Akt target sites of FOXO1 after phosphorylation, leading to masking the nuclear localization signal of FOXO1 and translocating it to the cytosol (2). However, it is not likely that 14-3-3 would be involved in translocating the phosphorylated FOXO1 by NLK because the motif recognized by 14-3-3 (RSXpSXP) is different from the NLK target sites on FOXO1. In this regard, there might be a novel mechanism of recognizing the phosphorylated FOXO1 by NLK and exporting it to the cytoplasm.
In this study, we revealed that the TAK1-NLK pathway is a novel negative regulator of FOXO1. Our findings will contribute to the expansion of our knowledge of FOXO1 regulation and the physiological role of the TAK1-NLK pathway.
Supplementary Material
Acknowledgments
We are grateful to Dr. W. H. Biggs III for cDNA constructs. We also thank members of the Chung laboratory for helpful discussions.
This work was supported by National Creative Research Initiatives Grant R16-2001-002-01001-0 from the Ministry of Education, Science and Technology and National Research Foundation of Korea.

The on-line version of this article (available at http://www.jbc.org) contains supplemental “Experimental Procedures” and Figs. 1–8.
- PI3K
- phosphatidylinositol 3-kinase
- NLK
- Nemo-like kinase
- MAPK
- mitogen-activated protein kinase
- PARP
- poly(ADP-ribose) polymerase
- HA
- hemagglutinin
- siRNA
- small interfering RNA
- WT
- wild type
- GST
- glutathione S-transferase.
REFERENCES
- 1.Calnan D. R., Brunet A. (2008) Oncogene 27, 2276–2288 [DOI] [PubMed] [Google Scholar]
- 2.Brunet A., Bonni A., Zigmond M. J., Lin M. Z., Juo P., Hu L. S., Anderson M. J., Arden K. C., Blenis J., Greenberg M. E. (1999) Cell 96, 857–868 [DOI] [PubMed] [Google Scholar]
- 3.Lin K., Hsin H., Libina N., Kenyon C. (2001) Nat. Genet. 28, 139–145 [DOI] [PubMed] [Google Scholar]
- 4.Huang H., Regan K. M., Lou Z., Chen J., Tindall D. J. (2006) Science 314, 294–297 [DOI] [PubMed] [Google Scholar]
- 5.von Groote-Bidlingmaier F., Schmoll D., Orth H. M., Joost H. G., Becker W., Barthel A. (2003) Biochem. Biophys. Res. Commun. 300, 764–769 [DOI] [PubMed] [Google Scholar]
- 6.Choi K. W., Benzer S. (1994) Cell 78, 125–136 [DOI] [PubMed] [Google Scholar]
- 7.Rocheleau C. E., Yasuda J., Shin T. H., Lin R., Sawa H., Okano H., Priess J. R., Davis R. J., Mello C. C. (1999) Cell 97, 717–726 [DOI] [PubMed] [Google Scholar]
- 8.Brott B. K., Pinsky B. A., Erikson R. L. (1998) Proc. Natl. Acad. Sci. U.S.A. 95, 963–968 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Miyata Y., Nishida E. (1999) Biochem. Biophys. Res. Commun. 266, 291–295 [DOI] [PubMed] [Google Scholar]
- 10.Ishitani T., Ninomiya-Tsuji J., Nagai S., Nishita M., Meneghini M., Barker N., Waterman M., Bowerman B., Clevers H., Shibuya H., Matsumoto K. (1999) Nature 399, 798–802 [DOI] [PubMed] [Google Scholar]
- 11.Meneghini M. D., Ishitani T., Carter J. C., Hisamoto N., Ninomiya-Tsuji J., Thorpe C. J., Hamill D. R., Matsumoto K., Bowerman B. (1999) Nature 399, 793–797 [DOI] [PubMed] [Google Scholar]
- 12.Ishitani T., Kishida S., Hyodo-Miura J., Ueno N., Yasuda J., Waterman M., Shibuya H., Moon R. T., Ninomiya-Tsuji J., Matsumoto K. (2003) Mol. Cell. Biol. 23, 131–139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ishitani T., Ninomiya-Tsuji J., Matsumoto K. (2003) Mol. Cell. Biol. 23, 1379–1389 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Smit L., Baas A., Kuipers J., Korswagen H., van de Wetering M., Clevers H. (2004) J. Biol. Chem. 279, 17232–17240 [DOI] [PubMed] [Google Scholar]
- 15.Thorpe C. J., Moon R. T. (2004) Development 131, 2899–2909 [DOI] [PubMed] [Google Scholar]
- 16.Ohkawara B., Shirakabe K., Hyodo-Miura J., Matsuo R., Ueno N., Matsumoto K., Shibuya H. (2004) Genes Dev. 18, 381–386 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kanei-Ishii C., Ninomiya-Tsuji J., Tanikawa J., Nomura T., Ishitani T., Kishida S., Kokura K., Kurahashi T., Ichikawa-Iwata E., Kim Y., Matsumoto K., Ishii S. (2004) Genes Dev. 18, 816–829 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kurahashi T., Nomura T., Kanei-Ishii C., Shinkai Y., Ishii S. (2005) Mol. Biol. Cell 16, 4705–4713 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zeng Y. A., Rahnama M., Wang S., Sosu-Sedzorme W., Verheyen E. M. (2007) Development 134, 2061–2071 [DOI] [PubMed] [Google Scholar]
- 20.Takada I., Mihara M., Suzawa M., Ohtake F., Kobayashi S., Igarashi M., Youn M. Y., Takeyama K., Nakamura T., Mezaki Y., Takezawa S., Yogiashi Y., Kitagawa H., Yamada G., Takada S., Minami Y., Shibuya H., Matsumoto K., Kato S. (2007) Nat Cell Biol. 9, 1273–1285 [DOI] [PubMed] [Google Scholar]
- 21.Kim Y., Park J., Kim S., Song S., Kwon S. K., Lee S. H., Kitada T., Kim J. M., Chung J. (2008) Biochem. Biophys. Res. Commun. 377, 975–980 [DOI] [PubMed] [Google Scholar]
- 22.Biggs W. H., 3rd., Meisenhelder J., Hunter T., Cavenee W. K., Arden K. C. (1999) Proc. Natl. Acad. Sci. U.S.A. 96, 7421–7426 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Qiao L., Shao J. (2006) J. Biol. Chem. 281, 39915–39924 [DOI] [PubMed] [Google Scholar]
- 24.Stahl M., Dijkers P. F., Kops G. J., Lens S. M., Coffer P. J., Burgering B. M., Medema R. H. (2002) J. Immunol. 168, 5024–5031 [DOI] [PubMed] [Google Scholar]
- 25.Puig O., Tjian R. (2005) Genes Dev. 19, 2435–2446 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rena G., Woods Y. L., Prescott A. R., Peggie M., Unterman T. G., Williams M. R., Cohen P. (2002) EMBO J. 21, 2263–2271 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yamagata K., Daitoku H., Takahashi Y., Namiki K., Hisatake K., Kako K., Mukai H., Kasuya Y., Fukamizu A. (2008) Mol. Cell 32, 221–231 [DOI] [PubMed] [Google Scholar]
- 28.Kortenjann M., Nehls M., Smith A. J., Carsetti R., Schüler J., Köhler G., Boehm T. (2001) Eur. J. Immunol. 31, 3580–3587 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.