Abstract
Replicative senescence in human fibroblasts is accompanied with alterations of various biological processes, including the impaired function of the proteasome. The proteasome is responsible for the removal of both normal and damaged proteins. Due to its latter function, proteasome is also considered a representative secondary antioxidant cellular mechanism. Nrf2 is a basic transcription factor responsible for the regulation of the cellular antioxidant response that has also been shown to regulate several proteasome subunits in mice. We have established in this study the proteasome-related function of Nrf2 in human fibroblasts undergoing replicative senescence. We demonstrate that Nrf2 has a declined function in senescence, whereas its silencing leads to premature senescence. However, upon its activation by a novel Nrf2 inducer, elevated levels of proteasome activity and content are recorded only in cell lines possessing a functional Nrf2. Moreover, treatment by the Nrf2 inducer results in the enhanced survival of cells following oxidative stress, whereas continuous treatment leads to lifespan extension of human fibroblasts. Importantly the Nrf2-proteasome axis is functional in terminally senescent cultures as these cells retain their responsiveness to the Nrf2 stimuli. In conclusion, these findings open up new directions for future manipulation of the senescence phenotype.
Keywords: Aging, Antioxidant, Proteasome, Transcription Factors, Transcription Regulation, Human Aging, Lifespan Extension, Nrf2, Proteasome Activation, Replicative Senescence
Introduction
Normal human fibroblasts undergo a limited number of divisions in culture and progressively reach a state of irreversible growth arrest, a process termed replicative senescence. Senescence has been attributed to a combination of genetic and environmental factors (1). Senescent fibroblasts are viable and fully functional, however, they exhibit several morphologic and biochemical changes as compared with their young/proliferating counterparts (2). We and others (3) have reported loss of proteasome function upon aging of several human tissues as well as in senescent primary cultures. Our previous work in human embryonic fibroblast cultures undergoing replicative senescence has shown that the reduced levels of proteasome activities are accompanied by lower proteasome content due to the down-regulation of its β-catalytic subunits (4). The fundamental link between cellular senescence and proteasome function is further supported by our additional studies (5, 6) demonstrating that when the proteasome is partially inhibited in young fibroblasts cultures, an irreversible senescence-like phenotype is triggered. Importantly, reconstitution of the levels of the “down-regulated” β-subunits by genetic means in human embryonic fibroblasts, results in enhanced rates of assembled, and functional proteasome and delays senescence by ∼15% (7).
The proteasome is the major cellular non-lysosomal threonine protease, implicated in the removal of normal as well as abnormal, denatured, or otherwise damaged proteins (8). It is a well organized protein complex that contains a proteolytic active 20 S core, composed of 28 subunits, 7 different α- and 7 different β-subunits, arranged as α1–7β1–7β1–7α1–7 structure (9). The β5, β1, and β2 subunits are the catalytic centers of chymotrypsin-like (CT-L),3 caspase-like/PGPH (peptidylglutamyl-peptide hydrolyzing), and trypsin-like activities, respectively, cleaving peptide bonds after hydrophobic, acidic, and basic residues, respectively (10). The 20 S proteasome is also central to the ATP/ubiquitin-dependent intracellular protein degradation pathway where it represents the proteolytic core of the 26 S complex (11).
Although the regulation of human proteasome is not yet entirely elucidated, reports in mice indicate that the expression of proteasome subunits and the relative proteasome activities can be modulated by exogenous stimuli. Specifically, Kensler and co-workers (12) have reported that the genes forming the 26 S proteasome complex are coordinately regulated by nuclear-erythroid factor 2 (Nrf2) in response to antioxidants such as 3H-1,2-dithiole-3-thione. Nrf2 is a basic leucine zipper (bZIP) transcription factor, featuring a Cap “n” collar structure, responsible for the regulation of the cellular antioxidant response. Nrf2 activates the transcription of multiple phase 2 genes encoding for proteins that protect cells against oxidative and electrophilic stress, such as glutathione S-transferases, γ-glutamylcysteine ligases, heme oxygenese 1, and NADPH quinone oxidoreductase (13) through a common DNA regulatory cis-acting element, called antioxidant response element (ARE) or electrophile responsive element (5′-TGA(C/T)NNNGC-3′) (14). Activation of the Nrf2 signaling pathway is regulated by Kelch-like ECH-associated protein 1 (Keap1) according to changes in the intracellular redox state. Under non-stimulated conditions, Nrf2 is sequestered in the cytoplasm, where it is associated with Keap1 (15). Keap1 does not just passively sequester Nrf2 in the cytoplasm but plays an active role in targeting Nrf2 for ubiquitination and proteasomal degradation (15). Depending on the type of stimulus, Nrf2 may became phosphorylated and/or Keap1 is modified, resulting in disruption of the Keap1-Nrf2 complex and the nuclear translocation of Nrf2 (16). Once in the nucleus, Nrf2 heterodimerizes with a small musculo-aponeurotic fibrosarcoma (Maf) protein and binds to AREs/electrophile responsive elements thereby promoting transcription of various cytoprotective genes (17, 18).
Given these findings, the aim of this study was establishing the proteasome-related function of Nrf2 during replicative senescence of human fibroblasts. We have studied the Nrf2 status in senescent cells and examined the effects of its activation on proteasome function as well as on the process of senescence.
EXPERIMENTAL PROCEDURES
Reagents and Antibodies
18α-Glycyrrhetinic acid (18α-GA), diethyl maleate (DEM), and dimethyl sulfoxide (DMSO) were obtained from Sigma. LLVY-AMC, LLE-NA, LSTR-AMC, MG132, as well as primary proteasomal antibodies against α4 (PW8120; 29 kDa), α6 (PW9390; 33 kDa), β5 (PW8895; 23 kDa), β1 (PW8140; 29 kDa), and β2 (PW8145; 28 kDa) subunits were purchased from BIOMOL. Primary antibodies against Nrf2 (sc-13032; 57 kDa), Keap1 (sc-1544; 68–75 kDa), lamin A/C (sc-20681; 69/62 kDa), p16 (sc467; 16 kDa), tubulin (sc-8035; 52 kDa), GAPDH (sc25778; 37 kDa), and secondary antibodies were purchased from Santa Cruz Biotechnology.
Cell Lines and Culture Conditions
HFL-1 human embryonic fibroblasts were obtained by the European Collection of Cell Cultures and maintained in Dulbecco's modified Eagle's medium (Invitrogen) supplemented with 10% fetal bovine serum (v/v; Invitrogen), 2 mm glutamine, and 1% non-essential amino acids (complete medium). HFL-1 cells subcultured at 37 °C, 5% CO2, and 95% humidity, were fed ∼16 h prior to each assay and the cell number was determined in duplicates using a Coulter Z2 counter (Beckman Coulter). HFL-1 cells were subcultured when they reached confluence at a split ratio of 1:2 until they entered senescence.
E8.T4 cells, a subclone of L929 cells (19), were kindly provided by Prof. J. Alam (20) and were cultured in the same medium supplemented with 200 mg/ml of geneticin (G418 sulfate). In the presence of 1 mg/ml of doxocycline, a tetracycline analogue, E8.T4 cells express the wild type and functional Nrf2, whereas upon doxocycline removal, cells express the mutated and non-functional form of Nrf2.
Proteasome Peptidase Assays
CT-L, PGPH, and trypsin-like proteasome activities were assayed in crude extracts with the hydrolysis of fluorogenic peptides, LLVY-AMC, LLE-NA, and LSTR-AMC, respectively, at 37 °C for 30 min, as described previously (21). Proteasome activity was determined as the difference between the total activity of crude extracts and the remaining activity in the presence of 20 μm MG132. Fluorescence was measured using a VersaFluorTM fluorescence spectrophotometer (Bio-Rad). Protein concentrations were determined using the Bradford method with bovine serum albumin as standard.
Real Time PCR Analysis
Young HFL-1 and E8.T4 cells were treated with 2 μg/ml of 18α-GA or DMSO for 2 and 16 h, respectively. Total RNA was extracted using TRIzol (Invitrogen) and converted into cDNA with the iScript cDNA synthesis kit (Bio-Rad). Real time PCR were performed in triplicate on an iCycler iQ using Gene Expression MacroTM version 1.1 (Bio-Rad). Primers used for real time PCR are summarized in supplemental Table S1. The GAPDH gene was used as normalizer.
Immunoblot Analysis and Detection of Oxidized Proteins
Cells were harvested at the indicated time points, lysed in reducing Laemmli buffer, and proteins were fractionated by SDS-PAGE according to standard procedures (22). Proteins were then transferred to nitrocellulose membranes for probing with appropriate antibodies. Secondary antibodies conjugated with horseradish peroxidase and enhanced chemiluminescence were used to detect the bound primary antibodies. Equal protein loading was verified by reprobing each membrane with a GAPDH antibody. The abundance of acidic acid residues found in Nrf2 causes the anomalous migration of Nrf2 in SDS-PAGE (23) and we report the appearance of four bands of ∼100 kDa (named as 1, 2, 3, and 4) along with a band at the predicted molecular mass of 57 kDa (band 5) (supplemental Fig. S1, A and B). These bands are Nrf2 specific as they all decrease following silencing of Nrf2 (supplemental Fig. S1A) and they all accumulate following proteasome inhibition by MG132, in accordance with previous studies (15), as revealed by immunoblot analysis (see supplemental Fig. S1B). Each immunoblot analysis was performed at least three times and representative blots are shown. For several blots a higher and a lower exposure are included in the figures and supplemental figures. Immunoblot detection of carbonyl groups into proteins (oxidized proteins) was performed with the “OxyblotTM protein oxidation kit” (Chemicon International) by using an anti-2,4-dinitrophenol antibody according to the manufacturer's instructions.
Immunoprecipitation of Proteasome
HFL-1 cells were treated with 2 μg/ml of 18α-GA or DMSO for 2 h and then lysed in lysis buffer containing 20 mm Tris-HCl, pH 7.5, 5 mm ATP, 10% glycerol, 0.2% Nonidet P-40, 10 mm phenylmethylsulfonyl fluoride, and 10 μg/ml of aprotinin. Protein A-agarose beads (Santa Cruz Biotechnology) equilibrated in lysis buffer were coupled with 1–2 μg of antibody against α6 subunit or 1–2 μg of normal mouse serum (for controls) at 4 °C for 3 h on a rotor. The same quantity of cell extracts was then added in the pre-coupled antibody against α6 proteasome subunit-protein A or in the pre-coupled normal mouse serum-protein A (for controls). Immunoprecipitation was performed on a rotor at 4 °C overnight. Immunoprecipitated protein complexes were collected, washed four times with 50 mm Tris/HCl buffer, pH 7.5, containing 5 mm ATP, 75 mm NaCl, 10% glycerol, and 0.2% Triton X-100 (washing buffer) and eluted from the agarose beads by boiling for 5 min in non-reducing Laemmli buffer. Controls were used to demonstrate the specificity of the observed immunoprecipitations. Following proteasome immunoprecipitation, the samples were processed for SDS-PAGE. For proteasome activity assays in immunoprecipitated proteasomes (to demonstrate that whole proteasomes are immunoprecipitated), elution of the complexes by boiling was omitted. Instead, immunoprecipitated protein complexes were diluted in proteasome assay buffer and equal volumes of different samples were assayed as described above.
Cell Fractionation
HFL-1 cells were treated with 2 μg/ml of 18α-GA or DMSO for 2 h and nuclear and cytosolic protein extracts were prepared according to the nuclear extract kit (Active motif). The cytosolic and nuclear fractions were then subjected to immunoblot analysis using anti-Nrf2, anti-tubulin, or anti-lamin A/C antibodies.
Immunofluorescence Analysis
HFL-1 cells were grown on coverslips and treated with 2 μg/ml of 18α-GA or DMSO for 2 h, then fixed in 2% paraformaldehyde followed by cell permeabilization with 0.2% Triton X-100 in phosphate-buffered saline (PBS). Cells were incubated with anti-Nrf2 antibody at room temperature for 1 h followed by incubation with rhodamine-conjugated secondary antibodies (Santa Cruz Biotechnology). Slides were mounted using Vectashield mounting medium for fluorescence with 4′,6-diamidino-2-phenylindole (Vector Laboratories) and analyzed using a Leica TCS SPE confocal laser scanning microscope (Leika Lasertechik). The LAS AF software was used for image acquisition.
Luciferase Activity
HFL-1 cells were co-transfected with pTi-Luc or pTi-ARE-Luc constructs and pCMX-β-galactosidase as internal control using EffecteneTM transfection reagent (Qiagen). pTi-ARE-Luc that contains a sequence of 41 bp ARE and its empty vector (pTi-Luc) were kindly provided by Prof. Fahl (14). Twenty four hours post-transfection, cells were treated with 2 μg/ml of 18α-GA or DMSO for 2 h. Cells were harvested and lysed; supernatants were used for luciferase and β-galactosidase assays, measured with the appropriate kits (Promega) according to manufacturer's instructions. Means and standard deviations were based on three independent transfections. DEM was used as positive control.
Measurement of Reactive Oxygen Species
2′,7′-Dichlorodihydrofluorescein diacetate (H2DCFDA) (Invitrogen) was used for detection of reactive oxygen species (ROS). HFL-1 cells treated with 18α-GA or DMSO for 2 and 24 h were resuspended in PBS ± the dye at a final concentration of 10 μm (loading buffer) and incubated at 37 °C for 30 min. The loading buffer was then removed; cells were resuspended in prewarmed complete medium and incubated at 37 °C for 5 min. The absorption and emission of the oxidation product were measured at 493 and 520 nm, respectively. Each sample was measured in triplicates.
Continuous Cell Treatment with 18α-GA
Five × 104 young HFL-1 cells were treated with 2 μg/ml of 18α-GA every day until they entered senescence. Upon 90% confluence, cell number was determined in triplicates using a Coulter Z2 counter and the population doublings were calculated using the formula: log (Nf/N0)/log 2, where Nf is the number of cells measured when each culture reached confluence and N0 represents the number of cells initially seeded. Control cultures were supplemented with 0.1% DMSO, the organic solvent of 18α-GA.
RNA Interference
A combination of four gene-specific small interfering RNAs (siRNA) against human Nrf2 was used (Dharmacon SMARTpool siRNA reagent; Dharmacon RNA Technologies). Applied siRNAs were complexed with liposome carrier Lipofectamine (Invitrogen) in serum-free Opti-MEM (Invitrogen). HFL-1 cells at 60–70% confluence were then transfected with 50 or 75 nm siRNA against Nrf2 (siNrf2) or an equal molar of mismatched scrambled siRNA (siCon) and incubated for 24 h. The medium was then changed and a further incubation in complete medium followed for another 24 h. Treatment with DMSO or 18α-GA was then performed for 2 h. To ensure functional and specific silencing, Nrf2 mRNA levels were revealed in siNrf2 and siCon groups before and after treatment in all experiments. Stable knockdown of the Nrf2 expression was performed by repeating Nrf2 siRNA transfection every 48 h for up to 17 days. Scrambled siRNA were used as control.
β-Galactosidase Staining
Staining for β-galactosidase activity was performed as described previously (24). Briefly, 1.5 × 105 cells were seeded in 6-well plates. After 24 h cells were washed with PBS, fixed in 0.2% glutaraldehyde and 2% formaldehyde for 5 min, washed again with PBS, and finally stained at 37 °C for 24 h in the absence of CO2, in staining solution (150 mm NaCl, 2 mm MgCl2, 5 mm K3Fe(CN)6, 40 mm citric acid, and 12 mm sodium phosphate, pH 6.0) containing 1 mg/ml of 5-bromo-4-chloro-3-indolyl-β-d-galactoside.
Survival Assays
Cell survival was monitored by counting the number of cells following treatment with the mentioned cytotoxic reagent. Briefly, 4 × 106 HFL-1 cells were plated in Petri dishes and treated with 2 μg/ml of 18α-GA or DMSO for 24 h, followed by incubation with 300 μm hydrogen peroxide (H2O2) for an additional 20 h with or without 20 μm MG132 (cells were treated for 6 h prior to H2O2 treatment). Cell number was determined in triplicates using a Coulter Z2 counter.
Statistical Analysis and Quantification
Statistics were performed using Microsoft Excel software. Statistical significance was evaluated using the one-way analysis of variance. Quantification was performed with Image Quant 5.2. All values including quantification data were reported as mean of three independent experiments ± S.D. and statistical significance at p < 0.05 or p < 0.01 are denoted in graphs by a single (*) or double (**) asterisk, respectively. Quantification of the Nrf2 immunoblots in the figures was performed including all specific Nrf2 bands together (1–5). Additionally, quantification of each specific band is shown separately in supplemental Fig. S1D.
RESULTS
The Transcription Factor Nrf2 Is Down-regulated during Replicative Senescence
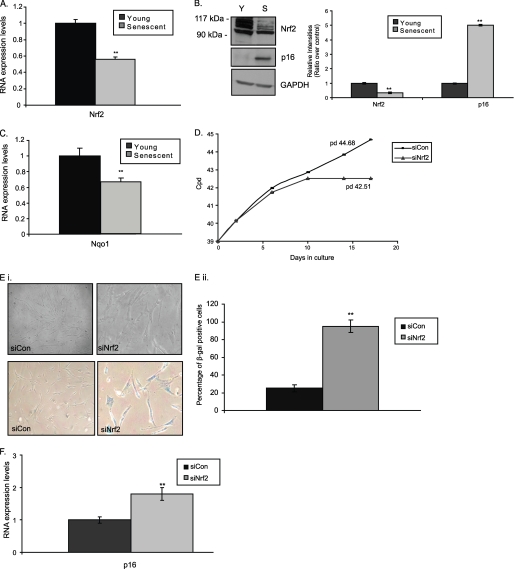
Initially, we aimed to reveal potential changes of the Nrf2 transcription factor status during replicative senescence. As shown in Fig. 1 both RNA (A) and protein (Fig. 1B and supplemental Fig. S1C for lower exposure) Nrf2 expression levels were found decreased by ∼45 and ∼65%, respectively, in HFL-1 senescent fibroblasts as compared with their young counterparts. In support, the transcriptional activity of Nrf2 was equally decreased because we detected lower RNA expression levels of its main gene target, namely NADPH quinone oxidoreductase (NQO1) (Fig. 1C).
FIGURE 1.
Lower expression levels of transcription factor Nrf2 during replicative senescence. Quantification of RNA expression levels of Nrf2 (A) and NADPH quinone oxidoreductase (Nqo1) (C) in young and senescent HFL-1 fibroblasts following real time PCR analysis. RNA expression levels in young fibroblasts were arbitrary set to 1. B, immunoblot analysis and the relative quantification of Nrf2 in young (Y) and senescent (S) HFL-1 fibroblasts. Lower exposure of the Nrf2 immunoblot to show the specific Nrf2 bands is included in supplemental Fig. S1C. p16 expression was used as a marker of senescence. GAPDH levels were used as loading control. Molecular mass markers are indicated on the left side of the Nrf2 blot. D, number of cpds of HFL-1 cells transfected with Nrf2 siRNA (siNrf2) or scrambled siRNA (siCon) constantly every 2 days for up to 17 days. Numbers on the graph show the population doublings (pd) measured on the 17th day of treatment. E, panel i, representative photographs of cells (upper panel) and cells following β-galactosidase activity staining (lower panel); panel ii, percentage of β-galactosidase positive cells; and F, quantification of p16 RNA expression levels in HFL-1 cells transfected with Nrf2 siRNA (siNrf2) or scrambled siRNA (siCon) constantly every 2 days for up to 17 days. RNA expression levels in HFL-1 cells transfected with scrambled siRNA were arbitrary set to 1. All values are reported as mean of three independent experiments ± S.D. Statistical significance at p < 0.01 is denoted by double asterisks.
To assess the effects of Nrf2 on cell proliferation, the expression of Nrf2 was knocked down using siRNA in HFL-1 cells. Our pilot experiments revealed a maximum ∼85% decrease of Nrf2 mRNA levels using 75 nm siNrf2 for 48 h (supplemental Fig. S2). Knockdown of Nrf2 resulted in inhibition of proliferation and induction of premature senescence in HFL-1 fibroblasts (Fig. 1D). The cells acquired the typical senescent morphology (Fig. 1E, panel i), exhibited a higher percentage of β-galactosidase positive staining (Fig. 1E, panel ii) and revealed increased levels of p16 protein (Fig. 1F), as compared with the HFL-1 cells transfected with scrambled siRNA.
Activation of Nrf2 by 18a-GA
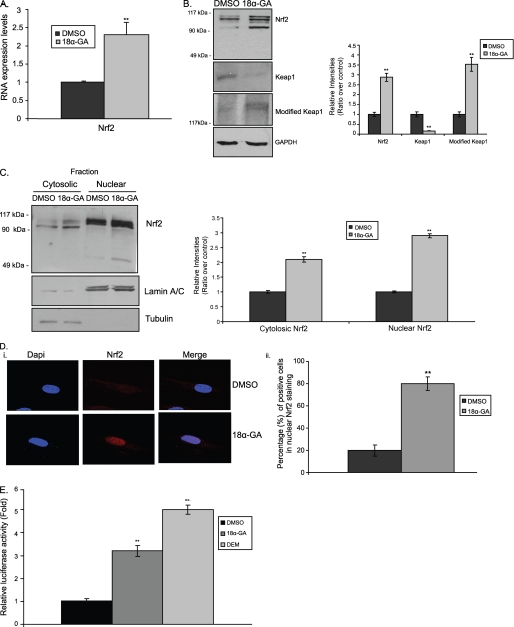
As it is known that many compounds are capable of activating Nrf2 (25), our screening analysis using a library of different compounds has identified two triterpenoids, namely 18α-GA and hederagenin (HED) as potential Nrf2 inducers. Extensive follow up analysis has revealed a dose- and time-dependent 18α-GA-mediated activation of Nrf2. Specifically, treatment of HFL-1 young cells with 2 μg/ml of 18α-GA for 2 h (optimal conditions, see next paragraph) induced both the RNA (Fig. 2A) and protein (Fig. 2B, top panel) expression levels of Nrf2. In support, we have observed reduced levels of Keap1 as well as increased levels of its modified isoform (a protein greater than 150 kDa). Moreover, cell fractionation experiments (Fig. 2C and supplemental Fig. S1D, iii for lower exposure) as well as immunofluorescence analysis (Fig. 2D) under the same conditions revealed that treatment with 18α-GA promoted Nrf2 translocation to the nucleus, a feature that concurs with its activation. Specifically, more than 80% of the cells treated with 18α-GA showed Nrf2 nuclear accumulation as compared with the control cells (Fig. 2D, panel ii). Finally, to demonstrate the binding of Nrf2 to the ARE elements upon 18α-GA treatment, a luciferase reporter plasmid containing the 41-bp murine ARE (pTi-ARE-Luc) and its relative empty vector (pTi-Luc) were used. As shown in Fig. 2E, 18α-GA treatment induced ARE-promoter activity, thus resulting in increased luciferase activity levels. DEM, a thiol-oxidizing agent and a known activator of the Nrf2 pathway (26), was used as a positive control. In total, our results demonstrate that 18α-GA treatment promotes Nrf2 activation.
FIGURE 2.
Activation of Nrf2 by 18α-GA. A, quantification of RNA expression levels of Nrf2 in HFL-1 cells treated with 2 μg/ml of 18α-GA or DMSO for 2 h. Nrf2 RNA expression levels in DMSO-treated cells were arbitrary set to 1. B, immunoblot analysis of Nrf2, Keap1, and modified Keap1 and the relative quantification in HFL-1 cells treated with 2 μg/ml of 18α-GA or DMSO for 2 h. GAPDH levels were used as loading control. C, cell fractionation and relative quantification of Nrf2 in HFL-1 cells treated with 2 μg/ml of 18α-GA or DMSO for 2 h. Lower exposure of the Nrf2 immunoblot to show the specific Nrf2 bands is included in supplemental Fig. 1D, iii. Tubulin and lamin A/C were used as markers of cytosolic and nuclear fractions, respectively. D, panel i, Nrf2 localization analyzed by confocal microscopy; and panel ii, percentage of cells with nuclear Nrf2 accumulation, in HFL-1 cells treated with 2 μg/ml of 18α-GA or DMSO for 2 h. 4′,6-Diamidino-2-phenylindole (DAPI) was used as a marker for nucleus visualization. E, luciferase activity in HFL-1 cells transfected with the pTi-ARE-Luc construct and treated with 2 μg/ml of 18α-GA or DMSO for 2 h. Treatment with DEM for 24 h was used as positive control. Luciferase activity in DMSO-treated cells was arbitrary set to 1. Molecular mass markers in B and C are indicated on the left side of the Nrf2 and modified Keap1 blots. All values are reported as mean of three independent experiments ± S.D. Statistical significance at p < 0.01 is denoted by double asterisks.
Proteasome Activation by 18α-GA
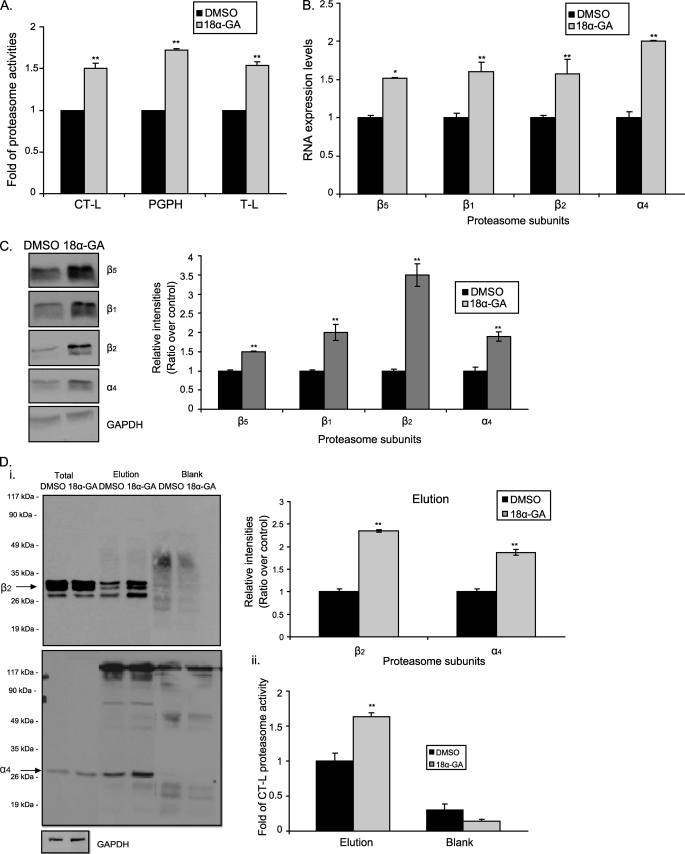
As Nrf2 has been related to proteasome regulation, next, we investigated the effects of 18α-GA on proteasome status. HFL-1 cells were treated with different concentrations of 18α-GA ranging from 0.5 to 10 μg/ml for 24 h and the CT-L proteasome activity was measured (supplemental Fig. S3A, i). The effective concentration was revealed at 2 μg/ml. Extensive time curve analysis was then conducted for all three proteasome activities (CT-L, PGPH, and trypsin-like) to conclude that the effective time treatment was at 2 h (supplemental Fig. S3B, i and Fig. 3A). More specifically, the activities were increased from 1.5- to 1.8-fold, with PGPH activity being the most affected. Moreover, we have also detected enhanced RNA (Fig. 3B) and protein (Fig. 3C) expression levels of representative proteasome subunits in the same cells and experimental conditions. Importantly, proteasome immunoprecipitation, followed by immunoblot analysis (Figs. 3D, panel I, and supplemental Fig. S1E for lower exposure of the β2 immunoblot in total cell lysates) and proteasome activity assay (Fig. 3D, panel ii) has verified an approximate ∼2-fold increase of the amount of assembled proteasome following treatment with 18α-GA, thus further verifying the activation of the proteasome by this substance. Finally to exclude the possibility of a single compound effect, we assayed the HED effects on proteasome status. As it is shown in supplemental Fig. S3, A, ii, and B, ii, HED treatment of HFL-1 cells results in a dose- and time-dependent activation of the proteasome that is similar to 18α-GA.
FIGURE 3.
Proteasome activation by 18α-GA. A, manifold of CT-L, PGPH, and trypsin-like proteasome activities in young HFL-1 cells treated with 2 μg/ml of 18α-GA or DMSO for 2 h. Use of proteasome inhibitor (MG132) in control reactions ensured the specificity of the enzymatic reaction. B, real time PCR; and C, immunoblot analysis and the relative quantification of proteasome subunits (β5, β1, β2, and α4) in HFL-1 cells treated with 2 μg/ml of 18α-GA or DMSO for 2 h. RNA expression levels of each gene in DMSO-treated cells were arbitrary set to 1. GAPDH levels were used as loading control. D, panel i, immunoblot analysis with the relative quantification of representative proteasome subunits (β2, α4) in total extracts and the relative elutions following proteasome immunoprecipitation in HFL-1 fibroblasts treated with 2 μg/ml of 18α-GA or DMSO for 2 h. Molecular mass markers are indicated on the left side of the blots. Panel ii, levels of CT-L proteasome activity in immunoprecipitated proteasomes of HFL-1 fibroblasts treated with 2 μg/ml of 18α-GA or DMSO for 2 h. Activity or protein levels in DMSO-treated cells were arbitrary set to 1. Blank samples represent the control immunoprecipitation samples. Immunoblot analysis of GAPDH (lower panel) in total extracts shows the equal starting protein quantity before immunoprecipitation. Lower exposure of the β2 immunoblot in total extracts is included in supplemental Fig. S1E. All values are reported as mean of three independent experiments ± S.D. Statistical significance at p < 0.05 or p < 0.01 are denoted by single or double asterisks, respectively.
Nrf2-mediated Proteasome Activation by 18α-GA
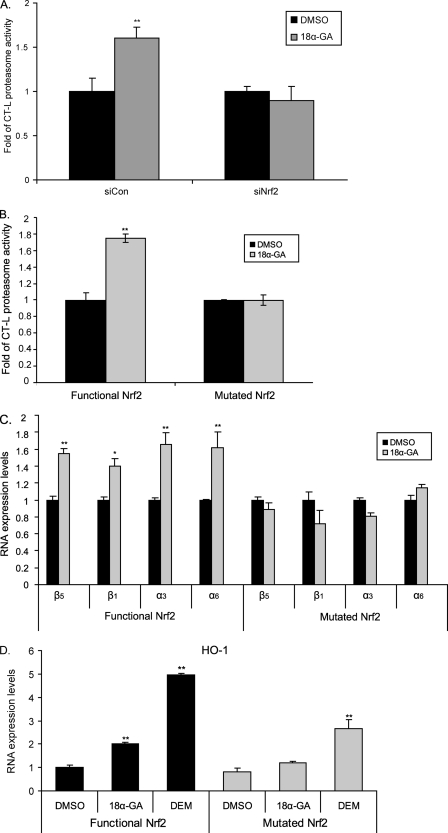
We then examined whether the observed 18α-GA-associated proteasome activation was mediated by Nrf2. To this end, we knocked down Nrf2 in HFL-1 human fibroblasts and the cells were then treated with 18α-GA. As shown in Fig. 4A, silencing Nrf2 inhibited the 18α-GA-mediated activation of the proteasome. We have also taken advantage of the E8.T4 cell line that conditionally expresses the functional Nrf2 (20). Specifically, in the presence of the mutated Nrf2, treatment with 18α-GA did not induce CT-L proteasome activity, whereas the same activity was increased upon 18α-GA treatment in the presence of a functional Nrf2 (Fig. 4B). This was also reflected in the RNA expression levels of representative proteasome subunits, because the levels of β5, β1, α3, and α6 were not affected following 18α-GA treatment in the presence of mutated Nrf2, in contrast to their induction in the presence of functional Nrf2 (Fig. 4C). Control experiments have shown a severe induction of an Nrf2 target gene, namely heme oxygenase-1, by 18α-GA only in the Nrf2 functional cell line (Fig. 4D). DEM was used as positive control. These data demonstrate that 18α-GA-mediated proteasome activation is directly linked to the function of Nrf2.
FIGURE 4.
Nrf2-mediated proteasome activation by 18α-GA. A, levels of CT-L proteasome activity in HFL-1 cells transfected with 75 nm Nrf2 siRNA (siNrf2) or scrambled siRNA (siCon) for 48 h and then treated with 2 μg/ml 18α-GA or DMSO for 2 h. B, levels of CT-L proteasome activity and C and D, RNA expression levels of (C) proteasome subunits (β5, β1, α3, α6), and (D) heme oxygenase-1 (HO-1) gene revealed by real time PCR analysis, in the presence or absence of functional Nrf2 in E8.T4 cells treated with 2 μg/ml 18α-GA or DMSO (and DEM in D) for 16 h. Mean value of activities and RNA expression levels in DMSO-treated cells were set to 1. All values are reported as mean of three independent experiments ± S.D. Statistical significance at p < 0.05 or p < 0.01 denoted by single or double asterisks, respectively.
We also examined the effects of HED on Nrf2 activation and proteasome status. Specifically, as shown in supplemental Fig. S4, treatment with 2 μg/ml HED for 2 h induced the protein expression levels of representative proteasome subunits and increased the amount of assembled proteasome. Finally, cell fractionation experiments revealed a HED-promoted translocation of Nrf2 in the nucleus. Thus both triterpenoids 18α-GA and HED, act similarly on proteasome and Nrf2 status.
Proteasome-mediated 18α-GA Protection against Oxidative Stress
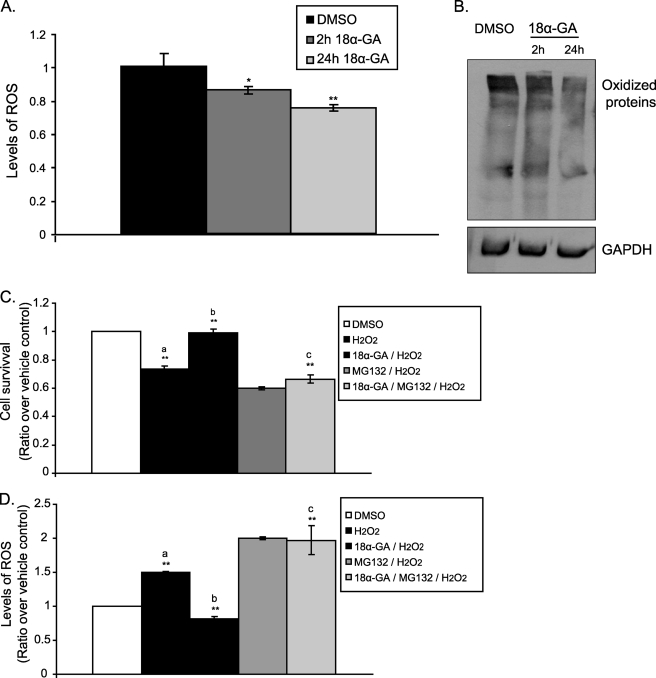
Next we tested the effects of 18α-GA on the intracellular levels of ROS and oxidized proteins. As shown in Fig. 5, A and B, we observed reduced levels of both ROS and oxidized proteins following cell treatment with 18α-GA for 2 h. A more pronounced effect was recorded when cells were treated with the compound for 24 h. Given these findings, we then addressed the question whether 18α-GA could protect the cells from oxidative stress. To this end HFL-1 cells were pretreated with 2 μg/ml of 18α-GA or DMSO for 24 h, then subjected to oxidative stress using 300 μm H2O2 for 20 h and their numbers and ROS levels were determined. Enhanced cytoprotection by ∼20% against H2O2 treatment was revealed in the presence of 18α-GA (Fig. 5C, compare the second and third columns); this protective effect was further verified by the recorded lower ROS levels in the 18α-GA-treated cells (Fig. 5D, compare the second and third columns). Importantly, the observed 18α-GA cytoprotection under these experimental conditions is linked highly to its proteasome activating properties because pretreatment of the cells with the proteasome inhibitor MG132 for 6 h abolished the protective effects of the compound (Fig. 5, C and D, compare the third and fifth columns).
FIGURE 5.
Cytoprotective effect of 18α-GA against oxidative stress. ROS levels (A) and immunoblot analysis (B) of oxidized proteins in HFL-1 cells treated with 2 μg/ml 18α-GA or DMSO for 2 and 24 h. Levels of ROS in DMSO-treated cells were set to 1. GAPDH levels were used as loading control. Cell survival (C) and ROS levels (D) following H2O2 treatment in the presence of 18α-GA with or without proteasome inhibition. HFL-1 cells were preincubated with 2 μg/ml 18α-GA or DMSO for 24 h, then challenged with 300 μm H2O2 for 20 h in the presence or not of 20 μm MG132, and the cell number and ROS levels were determined. Number of cells and levels of ROS in DMSO-treated cells were set to 1. a, p < 0.01 as compared with DMSO-treated cells; b, p < 0.01 as compared with H2O2-treated cells; c, p < 0.01 as compared with 18α-GA/H2O2-treated cells. All values are reported as mean of three independent experiments ± S.D. Statistical significance at p < 0.05 or p < 0.01 are denoted by single or double asterisks, respectively.
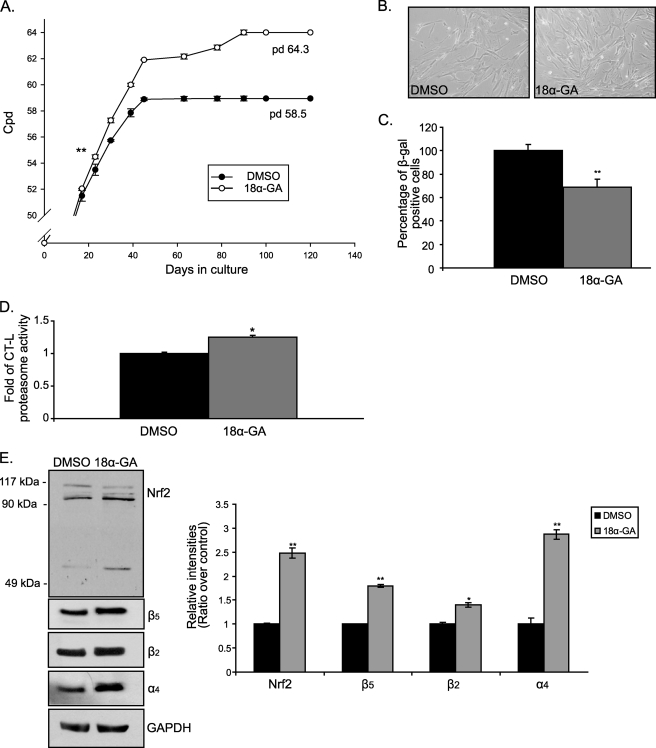
18α-GA Promotes Lifespan Extension in Human Fibroblasts
Given the beneficial cytoprotective effects of 18α-GA, along with its proteasome activating properties, its effects on the progression of senescence were examined. For these experiments, young HFL-1 cells were continuously treated with 2 μg/ml of 18α-GA throughout their lifespan. It was found that 18α-GA-treated cultures exhibited a 10 ± 1% increase of their lifespan as compared with the DMSO-treated cultures (Fig. 6A). The differences between the two groups were statistically significant beginning on the third week of treatment and maintained thereafter. Moreover, the young morphology of these cells (i.e. smaller size and nuclei, regular shape, and growth in parallel arrays) was maintained for longer as compared with the control cultures, further demonstrating the delay in the appearance of the senescent phenotype (Fig. 6B). In support, these cells exhibited a lower percentage of β-galactosidase positive staining, as compared with the DMSO-treated cells that coincides with their younger morphology (Fig. 6C).
FIGURE 6.
18α-GA treatment extends lifespan in human fibroblasts. A, number of cpds of HFL-1 cells treated with 2 μg/ml of 18α-GA or DMSO throughout their lifespan. Numbers on the graph show the population doublings (pd) performed at the end of treatment. Asterisks show the first statistically significant difference between the two different groups (on the 23rd day of treatment). This significance is maintained during all following time points. B, representative photographs of HFL-1 cells treated with 2 μg/ml of 18α-GA or DMSO throughout their lifespan at 120 days (terminally senescent cultures). C, percentage of β-galactosidase positive HFL-1 cells treated with 2 μg/ml of 18α-GA or DMSO throughout their lifespan at 120 days. D, levels of CT-L proteasome activity in senescent fibroblasts treated with 18α-GA or DMSO throughout their 120-day lifespan. Recorded levels in DMSO-treated cells were arbitrary set to 1. E, immunoblot analysis and the relative quantification of Nrf2 and proteasome subunits (β5, β2, α4) in HFL-1 cells treated with 2 μg/ml of 18α-GA or DMSO throughout their lifespan at 50 days (pre-senescent cultures). GAPDH levels were used as loading control. Molecular mass markers are indicated on the left side of the Nrf2 blot. All values are reported as mean of three independent experiments ± S.D. Statistical significance at p < 0.05 or p < 0.01 are denoted by single or double asterisks, respectively.
We were then asked whether the activating effects of 18α-GA on Nrf2 and proteasome still occur in cells during their progression to replicative senescence. Therefore, we tested the levels of CT-L proteasome activity and we revealed the protein expression levels of Nrf2 and representative proteasome subunits in pre-senescent cells that were serially passaged in the presence of 18α-GA. As shown in Fig. 6D, these cells possessed elevated CT-L proteasome activity by 1.2-fold as compared with control cells. Similarly, pre-senescent 18α-GA-treated cells had elevated protein levels of Nrf2 and proteasome subunits (β5, β2, and α4) as compared with the relative levels in DMSO-treated cells (Fig. 6E). Thus, although the increase of proteasome activity in senescent cultures was lower than the one observed in corresponding young cultures, the Nrf2-proteasome axis remains active as cells progress to senescence.
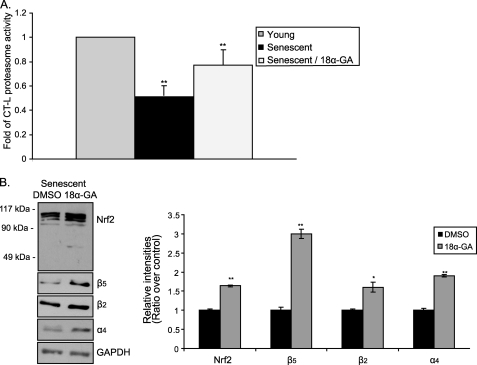
18a-GA Activates the Proteasome in Terminally Senescent Human Fibroblasts
Finally, we investigated the effect of 18α-GA in terminally senescent HFL-1 cells (cpd 64) that were irreversibly growth arrested after serial passaging in the absence of the substance. Treatment of these cells with 2 μg/ml of 18α-GA for 2 h still exhibited a positive effect on proteasome and Nrf2 status, as shown by the enhanced levels of CT-L proteasome activity by ∼1.5-fold (Fig. 7A, compare the second and third columns) as well as by the increased protein expression levels of Nrf2 and representative proteasome subunits (β5, β2, and α4; Fig. 7B). In conclusion, senescent fibroblasts retain their responsiveness to 18α-GA regarding the activation of the Nrf2-proteasome axis.
FIGURE 7.
18α-GA treatment induces proteasome activation in terminally senescent human fibroblasts. A, levels of CT-L proteasome activity; and B, immunoblot analysis and the relative quantification of Nrf2 and proteasome subunits (β5, β2, α4) in senescent HFL-1 cells treated with 2 μg/ml of 18α-GA or DMSO for 2 h. Mean value of CT-L proteasome activity in young cells was arbitrary set to 1. GAPDH levels were used as loading control. Molecular mass markers are indicated on the left side of the Nrf2 blot. All values are reported as mean of three independent experiments ± S.D. Statistical significance at p < 0.05 or p < 0.01 are denoted by single or double asterisks, respectively.
DISCUSSION
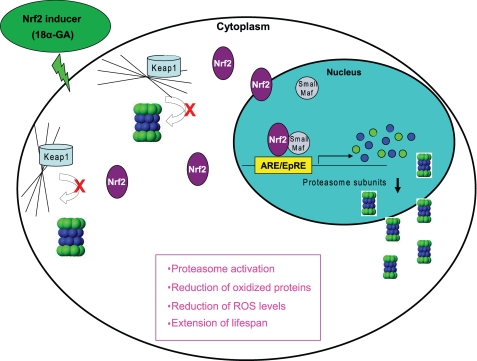
In this article we demonstrate that the transcription factor Nrf2 has an impaired function during replicative senescence of human fibroblasts and its silencing leads to premature senescence. Moreover, we have identified a novel Nrf2 inducer, 18α-GA, which by stimulating the Nrf2-mediated proteasome activation results in enhanced survival of the cells against oxidants and lifespan extension. Finally, we provide evidence that 18α-GA can stimulate the proteasome even in terminally senescent cultures. Fig. 8 summarizes the action and the overall effects of treatment of the cells with 18α-GA via the Nrf2 pathway and downstream proteasome activation.
FIGURE 8.
Overview of the Nrf2-mediated activation of the proteasome by 18α-GA. Model summarizing the effects of 18α-GA in human fibroblasts. 18α-GA induces Nrf2 dissociation from Keap1 as well as its de novo expression. Nrf2 translocates to the nucleus, binds to the ARE sequences of proteasome subunits, and activates their transcription. Increased transcription of proteasome subunits leads to increased levels of assembled proteasome, resulting in elevated proteasome activities and function, accompanied by decreased levels of ROS and oxidized proteins as well as lifespan extension.
Nrf2 factor plays a crucial role in transcriptional regulation and activation of phase 2 and antioxidant enzymes (27). We have observed reduced levels of Nrf2 expression and function in human senescent fibroblasts, in agreement with previous studies demonstrating a similar age-related decline of Nrf2 levels in rats (28), Caenorhabditis elegans (its homologue, SKN-1) (29) as well as in human subjects (30). Therefore, the well documented age-dependent decrease in the activity of the antioxidant defense systems (31) can be partly attributed to the decline of the function of Nrf2. For instance, the decline of Nrf2 transcriptional activity causes the age-associated loss of glutathione synthesis through the loss of expression of glutamylcysteine ligase in rats (32).
Proteasome expression has already been shown to be regulated by Nrf2 through AREs located on the proximal promoter region of several proteasome subunits in mouse cell lines (12, 33). Recently, it was also shown that silencing of Nrf2 promotes a decrease of the expression of proteasome subunits and a reduction of the relative proteasome activities (34). Given our previous findings where we have reported the down-regulation of several proteasome subunits in senescent human fibroblasts (4) as well as the currently reported senescence-related decline of Nrf2 expression and function, we have searched for Nrf2 inducers that could also serve as proteasome inducers. It is well established that several chemopreventive agents and natural compounds activate the Nrf2-dependent response at low doses. Examples of potent plant Nrf2 inducers include 3H-1,2-dithiole-3-thione, carnosol, curcumin, epigallocatechin-3-gallate, lycopene, quercetin, resveratrol, sulforaphane, and wasabi (25, 35). However, only a few of them have been correlated to proteasome regulation via Nrf2 in mice (12) and none of them have been studied regarding their downstream effects on replicative senescence. The identified Nrf2 inducers, 18α-GA and HED, are pentacyclic triterpenoids. Triterpenoids are known to activate the Nrf2 pathway. For instance, it has been demonstrated that they protect from lipopolysaccharide-induced inflammatory response through the Nrf2 pathway (36), whereas low concentrations of triterpenoids have been shown to activate an anti-inflammatory response correlated to Nrf2 activation (37).
Our data show that cell treatment with 18α-GA up-regulates Nrf2 levels per se, reduces the levels of Keap1, and increases the levels of modified Keap1, thus resulting in a higher ratio of Nrf2/Keap1 and a consequent Nrf2 activation via its translocation to the nucleus (27). Other compounds such as oltipraz (38) and quercetin (39) are also known to enhance Nrf2 transcription. Regarding Keap1, it mediates Nrf2 degradation by being a substrate adaptor protein for a Cul3-dependent E3 ubiquitin ligase that, in turn, targets Nrf2 for ubiquitin conjugation (40). A recent study suggests that oxidative signals are inhibitory for the ubiquitination activity of Keap1-Cul3, but not for the association activity of Keap1 with Nrf2, thus allowing newly synthesized Nrf2 to bypass Keap1-Cul3 and to accumulate in the nucleus (41). Alternatively, the interaction of Keap1 with oxidative inducers through its thiol groups triggers the disassociation of Nrf2 from the Nrf2-Keap1-Cul3 complex in the cytoplasm, thus protecting Nrf2 from ubiquitination (42). Moreover, several chemicals like ebselen (43), quercetin (39), sulforaphane (44), and tert-butylhydorquinone (45) have been shown to cause a reduction of the 70-kDa Keap1 band with a concomitant increase of a 150-kDa band that represents a modified form of Keap1. The exact nature of Keap1 modification is not clear but based on its molecular weight, it has been suggested that this form could be a covalently adducted protein from two different proteins (an intermolecular adduct), an intramolecular adduct of Keap1, or two covalently linked Keap1 proteins (a dimeric form of Keap1) (46). This modification has been shown to alter the Keap1-Cul3 interaction thus leading to Nrf2 accumulation and increase of Nrf2-mediated transcription (42). Regardless of the exact mechanism, the observed 18α-GA-mediated Nrf2 release from Keap1 is considered a critical step for Nrf2-driven gene transcription (47).
Activation of the Nrf2 pathway enhances the cellular antioxidant capacity through transcription of a panel of antioxidant and anti-inflammatory genes (27, 48). Our results indicate that 18α-GA is an antioxidant because it promotes reduced ROS levels and lower amounts of oxidized proteins. This is in full accordance with the action of other small molecule Nrf2 activators such as sulforaphane and dithiolethiones (27, 48, 49). However, our study additionally reveals that under our experimental conditions the cytoprotective effect of 18α-GA is mainly mediated through proteasome activation because it is severely attenuated upon proteasome inhibition, in accordance with previous studies (50). 18α-GA treatment also has a minor effect for better cellular survival and lower ROS levels even during proteasome inhibition (Fig. 5, C and D, compare the fourth and fifth columns). This could be attributed to the known induction of multiple antioxidant genes by Nrf2. Thus, although one cannot exclude the possibility that the observed results are due to the complementary and synergistic action of various pathways that are regulated by Nrf2, our study indicates the proteasome system is the major downstream mediator of the 18α-GA/Nrf2 cytoprotective effect.
Reduction of cellular oxidative cargo by various means has been shown to retard the rate of aging. Strategies include overexpression of antioxidant genes (51, 52) as well as delivery of various natural compounds possessing antioxidant properties (53, 54). Increased longevity achieved by proteasome manipulation is an emerging and novel area in anti-aging research. Few studies have shown that proteasome activation, via overexpression of either its catalytic subunits or its maturation protein POMP, delays the rate of aging in human fibroblasts (7) and in yeast (55), respectively. Moreover, proteasome activation through conformational changes induced by a natural compound also results in extended cellular lifespan (53). Our study envisages the transcriptional activation of proteasome as the reported 18α-GA effects on proteasome and human aging are Nrf2 mediated. The exact Nrf2 involvement in the aging process requires further investigation as it is implicated in the longevity of some species (56), but not in others (57). One possibility might be a species-specific activation of a different set of genes by Nrf2 that, in turn, influence diversely the rate of aging in different species. Regarding human aging, the Nrf2/proteasome axis belongs to this novel category of “healthspan” factors that act by maximizing cellular maintenance and homeodynamic balance. By definition, these factors may not result in a dramatic increase of lifespan, in contrast to those that affect directly the rate of cellular replication, but they rather provide longer “healthy aging,” which will indirectly affect lifespan. Importantly we have recorded Nrf2-mediated proteasome activation by 18α-GA even in terminally senescent cultures. This is one of the few cases where senescent cells retain their responsiveness, as in most cases there is a dramatic loss of response to different factors with advanced age (58). In this regard, the reported findings open up new directions for future manipulation of the senescence phenotype.
Supplementary Material
Acknowledgments
We are grateful to Prof. J. Alam at the Dept. of Molecular Genetics, Ochsner Medical Center, New Orleans, LA, for the E8.T4 cell line and Prof. T. Fahl at the McArdle Laboratory for Cancer Research, University of Wisconsin, Madison, WI, for the pTi-ARE-Luc plasmid. We also thank Dr. Varvara Trachana, Konstantinos Voutetakis, and Christina Sisoula for technical advice.
This work was supported by General Secretariat of Research and Technology Grant PENED 03EΔ9 (75% from the European Union & 25% from the Ministry of Development, Measure 8.3/Operational Program “Competitiveness”/3rd Community Support Framework) and European Union “Proteomage” Grant LSHM-CT-518230 (to E. S. G.). Data presented in this article are patent protected (patent submission number 2009-0100242).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1–S4 and Table S1.
We dedicate this article to the late Prof. Constantinos E. Sekeris.
- CT-L
- chymotrypsin-like
- 18α-GA
- 18α-glycyrrhetinic acid
- ARE
- antioxidant response element
- DEM
- diethyl maleate
- DMSO
- dimethyl sulfoxide
- HED
- hederagenin
- Keap1
- Kelch-like ECH-associated protein 1
- LLE-NA
- N-Cbz-Leu-Leu-Glu-β-naphthylamine
- LLVY-AMC
- Suc-Leu-Leu-Val-Tyr-7-amido-4-methylcoumarin
- LSTR-AMC
- N-t-Boc-Leu-Ser-Thr-Arg-7-amido-4-methylcoumarin
- Nrf2
- nuclear erythroid factor 2
- PBS
- phosphate-buffered saline
- PGPH
- peptidylglutamyl-peptide hydrolyzing
- ROS
- reactive oxygen species
- siRNA
- small interfering RNA
- GAPDH
- glyceraldehyde-3-phosphate dehydrogenase
- Cpd(s)
- cumulative population doubling(s).
REFERENCES
- 1.Petropoulou C., Chondrogianni N., Simões D., Agiostratidou G., Drosopoulos N., Kotsota V., Gonos E. S. (2000) Ann. N.Y. Acad. Sci. 908, 133–142 [DOI] [PubMed] [Google Scholar]
- 2.Collado M., Blasco M. A., Serrano M. (2007) Cell 130, 223–233 [DOI] [PubMed] [Google Scholar]
- 3.Chondrogianni N., Gonos E. S. (2005) Exp. Gerontol. 40, 931–938 [DOI] [PubMed] [Google Scholar]
- 4.Chondrogianni N., Stratford F. L., Trougakos I. P., Friguet B., Rivett A. J., Gonos E. S. (2003) J. Biol. Chem. 278, 28026–28037 [DOI] [PubMed] [Google Scholar]
- 5.Chondrogianni N., Gonos E. S. (2004) Biogerontology 5, 55–61 [DOI] [PubMed] [Google Scholar]
- 6.Chondrogianni N., Trougakos I. P., Kletsas D., Chen Q. M., Gonos E. S. (2008) Aging Cell 7, 717–732 [DOI] [PubMed] [Google Scholar]
- 7.Chondrogianni N., Tzavelas C., Pemberton A. J., Nezis I. P., Rivett A. J., Gonos E. S. (2005) J. Biol. Chem. 280, 11840–11850 [DOI] [PubMed] [Google Scholar]
- 8.Ciechanover A. (2005) Angew. Chem. Int. Ed. Engl. 44, 5944–5967 [DOI] [PubMed] [Google Scholar]
- 9.Demartino G. N., Gillette T. G. (2007) Cell 129, 659–662 [DOI] [PubMed] [Google Scholar]
- 10.Tanaka K. (1998) J. Biochem. 123, 195–204 [DOI] [PubMed] [Google Scholar]
- 11.Glickman M. H., Ciechanover A. (2002) Physiol. Rev. 82, 373–428 [DOI] [PubMed] [Google Scholar]
- 12.Kwak M. K., Wakabayashi N., Greenlaw J. L., Yamamoto M., Kensler T. W. (2003) Mol. Cell. Biol. 23, 8786–8794 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Nguyen T., Yang C. S., Pickett C. B. (2004) Free Radic. Biol. Med. 37, 433–441 [DOI] [PubMed] [Google Scholar]
- 14.Wasserman W. W., Fahl W. E. (1997) Proc. Natl. Acad. Sci. U.S.A. 94, 5361–5366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Furukawa M., Xiong Y. (2005) Mol. Cell. Biol. 25, 162–171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Surh Y. J., Kundu J. K., Na H. K. (2008) Planta Med. 74, 1526–1539 [DOI] [PubMed] [Google Scholar]
- 17.Itoh K., Chiba T., Takahashi S., Ishii T., Igarashi K., Katoh Y., Oyake T., Hayashi N., Satoh K., Hatayama I., Yamamoto M., Nabeshima Y. (1997) Biochem. Biophys. Res. Commun. 236, 313–322 [DOI] [PubMed] [Google Scholar]
- 18.Venugopal R., Jaiswal A. K. (1998) Oncogene 17, 3145–3156 [DOI] [PubMed] [Google Scholar]
- 19.Wei P., Ahn Y. I., Housley P. R., Alam J., Vedeckis W. V. (1998) J. Steroid Biochem. Mol. Biol. 64, 1–12 [DOI] [PubMed] [Google Scholar]
- 20.Alam J., Stewart D., Touchard C., Boinapally S., Choi A. M., Cook J. L. (1999) J. Biol. Chem. 274, 26071–26078 [DOI] [PubMed] [Google Scholar]
- 21.Chondrogianni N., Petropoulos I., Franceschi C., Friguet B., Gonos E. S. (2000) Exp. Gerontol. 35, 721–728 [DOI] [PubMed] [Google Scholar]
- 22.Harlow E., Lane D. (1999) Using Antibodies: A Laboratory Manual, Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- 23.Moi P., Chan K., Asunis I., Cao A., Kan Y. W. (1994) Proc. Natl. Acad. Sci. U.S.A. 91, 9926–9930 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Dimri G. P., Lee X., Basile G., Acosta M., Scott G., Roskelley C., Medrano E. E., Linskens M., Rubelj I., Pereira-Smith O., et al. (1995) Proc. Natl. Acad. Sci. U.S.A. 92, 9363–9367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Eggler A. L., Gay K. A., Mesecar A. D. (2008) Mol. Nutr. Food. Res. 52, Suppl. 1, S84–S94 [DOI] [PubMed] [Google Scholar]
- 26.Kobayashi M., Li L., Iwamoto N., Nakajima-Takagi Y., Kaneko H., Nakayama Y., Eguchi M., Wada Y., Kumagai Y., Yamamoto M. (2009) Mol. Cell. Biol. 29, 493–502 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kensler T. W., Wakabayashi N., Biswal S. (2007) Annu. Rev. Pharmacol. Toxicol. 47, 89–116 [DOI] [PubMed] [Google Scholar]
- 28.Shih P. H., Yen G. C. (2007) Biogerontology 8, 71–80 [DOI] [PubMed] [Google Scholar]
- 29.Przybysz A. J., Choe K. P., Roberts L. J., Strange K. (2009) Mech. Ageing Dev. 130, 357–369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Suzuki M., Betsuyaku T., Ito Y., Nagai K., Nasuhara Y., Kaga K., Kondo S., Nishimura M. (2008) Am. J. Respir. Cell Mol. Biol. 39, 673–682 [DOI] [PubMed] [Google Scholar]
- 31.Dröge W., Schipper H. M. (2007) Aging Cell 6, 361–370 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Suh J. H., Shenvi S. V., Dixon B. M., Liu H., Jaiswal A. K., Liu R. M., Hagen T. M. (2004) Proc. Natl. Acad. Sci. U.S.A. 101, 3381–3386 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kwak M. K., Kensler T. W. (2006) Biochem. Biophys. Res. Commun. 345, 1350–1357 [DOI] [PubMed] [Google Scholar]
- 34.Arlt A., Bauer I., Schafmayer C., Tepel J., Muerkoster S. S., Brosch M., Roder C., Kalthoff H., Hampe J., Moyer M. P., Folsch U. R., Schafer H. (2009) Oncogene 28, 3983–3996 [DOI] [PubMed] [Google Scholar]
- 35.Lau A., Villeneuve N. F., Sun Z., Wong P. K., Zhang D. D. (2008) Pharmacol. Res. 58, 262–270 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Thimmulappa R. K., Fuchs R. J., Malhotra D., Scollick C., Traore K., Bream J. H., Trush M. A., Liby K. T., Sporn M. B., Kensler T. W., Biswal S. (2007) Antioxid. Redox. Signal. 9, 1963–1970 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Dinkova-Kostova A. T., Liby K. T., Stephenson K. K., Holtzclaw W. D., Gao X., Suh N., Williams C., Risingsong R., Honda T., Gribble G. W., Sporn M. B., Talalay P. (2005) Proc. Natl. Acad. Sci. U.S.A. 102, 4584–4589 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kwak M. K., Itoh K., Yamamoto M., Kensler T. W. (2002) Mol. Cell. Biol. 22, 2883–2892 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tanigawa S., Fujii M., Hou D. X. (2007) Free Radic. Biol. Med. 42, 1690–1703 [DOI] [PubMed] [Google Scholar]
- 40.Kobayashi A., Kang M. I., Okawa H., Ohtsuji M., Zenke Y., Chiba T., Igarashi K., Yamamoto M. (2004) Mol. Cell. Biol. 24, 7130–7139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zhang D. D., Lo S. C., Sun Z., Habib G. M., Lieberman M. W., Hannink M. (2005) J. Biol. Chem. 280, 30091–30099 [DOI] [PubMed] [Google Scholar]
- 42.Dinkova-Kostova A. T., Holtzclaw W. D., Cole R. N., Itoh K., Wakabayashi N., Katoh Y., Yamamoto M., Talalay P. (2002) Proc. Natl. Acad. Sci. U.S.A. 99, 11908–11913 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sakurai T., Kanayama M., Shibata T., Itoh K., Kobayashi A., Yamamoto M., Uchida K. (2006) Chem. Res. Toxicol. 19, 1196–1204 [DOI] [PubMed] [Google Scholar]
- 44.Hong F., Freeman M. L., Liebler D. C. (2005) Chem. Res. Toxicol. 18, 1917–1926 [DOI] [PubMed] [Google Scholar]
- 45.Hong F., Sekhar K. R., Freeman M. L., Liebler D. C. (2005) J. Biol. Chem. 280, 31768–31775 [DOI] [PubMed] [Google Scholar]
- 46.Zhang D. D. (2006) Drug Metab. Rev. 38, 769–789 [DOI] [PubMed] [Google Scholar]
- 47.Itoh K., Wakabayashi N., Katoh Y., Ishii T., Igarashi K., Engel J. D., Yamamoto M. (1999) Genes Dev. 13, 76–86 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Fahey J. W., Haristoy X., Dolan P. M., Kensler T. W., Scholtus I., Stephenson K. K., Talalay P., Lozniewski A. (2002) Proc. Natl. Acad. Sci. U.S.A. 99, 7610–7615 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kraft A. D., Johnson D. A., Johnson J. A. (2004) J. Neurosci. 24, 1101–1112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kwak M. K., Cho J. M., Huang B., Shin S., Kensler T. W. (2007) Free Radic. Biol. Med. 43, 809–817 [DOI] [PubMed] [Google Scholar]
- 51.Orr W. C., Radyuk S. N., Prabhudesai L., Toroser D., Benes J. J., Luchak J. M., Mockett R. J., Rebrin I., Hubbard J. G., Sohal R. S. (2005) J. Biol. Chem. 280, 37331–37338 [DOI] [PubMed] [Google Scholar]
- 52.Dugan L. L., Quick K. L. (2005) Sci. Aging Knowledge Environ. 2005, pe20. [DOI] [PubMed] [Google Scholar]
- 53.Katsiki M., Chondrogianni N., Chinou I., Rivett A. J., Gonos E. S. (2007) Rejuvenation Res. 10, 157–172 [DOI] [PubMed] [Google Scholar]
- 54.Dhawan V., Jain S. (2004) Mol. Cell. Biochem. 266, 109–115 [DOI] [PubMed] [Google Scholar]
- 55.Chen Q., Thorpe J., Dohmen J. R., Li F., Keller J. N. (2006) Free Radic. Biol. Med. 40, 120–126 [DOI] [PubMed] [Google Scholar]
- 56.Bishop N. A., Guarente L. (2007) Nature 447, 545–549 [DOI] [PubMed] [Google Scholar]
- 57.Pearson K. J., Lewis K. N., Price N. L., Chang J. W., Perez E., Cascajo M. V., Tamashiro K. L., Poosala S., Csiszar A., Ungvari Z., Kensler T. W., Yamamoto M., Egan J. M., Longo D. L., Ingram D. K., Navas P., de Cabo R. (2008) Proc. Natl. Acad. Sci. U.S.A. 105, 2325–2330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Plisko A., Gilchrest B. A. (1983) J. Gerontol. 38, 513–518 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.