Abstract
O6-Alkylguanine-DNA alkyltransferase (AGT) plays a major role in repair of the cytotoxic and mutagenic lesion O6-methylguanine (m6G) in DNA. Unlike the Escherichia coli alkyltransferase Ogt that also repairs O4-methylthymine (m4T) efficiently, the human AGT (hAGT) acts poorly on m4T. Here we made several hAGT mutants in which residues near the cysteine acceptor site were replaced by corresponding residues from Ogt to investigate the basis for the inefficiency of hAGT in repair of m4T. Construct hAGT-03 (where hAGT sequence -V149CSSGAVGN157- was replaced with the corresponding Ogt -I143GRNGTMTG151-) exhibited enhanced m4T repair activity in vitro compared with hAGT. Three AGT proteins (hAGT, hAGT-03, and Ogt) exhibited similar protection from killing by N-methyl-N′-nitro-N-nitrosoguanidine and caused a reduction in m6G-induced G:C to A:T mutations in both nucleotide excision repair (NER)-proficient and -deficient Escherichia coli strains that lack endogenous AGTs. hAGT-03 resembled Ogt in totally reducing the m4T-induced T:A to C:G mutations in NER-proficient and -deficient strains. Surprisingly, wild type hAGT expression caused a significant but incomplete decrease in NER-deficient strains but a slight increase in T:A to C:G mutation frequency in NER-proficient strains. The T:A to C:G mutations due to O4-alkylthymine formed by ethylating and propylating agents were also efficiently reduced by either hAGT-03 or Ogt, whereas hAGT had little effect irrespective of NER status. These results show that specific alterations in the hAGT active site facilitate efficient recognition and repair of O4-alkylthymines and reveal damage-dependent interactions of base and nucleotide excision repair.
Keywords: Cancer, DNA/Damage, DNA/Enzymes, DNA/Repair, Mutagenesis Mechanisms, Nucleic Acid/Enzymology, Alkylation Damage
Introduction
Alkylating agents generate adducts at multiple sites in DNA. Among those, adducts at the O6-position of guanine and the O4-position of thymine have been shown to induce mutations and initiate the carcinogenic process (1–7). These adducts can be repaired by O6-alkylguanine-DNA alkyltransferase (AGT).3 All of the known AGTs can repair DNA adducts at guanine-O6, but the size of alkyl group greatly affects the efficiency of lesion repair (see Refs. 8–10). The human AGT (hAGT) has a larger substrate-binding pocket than the Escherichia coli Ada AGT (11, 12) and can rapidly repair some bulky adducts, such as benzyl. Ada can repair O6-methylguanine (m6G) effectively but is virtually inactive in repair of bulky O6-adducts (13, 14). The second AGT present in E. coli, Ogt, is slightly better than Ada in repair of the larger O6-alkyl adducts but is still much less efficient with bulky adducts than hAGT (14–16).
AGTs are also known to be able to repair alkyl adducts at the O4-position of thymine (m4T). Such adducts are formed at lower levels than those at guanine-O6, but some studies showed that m4T is a more potent inducer of mutations than m6G (6, 7, 17–19). The ability to repair m4T by AGTs varies widely. In in vitro assays, Ogt is highly proficient in this repair, whereas Ada is less effective (13, 15, 20), and hAGT has even less activity (21, 22). Reports of repair of these DNA adducts in vivo are inconsistent. In one study, expression of hAGT in E. coli actually retarded the removal of m4T by nucleotide excision repair (NER) (22). In contrast, expression of the AGTs from Drosophila melanogaster, mice, and rats was found to reduce the amount of m4T and incidence of T:A to C:G transition mutations caused by m4T in E. coli or mammalian cells (23, 24), and inactivation of rat AGT slowed the repair of m4T in rat liver (25).
At present, there is limited information on the features that allow efficient repair of m4T by AGTs. The repair of m6G is relatively well understood because structures of both Ada (11) and hAGT (12) are available as well as hAGT complexes with oligodeoxyribonucleotides containing m6G and other substrate analogs (26–28). These structures and biochemical studies support a model in which hAGT binds substrate DNA via the minor groove using a helix-turn-helix motif and the guanine deoxyribonucleotide substrate is flipped out from the base stack into the hAGT active site pocket via a 3′-phosphate rotation, which is promoted by the aromatic ring side chain of Tyr114 and stabilized by an Arg finger (Arg128). This rearrangement positions the adduct for repair by placing the alkyl group in close proximity to the Cys145 acceptor site. The cysteine residue has very high reactivity (29) due to its activation to a thiolate anion by a Glu172-His146-water-Cys145 hydrogen bond network (12, 27). In some species, the transfer reaction is facilitated by reduction of the negative charge on the repaired guanine via a hydrogen bond from Tyr114 to the guanine-N1. The description given above (and amino acid numbering) relates to the hAGT, but it is likely to be universally applicable because all of the key residues mentioned above are fully conserved except for Tyr114, which is Phe in some species. It seems very likely that repair of m4T can occur in a similar way and that the key difference leading to species specificity in this repair is in the positioning of the thymine adduct. However, currently, there are no structures available of either AGTs that are known to repair m4T well or other AGTs bound to m4T substrates that can be used to test this hypothesis.
Several explanations for the poor repair of m4T have been proposed. It is possible that, based on crystal structures of hAGT bound to a modified cytosine base (28), the interaction of Tyr114 is with the O2 atom rather than N3 of thymine, and this interaction may reduce the repair efficiency of m4T (28). Also, it is possible that the pairing of m4T with adenine in DNA causes larger local curvature, interfering with the repair of m4T (30). However, neither of these possibilities would explain the species specificity of m4T repair. A major factor appears likely to reside in the ability of amino acid residues in the protein to position accurately the target m4T in close enough proximity to the reactive cysteine acceptor site. Support for this concept was provided by studies from Encell and Loeb (31), who showed that some hAGT mutants obtained in a screen for resistance to inhibition by O6-benzylguanine (BG) repaired m4T at a faster rate. The most effective mutant showed a rate of repair ∼11 times greater and had eight substitutions near the active site. Its expression in E. coli produced a ∼4-fold reduction in T:A to C:G mutations caused by m4T, whereas the wild type hAGT produced only a 1.1-fold reduction (32).
The NER system can also repair alkyl adducts at O6-guanine and O4-thymine in in vitro assays, although recognition of non-bulky adducts is relatively poor (33, 34). Evidence for such repair in vivo has been published (35). However, it was reported that repair of O4-ethylthymine (e4T) in human cells was slow and not affected by NER or AGT status (36, 37), and the relative roles of NER and AGTs and their potential interactions in repair of alkyl adducts are unclear.
To address these questions regarding hAGT and Ogt DNA repair activities, we have constructed several hAGT mutants with inserts based on equivalent regions in Ogt and show that substitutions of a few residues in the active site pocket can significantly improve the repair of O4-alkylthymine in vitro and in vivo. The results suggest a unified understanding of repair activities by this protein family. The relative abilities of NER and AGT to repair methyl, ethyl, and propyl adducts were also examined. Remarkably, the repair of m4T (but not its ethyl- and propyl- analogs) was actually inhibited when both NER and hAGT were active, revealing damage-dependent pathway interactions.
EXPERIMENTAL PROCEDURES
Materials
Oligodeoxyribonucleotides and all of the primers were synthesized and purified at the Macromolecular Core Facility, Hershey Medical Center. E. coli XL1-Blue bacterial cells, Pfu Turbo hot start DNA polymerase, Pfu polymerase, and QuikChange site-directed mutagenesis kit were purchased from Stratagene (La Jolla, CA). DNA isolation kits, the pQE-30 plasmid, and penta-His antibody were obtained from Qiagen (Chatsworth, CA). All restriction enzymes were obtained from New England Biolabs (Ipswich, MA). Ampicillin, kanamycin, tetracycline, chloramphenicol, isopropyl β-d-thiogalactopyranoside, hemocyanin, calf thymus DNA, MNNG, ENNG, PNNG, phenylmethylsulfonyl fluoride, and most other biochemical reagents were purchased from Sigma. Talon metal affinity IMAC resin was obtained from BD Biosciences Clontech (Palo Alto, CA). The Rapid DNA ligation kit and urea were purchased from Roche Applied Science. Nitrocellulose filters (0.45 μm) were obtained from Millipore (Temecula, CA). The inhibitor BG was synthesized as described (38) and generously provided by Dr. G. Pauly (ABL-Basic Research program, NCI-Frederick Cancer Research and Development Center, Frederick, MD). [γ-35S]ATPγS (10mCi/ml) was obtained from ICN Biomedicals Inc. (Irvine, CA). Sephadex MicroSpin G-25 columns were purchased from Amersham Biosciences. Horseradish peroxidase-linked anti-mouse IgG and anti-rabbit IgG were purchased from Cell Signaling Technology (Danvers, MA).
Bacterial Strains and Media
The bacterial strains FC326, FC218, CJM1, and CJM2, in which the endogenous AGT genes Ada and Ogt were deleted (22, 39), were generously provided by Dr. L. Samson (Biological Engineering Division and Center for Environmental Health Sciences, MIT, Cambridge, MA). FC326 and FC218 carry a reporter gene that can be reverted to the wild type through T:A to C:G and G:C to A:T transition mutations, respectively (40). CJM1 and CJM2 are derived from FC326 and FC218, respectively, but are deficient in NER due to inactivation of the uvrB gene. Cells were grown in M9 minimal medium and plated in M9 minimal plates that contained either 0.025% glucose or lactose, 40 μg/ml methionine, 0.025% thiamine, 1 mm MgSO4, 0.1 mm CaCl2, and antibiotics (75 μg/ml ampicillin, 40 μg/ml kanamycin, 25 μg/ml tetracycline (CJM1 and CJM2 only)) as described (22, 39).
Construction of pQE Plasmids for Expression of Different AGT Mutants
Different hAGT mutants were constructed in which some or all of the residues around the Cys145 acceptor site were changed to those found in Ogt (see Table 1). Plasmids for the preparation of these mutants were constructed from previously described plasmids for hAGT expression, which have either an N-terminal His6 tag replacing the terminal M- with the sequence MRGS(H)6GS- or C-terminal His6-tag replacing the last 6 residues (41, 42). Mutants were constructed using PCR carried out with Pfu polymerase under conditions described previously (43). All plasmid constructs were verified by sequencing.
TABLE 1.
Sequences of the AGT mutants used
| Mutant | Sequence |
|---|---|
| Sequences from residues 137–172 in hAGT (active site pocket)a | |
| hAGT | -NPVPILIPCHRVVCSSGAVGNYSGGLAVKEWLLAHE- |
| Ogt | -NPISIVVPCHRVIGRNGTMTGYAGGVQRKEWLLRHE- |
| hAGT-01 | -NPISIVVPCHRVIGRNGTMTGYAGGVQRKEWLLAHE- |
| hAGT-02 | -NPVPILIPCHRVIGRNGTMTGYAGGVQRKEWLLAHE- |
| hAGT-03 | -NPVPILIPCHRVIGRNGTMTGYSGGLAVKEWLLAHE- |
| hAGT-04 | -NPVPILIPCHRVIGRNGAVGNYSGGLAVKEWLLAHE- |
| hAGT-05 | -NPVPILIPCHRVVCSSGTMTGYSGGLAVKEWLLAHE- |
| hAGT-06 | -NPVPILIPCHRVVCRNGTMGNYSGGLAVKEWLLAHE- |
| hAGT-07 | -NPVPILIPCHRVIGRNGAVGGYSGGLAVKEWLLAHE- |
| hAGT-08 | -NPVPILIPCHRVVCSNGTVTNYSGGLAVKEWLLAHE- |
| hAGT-(C145S) | -NPVPILIPSHRVVCSSGAVGNYSGGLAVKEWLLAHE- |
| hAGT-03-(C145S) | -NPVPILIPSHRVIGRNGTMTGYSGGLAVKEWLLAHE- |
| Sequences from residues 94–136 in hAGT (DNA binding domain) | |
| hAGT | -FTRQVLWKLLKVVKFGEVISYQQLAALAGNPKAARAVGGAMRG- |
| Ogt | -FQREV-WKTLRTIPCGQVMHYGQLAEQLGRPGAARAVGAANGS- |
| hAGT-(S113H) | -FTRQVLWKLLKVVKFGEVIHYQQLAALAGNPKAARAVGGAMRG- |
| Other mutants used | |
| hAGT-03-(S113H) | |
| hAGT-06-(S113H) | |
a Residues in hAGT that are changed to those in Ogt are shown underlined. The Cys145 acceptor site and its Ser145 mutant are shown in boldface type.
Creation of DraIII Restriction Site in Ogt Sequence Spanning Amino Acid Residues CHRV
The pQE30-Ogt plasmid construct described previously (14) was used as a template for PCR with the following primers: sense primer, 5′-d(CGTACCTTGCCACAGAGTGATTGGCCGAAAC)-3′; antisense primer, 5′-d(GTTTCGGCCAATCACTCTGTGGCAAGGTACG)-3′. The nucleotides that are different from the original Ogt sequence are underlined. The Ogt coding sequence upstream of the intended DraIII site (product 1) was amplified with the above antisense primer and 5′-d(CACACAGAATTCATTAAAG)-3′ (primer A). The Ogt coding sequence downstream of the intended DraIII site (product 2) was amplified with the above sense primer and 5′-d(AATTAAGCTTGGCTGCAGG)-3′ (primer B). The full-length Ogt sequence was then generated by PCR by adding primers A and B to the mixture of the purified products 1 and 2. The product was digested with BamHI and KpnI restriction enzymes and ligated into pQE30-Ogt plasmid digested with the same enzymes to generate pQE30-Ogt with a DraIII site in the Ogt coding sequence.
Construction of pQE-hAGT-01 and hAGT-02 Plasmids
Sense primer 5′-d(GCAGTGGGAGGCGCCATGAGAGGCAATCCCATCAGC)-3′ (sequence for the NarI site is in italic type, and the sequence matching Ogt is underlined) and antisense primer 5′-d(TTCCCCAACCGGTGGCCTTCATGGGCCAGAAGCCATTCTTTTCGCTGAACTCC)-3′ (sequence for the AgeI site is in italic type, and the sequence matching Ogt is underlined) were used to produce hAGT-01. The nucleotide sequence in the primers that is not underlined corresponds to the hAGT sequence. The Ogt insert was amplified with the above set of primers from the pQE30-Ogt-(DraIII) plasmid template. The purified PCR product containing the Ogt sequence with flanking hAGT sequence was digested with NarI and AgeI and ligated into pQE30-hAGT plasmid digested with the same enzymes.
Sense primer 5′-d(CCTTGCCACAGAGTGATTG)-3′ (sequence for the DraIII site is in italic type, and the sequence matching Ogt is underlined) and antisense primer 5′-d(TTCCCCAACCGGTGGCCTTCATGGGCCAGAAGCCATTCTTTTCGCTGAACTCC)-3′ (sequence for the AgeI site is in italic type and the sequence matching Ogt is underlined) were used to produce hAGT-02. The nucleotide sequence in the primer that is not underlined is the hAGT sequence. The Ogt insert was amplified with the above primers on pQE30-Ogt-(DraIII) plasmid template. The purified PCR product containing Ogt sequence with flanking hAGT sequence was digested with DraIII and AgeI restriction enzymes and ligated into pQE30-hAGT plasmid digested with the same enzymes.
Construction of pQE-hAGT-03 to hAGT-08 Plasmids
The antisense primer 5′-d(GGATCTATCAACAGGAGTCC)-3′ was used with the sense primers 5′-d(CCGTGCCACAGAGTGATCGGCCGAAACGGAACCATGACCGGATACTCCGGAGGGCTA)-3′, 5′-d(CCGTGCCACAGAGTGATCGGCCGAAACGGAGCCGTGGGCAAC)-3′, 5′-d(CCGTGCCACAGAGTGGTCTGCAGCAGCGGAACCATGACCGGCTACTCCGGAGGGCTA)-3′, 5′-d(CCGTGCCACAGAGTGGTCTGCCGAAACGGCACCATGGGCAACTACTCCGGA)-3′, 5′-d(CCGTGCCACAGAGTGATCGGCCGAAACGGAGCCGTGGGCGGCTACTCCGGAGGGCTA)-3′, and 5′-d(CCGTGCCACAGAGTGGTCTGCAGCAACGGAACCGTGACCAACTACTCCGGAGGG)-3′ (underlined sequences correspond to Ogt) to produce hAGT-03 to hAGT-08, respectively. The PCR products were digested with DraIII and KpnI, and the fragments containing the desired sequences were ligated into a pQE30-hAGT vector digested with the same enzymes. The resulting plasmids were transformed into E. coli XL1-Blue cells.
Construction of Plasmids for the Production of Mutants at Ser113 and Cys145
The plasmids for production of hAGT-(S113H), hAGT-03-(S113H), and hAGT-06-(S113H) were constructed from the template plasmids of hAGT, hAGT-03, and hAGT-06, respectively, with the QuikChange site-directed mutagenesis kit according to the manufacturer's instructions. Primers 5′-d(GGAGAAGTGATTCATTACCAGCAATTGGCC)-3′ and 5′-d(GGCCAATTGCTGGTAATGAATCACTTCTCC)-3′ (mismatches underlined) were used to construct the S113H point mutation.
Plasmids for the purification of hAGT-(C145S) and hAGT-03-(C145S) proteins were constructed similarly. The templates were hAGT and hAGT-03, and the primers were 5′-d(CCCATCCTCATCCCGAGCCACAGAGTGGTC)-3′ and 5′-d(GACCACTCTGTGGCTCGGGATGAGGATGGG)-3′ (mismatches underlined).
Protein Purification
The XL1-Blue cells containing plasmids encoding Ogt, hAGT, or mutant hAGTs were cultured for protein expression and purification using immobilized metal affinity chromatography essentially as previously described (43). Proteins hAGT, Ogt, hAGT-(S113H), and hAGT-(C145S) have an N-terminal His6 tag, and all hAGT mutants have a C-terminal His6 tag, which was used for the purification. Previous studies have shown that the tag has a minimal effect on the AGT activity (41–43). Because most of the mutant proteins were insoluble under native conditions, they were solubilized and purified in denaturing conditions in the presence of 8 m urea and refolded by sequential dialysis to remove the urea with buffer containing 50 μm Zn2+. The purification was carried out exactly as described (43) except that the proteins were not mixed with calf thymus DNA during the renaturation process. The purified proteins were analyzed by PAGE in the presence of SDS on 15% gels.
Alkyltransferase Activity Assays
Assays were conducted using double-stranded 11-mer-m6G/C (5′-d(GCGCm6GAAGTCG)-3′ paired with 3′-d(CGCGCTTCAGC)-5′), 11-mer-m4T-1/A (5′-d(GCGCAm4TAGTCG)-3′ paired with 3′-d(CGCGTATCAGC)-5′) or 11-mer-m4T-2/A (5′-d(CGTGAm4TCTGCG)-3′ paired with 3′-d(GCACTAGACGC)-5′).
In order to measure repair of m6G, 125 pmol of 11-mer-m6G/C in 15 μl of water was mixed with 200 pmol of the AGT protein in 15 μl of 50 mm Tris-HCl, pH 7.6, 5 mm dithiothreitol (DTT), and 0.1 mm EDTA. The reactions were performed in a RQF-3 rapid quench apparatus (KinTek Corp., Austin, TX). The incubation time varied from 1 to 16 s at 21 °C, and the reaction was stopped by the addition of 2% SDS.
To detect the repair activity of m4T for hAGT and mutant hAGTs, 125 pmol of 11-mer-m4T-1/A or 11-mer-m4T-2/A was mixed with 1000 pmol of AGT in 0.1 ml of 50 mm Tris-HCl, pH 7.6, 5 mm DTT, and 0.1 mm EDTA at 21 °C for 30 s to 2 h. The reaction was stopped by the addition of 2% SDS. Because the rate of m4T repair by Ogt is very fast, the reactions for Ogt were also carried out in the RQF-3 rapid quench for 1–16 s with 200 pmol of Ogt. In order to determine the amount of product, the oligonucleotides were separated by high performance liquid chromatography (HPLC) on a Beckman reverse-phase C-18 column at 40 °C using a flow rate of 1 ml/min and a gradient of 10–40% methanol for 60 min (43).
Inactivation of Alkyltransferase Activity by BG
Aliquots of hAGT-03, Ogt, or hAGT were incubated with different concentrations of BG at 37 °C for 30 min in 0.5 ml of 50 mm Tris-HCl, pH 7.6, 100 μg/ml hemocyanin, 2.5 μg/ml calf thymus DNA, 5 mm DTT, and 0.1 mm EDTA. The residual AGT activity was then determined, and the ED50 value representing the amount of inhibitor needed to produce a 50% loss of activity was calculated as described previously (41).
Mutagenesis Assays
E. coli strains FC326, FC218, CJM1, and CJM2 were transformed with plasmids pQE30, pQE30-hAGT, pQE30-Ogt, and pQE30-hAGT-03. The cells were grown overnight at 37 °C in 1 ml of M9 minimal medium containing appropriate antibiotics, and aliquots of 240 or 380 μl (for cells containing pQE30-hAGT-03) were added to 5 ml of fresh M9 minimal medium. After growth for 4 h or until A600 reached 0.6–0.8, FC326 and FC218 cells were treated with 0, 2.5, 5, and 10 μg/ml MNNG, ENNG, or PNNG for 40 min at 37 °C. CJM1 and CJM2 cells were treated with 0, 0.1, 0.5, 1, 2.5, 5 μg/ml MNNG, ENNG, or PNNG for the same time. Cells were collected by centrifugation at 5000 rpm for 10 min and washed and resuspended in M9 minimal medium. Cells were diluted 1–1,000,000-fold and then spread on M9 minimal plates containing either glucose (for measurements of survival) or lactose (for determination of mutation frequency). The plates were incubated for 72 h at 37 °C, and colonies were scored. The mutation frequency was expressed as the number of lacZ revertants/107 survivors (22). In order to study the effect of BG on the protection against MNNG-induced cytotoxicity and mutagenesis, these cell lines were treated with 100 μm BG for 1 h before treatment with different concentrations of MNNG.
Binding of AGTs to DNA
The association constant for the binding of the AGTs to oligonucleotides containing either m6G or m4T were determined using serial dilution analysis with electrophoretic mobility shift assays exactly as previously described (44, 45). A serial dilution method using a dilution coefficient of 0.8/step was used to obtain the monomeric association constant (Kmono) as described (44, 45). Briefly, the single-stranded 11-mer-m6G and 11-mer-m4T-1/A were 5′-end-labeled with [γ-35S]ATPγS and T4 polynucleotide kinase at 37 °C for 1 h, heated at 65 °C for 20 min, and passed through a Sephadex MicroSpin G-25 column to remove unincorporated [γ-35S]ATPγS. The labeled oligonucleotides were mixed with a 2-fold molar excess of reverse complement oligonucleotide at 100 °C and allowed to cool to room temperature slowly (46). Binding reactions were carried out in 10 mm Tris-HCl, pH 7.6, 1 mm DTT, 1 mm EDTA, and 10 μg/ml bovine serum albumin at 21 °C. Protein-DNA complexes were formed by adding appropriate amounts of hAGT-(C145S) or hAGT-03-(C145S) to solution containing 35S-labeled oligonucleotides. The mixtures were equilibrated at 21 °C for 45 min. PAGE was performed using 15% polyacrylamide gels (acrylamide/N,N′-methylene bisacrylamide ratio, 37.5:1) and run at 10 V/cm in buffer containing 90 mm Tris borate, 2 mm EDTA, pH 8.0, for 50 min. The gel was fixed, vacuum-dried, and developed with the Molecular Dynamics Storage Phosphor Screen for 24 h. The screen was quantified on a PhosphorImager using the program ImageQuant (Amersham Biosciences).
A binding competition assay was used to estimate the relative binding affinities. The distribution of AGT between reference and competitor DNAs depends on the equilibrium constant after the addition of competing DNA to a reference AGT-DNA complex, Kref/Kcomp = [PnD][C]/[D][PnC], where [D] is the free reference DNA concentration, [PnD] is the reference AGT-DNA complex concentration, [C] is the free competitor concentration, and [PnC] is the competitor AGT-DNA complex concentration (44). In the experiments, 11-mer-m4T-1/A served as reference DNA, and 11-mer-m6G/C or 11-mer-T/A served as competing DNA.
Determination of AGT Protein Expression
The expression levels of hAGT, Ogt, and hAGT-03 proteins in FC326, FC218, CJM1, and CJM2 cells were determined by Western blotting. Cells were grown in the same conditions as used for the mutagenesis assays. Aliquots of 10 ml of cell culture were centrifuged to collect cells. The cell pellets were suspended in 0.5 ml of buffer containing 50 mm Tris-HCl pH 7.6, 5 mm DTT, 0.1 mm EDTA, and 0.2 mm phenylmethylsulfonyl fluoride. Cells were sonicated, and cell lysate was quantified. Western blotting was carried out as described (47), using penta-His antibody as the primary antibody.
Generation of an hAGT-03-m4T-DNA Model
An initial model of hAGT-03 was built based on hAGT crystal structure Protein Data Bank entry 1t38, using the ESyPred3D Web server (48). Crystal structures of hAGT (Protein Data Bank entries 1yfh, 1t39, 1qnt, 1eh6, 1eh7, and 1eh8) were used as guides for Cys145 side chain placement, and hAGT-DNA complex crystal structures Protein Data Bank entries 1t38, 1t39, and 1yfh were used to model DNA. The initial model was subjected to conjugate gradient minimization, simulated annealing, and torsion angle dynamics by CNS (Crystallography and NMR System) (49), which employs molecular dynamics with CHARMM (Chemistry at Harvard Macromolecular Mechanics) (50, 51) force field parameters. One hundred steps of conjugate gradient minimization were followed by 40 steps of simulated annealing. Simulated annealing was carried out at a starting temperature of 1000 K and slow cooling with a drop in temperature of 25 K/cycle of dynamics. The model was validated using rigorous structure validation programs from the Joint Center for Structural Genomics Protein Structure Validation Suite and MolProbity. These programs, which are routinely used for checking the quality of models in theoretical studies, indicated that the model was acceptable and comparable with AGT crystal structures.
RESULTS
Repair of m4T in Vitro
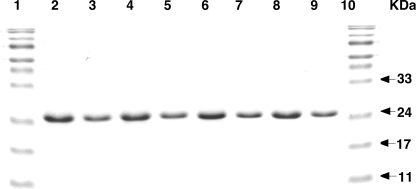
Various hAGT mutants in which some of the residues were replaced by the corresponding amino acids in Ogt (Table 1) were expressed in E. coli XL1-Blue cells for protein expression and purification. Because most of these hAGT mutants were insoluble, all of the proteins, including wild type hAGT were purified under denaturing conditions in 8 m urea and then refolded by dialysis against decreasing concentrations of urea in the presence of Zn2+, as described previously (43). All proteins purified under these denaturing conditions were soluble and obtained in pure form (Fig. 1). The wild type hAGT regains its full activity when purified under these conditions (43).
FIGURE 1.
Purity of AGT and hAGT mutants. The proteins were isolated under denaturing conditions, renatured, and analyzed by SDS-PAGE as described under “Experimental Procedures.” Lanes 1 and 10, molecular markers; lane 2–9, hAGT, hAGT-03, hAGT-02, hAGT-01, hAGT-05, hAGT-04, hAGT-03-(S113H), and hAGT-06, respectively. 10 μg of protein was loaded in lanes 2, 4, 6, and 8, and 5 μg of protein was loaded in lanes 3, 5, 7, and 9.
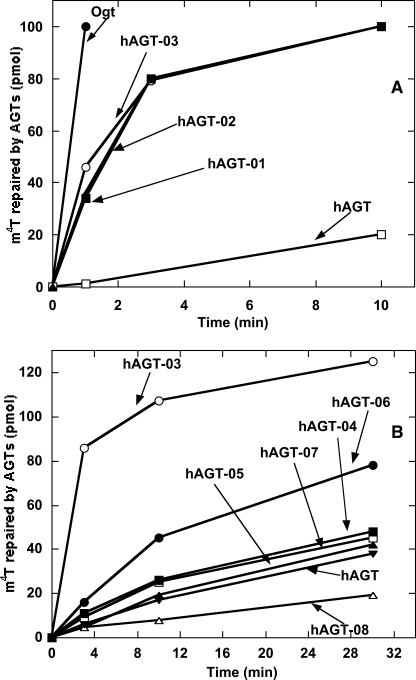
The purified proteins were assayed for their alkyltransferase activity on a double-stranded substrate 11-mer-m4T-1/A that contained a central m4T residue paired with A. The mutant proteins hAGT-01, hAGT-02, and hAGT-03, in which 16, 12, and 8 residues in the active site were altered respectively, showed major improvement in repair activity toward m4T compared with wild type hAGT (Fig. 2A). Since no significant differences in the rate of repair were observed for these three mutants, it can be concluded that the eight altered residues located in the Val149–Tyr158 region in the mutant hAGT-03 are critical for enhanced m4T repair. Attempts to narrow this region even further using mutants hAGT-04, hAGT-05, hAGT-06, hAGT-07, and hAGT-08, in which only subsets of these residues were varied, did not maintain the enhanced m4T repair, although the hAGT-06 mutant with four changes was better than hAGT (Fig. 2B). Therefore, hAGT-03 was chosen for the in vivo experiments.
FIGURE 2.
Repair of m4T by Ogt, hAGT, and hAGT mutants. Reactions were carried out using 11-mer-m4T-1/A as substrate, as described under “Experimental Procedures.” The products were separated by HPLC. A, the results for Ogt (●), hAGT-03 (○), hAGT-02 (■), hAGT-01 (▴), and hAGT (□). B, the results for hAGT-03 (○), hAGT-06 (●), hAGT-04 (■), hAGT-07 (□), hAGT-05 (▴), hAGT (▾), and hAGT-08 (▵).
We also examined the possibility that a change at residue 113 may influence the repair rate of hAGT. This residue is adjacent to the highly conserved residue Tyr114, which plays a key role in the alkyl transfer reaction by inducing rotation of the 3′-phosphate of the nucleotide substrate to promote the base-flipping process (12, 26). It was possible that replacement of the Ser113 of hAGT with His, the amino acid at this position in Ogt, might alter the flexibility and mobility of the side chain of Tyr114 in this base flipping. We therefore made the hAGT-(S113H), hAGT-03-(S113H), and hAGT-06-(S113H) mutants. However, the introduction of the S113H mutation caused only a slight change in m4T repair activity compared with hAGT (less than 2-fold) and gave about the same increase in the rate of m6G repair (results not shown).
Detailed kinetic studies of the hAGT-03 mutant and wild type hAGT and Ogt were carried out to determine the rate constant for repair of m6G or m4T contained in 11-mer double-stranded substrates. Assays were carried out with double-stranded 11-mer-m6G/C, 11-mer-m4T-1/A, or 11-mer-m4T-2/A. The two substrates containing m4T gave similar results. A RQF-3 rapid quench apparatus (KinTek Corp., Austin, TX) was used with 1–16 s assay times for the faster rates. Repair of m6G by hAGT-03 was ∼2-fold faster than hAGT but 3-fold slower than Ogt with a second order rate constant of 2.2 ± 0.4 × 107 m−1 min−1. Repair of m4T by hAGT-03 was increased about 47-fold over repair by wild type hAGT with a rate constant of 1.4 ± 0.5 × 105 m−1 min−1. This is still 92-fold slower than that for repair of m6G. Repair of m4T by Ogt had a rate constant of 1.3 ± 0.6 × 107 m−1 min−1, which is only slightly less than the repair rate for m6G (6.6 ± 0.6 × 107 m−1 min−1).
It is hard to measure the binding affinity of the AGT proteins to DNA containing substrate lesions due to the alkyl transfer reaction, which requires no cofactors and is thus difficult to prevent. We therefore mutated the Cys acceptor site to Ser and made the hAGT-(C145S) and hAGT-03-(C145S) mutants, which are known to maintain DNA-binding properties but cannot repair DNA (45, 52). These mutants were used in competitive binding assays to test the relative binding affinities of the mutants to double-stranded 11-mer DNA containing either m6G or m4T. The relative affinities were 0.07 ± 0.05 for hAGT-(C145S) and 0.8 ± 0.4 for hAGT-03-(C145S) when the 11-mer containing m4T was used for initial binding and then competed off with m6G containing duplex oligonucleotide. The corresponding values when an 11-mer oligonucleotide of the same sequence but containing T rather than m4T was used as competitor in the binding assay were 33 ± 15 and 169 ± 56, respectively. These results demonstrate that the 8-amino acid alteration significantly enhanced the relative affinity for binding to DNA containing m4T. We also determined the binding affinity to double-stranded 11-mer DNA containing an m4T adduct using a serial dilution electrophoretic mobility shift assay as previously described (45, 53). The calculated monomeric association constant Kmono was increased about 10-fold from ∼1 ± 0.4 × 106 m−1 for C145S to ∼10 ± 0.3 × 106 m−1 for hAGT-03-C145S. These values compare with the previously published values for hAGT binding to unmodified DNA with a Kmono of ∼0.6 × 106 m−1 and binding to DNA containing an m6G adduct with Kmono of ∼3 × 106 m−1 (45, 53). It should be noted that due to the cooperative nature of the binding of hAGT to DNA and the fact that there is only a relatively small increase in the association constant when DNA adducts are present, it is unlikely that initial DNA binding is the limiting step in alkyl transfer, and kinetic measurements of the hAGT reaction with substrates containing m6G confirm that alkyl transfer is the limiting step.
Some AGTs, such as hAGT, are very sensitive to inactivation by BG, whereas others, including Ogt, are resistant to this inhibitor (54, 55). The hAGT-03 mutant was also found to be resistant to BG (ED50 of 65 μm compared with 44 μm for Ogt and 0.2 μm for wild type hAGT).
Modeling of an hAGT-03-m4T-DNA Structure
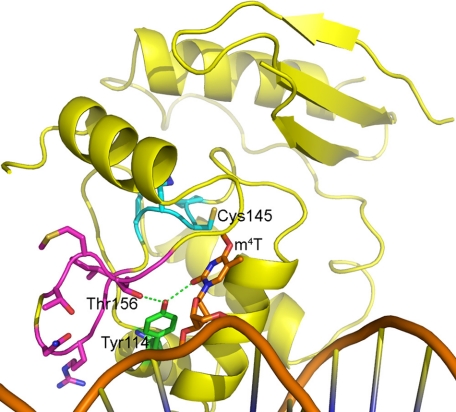
To determine how hAGT-03 might bind m4T and to test our hypothesis that m4T can be repaired similarly to m6G and that the species specificity of this repair results from the positioning of the thymine adduct, we made a model of hAGT-03 based on available AGT crystal structures. We then used AGT-DNA complex crystal structures to aid in modeling an m4T in the hAGT-03 active site. The finished model (Fig. 3) shows that many of the side chains of the mutated residues of hAGT-03 are too distant to directly interact with the m4T or with key residues Cys145, Tyr114, His146, or Glu172, with the exception that the Thr156 side chain (Gly in hAGT) could interact with the hydroxyl of Tyr114. However, the hAGT-03 mutations comprise a loop that is nearby the active site motif, both in the primary and the tertiary structure of the protein (Fig. 3). This loop in hAGT-03 has three glycines rather than two, as in hAGT, suggesting that it is more flexible in hAGT-03 than in hAGT. In addition, the Gly mutation at position 150 in hAGT-03 doubles the length of the flexible part of the loop from four residues (Gly-to-Gly) in hAGT to eight residues in hAGT-03, adding flexibility to the part of the loop that is closest to the DNA. The result of this added flexibility is likely to be significant in two ways. First, movement of this loop could help to shape the DNA slightly, allowing for better positioning of the m4T nearer the reactive cysteine acceptor site for repair. Second, a more flexible loop might allow for minor positional movements of active site motif residues to better optimize the distance between Cys145 and the methyl group of m4T, which is ∼1 Å further away in our model than the comparable Cys145-m6G methyl interaction derived from AGT crystal structures. Overall, this model supports the hypothesis that protein residues may aid in the correct positioning of the m4T for the methyl transfer reaction to occur. Coordinates for this model are included in the supplemental material.
FIGURE 3.
Model of hAGT-03 with m4T in the active site. Residues 149–157 are shown in magenta, the active site motif is shown in cyan, and the DNA is shown in orange.
Repair of O4-Methylthymine in Vivo
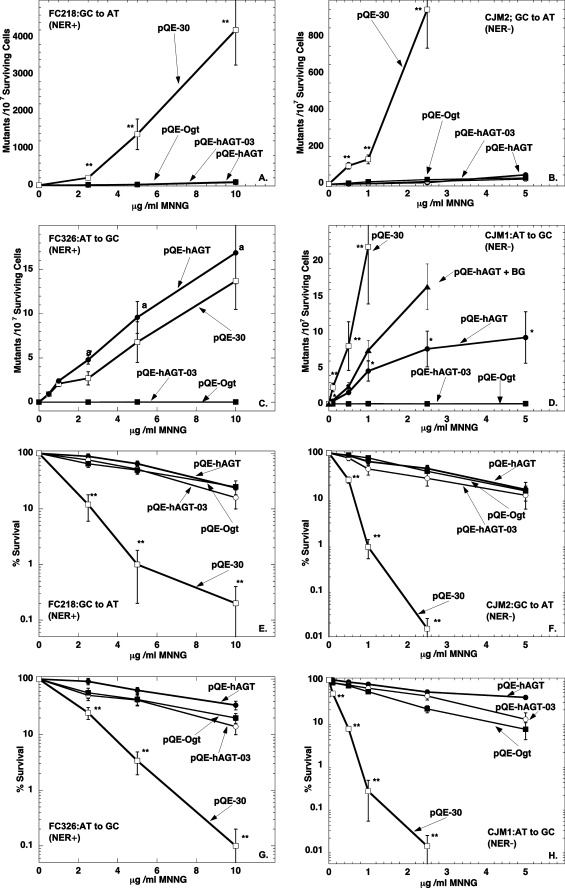
The ability of the wild type hAGT, hAGT-03, and Ogt to protect E. coli strains lacking endogenous AGTs from cytotoxicity and mutagenesis after exposure to methylation damage by MNNG was examined. All of the AGTs were expressed at similar levels (supplemental Fig. S1). Comparison was made in strains with wild type NER (Fig. 4, A, C, E, and G) and lacking NER (Fig. 4, B, D, F, and H). These results are summarized in Table 2.
FIGURE 4.
Ability of AGTs to protect from MNNG-induced cytotoxicity and mutagenicity in E. coli strains proficient and deficient in NER. Results are shown for cells with plasmids pQE-hAGT (●), pQE-hAGT-03 (○), pQE-Ogt (■), and pQE-30(□) in FC 218 cells (A and E), CJM2 cells (B and F), FC326 cells (C and G), and CJM1 cells (D and H), respectively. Results are shown for mutations in A–D and for survival in E–H. The effect of adding BG to cells expressing pQE-hAGT is also shown in D (▴). Experiments were performed as described under “Experimental Procedures.” Results are shown mean ± S.D. for at least three separate experiments. **, p < 0.01 compared with cells expressing hAGT, Ogt, or hAGT-03; *, p < 0.01 compared with cells expressing no AGT, Ogt, or hAGT-03; a, p < 0.01 compared with cells expressing Ogt or hAGT-03.
TABLE 2.
Summary of protection of E. coli cells from mutations and killing by MNNG, ENNG, and PNNG
| Agent | NER status | AGT expressed | ED20a | Mutations (percentage of “no AGT” valueb) |
|
|---|---|---|---|---|---|
| G:C to A:T | T:A to C:G | ||||
| μg/ml | % | % | |||
| MNNG | NER− | None | 0.4 | 100 | 100 |
| MNNG | NER− | hAGT | 4.5 | 3 | 19 |
| MNNG | NER− | Ogt | 3.4 | 6 | <2 |
| MNNG | NER− | hAGT-03 | 3.7 | 5 | <2 |
| ENNG | NER− | None | 0.6 | 100 | 100 |
| ENNG | NER− | hAGT | 4.6 | 2 | 123 |
| ENNG | NER− | Ogt | 2.7 | 4 | 3 |
| ENNG | NER− | hAGT-03 | 2.9 | 3 | 3 |
| PNNG | NER− | None | 0.6 | 100 | 100 |
| PNNG | NER− | hAGT | >5 | 7 | 62 |
| PNNG | NER− | Ogt | 5 | 9 | <2 |
| PNNG | NER− | hAGT-03 | >5 | 3 | <2 |
| MNNG | NER+ | None | 2.4 | 100c | 100c |
| MNNG | NER+ | hAGT | >10 | 2 | 125 |
| MNNG | NER+ | Ogt | 10 | 4 | <2 |
| MNNG | NER+ | hAGT-03 | 8.4 | 4 | <2 |
| ENNG | NER+ | None | 2.2 | 100d | 100d |
| ENNG | NER+ | hAGT | 9 | 16 | 89 |
| ENNG | NER+ | Ogt | 10 | 21 | 2 |
| ENNG | NER+ | hAGT-03 | 19 | 18 | 2 |
| PNNG | NER+ | None | 3.3 | 100e | 100e |
| PNNG | NER+ | hAGT | 10 | 53 | 83 |
| PNNG | NER+ | Ogt | 10 | 60 | <2 |
| PNNG | NER+ | hAGT-03 | 10 | 25 | <2 |
a There was no significant difference in survival between strains having different reporter genes but the same NER status (e.g. FC326 and FC218). Therefore, results for the survival are given as the mean of the two values. Results that are outside the range of doses tested are given as >5 or >10.
b The mean of the results obtained with each dose of the alkylating agent shown in Fig. 4 and in supplemental Figs. 2 and 3 was expressed as a percentage of the mutations obtained in the cells lacking AGT.
c In the absence of any AGT expression, the presence of NER reduced the MNNG-derived G:C to A:T mutations by 24% and the total A:T to C:G mutations by 22%.
d In the absence of any AGT expression, the presence of NER reduced the ENNG-derived G:C to A:T mutations by 79% and increased the A:T to C:G mutations insignificantly ∼1.2-fold.
e In the absence of any AGT expression, the presence of NER reduced the PNNG-derived G:C to A:T mutations by 92% and the total A:T to C:G mutations by 95%.
As shown in Fig. 4, A and B, all three AGTs tested prevented the formation of mutations caused by m6G and protected the cells from the cytotoxic effects of MNNG (Fig. 4, E–H). These results are consistent with the well established concept that the presence of m6G is a powerful inducer of cell death (56, 57). In the absence of AGT expression, NER was also able to provide partial protection against G:C to A:T mutations and MNNG cytotoxicity but was much less effective than AGT.
Expression of Ogt or hAGT-03 completely prevented the mutations caused by m4T, whereas wild type hAGT was only weakly active in the strain lacking NER (Fig. 4D) and inhibitory in the strain possessing NER (Fig. 4C). These results show that hAGT is very poorly active in repair of m4T but that the minor alterations in the protein sequence converting hAGT into hAGT-03 are sufficient to allow efficient repair of m4T. The increase in T:A to C:G transitions caused by m4T when wild type hAGT is expressed in cells with intact NER is in excellent agreement with previous reports by Samson et al. (22) that NER can repair m4T and that hAGT actually interferes with this process (22). However it is clear from the results with the cells lacking NER that hAGT does have the ability to provide some repair of m4T. Although this protective effect was quite modest compared with the robust protection provided by Ogt or hAGT-03, it is clearly due to repair of m4T because it was reduced when the cells were treated with the inhibitor BG (Fig. 4D). This inhibitor had no effect on the complete suppression of these mutations by hAGT-03 or Ogt, which, as described above, are much less effectively inhibited by BG.
Repair of O4-Ethylthymine and O4-Propylthymine in Vivo
Similar studies were carried out using ENNG and PNNG to examine the ability of the wild type hAGT, hAGT-03, and Ogt to protect from ethylation or propylation damage. Detailed graphs showing these responses are provided as supplemental material, and the results are summarized in Table 2. All three AGTs tested efficiently protected cells from mutations caused by O6-ethylguanine and O6-propylguanine. NER was also able to repair these DNA adducts. Expression of wild type hAGT had no effect on the incidence of T:A to C:G mutations, showing that this protein does not repair e4T and O4-propylthymine (p4T). However, expression of Ogt or the hAGT-03 mutant completely abolished these mutations. Thus, the active site alteration in hAGT-03 allows not only efficient repair of m4T but also the repair of e4T and p4T. NER was also able to repair p4T but was ineffective on e4T. However, NER did not interfere with repair of e4T and p4T by Ogt or hAGT-03.
DISCUSSION
Adducts at the exo-O atoms of guanine and thymine are of critical importance in the induction of mutations by N-nitroso- carcinogens (1, 58, 59). Although O4-alkylthymine is a minor adduct produced by alkylating agents and is formed in lower amounts compared with O6-alkylguanine, it is a highly persistent and mutagenic form of DNA damage (19, 36, 60, 61). Direct comparisons of mutagenicity in the absence of AGT-mediated repair in human and bacterial cells show that m4T is even more mutagenic than m6G (6, 7). Therefore, the ability to repair O4-alkylthymine is potentially an important mechanism in protection from the carcinogenic effects of nitrosamines and other alkylating agents that form such adducts.
There are contradictory reports of the ability of hAGT to repair m4T, although most suggest that hAGT activity for this substrate is much less than that of some microbial AGTs. Our studies with purified hAGT in in vitro assays and when expressed in E. coli cells confirm that m4T can be repaired by hAGT. These results also fully support previous observations that this repair is so slow that it may have little physiological significance. Our present studies carried out in the NER-deficient strain show clearly that hAGT does have a limited but measurable effect in repair of m4T. Mutations at T:A sites were reduced compared with cells transfected with the empty vector or cells in which the hAGT was inactivated with BG (see Fig. 4D). It is clear that these mutations are due to m4T because expression of Ogt or the hAGT-03 mutant abolished them (Table 2). When ENNG or PNNG were used as the alkylating agent, the protective effect of hAGT was not observed, although Ogt or the hAGT-03 mutant abolished these mutations. Thus, hAGT has a very poor ability to repair m4T in vivo even when expressed at high levels and no activity for repair of e4T or p4T. This is consistent with reports that there was little repair of e4T by hAGT in human cells treated with N-nitroso-N-ethylurea, whereas O6-ethylguanine was repaired (36, 62).
All characterized AGTs are highly active in the repair of m6G. They are also able to repair larger adducts at the O6-position of guanine and O4-thymine adducts, but such repair shows wide variation with species. The best substrate for hAGT is actually DNA containing O6-benzyldeoxyguanosine, whereas AGTs from yeast, E. coli, and many other microorganisms are virtually inactive with this substrate (14, 54). This is explained by the comparison of the crystal structures of the E. coli Ada-C and hAGT, which show that steric factors prevent acceptance of the benzyl group in the Ada-C active site pocket (12, 63), and by kinetic measurements with hAGT, which show that the benzyl group is more readily transferred than methyl (64). There is also striking variation in the ability to repair m4T by different AGTs, but there was no useful structural data to explain this because neither Ada-C nor hAGT repairs this product well and the only other available AGT structures are for proteins whose specificity in this respect has not been established. Our results provide strong evidence that changes of specific residues in the binding pocket have a profound effect on the ability to bind to and repair m4T. The substitution of eight residues (-VCSSGAVGN-) in the region 149–158 in hAGT with the corresponding sequence (-IGRNGTMTG-) from Ogt to form hAGT-03 allowed efficient repair of m4T, e4T, and p4T (Table 2). This change also increased the binding of the mutant hAGT-03 to m4T in DNA. It is likely that these changes not only improve binding of substrates containing m4T but also correctly position the alkyl group of O4-alkylthymine to allow attack by the highly reactive anion of Cys145. Our model (Fig. 3) furthermore suggests that the increased flexibility in the loop in the active site is an important factor in allowing the correct positioning.
A previous study observed that a mutant of hAGT selected in a screen for resistance to BG showed an increased ability to repair m4T (31, 32). This mutant had eight changes in the hAGT active site region from residue 150 to 170. It did not provide as high a level of protection from MNNG-induced T:A to C:G transitions as hAGT-03 and was somewhat impaired in repair of m6G. However, it also supports the concept that minor changes in the active site can allow recognition and repair of m4T. None of the amino acid changes that were reported for this BG-resistant mutant (31, 32) are the same as hAGT-03, although five changes are in the same region from 149 to 158.
Interestingly, a considerable enhancement of the rate of repair of m4T by hAGT can be achieved without loss of the capacity to repair m6G. This suggests an absence of strong evolutionary pressure to develop the activity toward m4T and that environmental exposure to this form of DNA damage is relatively insignificant. The alternative possibility that m4T is repaired well by some other process seems unlikely, because m4T is highly persistent in mammalian DNA after exposure to nitrosamines and 1,2-dimethylhydrazine (25, 65, 66). Another explanation might be that the changes allowing m4T repair render the hAGT protein less able to repair bulky adducts, such as benzyl or pyridyloxobutyl, and that these adducts have greater environmental significance. Although we did not examine the ability of the hAGT-03 mutant to repair such adducts in DNA directly, the fact that it is highly resistant to inactivation by the pseudosubstrate BG free base provides good evidence that it does have an impaired ability to deal with such bulky adducts. Our previous studies have shown an excellent correlation in reaction of hAGT mutants with BG as a free base and the ability to repair O6-benzyldeoxyguanosine present in DNA (14, 46). As mentioned above, other BG-resistant hAGT mutants have also been reported to be more effective in repair of m4T (31, 32).
Mutants of hAGT resistant to BG are under active investigation for use in gene therapy and gene expression studies in which an alkylating agent plus BG is used to select for cells transduced with vectors expressing such AGTs (67–69). Most of these studies are being carried out using the P140K hAGT mutant, but this mutation does not enhance m4T repair, and the hAGT-03 would be another viable candidate.
Our results and those previously published (22, 35) show that NER can repair m4T in E. coli. Our experiments show that p4T can also be repaired by NER but that NER had little effect on e4T repair, where there was virtually no difference in mutations between the two strains in the absence of AGT expression (Table 2). These results differ from those seen for the repair by NER of O6-alkyl adducts, where the efficiency of repair of these adducts by NER increases with the size of the adduct. This could be explained by an increased distortion of the DNA leading to better recognition of the larger adducts. Notably, recent studies have indicated that repair of some small alkyl adducts, such as O6-alkylguanine, by NER is facilitated by the binding of a ATL (alkyltransferase-like protein) (70, 71). This small protein has a structure similar to that of the DNA binding domain/active site of AGT and binds to DNA in a similar way although causing a greater bending of the DNA (71). ATL binds to the components of the NER, and it suppresses mutations caused by MNNG, PNNG, and hydroxyethylating agents, such as N-(2-hydroxyethyl)-N-nitrosourea (70, 71).4 All of the strains used in our studies contain ATL, and although it has not yet been established that ATL can interact with O4-alkylthymine, it is possible that unproductive competition between these pathways accounts for inhibition of m4T repair by the combination of NER and hAGT.
In general, despite many structural analyses of DNA base repair enzymes and their DNA interactions, questions still remain regarding the molecular basis for repair specificity and the existence of possible pathway interactions (72–74). The results presented here help establish the basis for differential repair specificity by hAGT and Ogt by specific amino acids in the active site pocket. These results test and extend the proposal from the uracil-DNA glycosylase-DNA complex, which identified flipping of the nucleotide and damaged base, that interactions within the active site pocket provide the damaged base specificity (75). Our new results furthermore reveal an unexpected damage-dependent interaction of base and nucleotide excision repair pathways that impact repair efficiency for alkylated base damage. Taken together, these results thus broaden our understanding of damaged base recognition and repair for DNA alkylation damage of biological and medical importance.
Supplementary Material
Acknowledgments
We thank Drs. Natalia A. Loktionova, Douglas S. Daniels, and Thomas E. Spratt for suggestions and for help with the experiments.
This work was supported, in whole or in part, by National Institutes of Health United States Public Health Service Grants R01-CA018137, CA-097209, and CA-071976.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1–S3 and supplemental material.
S. Kanugula and A. E. Pegg, unpublished observations.
- AGT
- O6-alkylguanine-DNA alkyltransferase
- hAGT
- human wild type AGT
- m6G
- O6-methylguanine
- m4T
- O4-methylthymine
- NER
- nucleotide excision repair
- BG
- O6-benzylguanine
- e4T
- O4-ethylthymine
- MNNG
- N-methyl-N′-nitro-N-nitrosoguanidine
- ENNG
- N-ethyl-N′-nitro-N-nitrosoguanidine
- PNNG
- N-propyl-N′-nitro-N-nitrosoguanidine
- HPLC
- high performance liquid chromatography
- DTT
- dithiothreitol
- p4T
- O4-propylthymine
- ATPγS
- adenosine 5′-O-thiotriphosphate.
REFERENCES
- 1.Lawley P. D. (1976) in Chemical Carcinogens (Searle C. E. ed) Vol. 173, pp. 83–244, American Chemical Society, Washington, D. C. [Google Scholar]
- 2.Loechler E. L., Green C. L., Essigmann J. M. (1984) Proc. Natl. Acad. Sci. U.S.A. 81, 6271–6275 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Preston B. D., Singer B., Loeb L. A. (1986) Proc. Natl. Acad. Sci. U.S.A. 83, 8501–8505 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Basu A. K., Essigmann J. M. (1990) Mutat. Res. 233, 189–201 [DOI] [PubMed] [Google Scholar]
- 5.Pauly G. T., Hughes S. H., Moschel R. C. (1995) Biochemistry 34, 8924–8930 [DOI] [PubMed] [Google Scholar]
- 6.Pauly G. T., Hughes S. H., Moschel R. C. (1998) Carcinogenesis 19, 457–461 [DOI] [PubMed] [Google Scholar]
- 7.Pauly G. T., Moschel R. C. (2001) Chem. Res. Toxicol. 14, 894–900 [DOI] [PubMed] [Google Scholar]
- 8.Pegg A. E., Dolan M. E., Moschel R. C. (1995) Prog. Nucleic Acid Res. Mol. Biol. 51, 167–223 [DOI] [PubMed] [Google Scholar]
- 9.Margison G. P., Santibáñez-Koref M. F. (2002) BioEssays 24, 255–266 [DOI] [PubMed] [Google Scholar]
- 10.Pegg A. E. (2000) Mutat. Res. 462, 83–100 [DOI] [PubMed] [Google Scholar]
- 11.Wibley J. E., Pegg A. E., Moody P. C. (2000) Nucleic Acids Res. 28, 393–401 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Daniels D. S., Mol C. D., Arvai A. S., Kanugula S., Pegg A. E., Tainer J. A. (2000) EMBO J. 19, 1719–1730 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Graves R. J., Li B. F., Swann P. F. (1989) Carcinogenesis 10, 661–666 [DOI] [PubMed] [Google Scholar]
- 14.Goodtzova K., Kanugula S., Edara S., Pauly G. T., Moschel R. C., Pegg A. E. (1997) J. Biol. Chem. 272, 8332–8339 [DOI] [PubMed] [Google Scholar]
- 15.Harris L. C., Margison G. P. (1993) Br. J. Cancer 67, 1196–1202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wilkinson M. C., Potter P. M., Cawkwell L., Georgiadis P., Patel D., Swann P. F., Margison G. P. (1989) Nucleic Acids Res. 17, 8475–8484 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dosanjh M. K., Essigmann J. M., Goodman M. F., Singer B. (1990) Biochemistry 29, 4698–4703 [DOI] [PubMed] [Google Scholar]
- 18.Singer B., Dosanjh M. K. (1990) Mutat. Res. 233, 45–51 [DOI] [PubMed] [Google Scholar]
- 19.Preston B. D., Singer B., Loeb L. A. (1987) J. Biol. Chem. 262, 13821–13827 [PubMed] [Google Scholar]
- 20.Sassanfar M., Dosanjh M. K., Essigmann J. M., Samson L. (1991) J. Biol. Chem. 266, 2767–2771 [PubMed] [Google Scholar]
- 21.Paalman S. R., Noll D. M., Clarke N. D. (1997) Nucleic Acids Res. 25, 1795–1801 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Samson L., Han S., Marquis J. C., Rasmussen L. J. (1997) Carcinogenesis 18, 919–924 [DOI] [PubMed] [Google Scholar]
- 23.Kooistra R., Zonneveld J. B., Watson A. J., Margison G. P., Lohman P. H., Pastink A. (1999) Nucleic Acids Res. 27, 1795–1801 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kawate H., Ihara K., Kohda K., Sakumi K., Sekiguchi M. (1995) Carcinogenesis 16, 1595–1602 [DOI] [PubMed] [Google Scholar]
- 25.O'Toole S. M., Pegg A. E., Swenberg J. A. (1993) Cancer Res. 53, 3895–3898 [PubMed] [Google Scholar]
- 26.Daniels D. S., Woo T. T., Luu K. X., Noll D. M., Clarke N. D., Pegg A. E., Tainer J. A. (2004) Nat. Struct. Mol. Biol. 11, 714–720 [DOI] [PubMed] [Google Scholar]
- 27.Tubbs J. L., Pegg A. E., Tainer J. A. (2007) DNA Repair 6, 1100–1115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Duguid E. M., Rice P. A., He C. (2005) J. Mol. Biol. 350, 657–666 [DOI] [PubMed] [Google Scholar]
- 29.Guengerich F. P., Fang Q., Liu L., Hachey D. L., Pegg A. E. (2003) Biochemistry 42, 10965–10970 [DOI] [PubMed] [Google Scholar]
- 30.Cruzeiro-Hansson L., Goodfellow J. M. (1994) Carcinogenesis 15, 1525–1533 [DOI] [PubMed] [Google Scholar]
- 31.Encell L. P., Loeb L. A. (1999) Biochemistry 38, 12097–12103 [DOI] [PubMed] [Google Scholar]
- 32.Encell L. P., Loeb L. A. (2000) Carcinogenesis 21, 1397–1402 [PubMed] [Google Scholar]
- 33.Voigt J. M., Van Houten B., Sancar A., Topal M. D. (1989) J. Biol. Chem. 264, 5172–5176 [PubMed] [Google Scholar]
- 34.Sancar A. (1996) Annu. Rev. Biochem. 65, 43–81 [DOI] [PubMed] [Google Scholar]
- 35.Samson L., Thomale J., Rajewsky M. F. (1988) EMBO J. 7, 2261–2267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bronstein S. M., Skopek T. R., Swenberg J. A. (1992) Cancer Res. 52, 2008–2011 [PubMed] [Google Scholar]
- 37.Engelbergs J., Thomale J., Galhoff A., Rajewsky M. F. (1998) Proc. Natl. Acad. Sci. U.S.A. 95, 1635–1640 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Dolan M. E., Moschel R. C., Pegg A. E. (1990) Proc. Natl. Acad. Sci. U.S.A. 87, 5368–5372 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mackay W. J., Han S., Samson L. D. (1994) J. Bacteriol. 176, 3224–3230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Cupples C. G., Miller J. H. (1989) Proc. Natl. Acad. Sci. U.S.A. 86, 5345–5349 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Fang Q., Loktionova N. A., Moschel R. C., Javanmard S., Pauly G. T., Pegg A. E. (2008) Biochem. Pharmacol. 75, 618–626 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Liu L., Xu-Welliver M., Kanugula S., Pegg A. E. (2002) Cancer Res. 62, 3037–3043 [PubMed] [Google Scholar]
- 43.Fang Q., Kanugula S., Pegg A. E. (2005) Biochemistry 44, 15396–15405 [DOI] [PubMed] [Google Scholar]
- 44.Fried M. G., Kanugula S., Bromberg J. L., Pegg A. E. (1996) Biochemistry 35, 15295–15301 [DOI] [PubMed] [Google Scholar]
- 45.Rasimas J. J., Pegg A. E., Fried M. G. (2003) J. Biol. Chem. 278, 7973–7980 [DOI] [PubMed] [Google Scholar]
- 46.Luu K. X., Kanugula S., Pegg A. E., Pauly G. T., Moschel R. C. (2002) Biochemistry 41, 8689–8697 [DOI] [PubMed] [Google Scholar]
- 47.Fang Q., Noronha A. M., Murphy S. P., Wilds C. J., Tubbs J. L., Tainer J. A., Chowdhury G., Guengerich F. P., Pegg A. E. (2008) Biochemistry 47, 10892–10903 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lambert C., Léonard N., De Bolle X., Depiereux E. (2002) Bioinformatics 18, 1250–1256 [DOI] [PubMed] [Google Scholar]
- 49.Brünger A. T., Adams P. D., Clore G. M., DeLano W. L., Gros P., Grosse-Kunstleve R. W., Jiang J. S., Kuszewski J., Nilges M., Pannu N. S., Read R. J., Rice L. M., Simonson T., Warren G. L. (1998) Acta Crystallogr. D Biol. Crystallogr. 54, 905–921 [DOI] [PubMed] [Google Scholar]
- 50.Brooks B. R., Bruccoleri R. E., Olafson B. D., States D. J., Swaminathan S., Karplus M. (1983) J. Comp. Chem. 4, 187–217 [Google Scholar]
- 51.MacKerell A. D., Brooks B. P., Brooks I. C. L., Nilsson L., Roux B., Won Y., Karplus M. (1998) in The Encyclopedia of Computational Chemistry (Schleyer P. V. R., Allinger N. L., Clark T., Gasteiger J., Kollaman P. A., Schaefer H. F., Schreiner P. R. eds) pp. 271–277, John Wiley & Sons, Chichester, UK [Google Scholar]
- 52.Melikishvili M., Rasimas J. J., Pegg A. E., Fried M. G. (2008) Biochemistry 47, 13754–13763 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Rasimas J. J., Kar S. R., Pegg A. E., Fried M. G. (2007) J. Biol. Chem. 282, 3357–3366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Pegg A. E., Boosalis M., Samson L., Moschel R. C., Byers T. L., Swenn K., Dolan M. E. (1993) Biochemistry 32, 11998–12006 [DOI] [PubMed] [Google Scholar]
- 55.Elder R. H., Margison G. P., Rafferty J. A. (1994) Biochem. J. 298, 231–235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Karran P., Bignami M. (1994) BioEssays 16, 833–839 [DOI] [PubMed] [Google Scholar]
- 57.Kaina B., Ochs K., Grösch S., Fritz G., Lips J., Tomicic M., Dunkern T., Christmann M. (2001) Prog. Nucleic Acid Res. Mol. Biol. 68, 41–54 [DOI] [PubMed] [Google Scholar]
- 58.Pegg A. E. (1984) Cancer Invest. 2, 223–231 [DOI] [PubMed] [Google Scholar]
- 59.Saffhill R., Margison G. P., O'Connor P. J. (1985) Biochim. Biophys. Acta 823, 111–145 [DOI] [PubMed] [Google Scholar]
- 60.Dosanjh M. K., Singer B., Essigmann J. M. (1991) Biochemistry 30, 7027–7033 [DOI] [PubMed] [Google Scholar]
- 61.Singer B., Essigmann J. M. (1991) Carcinogenesis 12, 949–955 [DOI] [PubMed] [Google Scholar]
- 62.Bronstein S. M., Cochrane J. E., Craft T. R., Swenberg J. A., Skopek T. R. (1991) Cancer Res. 51, 5188–5197 [PubMed] [Google Scholar]
- 63.Moore M. H., Gulbis J. M., Dodson E. J., Demple B., Moody P. C. (1994) EMBO J. 13, 1495–1501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Zang H., Fang Q., Pegg A. E., Guengerich F. P. (2005) J. Biol. Chem. 280, 30873–30881 [DOI] [PubMed] [Google Scholar]
- 65.Den Engelse L., Menkveld G. J., De Brij R. J., Tates A. D. (1986) Carcinogensis 7, 393–403 [DOI] [PubMed] [Google Scholar]
- 66.Devereux T. R., Anderson M. W., Belinsky S. A. (1991) Carcinogenesis 12, 299–303 [DOI] [PubMed] [Google Scholar]
- 67.Milsom M. D., Jerabek-Willemsen M., Harris C. E., Schambach A., Broun E., Bailey J., Jansen M., Schleimer D., Nattamai K., Wilhelm J., Watson A., Geiger H., Margison G. P., Moritz T., Baum C., Thomale J., Williams D. A. (2008) Cancer Res. 68, 6171–6180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Reese J. S., Roth J. C., Gerson S. L. (2008) Stem Cells 26, 675–681 [DOI] [PubMed] [Google Scholar]
- 69.Zhao H., Pestina T. I., Nasimuzzaman M., Mehta P., Hargrove P. W., Persons D. A. (2009) Blood 113, 5747–5756 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Mazon G., Philippin G., Cadet J., Gasparutto D., Fuchs R. P. (2009) DNA Repair 8, 697–703 [DOI] [PubMed] [Google Scholar]
- 71.Tubbs J. L., Butt A., Kanugula S., Melikishvili M., Marriott A., Watson A. J., Verbeek B., Santibanez-Koref M. F., Kraehenbuehl R., Fleck O., Millington C., Arvai A. S., Kroeger M. D., Peterson L. A., Williams D. M., Fried M. G., Margison G. P., Pegg A. E., Tainer J. A. (2009) Nature 458, 808–813 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Mol C. D., Parikh S. S., Putnam C. D., Lo T. P., Tainer J. A. (1999) Annu. Rev. Biophys. Biomol. Struct. 28, 101–128 [DOI] [PubMed] [Google Scholar]
- 73.Huffman J. L., Sundheim O., Tainer J. A. (2005) Mutat. Res. 577, 55–76 [DOI] [PubMed] [Google Scholar]
- 74.Hitomi K., Iwai S., Tainer J. A. (2007) DNA Repair 6, 410–428 [DOI] [PubMed] [Google Scholar]
- 75.Slupphaug G., Mol C. D., Kavli B., Arvai A. S., Krokan H. E., Tainer J. A. (1996) Nature 384, 87–92 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.