Abstract
Idiopathic pulmonary fibrosis (IPF) is a poorly understood progressive disease characterized by the accumulation of scar tissue in the lung interstitium. A hallmark of the disease is areas of injury to type II alveolar epithelial cells with attendant accumulation of fibroblasts in areas called fibroblastic foci. In an effort to better characterize the lung fibroblast phenotype in IPF patients, we isolated fibroblasts from patients with IPF and looked for activation of signaling proteins, which could help explain the exaggerated fibrogenic response in IPF. We found that IPF fibroblasts constitutively expressed increased basal levels of SPARC, plasminogen activator inhibitor-1 (PAI-1), and active β-catenin compared with control cells. Control of basal PAI-1 expression in IPF fibroblasts was regulated by SPARC-mediated activation of Akt, leading to inhibition of glycogen synthase kinase-3β and activation of β-catenin. Additionally, IPF fibroblasts (but not control fibroblasts) were resistant to plasminogen-induced apoptosis and were sensitized to plasminogen-mediated apoptosis by inhibition of SPARC or β-catenin. These findings uncover a newly discovered regulatory pathway in IPF fibroblasts that is characterized by elevated SPARC, giving rise to activated β-catenin, which regulates expression of downstream genes, such as PAI-1, and confers an apoptosis-resistant phenotype. Disruption of this pathway may represent a novel therapeutic target in IPF.
Keywords: Cell/Apoptosis, Extracellular Matrix, Organisms/Mammal, Tissue/Organ Systems/Lung, Signal Transduction, Secreted Protein Acidic and Rich in Cysteine, β-Catenin, Idiopathic Pulmonary Fibrosis, Plasminogen Activator Inhibitor 1
Introduction
Idiopathic pulmonary fibrosis (IPF)2 is a progressive and fatal lung disease of unknown cause. Current estimates of disease incidence are 40–50 per 100,000 and ∼125,000 cases in the United States. Most patients are 50–70 years old, but patients with familial IPF tend to present earlier. Patients are usually symptomatic for 6–24 months before diagnosis but often present with advanced fibrotic disease. Despite therapy, IPF has a median survival of 4–5 years (1). Most of the current therapy targeted at IPF is anti-inflammatory. These treatments have yielded few durable responses. Their failure is due in great part to the unique pathogenesis of IPF. Our understanding of the pathogenesis of IPF is evolving, with more recent evidence suggesting a process of alveolar epithelial injury, possibly ongoing, and dysregulated repair, leading to proliferation of myofibroblasts and fibrotic scarring in the lung.
Myofibroblasts in IPF are thought to arise, at least in part, from differentiation of fibroblasts in response to transforming growth factor-β (TGF-β). They are characterized by expression of α-smooth muscle actin (α-SMA) and synthesis of matrix-modifying factors such as collagen and fibronectin. The proliferation of myofibroblasts is a critical step in the generation of fibrotic scarring in IPF (2). This process is very similar to what occurs during normal wound healing. Myofibroblasts are responsible for pulling on a wound in order for its edges to approximate and close. They also lay down the extracellular matrix (ECM) that is part of the wound scar. Once wound repair is complete, myofibroblasts normally undergo apoptosis, and other cells in the vicinity break down the ECM. In IPF, however, apoptosis of myofibroblasts is impaired, and they maintain an environment that is non-degradative, thereby preserving and extending the ECM. A critical regulator of fibroblast differentiation into myofibroblasts and the genesis of matrix components is isoform 1 of TGF-β, a powerful mitogen that is secreted by type II alveolar epithelial cells (AEC), macrophages, platelets, and myofibroblasts themselves in response to injury (3).
TGF-β regulates multiple signaling pathways that coordinate cellular responses to injury, tissue repair, and fibrosis. Recent studies have elucidated that TGF-β activates Wnt/β-catenin. The Wnt/β-catenin pathway has been shown to organize diverse regulatory pathways during development, cell growth and differentiation, tissue remodeling, and tumorigenesis (4). The Wnt/β-catenin or canonical Wnt signaling pathway is characterized by the nuclear accumulation of β-catenin, which forms a complex with members of the T cell factor/lymphoid enhancer factor-1 family of transcription factors (4). Many of these genes are involved in matrix remodeling and fibrogenesis, such as SPARC (secreted protein acidic and rich in cysteine); matrix metalloproteinase-2, -3, and -9; cyclin D1; matrilysin; and fibronectin. In recent studies, TGF-β has been shown to induce rapid nuclear translocation of β-catenin in mesenchymal stem cells in a Smad-3-dependent manner (5). Also, a recent study showed that TGF-β activates the β-catenin pathway in lung fibroblasts through inhibition of glycogen synthase kinase-3β (GSK-3β) (6). In the lung, a Gata6-Wnt/β-catenin pathway was recently shown to be required for epithelial stem cell development and airway regeneration (7). Recent studies also point to a role for the Wnt/β-catenin pathway in pulmonary fibrosis (8). Chilosi et al. (9) reported the accumulation of nuclear β-catenin in damaged type II AEC and myofibroblasts in fibroblastic foci in IPF lung but not in other idiopathic interstitial pneumonias such as nonspecific interstitial pneumonia. A more recent study revealed activation of the Wnt/β-catenin pathway in type II AEC from IPF patients, which appeared to play a role in mediating epithelial injury and hyperplasia (8). Furthermore, it was suggested that activation of Wnt signaling in adjacent lung mesenchyme may prevent the proper differentiation of the alveolar epithelium (10).
SPARC, a matricellular protein that regulates tissue repair and wound healing, is a known target of TGF-β (reviewed in Ref. 36). It is known to accumulate in myofibroblasts in fibroblastic foci in IPF (11). It also plays a role in the assembly of fibrillar collagen in the ECM (44). Recently, a study by Nie and Sage (12) demonstrated that SPARC induces the accumulation and activation of β-catenin in preadipocytes, leading to an enhanced association of β-catenin with T cell factor/lymphoid enhancer factor and inhibition of adipogenesis. They showed that integrin-linked kinase (ILK), but not Akt, is required for SPARC activation of β-catenin and that LiCl mimics the effects of SPARC. They also revealed that SPARC regulates expression of α5- and α6-integrins through β-catenin. Their laboratory also recently showed that SPARC mediates cell survival through its interaction with β1-integrin and activation of ILK (45).
We were interested in identifying genes and signaling pathways in lung fibroblasts from IPF patients that contribute to the fibrogenic phenotype through promoting ECM deposition and/or inhibition of apoptosis. Recently published supporting studies reveal that TGF-β induces protection from serum starvation-mediated apoptosis through the p38 MAPK (mitogen-activated protein kinase) and phosphoinositide 3-kinase (PI3K)/Akt pathways (13, 14). Also, endothelin-1 and TGF-β appear to independently protect lung fibroblasts from apoptosis via these pathways (13, 14). The same laboratory also showed that TGF-β-mediated induction of plasminogen activator inhibitor-1 (PAI-1) protects lung fibroblasts from plasminogen-induced apoptosis (15). Several previous studies show that PAI-1 promotes fibrosis in lung and other tissues and that plasminogen activation is anti-fibrogenic (16, 17). Additionally, PAI-1 has been shown to impair alveolar epithelial repair through binding to vitronectin, and TGF-β requires PAI-1 for its cytostatic effect on epithelial cells (18, 19).
To further characterize the interplay between these matrix-modifying pathways in IPF fibroblasts, we isolated fibroblasts from IPF patients and used fibroblasts isolated from patients undergoing resection for lung cancer as our control. Like others previously, we observed increased expression of total α-SMA in IPF fibroblast samples versus control fibroblasts, which is consistent with an increase in the number of myofibroblasts in IPF, as has been observed in IPF. However, there was significantly more heterogeneity in the level of total α-SMA expression in IPF fibroblasts versus control fibroblasts. We report, for the first time, that IPF fibroblasts constitutively express significantly more SPARC and nuclear, i.e. active, β-catenin than control fibroblasts. Because the resistance of IPF fibroblasts to normal apoptotic signals may play a role in disease pathogenesis, we looked for targets of SPARC/β-catenin, which may mediate this protection. Because PAI-1 has been shown to mediate apoptotic resistance in lung fibroblasts, we compared the levels of PAI-1 in control fibroblasts versus IPF fibroblasts and found significantly higher basal PAI-1 expression in IPF fibroblasts. Also, IPF fibroblasts were significantly more resistant than control fibroblasts to plasminogen-induced apoptosis. Finally, we show that SPARC mediates activation of β-catenin through activation of Akt, leading to inhibition of GSK-3β. Also, this pathway regulates PAI-1 expression in IPF fibroblasts. These data describe, for the first time, a constitutive signaling pathway regulated by SPARC/β-catenin in IPF fibroblasts that leads to increased basal expression of PAI-1, which mediates resistance to plasminogen-induced apoptosis. This may contribute to both impairment of epithelial repair and fibrosis in IPF.
EXPERIMENTAL PROCEDURES
Cell Culture and Reagents
Lung tissue was obtained from patients undergoing surgical biopsy for the diagnosis of interstitial lung disease or lung transplant for IPF, and non-neoplastic tissue was obtained from patients undergoing surgical lung cancer resection. The tissue was minced into small pieces with a scalpel and digested with type I collagenase (1 mg/ml; Invitrogen) and hyaluronidase (125 units/ml; Sigma) at 37 °C with agitation for 18 h in Dulbecco's modified Eagle's medium (DMEM; Invitrogen) supplemented with 10% fetal bovine serum. The dissociated tissues were incubated without shaking for 5 min at room temperature, followed by the separation of cell-enriched supernatant into a new tube. The cell fraction was centrifuged at 250 × g for 5 min, and the pellet was then resuspended in DMEM with 10% fetal bovine serum. Epithelial cells did not, in general, survive more than one passage and were in large part eliminated through trypsinization. Surviving fibroblasts were cultured in DMEM supplemented with 10% fetal bovine serum at 5% CO2 at 37 °C. Each fibroblast culture was frozen at its earliest available passage and was used for studies for up to five passages. For each experiment, cells were plated in culture vessels in DMEM and cultured until 70–80% confluent, unless indicated otherwise. Cells were subjected to starvation by washing cells twice with 1× phosphate-buffered saline (PBS), followed by the addition of DMEM and 0.1% serum to each well and incubation for an additional 24 h. In the experiments for cell death induced by Glu-plasminogen (American Diagnostica Inc., Stamford, CT), phenol red-free DMEM was used.
Western Blot Analysis
Western blot analysis and band intensity quantitation were performed as described previously (20). Briefly, the protein concentration was measured by Bradford assay (Bio-Rad) according to the manufacturer's instructions. An equal amount of protein was separated by SDS-PAGE and transferred to polyvinylidene difluoride membrane. Immunoblotting was performed using primary antibodies for α-SMA (American Research Products, Inc., Belmont, MA); SPARC (Biodesign International, Saco, ME); cleaved caspase-3 (Asp175), phospho-Akt (Ser473), phospho-GSK-3β (Ser9), and GSK-3β (Cell Signaling Technology, Danvers, MA); active β-catenin (8E7; Millipore, Billerica, MA); total β-catenin (BD Biosciences); PAI-1 (Santa Cruz Biotechnology, Santa Cruz, CA); and α-tubulin (used as a loading control; Sigma) overnight at 4 °C, followed by incubation with the appropriate horseradish peroxidase-conjugated secondary antibody (GE Healthcare). The blot was visualized by enhanced chemiluminescence (GE Healthcare) and analyzed using a Kodak Image Station 4000R system (Carestream Health, Rochester, NY).
To detect secreted PAI-1, the culture medium was first cleared by centrifugation, and proteins were precipitated in the presence of ammonium sulfate at 50% saturation overnight at 4 °C with gentle agitation. The excess salts were removed by dialysis against lysis buffer used for total lysate preparation, and protein quantitation was performed by the Bradford assay as described above. To suppress endogenous PI3K activity, 10 μm LY293002 or 1 μm wortmannin (Sigma) was used, and dimethyl sulfoxide (Sigma) was used as a control.
Immunofluorescent Staining and Nuclear Localization of β-Catenin
Fibroblasts were grown on 2-well chamber slides (Thermo Fisher Scientific) in complete culture medium until 50–60% confluent. Cells were subjected to starvation (0.1% serum) for 48 h. After washing with 1× PBS, cells were treated with 100% methanol at −20 °C for 5 min. Cells were then blocked in 1× PBS and 3% normal goat serum (Sigma) for 30 min at room temperature, followed by incubation with anti-active β-catenin at 1:100 dilution in blocking solution for 16 h at 4 °C. The target proteins were visualized with fluorescein isothiocyanate-conjugated secondary antibody (Calbiochem), and 4′,6-diamidino-2-phenylindole (Molecular Probes, Eugene, OR) was used for nuclear counterstaining. Fluorescent images were taken and processed using a Labophot-2 microscope equipped with an episcopic fluorescence attachment (EFD-3; Nikon Instruments Inc., Melville, NY). For the quantitation of nuclear localized β-catenin, at least total 100 cells were randomly selected from five fields of each stained sample to obtain the percentage of nuclear localization of β-catenin.
Nuclear Isolation, Staining, and Fluorescence-activated Cell Sorting
To isolate stable cell nuclei from fibroblasts for the staining of intranuclear β-catenin and for analyzing on a flow cytometer, a protocol disrupting cell membrane by detergent (Triton X-100) and maintaining nuclear membrane integrity by magnesium was adopted from the Flow Cytometry Core Laboratory at the NCI ETI Branch (home.ncifcrf.gov/ccr/flowcore/nuclei.pdf). Briefly, cells were collected by gentle scraping and washed twice with cold PBS. Cells were then resuspended in cold nuclear isolation buffer (320 mm sucrose, 5 mm MgCl2, 10 mm HEPES, and 1% Triton X-100, pH 7.4) and allowed to incubate on ice for 10 min. Nuclear yield and integrity were confirmed by microscopic examination with trypan blue staining. We routinely observed >98% nuclear isolation efficiency (data not shown). Nuclei were pelleted by centrifugation at 2000 × g and washed twice with nuclear wash buffer (320 mm sucrose, 5 mm MgCl2, and 10 mm HEPES, pH 7.4). Isolated nuclei were then incubated overnight with anti-β-catenin antibody (5 μg/ml) or normal mouse IgG (Santa Cruz Biotechnology), followed by a 1-h incubation with fluorescein isothiocyanate-conjugated anti-mouse IgG (2 μg/ml) in nuclear wash buffer plus 1% bovine serum albumin and 0.1% sodium azide. All steps described above were done at 4 °C. After extensive washing, nuclei were resuspended in 250 μl of nuclear wash buffer before flow cytometry analysis. Flow cytometry was performed on an Accuri C6 flow cytometer system (Accuri Cytometers, Inc., Ann Arbor, MI) using 488 nm excitation and standard fluorescein isothiocyanate emission optics with 10,000 events from each sample, and analysis was performed using FlowJo software (Tree Star Inc., Ashland, OR).
Analysis of Gene Expression
RNA was extracted from fibroblasts using TRIzol (Invitrogen), and cDNA converted from 5 μg of total RNA was obtained using a SuperScript first-strand synthesis system for reverse transcription-PCR kit (Invitrogen). To control for genomic DNA contamination, additional RNA samples were processed without reverse transcriptase. The reverse transcription product equivalent to 25 ng of total RNA was then added to a real-time quantitative PCR (qPCR) using a Dynamo SYBR Green qPCR kit (Finnzymes, Espoo, Finland) according to the manufacturer's protocol. The following primers were used: human PAI-1, 5′-TGGAACAAGGATGAGATCAG-3′ (sense) and 5′-CCGTTGAAGTAGAGGGCATT-3′ (antisense); human α-SMA, 5′-CTGTTCCAGCCATCCTTCAT-3′ (sense) and 5′-CCGTGATCTCCTTCTGCATT-3′ (antisense); and glyceraldehyde-3-phosphate dehydrogenase, 5′-GACCCCTTCATTGACCTCAAC-3′ (sense) and 5′-CTTCTCCATGGTGGTGAAGA-3′ (antisense). The annealing and amplification temperature was 60 °C. Real-time qPCR was performed in strip tubes using a StepOne PCR system (Applied Biosystems, Foster City, CA) according to the manufacturer's instructions. The specificity of amplified products was suggested by a melting curve resulting in only one peak. This was further confirmed by agarose gel electrophoresis of the PCR products visualized under ethidium bromide/UV illumination. Target amplifications were compared with the reference amplifications (glyceraldehyde-3-phosphate dehydrogenase) in the same experiment for each reverse transcription product tested. All reactions were carried out in duplicate, and the threshold cycle (Ct) values were determined by automated threshold analysis with the StepOne software. The final results are presented as relative -fold change in target gene expression compared with the reference based on the comparative or ΔΔCt method. The efficiency of each primer pair was determined by the qPCR procedure from standard dilutions of cDNA (equivalent to 10 pg to 10 ng of total RNA in the reverse transcription reaction). To assess the effect of an inhibitor of the TGF-β1 receptor/ALK5 in suppressing PAI-1 expression, 10 μm SB431542 (Tocris Bioscience, Ellisville, MO) was added 1 h prior to the addition of human recombinant TGF-β1 at 2 ng/ml overnight.
Enzyme-linked Immunosorbent Assay for Secreted PAI-1
To determine the concentration of PAI-1 in the culture medium of human lung fibroblasts, cells were plated in a 24-well plate at 5 × 105 cells/well overnight in DMEM and 10% fetal bovine serum. After serum deprivation for 24 h, the culture medium was collected and spun at 10,000 × g for 5 min at 4 °C to remove any cell debris. The PAI-1 concentration of each supernatant was determined using a Quantikine human serpin E1/PAI-1 immunoassay kit (R&D Systems, Minneapolis, MN) according to the manufacturer's instructions.
Determination of TGF-β Bioactivity
The active TGF-β concentration in the cultured fibroblasts was assayed using the co-culture method with mink lung epithelial cells transfected with a truncated PAI-1 promoter fused to a firefly luciferase gene (referred to as MLEC-PAI-1-Lux cells; a kind gift from Dr. George Yang, Stanford University) (21). Fibroblasts were seeded in 96-well plates at 2.5 × 104 cells/well in triplicates, along with MLEC-PAI-1-Lux cells at 1.5 × 104 cells/well in low serum (1%) medium. The use of 1% serum instead of starvation medium with 0.1% serum was needed to support survival of the MLEC-PAI-1-Lux cells. A standard curve of active TGF-β1 was generated in MLEC-PAI-1-Lux cells with serial dilutions of human recombinant TGF-β1 (0–10 ng/ml; Sigma). After incubation for 24 h, the viability of cells was checked microscopically before washing twice with 1× PBS. Cells were then lysed in 1× passive lysis buffer at 50 μl/well (luciferase assay kit, Promega, Madison, WI) and incubated with agitation at room temperature for 20 min. 10 μl of cell lysate was analyzed for luciferase activity according to the manufacturer's instructions. Cells from fibroblast-only wells were used for the cell count. The mean values of luciferase activity from triplicates were then converted into concentrations of TGF-β in picograms/number of cells using a standard curve obtained with human recombinant TGF-β1, normalized with cell numbers. The concentration of total TGF-β1 in the culture medium for corresponding fibroblasts was determined using a Quantikine human TGF-β1 immunoassay kit (R&D Systems) according to the manufacturer's instructions.
RNA Interference
We constructed a lentivirus-driven β- catenin small hairpin RNA (shRNA) expression plasmid from the pLKO.1 vector (22), targeting to β-catenin (5′-CGGGATGTTCACAACCGAATT-3′; pLKO.1-shβ-Cat), SPARC (5′- AACAAGACCTTCGACTCTTCC-3′; pLKO.1-shSPARC), or a scrambled sequence (5′-GTTCTCCGAACGTGTCACGTT-3′; pLKO.1-Scr). pLKO.1-shβ-Cat, pLKO.1-shSPARC, or pLKO.1-Scr was transduced into cells, followed by puromycin selection at 2 μg/ml for at least 48 h. The efficiency of shRNA knockdown of endogenous β-catenin or SPARC was assessed by Western blot analysis.
Quantitation of Cell Viability and Caspase-3 Activity Assay
Cell viability was determined by an alamarBlue assay. Briefly, primary fibroblasts were seeded overnight in 96-well plates in triplicates and then subjected to serum starvation (0.1% serum) for 24 h. Cells were left untreated or were treated with Glu-plasminogen at the indicated concentrations for 48 h. alamarBlue (resazurin from Sigma) was added to each well at 1.25 μg/ml for 2–4 h, and the fluorometric assay was done with excitation wavelength at 560 nm and emission wavelength at 590 nm with a fluorescence plate reader (FLUOstar Omega, BMG Labtech, Durham, NC). For each assay, data were collected from triplicates and analyzed and are represented as the percentage of viable cells relative to the untreated sample.
Caspase-3 protease activity in Glu-plasminogen-treated lung fibroblasts was determined by the fluorometric reaction using acetyl-Asp-Glu-Val-Asp-7-amino-4-trifluoromethylcoumarin (DEVD-AFC; R&D Systems) as substrate according to the manufacturer's instructions. Briefly, following the induction of cell death by Glu-plasminogen for 24 h, cells were collected by centrifugation and lysed in lysis buffer on ice for 10 min. 50 μl of cell lysate (from 2 × 106 cells) was mixed with 50 μl of 2× reaction buffer and 10 mm dithiothreitol and 5 μl of 1 mm DEVD-AFC in a 96-well plate. After incubation at 37 °C for 90 min, the release of free AFC cleaved by active caspase-3 proteases was determined using the fluorescence microplate reader with excitation at 400 nm and emission at 505 nm. The level of caspase-3 enzymatic activity in the cell lysate is directly proportional to the fluorescent signal of cleaved AFC.
Statistical Analysis
Data are expressed as the mean ± S.D. One-way analysis of variance and Student's t test were used for intergroup comparison. A probability level of 0.05 (p < 0.05) was considered significant.
RESULTS
Increased Expression of SPARC in IPF Fibroblasts Leads to Activation of β-Catenin
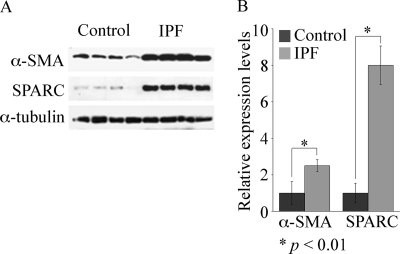
To further elucidate the phenotype of fibroblasts originating from IPF lung, we isolated fibroblasts from IPF patients and compared them with control fibroblasts, which were isolated from tissue taken at the time of lung cancer resection surgery and remote from the cancer. As others have reported previously, we found an increase in total α-SMA in IPF fibroblasts versus control fibroblasts (Fig. 1). This agrees with the observation that increased numbers of myofibroblasts are present in IPF, but it is noteworthy that they also retain this phenotypic identity in culture (23). There was also greater heterogeneity in the expression of α-SMA in IPF fibroblasts compared with control fibroblasts, which likely reflects varying numbers of myofibroblasts in IPF samples. While examining the expression of known matrix regulatory proteins in fibroblasts isolated from control or IPF lung, which have been shown to regulate the wound-healing response, we found SPARC expression increased 8-fold in fibroblasts from IPF lung fibroblasts versus control fibroblasts (Fig. 1). SPARC was more consistently expressed at higher levels in IPF fibroblasts versus control fibroblasts than fibronectin. Type I collagen was consistently expressed at higher levels in IPF fibroblasts, but unlike SPARC, levels varied more with serial passage (data not shown). Also, serial passage of cells did not diminish the percentage of fibroblasts in the culture, but we found that both populations of fibroblasts began to senesce after nine passages (data not shown). There was no significant difference, however, in proliferation or survival of control fibroblasts versus IPF fibroblasts (supplemental Fig. 1S).
FIGURE 1.
α-SMA and SPARC are highly expressed in IPF fibroblasts. A, Western blot analysis of lysate from control (n = 4) and IPF (n = 4) fibroblast cultures showing expression of α-SMA and SPARC, with α-tubulin as a loading control. B, quantitation of α-SMA and SPARC in control (n = 4) and IPF (n = 4) fibroblasts.
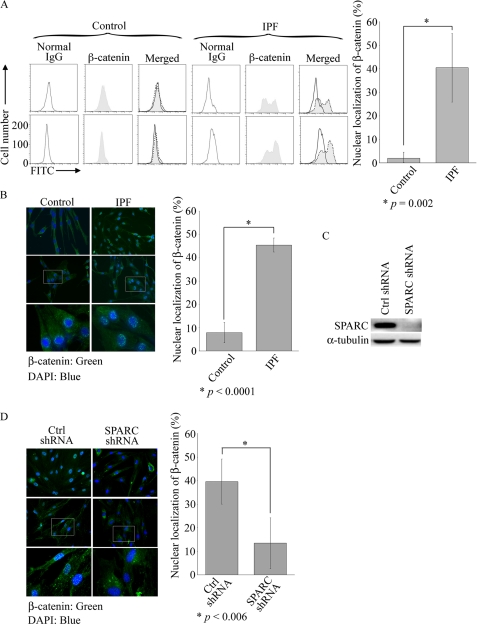
In view of recent studies demonstrating the activation of β-catenin in lung epithelial cells and fibroblasts in IPF lung and a study showing that SPARC activates β-catenin (8, 12), we examined whether β-catenin is activated in IPF fibroblasts. Because β-catenin regulates transcriptional responses in the nucleus, nuclear localization of β-catenin is considered synonymous with activated β-catenin. Using fluorescence-activated cell sorter (FACS) analysis to detect nuclear β-catenin, we discovered that 25–55% of IPF fibroblasts expressed nuclear β-catenin compared with <5% of control fibroblasts (Fig. 2A). The same four IPF and control samples were used at early passage (lower than six) for this and all subsequent experiments shown. There was heterogeneity in the number of fibroblasts expressing nuclear β-catenin, like α-SMA, between IPF patients, but control fibroblasts from different patients uniformly expressed little nuclear β-catenin (Fig. 2A). In Fig. 2B, a representative immunostaining from two of these IPF patients and two control patients shows nuclear localization of β-catenin in IPF fibroblasts versus cytoplasmic localization in control fibroblasts. Analysis of nuclear β-catenin expression by immunostaining in IPF and control fibroblasts revealed similar results as seen with FACS analysis (Fig. 2B). Because SPARC can activate β-catenin in adipocytes (12), we investigated whether this was also the case in IPF fibroblasts. Following successful down-regulation of SPARC by RNA interference (Fig. 2C), we observed that 40% of control shRNA cells versus 12% of SPARC shRNA cells expressed nuclear β-catenin as detected by immunohistochemical analysis (Fig. 2D).
FIGURE 2.
Increased active β-catenin in IPF fibroblasts and its regulation by SPARC. A, quantitation of nuclear, i.e. active, β-catenin in control and IPF fibroblast cultures by FACS analysis with anti-β-catenin antibody. Results are shown from two control and two IPF samples. Statistical analysis of IPF (n = 4) and control (n = 4) fibroblast cultures was performed using Student's t test and is presented as the mean ± S.D. in the right panel. FITC, fluorescein isothiocyanate. B, quantitation of nuclear localization of active β-catenin in control and IPF fibroblasts by immunostaining with β-catenin antibody and 4′,6-diamidino-2-phenylindole (DAPI) for nuclear staining. Representative control cells show peripheral cytoplasmic localization of β-catenin, whereas β-catenin is expressed in the nuclei of IPF cells. Results are shown in higher magnification (lower panels). The percentages of nuclear localized β-catenin in control (n = 4) and IPF (n = 4) fibroblast cultures are presented as the mean ± S.D. in the right panel. Statistical analysis was performed using Student's t test. C and D, SPARC regulates nuclear localization of β-catenin. Following lentiviral transduction of control (Ctrl; scrambled) or SPARC shRNA, IPF fibroblasts were harvested for Western blot analysis with anti-SPARC antibody (C). The nuclear localization of active β-catenin in control or SPARC shRNA of four IPF samples was assessed as described for B. Representative results from two samples are shown in the left panel of D. In the right panel, the percentage of nuclear localized β-catenin in control shRNA (n = 4) versus SPARC shRNA in IPF fibroblasts is presented as the mean ± S.D. Statistical analysis was performed using Student's t test.
PAI-1 Expression Is Increased in IPF Fibroblasts and Is Regulated by SPARC/β-Catenin
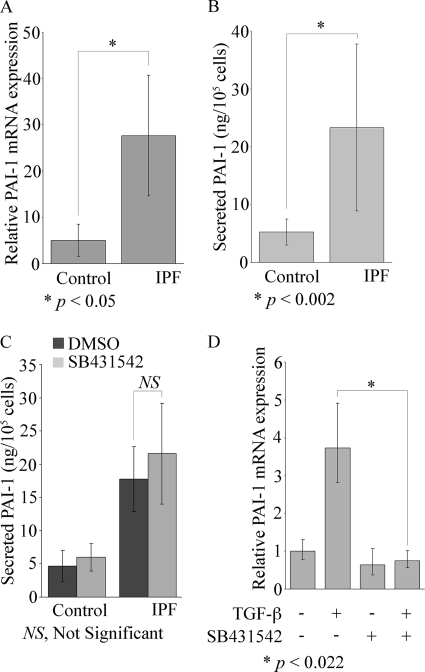
We then sought to identify potential targets of SPARC and β-catenin, which would play a role in promoting fibrosis by inducing matrix production or reducing matrix turnover and/or suppressing apoptosis of IPF fibroblasts. We identified PAI-1 as a potential target because the balance of expression of plasminogen versus PAI-1 regulates matrix production and turnover during wound healing, PAI-1 expression is increased in IPF lung, deficiency in PAI-1 protects mice from bleomycin-induced fibrosis, and it has been shown to mediate resistance of lung fibroblasts to plasminogen-induced apoptosis in response to TGF-β and endothelin-1 (Ref. 15; reviewed in Ref. 17). We found mean basal PAI-1 mRNA to be increased 6-fold in IPF fibroblasts versus control fibroblasts, but again there was more heterogeneity in its level of expression in IPF fibroblasts versus control fibroblasts (Fig. 3A). Additionally, basal levels of extracellular secreted PAI-1 were 4.7 times higher in IPF cells versus control cells (Fig. 3B). Interestingly, SPARC, α-SMA, PAI-1, and β-catenin are all targets of TGF-β and are induced or activated during the differentiation of fibroblasts into myofibroblasts. It was therefore possible that the increased expression or activity of PAI-1 resulted from an increase in basal TGF-β levels and/or activity, which led to autocrine induction of PAI-1. However, SB431542, a TGF-β1 receptor/ALK5-selective inhibitor, did not suppress basal levels of secreted PAI-1 in either IPF or control fibroblasts (Fig. 3C). It did, however, block TGF-β-mediated induction of PAI-1 in control fibroblasts (Fig. 3D), suggesting that basal PAI-1 in IPF and control fibroblasts is not regulated by the TGF-β1 receptor pathway. However, the addition of exogenous TGF-β1 still requires its cognate receptor to induce PAI-1. Also, we examined whether total or active secreted TGF-β levels were different in IPF fibroblasts versus control fibroblasts, but neither total nor active TGF-β differed significantly in control or IPF fibroblasts (Table 1).
FIGURE 3.
Elevated basal PAI-1 expression in IPF fibroblasts does not require the TGF-β1 receptor/ALK5. A, PAI-1 mRNA expression was measured by real-time qPCR in control and IPF cells after serum starvation. B, secreted PAI-1 was determined by enzyme-linked immunosorbent assay (average of triplicates) in serum-starved control or IPF cells. Data represent the mean ± S.D. (n = 4, control and IPF samples). Statistical analysis was performed using Student's t test. C, human lung fibroblasts from control (n = 4) and IPF (n = 4) cultures were incubated overnight with the TGF-β1 receptor/ALK5 inhibitor SB431542 at 10 μm. Secreted PAI-1 was determined by enzyme-linked immunosorbent assay as described for B. Data represent the mean ± S.D. Statistical analysis was performed using Student's t test. D, SB431542 suppressed the induction of PAI-1 mRNA expression by exogenous TGF-β1 as measured by real-time qPCR. Human control fibroblasts (n = 3) were preincubated with SB431542 at 10 μm for 1 h before the addition of TGF-β1 overnight at 2 ng/ml. The PAI-1 mRNA levels were measured by real-time qPCR. Results are shown as the mean ± S.D. with statistical analysis by Student's t test.
TABLE 1.
TGF-β activity in control and IPF fibroblasts
| Fibroblasts |
p valuea | ||
|---|---|---|---|
| Control (n = 4) | IPF (n = 4) | ||
| Active TGF-β (pg/250,000 cells) | 49.8 ± 29.7 | 63.9 ± 41.5 | 0.12 (NSb) |
| Total TGF-β (pg/100,000 cells) | 523.8 ± 166.6 | 570.6 ± 177.0 | 0.35 (NS) |
a Statistical analysis was performed using Student's t test.
b NS, not significant.
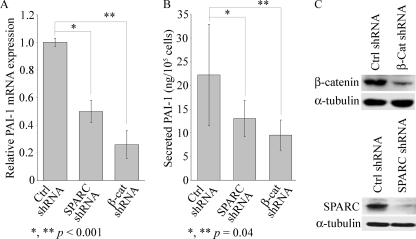
Because basal SPARC was elevated in IPF fibroblasts and accompanied by activated β-catenin, we hypothesized that PAI-1 is a target of SPARC/β-catenin. To examine this, we first down-regulated SPARC or β-catenin by RNA interference in IPF fibroblasts. This resulted in significantly reduced basal PAI-1 mRNA and secreted PAI-1 compared with the scrambled control (Fig. 4, A and B). Total β-catenin was reduced by 70% following transduction of the β-catenin shRNA compared with the scrambled control shRNA, and SPARC was reduced by 90% following transduction of a SPARC shRNA (Fig. 4C). Also, a similar result was observed in 293T cells, where we first down-regulated endogenous PAI-1 in 293T cells by transduction of a β-catenin shRNA, followed by overexpression of constitutively active β-catenin (supplemental Fig. 2S). This construct displayed nuclear localization of the fusion protein, in contrast to the diffused cytoplasmic distribution of green fluorescent protein alone (supplemental Fig. 2S).
FIGURE 4.
SPARC and β-catenin regulate expression of PAI-1. Down-regulation of SPARC or β-catenin suppressed PAI-1 in IPF fibroblasts. A, PAI-1 mRNA expression was assessed by real-time qPCR in IPF fibroblasts following transduction of control (Ctrl; scrambled), SPARC, or β-catenin (β-cat) shRNA. Data are from duplicate samples (mean ± S.D., n = 4). B, secreted PAI-1 in the culture medium from IPF fibroblasts was determined by enzyme-linked immunosorbent assay (done in triplicate), and data are the mean ± S.D. (n = 4). Statistical analysis was performed using Student's t test. C, Western blot analysis shows down-regulation of endogenous SPARC (upper panel) and β-catenin (lower panel) proteins in IPF cells following transduction of the respective shRNA constructs.
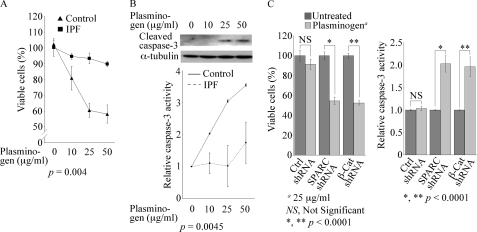
Elevated PAI-1 in IPF Fibroblasts Confers Resistance to Plasminogen-induced Apoptosis
A prevailing hypothesis is that fibrosis in IPF is due, at least in part, to inadequate apoptosis of interstitial fibroblasts. A recent study showed, for example, that TGF-β induction of PAI-1 in lung fibroblasts inhibits plasminogen-induced apoptosis of normal lung fibroblasts (15). Following our observation that IPF fibroblasts express significantly higher basal PAI-1 than control fibroblasts, we speculated that IPF fibroblasts would be more resistant than control fibroblasts to plasminogen-induced apoptosis. To examine this, exogenous plasminogen (10–50 μg/ml) was added to control or IPF fibroblasts, followed by an alamarBlue assay to analyze cell viability. Plasminogen caused a dose-dependent reduction in cell viability, with a maximal 42% reduction in cell viability at 50 μg/ml in control fibroblasts; this compared with a 10% decrease in cell viability of IPF fibroblasts at the same concentration (Fig. 5A). Cell death following exposure to plasminogen was caspase-mediated in control fibroblasts, as plasminogen induced a dose-dependent activation of caspase-3 as evidenced by the appearance of cleaved caspase-3. The activation of caspase-3 was not observed in plasminogen-treated IPF fibroblasts (Fig. 5B and data not shown). Using a more sensitive caspase-3 activity assay, we did detect a 1.5-fold increase in caspase-3 activity in IPF fibroblasts after exposure to the maximal plasminogen concentration (50 μg/ml), but this was significantly less than the 3.5-fold increase in control fibroblasts (Fig. 5B, lower panel). Because IPF fibroblasts are relatively resistant to plasminogen-induced apoptosis, which is likely a result of elevated PAI-1, and SPARC and β-catenin regulate PAI-1 expression, it is possible that down-regulation of SPARC or β-catenin would sensitize IPF fibroblasts to plasminogen-induced apoptosis. This was, in fact, what we observed, as lentiviral transduction of either SPARC shRNA or β-catenin shRNA into IPF fibroblasts caused a significant 46% (SPARC shRNA) or 48% (β-catenin shRNA) reduction in cell viability, compared with a 5–7% reduction in viability in the scrambled control, and a 2-fold increase in caspase-3 activity following exposure to plasminogen (Fig. 5C).
FIGURE 5.
IPF fibroblasts are resistant to plasminogen-induced apoptosis. A, cell survival determined by the alamarBlue assay following overnight incubation with various concentrations of plasminogen is shown. Data are from experiments repeated twice with control (n = 4) and IPF (n = 4) fibroblast cultures and are shown as the mean ± S.D. Statistical analysis was performed using a one-way analysis of variance. B, Western blot analysis using an antibody specific for cleaved caspase-3 shows that plasminogen caused a concentration-dependent increase in cleaved caspase-3 in control cells. Results are representative of three independent experiments from control cell lines (n = 4). In the lower panel, a significant increase in caspase-3 activity is shown in control fibroblasts (n = 4) treated with various concentrations of plasminogen for 24 h but not in IPF fibroblasts (n = 4). Data are presented as the mean ± S.D. from triplicates for each sample. Statistical analysis was performed using a one-way analysis of variance. C, SPARC and β-catenin regulate plasminogen-induced apoptosis. Endogenous SPARC or β-catenin in IPF cells was attenuated by transduction of SPARC or β-catenin (β-Cat) shRNA. Left panel, following the addition of plasminogen, an alamarBlue assay revealed a significant decrease in cell viability in SPARC or β-catenin shRNA cells but not in control (Ctrl) shRNA cells. Right panel, under the same treatment conditions, a significant increase in caspase-3 activity was observed following attenuation of SPARC or β-catenin. Data are the mean ± S.D. (n = 4). Statistical analysis was performed using Student's t test.
To further validate that PAI-1 is regulated by β-catenin, we examined whether LiCl, a known activator of the β-catenin pathway through phosphorylation and inactivation of GSK-3β (reviewed in Ref. 24), induces PAI-1 in control fibroblasts. The addition of LiCl to control fibroblasts caused a dose-dependent increase in PAI-1 mRNA, with a 4-fold maximal increase at 50 mm LiCl and coincident nuclear translocation of β-catenin (supplemental Fig. 3S). Additionally, LiCl (25 or 50 mm) protected control fibroblasts from plasminogen-induced apoptosis (supplemental Fig. 4S).
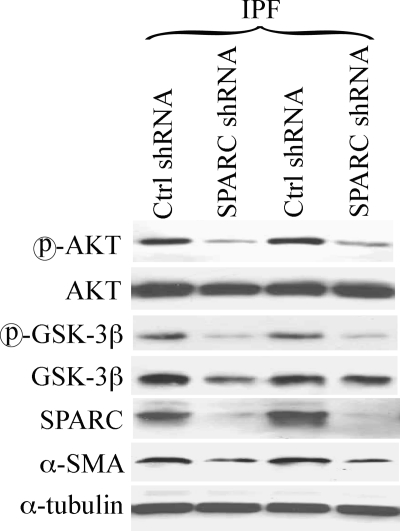
SPARC Activates β-Catenin in IPF Fibroblasts through Akt and GSK-3β
To elucidate the mechanism of SPARC-mediated activation of β-catenin in IPF fibroblasts, we examined the effect of attenuating SPARC in IPF fibroblasts on Akt and GSK-3β, known signaling targets of SPARC. Fig. 6 shows that down-regulation of SPARC suppressed both basal phospho-Akt and phospho-GSK-3β. Interestingly, SPARC down-regulation also attenuated α-SMA in IPF fibroblasts (Fig. 6), which suggests that SPARC may be required to maintain a myofibroblast phenotype.
FIGURE 6.
SPARC regulates β-catenin activity thorough Akt. Akt and GSK-3β activity was reduced in SPARC shRNA IPF fibroblasts with a coincident decrease in expression of PAI-1 and α-SMA. IPF fibroblast cultures (n = 4) were transduced with control (Ctrl) or SPARC shRNA, followed by Western blot analysis for the indicated proteins. Representative results from three independent experiments are shown.
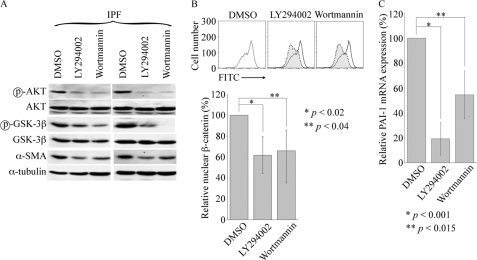
Our results revealing a mechanism of β-catenin activation by SPARC through Akt differ somewhat from those of Nie and Sage (12), who found that SPARC activation of β-catenin in adipocytes does not involve Akt but instead is regulated by integrin-linked kinase. It is possible that integrin-linked kinase also regulates β-catenin activity in IPF fibroblasts, and studies to address this are planned. To validate that Akt mediates, at least in part, the activation of β-catenin in IPF fibroblasts, we examined the effect of LY294002 and wortmannin, PI3K/Akt-selective inhibitors, on β-catenin activation in IPF fibroblasts. Both inhibitors functioned as expected to suppress basal phospho-Akt and phospho-GSK-3β levels. Additionally, each reduced nuclear β-catenin to a similar extent in IPF fibroblasts (Fig. 7). Both also inhibited PAI-1 mRNA and reduced basal α-SMA in IPF fibroblasts, albeit LY294002 to a greater extent (Fig. 7, A and C). These data suggest that SPARC causes activation of β-catenin in IPF fibroblasts by first activating Akt, which in turn phosphorylates, i.e. inactivates, GSK-3β and is then accompanied by the nuclear import of β-catenin (see schematic in Fig. 8).
FIGURE 7.
Reduced nuclear β-catenin and PAI-1 expression in the presence of PI3K/Akt inhibitors. A, Akt and GSK-3β activity was reduced in IPF fibroblasts by PI3K inhibitor LY294002 (10 μm) or wortmannin (1 μm) treatment for 24 h, in concert with a decrease in α-SMA expression. α-Tubulin was used as a loading control. Representative results of Western blot analysis from four IPF samples are shown with specific antibodies against the indicated proteins. DMSO, dimethyl sulfoxide. B, shown is the decrease in nuclear β-catenin localization in IPF fibroblasts treated with PI3K inhibitors. A decrease in nuclear β-catenin in inhibitor-treated IPF fibroblasts was assessed by FACS analysis as described in the legend to Fig. 2. Representative results are shown in the upper panel, and the results from statistical analysis of four IPF samples performed using Student's t test shown are shown in the lower panel. FITC, fluorescein isothiocyanate. C, suppression of basal PAI-1 mRNA expression in IPF fibroblasts by PI3K inhibitors was demonstrated by real-time qPCR. Data are from duplicate four IPF samples and are represented as the mean ± S.D.
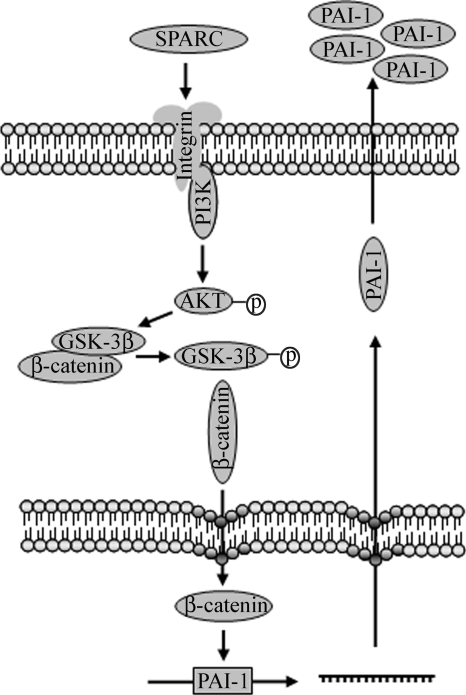
FIGURE 8.
Schematic of signaling through SPARC in IPF fibroblasts. Enhanced expression of SPARC in IPF fibroblasts stimulates PI3K activity, followed by Akt activation. Akt-mediated phosphorylation of GSK-3β releases β-catenin for nuclear translocation and transactivation of downstream gene expression, e.g. PAI-1. The accumulation of secreted PAI-1 protects IPF fibroblasts from plasminogen-induced apoptosis.
DISCUSSION
IPF is a devastating progressive fibrotic disease of the lung. The pathogenesis is unknown, although some recent studies have identified telomerase mutations in almost 10% of patients with familial IPF, which suggests that telomere shortening in epithelial and mesenchymal cell compartments may limit lung regenerative capacity (25–28). A recent study by Larsson et al. (23) showed that myofibroblasts are in greater abundance in IPF fibroblast populations versus control fibroblast populations and exhibit genome-wide abnormalities in translational control, which may confer an anti-apoptotic phenotype. We, too, found an increase in α-SMA expression in IPF fibroblasts versus control fibroblasts, consistent, at least in part, with the existence of an increased number of myofibroblasts in IPF samples versus control samples. There was, however, significantly more heterogeneity in the level of expression of markers of fibroblast activation, i.e. SPARC, type I collagen, and α-SMA, and activation of signaling pathways, i.e. Akt and β-catenin, in IPF samples versus control samples, which in part likely reflects differences in the numbers of myofibroblasts. This is not surprising because our patient samples were taken from patients at various stages of disease, and IPF patients exhibit profound variability in how they present and progress. Also, we found no significant differences in base-line proliferation or apoptosis in control fibroblasts versus IPF myofibroblasts, which agrees with the findings of Huang et al. (29). For the first time, we have shown that IPF fibroblasts express higher levels of SPARC compared with control fibroblasts, which mediates activation of β-catenin through activation of Akt and inhibition of GSK-3β. This results in an increase in basal PAI-1 expression, which renders IPF myofibroblasts resistant to plasminogen-induced apoptosis. SPARC has been shown to induce PAI-1 expression in aortic endothelial cells, but the mechanism was not elucidated (30). Our findings of elevated basal PAI-1 in IPF fibroblasts complement those of Horowitz et al. (15), who found that TGF-β induces PAI-1 in lung fibroblasts, which confers resistance to anoikis and plasminogen-induced apoptosis. Our findings suggest that SPARC, but not TGF-β, regulates basal expression of PAI-1; we did find, however, that TGF-β-dependent induction of PAI-1 in control fibroblasts requires the TGF-β1 receptor/ALK5 (Fig. 3). Also, TGF-β induced PAI-1 in IPF fibroblasts, but down-regulation of SPARC did not abrogate TGF-β-mediated induction of PAI-1 in control or IPF fibroblasts (data not shown). This suggests that regulation of basal PAI-1 in lung fibroblasts is controlled by a pathway distinct from the TGF-β-mediated induction of PAI-1. Our findings and published reports suggest that myofibroblasts, which are increased in number in IPF, acquire an apoptosis-resistant phenotype, which is characterized in part by elevated basal expression of PAI-1.
A recent study revealed that IPF fibroblasts are more resistant to prostaglandin E2-induced apoptosis compared with control fibroblasts, which, as speculated, may be due to reduced levels of prostaglandin E2 receptors, diminished protein kinase A levels and activity, and reduced expression of PTEN (phosphate and tensin homologue) in IPF lung (31). The authors revealed that prostaglandin E2 requires PTEN-mediated inhibition of Akt to induce apoptosis in fibroblasts. Also, pathologic signaling through β1-integrin in IPF fibroblasts has been shown to generate defective PTEN function (32). Moreover, a recent study showed that both TGF-β and endothelin-1 activate Akt in lung fibroblasts and that Akt promotes fibroblast resistance to apoptosis (14); endothelin-1 levels are known to be increased in IPF, and clinical trials in IPF with endothelin receptor antagonists are ongoing (33). Increased levels of SPARC in IPF lung and lung fibroblasts would activate Akt in IPF lung fibroblasts, which may also play a role in the resistance of IPF fibroblasts to prostaglandin E2-induced apoptosis.
We identified a constitutively active SPARC/Akt/β-catenin signaling cascade in IPF fibroblasts, which does not appear to be regulated by autocrine activation by TGF-β signaling because neither total nor active TGF-β was significantly increased in IPF myofibroblasts compared with control fibroblasts. In a previous study by Koli et al. (34), cultured IPF fibroblasts secreted ∼3-fold higher levels of active TGF-β, whereas the total TGF-β levels increased only ∼1.5-fold. Only two IPF fibroblast samples and one control fibroblast sample were used in this study, and the activity of TGF-β was measured in conditioned medium at 24 h with a mink lung epithelial cell reporter assay. Because active TGF-β binds to its receptors only after being liberated from the latent complex, TGF-β is probably not formed in solution and may not be stable in its soluble phase (35). Therefore, the measurement of the activity of TGF-β in conditioned medium at 24 h may underestimate its biological activity, whereas our study is based on the co-culture of mink lung epithelial cell reporter cells with fibroblasts, which yields a continuous measurement of the basal activity of TGF-β. A plausible model in IPF may be that lung epithelial injury is followed by a TGF-β-dependent phase, which causes differentiation of fibroblasts into myofibroblasts and contributes to persistent epithelial injury and possibly epithelial-mesenchymal transition (EMT). Pathways that become active in myofibroblasts, such as SPARC/Akt/β-catenin, may be autonomous and no longer require TGF-β to perpetuate the fibrotic response, but fibrosis could be further exacerbated by exposure of myofibroblasts to TGF-β, e.g. accentuated production of PAI-1 and other fibrogenic factors. In the absence of augmented TGF-β signaling, it is unclear why SPARC levels are elevated in IPF fibroblasts, and studies are ongoing to investigate this further. In an intratracheal bleomycin model, Strandjord et al. (37) showed that collagen accumulation is decreased in SPARC-null mice. Strandjord et al. showed a decrease in collagen accumulation in the lungs of SPARC-null mice following bleomycin injury, a finding supported by recent studies such as those of Zhou et al. (38) showing that SPARC regulates collagen expression. The bleomycin model in mice clearly does not completely mirror IPF, especially in regard to chronicity and the presence of fibroblastic foci in IPF but not in the bleomycin model. Myofibroblasts do accumulate in bleomycin injury but not in defined fibroblastic foci. Also, SPARC is clearly expressed in myofibroblasts in fibroblastic foci in IPF lung (11). Chronic models of lung injury and fibrosis may be more informative in discerning a role for SPARC in the regulation of lung fibrosis.
It is interesting to speculate that, because type II AEC in fibroblastic foci constitutively express active β-catenin, SPARC secretion from myofibroblasts in these foci may contribute to activation of β-catenin in AEC. It is also possible that trophic factors secreted from IPF myofibroblasts, in addition to SPARC, are responsible for the constitutive β-catenin activity. For example, Königshoff et al. (8) demonstrated recently that Wnt1, Wnt7b, Wnt10b, Fzd2, Fzd3, β-catenin, and lymphoid enhancer factor-1 are significantly increased in IPF lung tissue with similar elevations in cultured type II AEC from IPF patients. Interestingly, in a recent study, the same laboratory showed that WISP1 (Wnt1-inducible signaling protein-1) is increased in type II AEC in bleomycin lung injury and in patients with IPF (39). They also showed that WISP1 activates Akt, induces PAI-1, and promotes EMT in cultured type II AEC and enhances myofibroblast activation and ECM expression in lung fibroblasts; neutralization of WISP1 inhibited WISP1-induced EMT in cultured type II AEC and reduced expression of matrix proteins in lung fibroblasts, suggesting that both WISP-1 and SPARC may contribute to increased Akt activity in IPF lung fibroblasts and Akt-mediated resistance of lung fibroblasts to apoptosis by TGF-β and endothelin (13, 14, 23, 32). Also, the inhibition of WISP1 in a murine bleomycin lung injury model suppressed lung fibrosis and improved survival (39). We did not observe, however, increased expression of WISP1 mRNA in IPF fibroblasts versus control fibroblasts (data not shown). A preliminary analysis in our laboratory also did not reveal elevation of Wnt1b, Wnt3a, or Wnt antagonist Fzd2 or Fzd3 mRNA in IPF fibroblasts (data not shown). However, Wnt proteins and WISP1 originating from type II AEC could act in a paracrine manner to activate β-catenin in IPF fibroblasts; selective perturbation of Wnt proteins or β-catenin in murine models of lung injury will help to decipher the contribution of β-catenin to lung injury and fibrosis.
The cellular origin of myofibroblasts in IPF is as yet unknown. Fibrocytes, circulating progenitors of fibroblasts, have been identified in fibroblastic foci in IPF lung but not in normal lung (40). In a recent study by Larsson et al. (23), fibroblasts from IPF patients were shown to express keratin 18, an epithelium-specific marker, suggesting that these fibroblasts may arise through EMT. The Wnt/β-catenin pathway is known to regulate EMT in other cell types (41, 42); given that both type II AEC and myofibroblasts constitutively express β-catenin in IPF, it is possible that β-catenin mediates a transition of type II AEC to myofibroblasts in IPF.
Our results in IPF fibroblasts reveal that SPARC activates Akt, which then phosphorylates GSK-3β, leading to its inactivation, which results in nuclear translocation and activation of β-catenin. GSK-3β has recently been shown to regulate heart development through a β-catenin-dependent but Wnt-independent pathway (43). Kerkela et al. (43) showed that deletion of GSK-3β leads to cardiac hypertrophy. Our findings reveal a central role for SPARC/β-catenin in mediating the maintenance of a myofibroblast phenotype, which is supported by our observation that down-regulation of either SPARC or β-catenin suppresses α-SMA and confers resistance to plasminogen-induced apoptosis. Coupled with recently published data (8), our findings suggest that inhibition of SPARC/β-catenin may represent a novel therapeutic target in IPF.
Supplementary Material
Acknowledgments
We are very grateful to Dr. Helene Sage (Benaroya Research Institute, Seattle, WA) for critical reading of the manuscript. The lentiviral construct expressing constitutively active β-catenin fused with a 3′-green fluorescent protein was a kind gift from Dr. Irving Weissman (Stanford University).
This work was supported by SAMFUND, Division of Pulmonary and Critical Care, Department of Medicine, Stanford University.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. 1S–4S.
- IPF
- idiopathic pulmonary fibrosis
- TGF-β
- transforming growth factor-β
- α-SMA
- α-smooth muscle actin
- ECM
- extracellular matrix
- AEC
- alveolar epithelial cells
- GSK-3β
- glycogen synthase kinase-3β
- PI3K
- phosphoinositide 3-kinase
- PAI-1
- plasminogen activator inhibitor-1
- DMEM
- Dulbecco's modified Eagle's medium
- PBS
- phosphate-buffered saline
- qPCR
- quantitative PCR
- shRNA
- small hairpin RNA
- DEVD-AFC
- acetyl-Asp-Glu-Val-Asp-7-amino-4-trifluoromethylcoumarin
- FACS
- fluorescence-activated cell sorter
- EMT
- epithelial-mesenchymal transition.
REFERENCES
- 1.Zisman D. A., Keane M. P., Belperio J. A., Strieter R. M., Lynch J. P., 3rd (2005) Methods Mol. Med. 117, 3–44 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Phan S. H. (2002) Chest 122, 286S–289S [DOI] [PubMed] [Google Scholar]
- 3.Kapanci Y., Desmouliere A., Pache J. C., Redard M., Gabbiani G. (1995) Am. J. Respir. Crit. Care Med. 152, 2163–2169 [DOI] [PubMed] [Google Scholar]
- 4.Cadigan K. M. (2008) Dev. Cell 14, 322–323 [DOI] [PubMed] [Google Scholar]
- 5.Jian H., Shen X., Liu I., Semenov M., He X., Wang X. F. (2006) Genes Dev. 20, 666–674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Caraci F., Gili E., Calafiore M., Failla M., La Rosa C., Crimi N., Sortino M. A., Nicoletti F., Copani A., Vancheri C. (2008) Pharmacol. Res. 57, 274–282 [DOI] [PubMed] [Google Scholar]
- 7.Zhang Y., Goss A. M., Cohen E. D., Kadzik R., Lepore J. J., Muthukumaraswamy K., Yang J., DeMayo F. J., Whitsett J. A., Parmacek M. S., Morrisey E. E. (2008) Nat. Genet. 40, 862–870 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Königshoff M., Balsara N., Pfaff E. M., Kramer M., Chrobak I., Seeger W., Eickelberg O. (2008) PLoS ONE 3, e2142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chilosi M., Poletti V., Zamò A., Lestani M., Montagna L., Piccoli P., Pedron S., Bertaso M., Scarpa A., Murer B., Cancellieri A., Maestro R., Semenzato G., Doglioni C. (2003) Am. J. Pathol. 162, 1495–1502 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ramasamy S. K., Mailleux A. A., Gupte V. V., Mata F., Sala F. G., Veltmaat J. M., Del Moral P. M., De Langhe S., Parsa S., Kelly L. K., Kelly R., Shia W., Keshet E., Minoo P., Warburton D., Bellusci S. (2007) Dev. Biol. 307, 237–247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kuhn C., Mason R. J. (1995) Am. J. Pathol. 147, 1759–1769 [PMC free article] [PubMed] [Google Scholar]
- 12.Nie J., Sage E. H. (2009) J. Biol. Chem. 284, 1279–1290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Horowitz J. C., Lee D. Y., Waghray M., Keshamouni V. G., Thomas P. E., Zhang H., Cui Z., Thannickal V. J. (2004) J. Biol. Chem. 279, 1359–1367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kulasekaren P., Scavone C. A., Rogers D. S., Arenberg D. A., Thannickal V. J., Horowitz J. C. (2009) Am. J. Respir. Cell Mol. Biol. 42, 484–493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Horowitz J. C., Rogers D. S., Simon R. H., Sisson T. H., Thannickal V. J. (2008) Am. J. Respir. Cell Mol. Biol. 38, 78–87 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kortlever R. M., Bernards R. (2006) Cell Cycle 5, 2697–2703 [DOI] [PubMed] [Google Scholar]
- 17.Liu R. M. (2008) Antioxid. Redox Signal. 10, 303–319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lazar M. H., Christensen P. J., Du M., Yu B., Subbotina N. M., Hanson K. E., Hansen J. M., White E. S., Simon R. H., Sisson T. H. (2004) Am. J. Respir. Cell Mol. Biol. 31, 672–678 [DOI] [PubMed] [Google Scholar]
- 19.Kortlever R. M., Nijwening J. H., Bernards R. (2008) J. Biol. Chem. 283, 24308–24313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chang W. T., Kang J. J., Lee K. Y., Wei K., Anderson E., Gotmare S., Ross J. A., Rosen G. D. (2001) J. Biol. Chem. 276, 2221–2227 [DOI] [PubMed] [Google Scholar]
- 21.Abe M., Harpel J. G., Metz C. N., Nunes I., Loskutoff D. J., Rifkin D. B. (1994) Anal. Biochem. 216, 276–284 [DOI] [PubMed] [Google Scholar]
- 22.Moffat J., Grueneberg D. A., Yang X., Kim S. Y., Kloepfer A. M., Hinkle G., Piqani B., Eisenhaure T. M., Luo B., Grenier J. K., Carpenter A. E., Foo S. Y., Stewart S. A., Stockwell B. R., Hacohen N., Hahn W. C., Lander E. S., Sabatini D. M., Root D. E. (2006) Cell 124, 1283–1298 [DOI] [PubMed] [Google Scholar]
- 23.Larsson O., Diebold D., Fan D., Peterson M., Nho R. S., Bitterman P. B., Henke C. A. (2008) PLoS ONE 3, e3220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Medina M., Castro A. (2008) Curr. Opin. Drug Discov. Devel. 11, 533–543 [PubMed] [Google Scholar]
- 25.Alder J. K., Chen J. J., Lancaster L., Danoff S., Su S. C., Cogan J. D., Vulto I., Xie M., Qi X., Tuder R. M., Phillips J. A., 3rd, Lansdorp P. M., Loyd J. E., Armanios M. Y. (2008) Proc. Natl. Acad. Sci. U.S.A. 105, 13051–13056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Armanios M. Y., Chen J. J., Cogan J. D., Alder J. K., Ingersoll R. G., Markin C., Lawson W. E., Xie M., Vulto I., Phillips J. A., 3rd, Lansdorp P. M., Greider C. W., Loyd J. E. (2007) N. Engl. J. Med. 356, 1317–1326 [DOI] [PubMed] [Google Scholar]
- 27.Cronkhite J. T., Xing C., Raghu G., Chin K. M., Torres F., Rosenblatt R. L., Garcia C. K. (2008) Am. J. Respir. Crit. Care Med. 178, 729–737 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tsakiri K. D., Cronkhite J. T., Kuan P. J., Xing C., Raghu G., Weissler J. C., Rosenblatt R. L., Shay J. W., Garcia C. K. (2007) Proc. Natl. Acad. Sci. U.S.A. 104, 7552–7557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Huang S. K., Wettlaufer S. H., Hogaboam C. M., Flaherty K. R., Martinez F. J., Myers J. L., Colby T. V., Travis W. D., Toews G. B., Peters-Golden M. (2008) Am. J. Respir. Crit. Care Med. 177, 66–74 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hasselaar P., Loskutoff D. J., Sawdey M., Sage E. H. (1991) J. Biol. Chem. 266, 13178–13184 [PubMed] [Google Scholar]
- 31.Huang S. K., White E. S., Wettlaufer S. H., Grifka H., Hogaboam C. M., Thannickal V. J., Horowitz J. C., Peters-Golden M. (2009) FASEB J. 23, 4317–4326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Xia H., Diebold D., Nho R., Perlman D., Kleidon J., Kahm J., Avdulov S., Peterson M., Nerva J., Bitterman P., Henke C. (2008) J. Exp. Med. 205, 1659–1672 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Shi-Wen X., Rodríguez-Pascual F., Lamas S., Holmes A., Howat S., Pearson J. D., Dashwood M. R., du Bois R. M., Denton C. P., Black C. M., Abraham D. J., Leask A. (2006) Mol. Cell. Biol. 26, 5518–5527 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Koli K., Myllärniemi M., Vuorinen K., Salmenkivi K., Ryynänen M. J., Kinnula V. L., Keski-Oja J. (2006) Am. J. Pathol. 169, 61–71 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Jurukovski V., Dabovic B., Todorovic V., Chen Y., Rifkin D. B. (2005) Methods Mol. Med. 117, 161–175 [DOI] [PubMed] [Google Scholar]
- 36.Clark C. J., Sage E. H. (2008) J. Cell. Biochem. 104, 721–732 [DOI] [PubMed] [Google Scholar]
- 37.Strandjord T. P., Madtes D. K., Weiss D. J., Sage E. H. (1999) Am. J. Physiol. Lung Cell. Mol. Physiol. 277, L628–L635 [DOI] [PubMed] [Google Scholar]
- 38.Zhou X., Tan F. K., Guo X., Wallis D., Milewicz D. M., Xue S., Arnett F. C. (2005) Arthritis Rheum. 52, 257–261 [DOI] [PubMed] [Google Scholar]
- 39.Königshoff M., Kramer M., Balsara N., Wilhelm J., Amarie O. V., Jahn A., Rose F., Fink L., Seeger W., Schaefer L., Günther A., Eickelberg O. (2009) J. Clin. Invest. 119, 772–787 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Andersson-Sjöland A., de Alba C. G., Nihlberg K., Becerril C., Ramírez R., Pardo A., Westergren-Thorsson G., Selman M. (2008) Int. J. Biochem. Cell Biol. 40, 2129–2140 [DOI] [PubMed] [Google Scholar]
- 41.Medici D., Hay E. D., Olsen B. R. (2008) Mol. Biol. Cell 19, 4875–4887 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Dissanayake S. K., Wade M., Johnson C. E., O'Connell M. P., Leotlela P. D., French A. D., Shah K. V., Hewitt K. J., Rosenthal D. T., Indig F. E., Jiang Y., Nickoloff B. J., Taub D. D., Trent J. M., Moon R. T., Bittner M., Weeraratna A. T. (2007) J. Biol. Chem. 282, 17259–17271 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kerkela R., Kockeritz L., Macaulay K., Zhou J., Doble B. W., Beahm C., Greytak S., Woulfe K., Trivedi C. M., Woodgett J. R., Epstein J. A., Force T., Huggins G. S. (2008) J. Clin. Invest. 118, 3609–3618 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bradshaw A. D. (2009) J. Cell Commun. Signal. 3, 239–246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Weaver M. S., Workman G., Sage E. H. (2008) J. Biol. Chem. 283, 22826–22837 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.