Abstract
The molecular mechanisms governing breast tumor cellular self-renewal contribute to breast cancer progression and therapeutic resistance. The ErbB2 oncogene is overexpressed in ∼30% of human breast cancers. c-Jun, the first cellular proto-oncogene, is overexpressed in human breast cancer. However, the role of endogenous c-Jun in mammary tumor progression is unknown. Herein, transgenic mice expressing the mammary gland-targeted ErbB2 oncogene were crossed with c-junf/f transgenic mice to determine the role of endogenous c-Jun in mammary tumor invasion and stem cell function. The excision of c-jun by Cre recombinase reduced cellular migration, invasion, and mammosphere formation of ErbB2-induced mammary tumors. Proteomic analysis identified a subset of secreted proteins (stem cell factor (SCF) and CCL5) induced by ErbB2 expression that were dependent upon endogenous c-Jun expression. SCF and CCL5 were identified as transcriptionally induced by c-Jun. CCL5 rescued the c-Jun-deficient breast tumor cellular invasion phenotype. SCF rescued the c-Jun-deficient mammosphere production. Endogenous c-Jun thus contributes to ErbB2-induced mammary tumor cell invasion and self-renewal.
Keywords: Breast Cancer, Cell Migration, Chemokines, Cytokine, Jun Transcription Factor, Stem Cell
Introduction
The mechanisms governing the invasive phenotype of cancer cells are currently thought to involve either autonomous changes in which alterations in the cellular genome contribute to the invasive phenotype, or alternatively, non-cell autonomous mechanisms in which heterotypic paracrine signals may determine the metastatic behaviors of the tumor (1, 2). In this regard, the heterotypic signals derived from mesenchymal cells within the tumor associated stromal microenvironment are known to promote tumor growth and invasiveness (3). An alternative hypothesis considers that tumors may grow and invade through a process of self-seeding via the expansion of a self-renewing population of cells within the tumor at the leading edge, known as cancer stem cells (4, 5).
c-Jun has been identified in the invasive front of breast cancers and correlates with increased microvessel density (6). The c-jun oncogene encodes a member of the activator protein-1 (AP-1) transcription factor family that heterodimerizes through a leucine zipper motif with members of the Jun, Fos, activating transcription factor (ATF), and Maf families (7). Induction of c-Jun abundance regulates activity of downstream target genes involved in processes governing cellular growth, proliferation, and development (8). Phosphorylation of the c-Jun by the c-Jun N-terminal kinase subgroup of mitogen-activated protein kinases contributes to cellular apoptosis in a cell type-specific manner and the regulation of cellular migration (9).
An analysis of the role of c-Jun in vivo requires the use of transgenic animals carrying floxed c-jun alleles (c-junf/f) in which the c-jun gene is flanked by lox P-sites as c-jun−/− mice die from cardiovascular and hepatic defects during gestation (10). Analysis of mice embryo fibroblasts (MEFs)2 derived from either c-jun−/− or c-junf/f mice identified a role for c-Jun in regulating expression of the epidermal growth factor receptor, cellular proliferation, and migration (11, 12). c-jun−/− MEFs undergo premature senescence associated with a proliferative defect due to the induction of p53/p21CIP1 (13, 14). Although in tissue culture experiments, overexpression of either the dominant negative c-Jun or the wild type c-Jun in transformed mammary epithelial cells has suggested the importance of c-Jun in promoting breast cancer cellular proliferation (15), the role of endogenous c-Jun in mammary epithelial cell invasion and progenitor cell expansion was previously unknown.
The potential role of epithelial stem cells in mammary tumor growth and invasion is of fundamental importance (1, 5). Stem cell factor (SCF), through its receptor c-Kit, regulates hematopoietic stem cell proliferation and migration (16). SCF is secreted in a soluble form and as a membrane-associated glycoprotein. Intracellular kinases activated upon dimerization induced by ligand binding include trans-phosphorylation of c-Kit, a type III receptor tyrosine kinase (17), but other cytokines and growth factors may also contribute to cellular migration (18). CCL5 expression correlates with poor prognosis in human breast cancer (19, 20) and is known to induce expression of matrix metalloproteases, which enhance cellular invasiveness (21). Human breast milk contains high concentrations of CCL5 (22), and CCL5 is produced by human tumors (23, 24). CCL5 acts through the three G-protein-coupled receptors CCR1, CCR3, and CCR5 where CCR5 is the main receptor for CCL5 in MDA-MB-231 cells (25). The molecular mechanisms governing CCR5 expression and its role in tumorigenesis are poorly understood.
Given the clinical studies demonstrating c-Jun overexpression in human breast cancer and its distribution at the leading edge of breast tumors (6), we examined the role of endogenous c-Jun in mammary epithelial cell migration and its role in mammary tumor cellular migration, invasion, and stem cell expansion. Using mammary epithelial cells and mammary tumors derived through intercrossing transgenic mice expressing MMTV-ErbB2 with c-junf/f, we show that endogenous c-Jun plays a key role in ErbB2-induced migration and invasion of mammary epithelial cells. c-Jun induced the abundance of SCF and CCL5 through transcriptional activation of the gene promoters. c-Jun-mediated induction of SCF and CCL5 promoted the expansion of a self-renewing mammary epithelial cell population and promoted cellular invasiveness. Thus, c-Jun mediates the expansion of a self-renewing population of mammary tumor stem cells via the production of CCL5 and SCF to enhance tumor invasiveness.
EXPERIMENTAL PROCEDURES
Transgenic Mice, Expression Plasmids, and Promoter Cloning
Transgenic animals carrying floxed c-jun alleles, c-junf/f, and the MMTV-ErbB2 transgenic mice were previously described (12, 26, 27). All experimental procedures performed using these mice were approved by the Institutional Animal Care and Use Committee (IACUC) of Thomas Jefferson University. The methods for derivation and culture of mammary epithelial cell (MEC) (28) and mammary tumor cell lines (29) were previously described.
The expression plasmids encoding adenovirus directing Cre (Ad-Cre) expression or control virus (Ad-null) were previously described (11). The retroviral expression plasmid encoding Cre was cloned through the insertion of the cDNA from the vector pMC-Cre-PGK-Hyg (from Dr. P. Stanley) as an EcoRI fragment into the retroviral expression plasmid pMSCV-IRES-GFP (30). Expression of Cre from pMSCV-IRES-GFP vector was confirmed by Western blot using an antibody directed to Cre (MMS-106) (12). The EcoRI fragment of the rat c-Jun DNA (31) was subcloned into the retroviral expression vector MSCV-IRES-GFP to form MSCV-c-Jun-IRES-GFP. The murine SCF promoter was cloned by amplifying a 2-kb fragment from the 5′-flanking region of the kit ligand (Kitl) gene followed by its insertion into the SmaI site of pGL3-basic luciferase reporter vector (11). The CCL5 luciferase reporter was previously described (11).
Reagents and Antibodies
SCF was from Peprotech Inc. (Rocky Hill, NJ). CCL5 was from R&D Systems (Minneapolis, MN). Anti-c-Jun (H-79) rabbit polyclonal antibody was from Santa Cruz Biotechnology (Santa Cruz, CA). Anti-paxillin (5H11, 05-417) mouse monoclonal antibody was from Millipore (Billerica, MA). Anti-phospho-paxillin (Tyr-118, 44-722G) rabbit polyclonal antibody was from BIOSOURCE (Camarillo, CA). Anti-GDI rabbit polyclonal antibody was from RTG Sol (Gaithersburg, MD). Rhodamine-phalloidin and 4′,6′-diamino-2-phenylindole were from Sigma-Aldrich. BODIPY 650/665-phalloidin was from Molecular Probes Inc. (Eugene, OR).
Cell Culture, Viral Cell Transduction, and Reporter Gene Assays
Cells were maintained in Ham's F12 supplemented with 10% fetal bovine serum, 100 μg/ml each of penicillin and streptomycin, 4 μg/ml insulin, 10 ng/ml epidermal growth factor, 10 ng/ml cholera toxin, 1 μg/ml hydrocortisone and were cultured in 5% CO2 at 37 °C. Mammosphere cultures were conducted as described previously (32). Aldehyde dehydrogenase (ALDH) activity was determined using the ALDEFLUOR assay and Sca-1 staining as described previously (33, 34). Adenovirus propagation was previously described (35). Infection was done at a multiplicity of infection of 20, cells were cultured overnight, and medium was changed prior to experimental analysis. Retroviral infections were conducted as described (35). Transfections were conducted using GeneJuice transfection reagent (EMD Biosciences, San Diego, CA) and Lipofectamine (Invitrogen) as described (36). Statistical analysis was conducted using the Student's t test.
Microscopy and Phalloidin Staining for F-actin Quantitation
Immunopositive MSCV-IRES-GFP- and MSCV-Cre-IRES-GFP-transduced cells were examined in 6-well plates. Phase contrast and fluorescent imaging were carried out using the ×20 and ×40 objectives of a Zeiss LSM 510 Meta laser confocal scanning microscope. Rhodamine-phalloidin F-actin staining was conducted as described previously (30).
Cell Adhesion Assay
96-well cell surface matrix-coated strip well tissue culture plates (no coating, bovine serum albumin, poly-l-lysine, collagen I, collagen IV, fibronectin, and laminin) were used for cell adhesion assays. An equal number of cells were seeded at the bottom of each coated well and allowed to adhere by incubating the plates at 37 °C, 5% CO2 for planned intervals. Strip wells containing adherent cells were removed at 1 h, fixed in 1% glutaraldehyde for 10 min, and stained with 0.1% crystal violet for 30 min. Following phosphate-buffered saline washes, 100 μl of 0.5% Triton X-100 was added to each well to lyse the cells and extract dye by incubating the plates overnight at room temperature with gentle shaking. Quantitation of extracted dye was conducted by measuring the absorbance at 595 nm. For each cell surface matrix, the background was noted from a coated well with no cells seeded in them.
Assays of Cell Motility, Migration, and Invasion
Cells were plated on plastic dishes and cultured overnight in Ham's F12 medium containing 10% fetal bovine serum, 100 μg/ml each of penicillin and streptomycin, 4 μg/ml insulin, 10 ng/ml epidermal growth factor, 10 ng/ml cholera toxin, 1 μg/ml hydrocortisone. Cell movements were monitored using a Zeiss inverted microscope. Video images were collected with a CCD camera (model 2400) at planned intervals, digitized, and stored as images using the MetaMorph 3.5 software (38). The position of nuclei was tracked to quantify cell motility. Cellular velocity was calculated in micrometers using MetaMorph software. Prior to examination for effects on cell motility, analyses were conducted for 3 h. Net displacements were measured every 20 min from start point to end point. Data from at least 100 cells were collected for each point.
Migration of cells across a membrane was determined using the Boyden chamber, as described previously (28, 39). A gradient of CCL5 or SCF was created through the addition of CCL5 (RANTES (regulated on activation normal T cell expressed and secreted)) (concentration 10 ng/ml) or SCF (0.5 ng/ml) to the lower chamber.
Three-dimensional Invasion Assay
The three-dimensional invasion assay was adapted from Vial et al. (40). Briefly, 100 μl of 1.67 mg/ml rat tail collagen type I (BD Biosciences) was pipetted in the top chamber of a 24-well 8-μm pore Transwell (Corning). The Transwell was incubated at 37 °C overnight to allow the collagen to solidify. 30,000 cells were then seeded on the bottom of the Transwell membrane and allowed to attach. Serum-free growth medium was placed in the bottom chamber, whereas 5% serum was used as a chemoattractant in the growth medium of the upper chamber. The cells were then chemoattracted across the filter through the collagen above for 3 days. Cells were fixed in 4% formaldehyde and permeabilized with 0.2% Triton-X in phosphate-buffered saline and then stained with 40 μg/μl propidium iodide for 2 h. Fluorescence was analyzed by confocal z-sections (1 section every 4 μm) at ×10 magnification from the bottom of the filter. Three-dimensional reconstructions of the propidium iodide-stained cells were done using the Carl Zeiss ZEN software (2007 Light Edition).
Cytokine Array Analysis
Mouse cytokine arrays spotted on nitrocellulose membranes were obtained from RayBiotech (Norcross, GA). Conditioned medium from either c-junPf/f or c-jun−/− cells was prepared by culturing cells in serum-free Ham's F12 for 24–48 h. Membranes were then processed according to the manufacturer's instructions for assessment of secreted cytokines and growth factors present in conditioned medium.
Real-time PCR and ELISA
All gel-based PCRs and RT-PCRs were done with an Ex Taq DNA polymerase kit (Takara Shuzo, Shiga, Japan) using oligonucleotide primers mentioned in Table 1. RNA was extracted using the standard guanidinium-isothiocyanate method, RQ1 DNase I (Promega, Madison, WI)-treated, and phenol-chloroform-extracted. RNA quantitation was done in an Agilent 2100 bioanalyzer (Agilent Technologies, Palo Alto, CA), and equal quantities were used for the reverse transcription reactions. Primers for all the genes were either designed using Primer Express 5.1 (Applied Biosystems Inc., Foster City, CA) (Table 1) or as described in Ref. 41.
TABLE 1.
List of oligonucleotide primers used PCR, RT-PCR, and real-time quantitative RT-PCR analysis (41)
| Gene | Orientation | Sequence 5′ → 3′ |
|---|---|---|
| c-jun genotyping | Forward | CTC ATA CCA GTT CGC ACA GGC GGC |
| Reverse | CCG CTA GCA CTC ACG TTG GTA GGC | |
| Reverse | CAG GGC GTT GTG TCA CTG AGC T | |
| RPL-19 (DNA PCR) | Forward | AAT GCT CGG ATG CCT GAG AA |
| Reverse | CTC CAT GAG GAT GCG CTT GT | |
| Cre recombinase | Forward | TGC TCT GTC CGT TTG CCG |
| Reverse | ATC GTG TCC AGA CCA GGC | |
| RPL-19 (for RT-PCR) | Forward | CTGAAGGTCAAAGGGAATGTG |
| Reverse | GGACAGAGTCTTGATGATCTC | |
| c-jun (for RT-PCR) | Forward | AGA GCG GTG CCT ACG GCT ACA GTA A |
| Reverse | CGA CGT GAG AAG GTC CGA GTT CTT G | |
| 18 S rRNA | Forward | AGGAATTCCCAGTAAGTGCG |
| Reverse | GCCTCACTAAACCATCCAA |
For ELISA, cells were seeded at 80% of confluence, and the growth medium was changed 24 h later to basal medium containing 0.1% bovine serum albumin after washing with phosphate-buffered saline. 48 h later, the conditioned media were collected, and supernatant were obtained by centrifugation at 2000 rpm for 5 min followed by filtration through 0.45-μm membrane filter. SCF and CCL5 in the conditioned media were measured using the mouse SCF and CCL5 ELISA kits (RayBiotech) in triplicate as per the manufacturer's recommendations and normalized by the total protein levels in the media of each individual sample. Experiments were conducted at least three separate times.
FACS Analysis
Cell labeling and FACS analysis were based on Refs. 34, 42, and 43 with modification. Before labeling, the cells were blocked with normal rat IgG (1/100) and purified rat anti-mouse Fcγ III/II receptor antibody (1/100) (clone 2.4G2, BD Pharmingen) for 30 min and then incubated with phycoerythrin labeled rat anti-mouse CD24 (1/50) (clone M1/69, BD Pharmingen), Sca-1 (1/50) (clone E13-161.7, BD Pharmingen), and/or phycoerythrin/Cy5-labeled rat anti-human/mouse CD44 (1/50) (clone IM7, BioLegend, San Diego, CA) for 1 h. All experiments were carried on at 4 °C. Cell sorting was performed on a FACSCalibur cell sorter (BD Biosciences). The data were analyzed with the FlowJo software (Tree Star, Inc., Ashland, OR).
RESULTS
Endogenous c-Jun Promotes Mammary Tumor Cell Persistence of Migratory Directionality and Velocity in Bitransgenic Mice Encoding c-junf/f
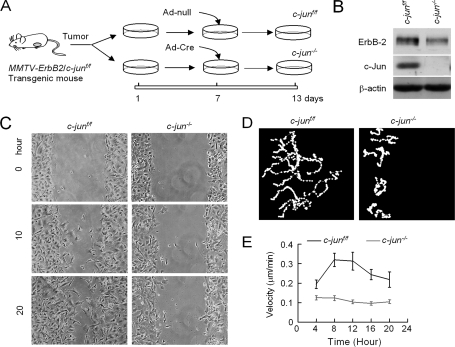
c-Jun and JNK contribute to fibroblast cellular migration (9, 11, 12). To determine the role of endogenous c-Jun in oncogene-induced MEC cellular migration and invasion, transgenic mice expressing mammary gland-targeted ErbB2 (MMTV-ErbB2)were crossed with c-junf/f mice. MEC and mammary epithelial cell tumors (MET) were derived from the offspring of these mice. The mammary tumor-derived epithelial cell lines were cultured and treated with either adenoviral vector or a vector expressing Cre recombinase (Fig. 1A). c-Jun abundance was reduced in c-junf/f MET treated with Ad-Cre (Fig. 1B). Cells deleted of c-jun appeared more spread by phase-contrast microscopy (Fig. 1C and supplemental Fig. 1A). The addition of adenovirus expressing Cre resulted in the presence of the 600-bp fragment, reflecting the excised c-jun allele (supplemental Fig. 1B).
FIGURE 1.
Endogenous c-Jun determines mammary epithelial tumor cell migration velocity. A, schematic representation of experimental protocol in which MMTV-ErbB2-c-junf/f double transgenic mice tumors were analyzed. B, Western blot analysis of c-junf/f METs demonstrating reduction in c-Jun abundance upon transduction with Ad-Cre. C, mammary epithelial tumor cells from c-junf/f cells transduced with either Ad-Cre or Ad-null were assessed in a wound healing assay (B). D, video microscopy of either c-junf/f or c-jun−/− cells. E, video microscopy data were used to determine the cellular velocity of the MET cells. Error bars indicate S.E.
The migratory properties of mammary epithelial cells expressing c-Jun or deleted of c-Jun was then examined using video microscopy. When c-jun+/+ cells migrated into the wound, migration was reduced 61.5% in the c-jun−/− cells (Fig. 1C). Mammary epithelial cells expressing or deleted of c-Jun were compared using video microscopy. Cellular velocity of migration was determined from video microscopy including at least 50 cells (Fig. 1D). The excision of c-Jun was associated with a 71.5% reduction in cellular migratory velocity at 8–12 h after plating (Fig. 1E).
Mammary Epithelial Tumor Cell c-Jun Promotes Cellular Invasiveness
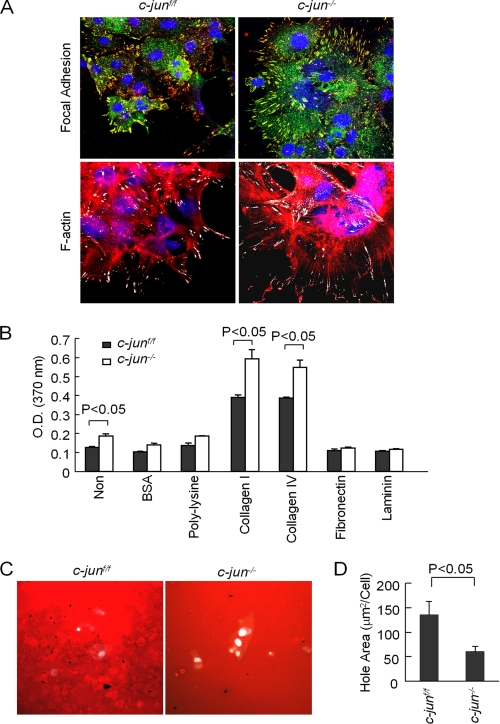
Analyses were conducted of the spread cellular phenotype including F-actin staining. MECs from c-junf/f were stained for focal adhesions using immunohistochemical staining to phospho-paxillin (yellow color). Cellular nuclei were identified by 4′,6′-diamino-2-phenylindole staining in blue (Fig. 2A). Cells deleted of c-Jun showed a centripetal distribution of focal contacts. The cells were increased in diameter and showed a more spread morphology. F-actin staining demonstrated the presence of F-actin at the focal contacts in c-jun+/+ mammary tumor cells; however, in c-jun−/−, MEC F-actin was dispersed in a centripetal distribution consistent with the centripetal distribution of focal contact staining (Fig. 2A).
FIGURE 2.
c-Jun reduces mammary epithelial cell adhesion and enhances invadopodia. A, confocal microscopy for focal contacts (tyrosine phosphorylated paxillin in yellow, with nuclei marked by 4′,6′-diamino-2-phenylindole in blue). Stress fiber formation is demarcated by F-actin distribution in cells. Note: Points of focal contact are shown in white. B, cellular adhesion assays comparing c-jun+/+ and c-jun−/− cells plated on distinct substrates. Non, non-cell; BSA, bovine serum albumin. C and D, invadopodia assays were conducted on c-jun+/+ and c-jun−/− MET. Holes indicating active invadapodia are shown in black. The hole area is shown as mean ± S.E. for n = 10 separate images.
Cellular adhesion was determined on distinct surfaces to investigate the role of integrin engagement in cellular adhesion. The ErbB2-c-junf/f tumor-derived cells plated on distinct surfaces were compared. The adhesion of c-jun−/− MET was increased on collagen I and collagen IV but not on fibronectin, poly-l-lysine, or laminin. To determine where the ErbB2 mammary tumor cellular invasion was c-Jun-dependent, phagocytosis assays were conducted (Fig. 2C). Areas of invasion were quantified as holes in the gelatin measured in square micrometer per cell (μm2/cell). Deletion of c-jun was associated with a reduction in the phagocytosis of red fluorescent Matrigel (Fig. 2C), consistent with the reduction in cellular invasive properties.
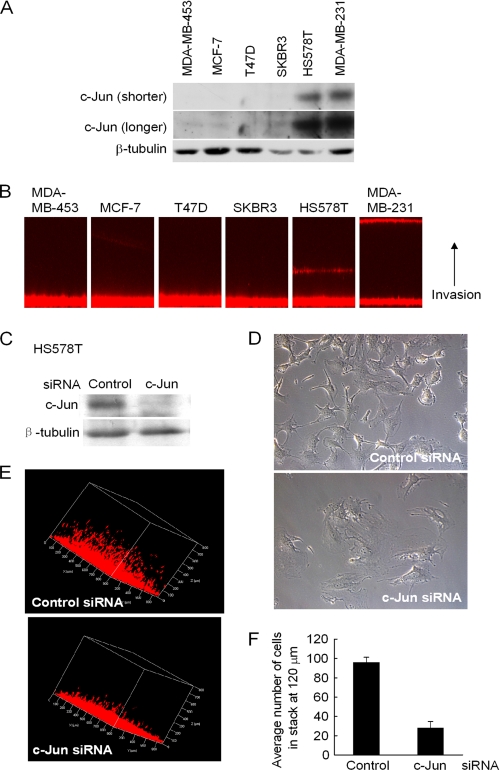
To examine further the role of endogenous c-Jun in breast tumor cellular invasiveness, Matrigel invasion assays were conducted. Western blot analysis was conducted on six human breast cancer cell lines (Fig. 3A). The relative abundance of c-Jun was greater in HS578T and MDA-MB-231 cells. Matrigel invasion assays demonstrated greater invasiveness in the HS578T and MDA-MB-231 cell lines (Fig. 3B) among a total of six mammary cancer cell lines that were examined. To determine the role of endogenous c-Jun in Matrigel invasion, HS578T cells were treated with either scrambled control small interfering RNA or small interfering RNA directed against c-Jun (Fig. 3C). The reduction in c-Jun protein abundance was associated with a less polarized morphology by phase-contrast microscopy (Fig. 3D) and reduced invasiveness in Matrigel (Fig. 3, E and F).
FIGURE 3.
Endogenous c-Jun promotes breast tumor cellular invasiveness. A, Western blot of human breast cancer cell lines with antibodies as indicated. B, three-dimensional Matrigel invasion assay in which invading cells are indicated in red as migrating toward the upper surface. C–F, Western blot of HS578T cells treated with c-Jun small interfering RNA (siRNA) (C) and assessed for morphology by phase-contrast microscopy (D) or three-dimensional reconstruction of cellular invasion (E) shown quantitated as mean ± S.E. data for relative invasion (F).
c-Jun Induces CCL5 and SCF Expression
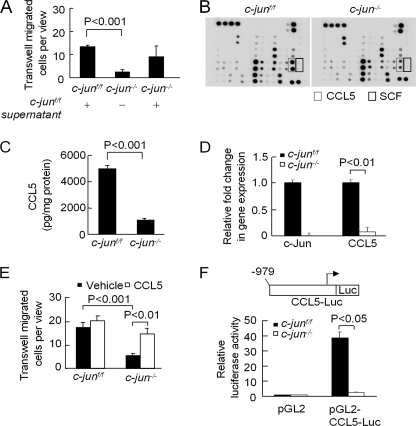
Previous studies performed on fibroblasts derived from c-jun−/− MEFs demonstrated reduced cellular migration that was rescued by the addition of secreted cellular factors from wild type fibroblasts (11). To further examine the mechanism governing the reduced cellular migration phenotype in c-Jun-deficient ErbB2 mammary tumor lines, the supernatant of ErbB2-c-jun+/+ cells was used to assess whether c-Jun-mediated mammary epithelial tumor cell migration was via a secreted factor (Fig. 4A). The conditioned medium (supernatant) from ErbB2-c-jun+/+ cells rescued the migration defect of ErbB2-c-jun−/− cells. To determine candidate secreted proteins governing c-Jun-mediated cellular migration, unbiased proteomic analyses were performed using cytokine arrays. Although the majority of cytokines and chemokines secreted by the mammary tumor cells remained unchanged between wild type and c-jun−/− MET cells, a subset of cytokines and chemokines were altered upon deletion of c-Jun (Fig. 4B and supplemental Fig. 3). CCL5 and SCF were substantially reduced upon c-jun deletion (Fig. 4B and data not shown). Further analyses were conducted to quantify the relative expression and secretion of CCL5. The excision of c-jun was associated with a 79% reduction in the abundance of CCL5 secreted by the MET cells (Fig. 4C). The mRNA abundance of CCL5 assessed by quantitative PCR was reduced 92.5% (Fig. 4D).
FIGURE 4.
Endogenous c-Jun induces CCL5 and SCF. A, Transwell migration of MET in response to conditioned medium (supernatant) from either MET (c-jun+/+) or MET (c-jun−/−). Data are mean ± S.E. B, cytokine array of proteins secreted by ErbB2 MET derived from c-jun+/+ and c-jun−/− mice (n = 2). Key differences in the relative abundance of cytokines and chemokines identify SCF and CCL5. Quantitative analysis is shown in supplemental Fig. 3. C, ELISA quantitating the relative abundance of CCL5 secreted by ErbB2 mammary tumor cells (n = 6 for c-jun+/+ and n = 9 for c-jun−/−). D, relative mRNA abundance of CCL5 determined by quantitative PCR shown as mean ± S.E. for n = 4. E, Transwell migration assays in response to the addition of CCL5. F, activity of the CCL5 promoter linked to a luciferase reporter gene. Luciferase activity was normalized to a co-transfected β-galactosidase report gene and luciferase reporter control vector. Data are mean ± S.E. for three separate transfection.
To determine the contribution of CCL5 and SCF to mammary epithelial tumor cellular migration, Transwell migration assays were conducted. A comparison was made between the c-junf/f and c-jun−/− MET. The defect in migration of c-jun−/− MET was rescued by the addition of CCL5 (Fig. 4E). The addition of SCF to the c-Jun−/− MEC had no effect on the migration defect of c-jun−/− MET cells (data not shown).
To determine whether endogenous c-Jun induced CCL5, the CCL5 promoter, linked to a luciferase reporter gene, was assessed. Comparison was made of CCL5 promoter activity by contrasting c-junf/f and c-jun−/− 3T3 fibroblast (Fig. 4F). When normalized to transfection efficiency, CCL5 promoter activity was decreased 93% by the absence of endogenous c-Jun (Fig. 4F).
c-Jun-mediated Contact-independent Growth of MET Cells Occurs via c-Jun without Affecting Proliferative Indices
To determine the role of c-Jun in the key properties determining cellular growth and renewal, mammary epithelial cells from the MMTV-ErbB2 tumors were examined further. The MMTV-ErbB2-MEC cell line (MET) was treated with Ad-Cre or control vector (Ad-null), and cellular proliferation assays were conducted using a450 and cell counting. The proliferation rate was unchanged at days 1 and 5 (supplemental Fig. 2A). The proliferation marker Ki-67 showed no significant reduction in abundance (supplemental Fig. 2, B and C). In contrast, endogenous c-Jun enhanced contact-independent growth as MEC wild type colony size was increase ∼2-fold assessed using colony forming assays (supplemental Fig. 1C).
Endogenous c-Jun Promotes Mammary Epithelial Tumor Cell Self-renewal
The expansion of cell populations through self-renewal may contribute to tumor growth (1, 5). In view of the insignificant change in proliferative index associated with the growth advantage contributed by c-Jun, we examined the possibility that endogenous c-Jun may enhance mammary epithelial progenitor cell expansion. Mammosphere analysis was conducted on MMTV-ErbB2-derived MET cells. There was a corresponding reduction in the number of mammospheres produced from METc-jun+/+ when compared with c-jun−/− (Fig. 5A).
FIGURE 5.
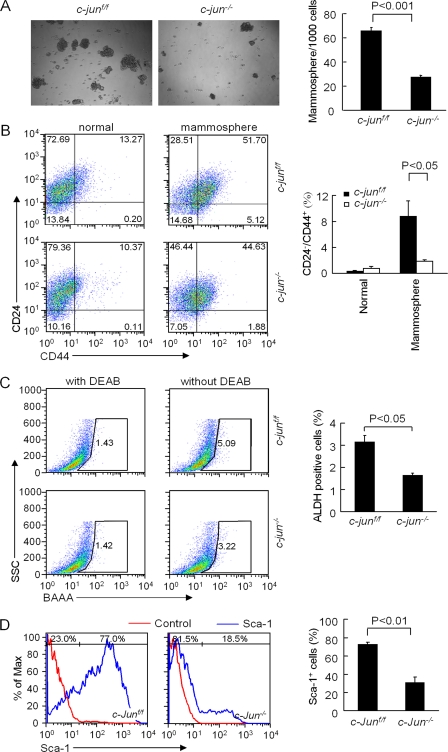
c-Jun promotes mammary epithelial tumor stem cell expansion. A, mammosphere production of ErbB2 tumor cell lines comparing c-jun+/+ with c-jun−/−. B, the proportion of CD24−/CD44+ cells was determined by FACS analysis and used as a surrogate marker of MEC progenitor cells. Comparison is shown of METs grown under normal or mammosphere culture conditions. C, ALDH1 activity was determined by FACS analysis of ErbB2-c-jun+/+ versus ErbB2-c-jun−/− MET cells. DEAB, dethylaminobenzaldehyde; BAAA, BODIPY-aminoacetaldehyde. D, Sca-1 staining in c-jun+/+ versus c-jun−/− MET cells. Error bars indicate S.E.
A subpopulation of CD44+/CD24− breast cancer cells has stem/progenitor cell properties (42). Although an area of controversy, CD44highCD24low murine breast cancer cells are enriched for breast tumor-initiating cells in non-obese diabetic/severe combined immunodeficient (NOD/SCID) mice, express stem cell surface antigens, and form mammospheres (44). The relative proportion of CD44+/CD24− cells increased in mammosphere culture (0.30 versus 8.67%) (Fig. 5B). Excision of c-jun reduced the proportion of CD44+/CD24− MET by 79.5% (8.67% versus 1.78%) (Fig. 5B). Normal and tumorous human mammary epithelial cells with increased ALDH activity have stem/progenitor properties (33). ALDH activity was measured as described previously (33). Tumor cells suspended in buffer containing ALDH1 substrate (BAAA) were compared with the negative control cells incubated with dethylaminobenzaldehyde (Fig. 5C, DEAB), a specific ALDH inhibitor. Mammary tumor stem cells demonstrated an increased ALDH1 activity. The relative proportion of ALDH1-positive MET was reduced 48.3% upon excision of c-Jun (Fig. 5C). Finally Sca-1 has been used as a marker enriched in breast cancer stem cells of ErbB2 transgenic mice (34). The deletion of the endogenous c-jun gene in MET reduced the proportion of Sca-1+ cells from 72.5 to 30.4% (Fig. 5D).
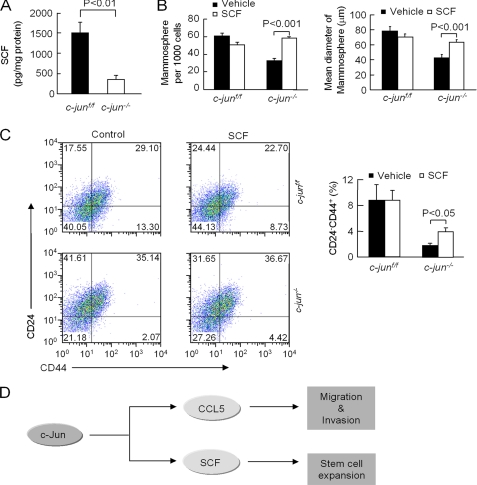
The excision of c-jun in the MET was associated with 76.3% reduction in the abundance of SCF (Fig. 6A). Although SCF did not affect Transwell migration, the addition of SCF rescued the defect in c-jun−/− MET mammosphere expansion (Fig. 6B) and increased the proportion of CD24−/CD44+ cells 2.2-fold (Fig. 6C). Collectively these studies are consistent with a model in which endogenous c-Jun in mammary epithelial tumors induces the expression and secretion of SCF and CCL5, which promotes mammary epithelial tumor progenitor cell expansion and tumor cell migration respectively (Fig. 6D).
FIGURE 6.
SCF rescued the defect in c-jun−/− MET stem cell expansion. A, ELISA of conditioned medium from MET (c-jun+/+) versus MET (c-jun−/−) for SCF. B, mammosphere production assays in the presence or absence of SCF indicating the induction of mammosphere formation upon the addition of SCF to c-jun−/− METs. (Data are shown throughout for n = 4.) C, fluorescence activated cell sorting for the relative proportion of CD44highCD24low. The cells were from mammospheres and treated with SCF (10 ng/ml) or vehicle. Error bars indicate S.E. in panels A–C. D, schematic representation of c-Jun-mediated cellular migration and mammosphere expansion via induction of SCF and CCL5 (RANTES) production.
DISCUSSION
The current studies demonstrate for the first time that endogenous c-Jun enhances MET cell growth, invasion, and tumor stem cell expansion. Mammary tumors secreted SCF and CCL5 in a manner that was dependent upon the abundance of endogenous c-Jun. CCL5, but not SCF, additionally rescued the defective migration of c-jun−/− mammary tumor cells. SCF addition rescued the defect in mammosphere production of c-jun−/− mammary tumors. The CCL5 and SCF gene promoters were direct transcriptional targets induced by c-Jun in breast tumor cells in culture (Fig. 4F) (11). Inhibitors of JNK (SP600125, 5 μm) reduced CCL5 promoter activity 26.8% and SCF promoter activity 25.0% in c-jun+/+ cells (supplemental Fig. 2E). The ErbB2 inhibitor (AG-825, 50 μm) reduced SCF promoter activity but not CCL5 promoter activity in c-jun+/+ cells (supplemental Fig. 2E). These findings are consistent with a role for c-Jun/JNK in activating the SCF and CCL5 promoters in c-jun+/+ cells. Collectively these studies are consistent with a novel model in which c-Jun may enhance mammary tumor growth through enhancing tumor stem cell expansion via heterotypic secretion of SCF.
Herein, endogenous c-Jun reduced cell adhesion and enhanced mammary epithelial tumor cellular migration and invasion. c-Jun enhanced the persistence of migratory directionality. c-Jun promotes fibroblast and keratinocyte migration (45, 46). c-Jun enhanced fibroblast migration via the induction of SCF secretion. In contrast, in mammary tumor cells, CCL5, but not SCF, governed cellular migration. CCL5 is a chemoattractant for stromal cells, including macrophages that express the CCR5 receptor (22–24, 47, 48). The chemokine CCL5 is secreted by T-47D, MCF-7, MDA-MB-435, and BT-20 cells (19, 49). High levels of tumor-associated macrophages are correlated with poor prognosis (50, 51). The CC family of chemokines has been characterized as major mediators of monocyte and T-cell migration. CCL5 has been shown to produce mesenchymal stem cells in a heterotypic manner and to enhance the expression of CCR5, thereby inducing breast cancer cellular motility, invasion, and metastasis (3). Heterotypic signals contribute to the onset and progression of breast cancer derived in part from an infiltrating set of T-cells and tumor-associated macrophages. The CC chemokine CCL5 has been implicated in breast cancer progression. Stress fiber formation was increased in c-jun−/− MET cells associated with a centripetal distribution of focal contacts resembling findings in c-jun−/− MEF cells (12). The induction of migration by c-Jun in MEFs involves increased expression of c-Src, a direct target of c-Jun, consistent with the current study in which Src abundance was reduced in the c-jun−/− MECs.
Several lines of evidence herein demonstrate an important role for c-Jun in promoting MET progenitor cell expansion. Mammary tumor stem cells are characterized by enrichment for the cell surface markers CD24−/CD44high, production of ALDH1, expression of Sca-1, and growth as mammospheres under serum-deprived conditions (52). Endogenous c-Jun enhanced each of these characteristic features in ErbB2-derived mammary epithelial tumor cell lines. The mammosphere assay determines the number and size of free floating aggregates, which have been previously shown to maintain the potential for self-renewal and to differentiate into all cell types of the mammary gland (53). This assay of self-renewal is based on the idea that stem cells may survive in anchorage-independent conditions. Differentiated cells, by contrast, need attachment to survive. Cancer stem cells have self-renewal capacity driving tumorigenicity recurrence and metastasis. Murine and human hematopoietic and neural stem and progenitor cells have high ALDH1 activity (54–56). ALDH-positive cells isolated from human breast tumor contain a cancer stem cell population (33). ALDH staining on human breast cancer correlates with poor prognosis (33).
An unbiased proteomic approach demonstrated that the mechanism by which c-jun mediates mammary epithelial stem cell expansion involves c-Jun-mediated induction of SCF secretion. SCF production was reduced in ErbB2-c-jun−/− cells, and SCF addition rescued the defective mammosphere production. Breast tumor stem cells contribute to therapy resistance and tumor recurrence (37). The finding that c-Jun induced tumor stem cells via SCF raises the possibility that SCF may be a useful target for c-Jun-expressing breast tumors.
Supplementary Material
Acknowledgment
We thank Atenssa L. Cheek for the preparation of this manuscript. The Kimmel Cancer Center was supported by the National Institutes of Health Cancer Center Core grant P30CA56036 (to R. G. P.).
This work was supported, in whole or in part, by National Institutes of Health Grants R01CA70896, R01CA75503, and R01CA107382 (to R. G. P.) and R01CA120876 (to M. P. L.). This project was also funded in part by grants from the Dr. Ralph and Marian C. Falk Medical Research Trust and the Pennsylvania Department of Health (to R. G. P.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. 1–3.
- MEF
- mouse embryonic fibroblast
- SCF
- stem cell factor
- MEC
- mammary epithelial cell
- MET
- mammary epithelial cell tumors
- JNK
- c-Jun N-terminal kinase
- RANTES
- regulated on activation normal T cell expressed and secreted
- ALDH
- aldehyde dehydrogenase
- ELISA
- enzyme-linked immunosorbent assay
- FACS
- fluorescence-activated cell sorting
- RT-PCR
- real-time PCR
- MSCV
- murine stem cell virus
- MMTV
- murine mammary tumor virus
- IRES
- internal ribosomal entry site
- GFP
- green fluorescent protein
- Ad
- adenovirus.
REFERENCES
- 1.Gupta G. P., Massagué J. (2006) Cell 127, 679–695 [DOI] [PubMed] [Google Scholar]
- 2.Nguyen D. X., Massagué J. (2007) Nat Rev Genet 8, 341–352 [DOI] [PubMed] [Google Scholar]
- 3.Karnoub A. E., Dash A. B., Vo A. P., Sullivan A., Brooks M. W., Bell G. W., Richardson A. L., Polyak K., Tubo R., Weinberg R. A. (2007) Nature 449, 557–563 [DOI] [PubMed] [Google Scholar]
- 4.Polyak K. (2007) J. Clin. Invest. 117, 3155–3163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Li F., Tiede B., Massagué J., Kang Y. (2007) Cell Res. 17, 3–14 [DOI] [PubMed] [Google Scholar]
- 6.Vleugel M. M., Greijer A. E., Bos R., van der Wall E., van Diest P. J. (2006) Hum. Pathol. 37, 668–674 [DOI] [PubMed] [Google Scholar]
- 7.Karin M., Liu Z., Zandi E. (1997) Curr. Opin. Cell Biol. 9, 240–246 [DOI] [PubMed] [Google Scholar]
- 8.Shaulian E., Karin M. (2002) Nature Cell Biol. 4, E131–136 [DOI] [PubMed] [Google Scholar]
- 9.Xia Y., Karin M. (2004) Trends Cell Biol. 14, 94–101 [DOI] [PubMed] [Google Scholar]
- 10.Eferl R., Sibilia M., Hilberg F., Fuchsbichler A., Kufferath I., Guertl B., Zenz R., Wagner E. F., Zatloukal K. (1999) J. Cell Biol. 145, 1049–1061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Katiyar S., Jiao X., Wagner E., Lisanti M. P., Pestell R. G. (2007) Mol. Cell. Biol. 27, 1356–1369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jiao X., Katiyar S., Liu M., Mueller S. C., Lisanti M. P., Li A., Pestell T. G., Wu K., Ju X., Li Z., Wagner E. F., Takeya T., Wang C., Pestell R. G. (2008) Mol. Biol. Cell 19, 1378–1390 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Schreiber M., Kolbus A., Piu F., Szabowski A., Möhle-Steinlein U., Tian J., Karin M., Angel P., Wagner E. F. (1999) Genes Dev. 13, 607–619 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Johnson R. S., van Lingen B., Papaioannou V. E., Spiegelman B. M. (1993) Genes Dev. 7, 1309–1317 [DOI] [PubMed] [Google Scholar]
- 15.Liu Y., Lu C., Shen Q., Munoz-Medellin D., Kim H., Brown P. H. (2004) Oncogene 23, 8238–8246 [DOI] [PubMed] [Google Scholar]
- 16.Zsebo K. M., Williams D. A., Geissler E. N., Broudy V. C., Martin F. H., Atkins H. L., Hsu R. Y., Birkett N. C., Okino K. H., Murdock D. C., et al. (1990) Cell 63, 213–224 [DOI] [PubMed] [Google Scholar]
- 17.Yarden Y., Kuang W. J., Yang-Feng T., Coussens L., Munemitsu S., Dull T. J., Chen E., Schlessinger J., Francke U., Ullrich A. (1987) EMBO J. 6, 3341–3351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lin W. W., Karin M. (2007) J. Clin. Invest. 117, 1175–1183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Luboshits G., Shina S., Kaplan O., Engelberg S., Nass D., Lifshitz-Mercer B., Chaitchik S., Keydar I., Ben-Baruch A. (1999) Cancer Res. 59, 4681–4687 [PubMed] [Google Scholar]
- 20.West R. B., Nuyten D. S., Subramanian S., Nielsen T. O., Corless C. L., Rubin B. P., Montgomery K., Zhu S., Patel R., Hernandez-Boussard T., Goldblum J. R., Brown P. O., van de Vijver M., van de Rijn M. (2005) PLoS Biol. 3, e187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Azenshtein E., Luboshits G., Shina S., Neumark E., Shahbazian D., Weil M., Wigler N., Keydar I., Ben-Baruch A. (2002) Cancer Res. 62, 1093–1102 [PubMed] [Google Scholar]
- 22.Michie C. A., Tantscher E., Schall T., Rot A. (1998) Eur. Cytokine Netw. 9, 123–129 [PubMed] [Google Scholar]
- 23.Negus R. P., Stamp G. W., Hadley J., Balkwill F. R. (1997) Am. J. Pathol. 150, 1723–1734 [PMC free article] [PubMed] [Google Scholar]
- 24.Mrowietz U., Schwenk U., Maune S., Bartels J., Küpper M., Fichtner I., Schröder J. M., Schadendorf D. (1999) Br. J. Cancer 79, 1025–1031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tanaka T., Bai Z., Srinoulprasert Y., Yang B. G., Hayasaka H., Miyasaka M. (2005) Cancer Sci. 96, 317–322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.D'Amico M., Wu K., Di Vizio D., Reutens A. T., Stahl M., Fu M., Albanese C., Russell R. G., Muller W. J., White M., Negassa A., Lee H. W., DePinho R. A., Pestell R. G. (2003) Cancer Res. 63, 3395–3402 [PubMed] [Google Scholar]
- 27.Hulit J., Lee R. J., Li Z., Wang C., Katiyar S., Yang J., Quong A. A., Wu K., Albanese C., Russell R., Di Vizio D., Koff A., Thummala S., Zhang H., Harrell J., Sun H., Muller W. J., Inghirami G., Lisanti M. P., Pestell R. G. (2006) Cancer Res. 66, 8529–8541 [DOI] [PubMed] [Google Scholar]
- 28.Li Z., Jiao X., Wang C., Ju X., Lu Y., Yuan L., Lisanti M. P., Katiyar S., Pestell R. G. (2006) Cancer Res. 66, 9986–9994 [DOI] [PubMed] [Google Scholar]
- 29.Ju X., Katiyar S., Wang C., Liu M., Jiao X., Li S., Zhou J., Turner J., Lisanti M. P., Russell R. G., Mueller S. C., Ojeifo J., Chen W. S., Hay N., Pestell R. G. (2007) Proc. Natl. Acad. Sci. U.S.A. 104, 7438–7443 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Neumeister P., Pixley F. J., Xiong Y., Xie H., Wu K., Ashton A., Cammer M., Chan A., Symons M., Stanley E. R., Pestell R. G. (2003) Mol. Biol. Cell 14, 2005–2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sonnenberg J. L., Rauscher F. J., 3rd, Morgan J. I., Curran T. (1989) Science 246, 1622–1625 [DOI] [PubMed] [Google Scholar]
- 32.Lindsay J., Jiao X., Sakamaki T., Casimiro M. C., Shirley L. A., Tran T., Ju X., Liu M., Li Z., Wang C., Katiyar S., Rao M., Allen K. G., Glazer R. I., Ge C., Stanley P., Lisanti M., Rui H., Pestell R. G. (2008) Clin. Transl. Sci. 1, 107–115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ginestier C., Hur M. H., Charafe-Jauffret E., Monville F., Dutcher J., Brown M., Jacquemier J., Viens P., Kleer C. G., Liu S., Schott A., Hayes D., Birnbaum D., Wicha M. S., Dontu G. (2007) Cell Stem. Cell 1, 555–567 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Grange C., Lanzardo S., Cavallo F., Camussi G., Bussolati B. (2008) Neoplasia 10, 1433–1443 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wang C., Pattabiraman N., Zhou J. N., Fu M., Sakamaki T., Albanese C., Li Z., Wu K., Hulit J., Neumeister P., Novikoff P. M., Brownlee M., Scherer P. E., Jones J. G., Whitney K. D., Donehower L. A., Harris E. L., Rohan T., Johns D. C., Pestell R. G. (2003) Mol. Cell. Biol. 23, 6159–6173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bouras T., Fu M., Sauve A. A., Wang F., Quong A. A., Perkins N. D., Hay R. T., Gu W., Pestell R. G. (2005) J. Biol. Chem. 280, 10264–10276 [DOI] [PubMed] [Google Scholar]
- 37.Jamieson C. H., Ailles L. E., Dylla S. J., Muijtjens M., Jones C., Zehnder J. L., Gotlib J., Li K., Manz M. G., Keating A., Sawyers C. L., Weissman I. L. (2004) N. Engl. J. Med. 351, 657–667 [DOI] [PubMed] [Google Scholar]
- 38.Gu J., Tamura M., Pankov R., Danen E. H., Takino T., Matsumoto K., Yamada K. M. (1999) J. Cell Biol. 146, 389–403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Li Z., Wang C., Jiao X., Lu Y., Fu M., Quong A. A., Dye C., Yang J., Dai M., Ju X., Zhang X., Li A., Burbelo P., Stanley E. R., Pestell R. G. (2006) Mol. Cell. Biol. 26, 4240–4256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Vial E., Sahai E., Marshall C. J. (2003) Cancer Cell 4, 67–79 [DOI] [PubMed] [Google Scholar]
- 41.Sugimoto Y., Koji T., Miyoshi S. (1999) J. Cell. Physiol. 181, 285–294 [DOI] [PubMed] [Google Scholar]
- 42.Al-Hajj M., Wicha M. S., Benito-Hernandez A., Morrison S. J., Clarke M. F. (2003) Proc. Natl. Acad. Sci. U.S.A. 100, 3983–3988 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Shackleton M., Vaillant F., Simpson K. J., Stingl J., Smyth G. K., Asselin-Labat M. L., Wu L., Lindeman G. J., Visvader J. E. (2006) Nature 439, 84–88 [DOI] [PubMed] [Google Scholar]
- 44.Wright M. H., Calcagno A. M., Salcido C. D., Carlson M. D., Ambudkar S. V., Varticovski L. (2008) Breast Cancer Res. 10, R10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Eferl R., Wagner E. F. (2003) Nat. Rev. Cancer 3, 859–868 [DOI] [PubMed] [Google Scholar]
- 46.Maeda S., Karin M. (2003) Cancer Cell 3, 102–104 [DOI] [PubMed] [Google Scholar]
- 47.Böttcher M. F., Jenmalm M. C., Björkstén B., Garofalo R. P. (2000) Pediatr. Res. 47, 592–597 [DOI] [PubMed] [Google Scholar]
- 48.von Luettichau I., Nelson P. J., Pattison J. M., van de Rijn M., Huie P., Warnke R., Wiedermann C. J., Stahl R. A., Sibley R. K., Krensky A. M. (1996) Cytokine 8, 89–98 [DOI] [PubMed] [Google Scholar]
- 49.Ali S., Kaur J., Patel K. D. (2000) Am. J. Pathol. 157, 313–321 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.van Netten J. P., George E. J., Ashmead B. J., Fletcher C., Thornton I. G., Coy P. (1993) Lancet 342, 872–873 [PubMed] [Google Scholar]
- 51.Leek R. D., Lewis C. E., Whitehouse R., Greenall M., Clarke J., Harris A. L. (1996) Cancer Res. 56, 4625–4629 [PubMed] [Google Scholar]
- 52.Charafe-Jauffret E., Ginestier C., Iovino F., Wicinski J., Cervera N., Finetti P., Hur M. H., Diebel M. E., Monville F., Dutcher J., Brown M., Viens P., Xerri L., Bertucci F., Stassi G., Dontu G., Birnbaum D., Wicha M. S. (2009) Cancer Res. 69, 1302–1313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Velasco-Velázquez M. A., Yu Z., Jiao X., Pestell R. G. (2009) Expert Rev. Anticancer Ther. 9, 275–279 [DOI] [PubMed] [Google Scholar]
- 54.Armstrong L., Stojkovic M., Dimmick I., Ahmad S., Stojkovic P., Hole N., Lako M. (2004) Stem. Cells 22, 1142–1151 [DOI] [PubMed] [Google Scholar]
- 55.Matsui J., Wakabayashi T., Asada M., Yoshimatsu K., Okada M. (2004) J. Biol. Chem. 279, 18600–18607 [DOI] [PubMed] [Google Scholar]
- 56.Hess D. A., Wirthlin L., Craft T. P., Herrbrich P. E., Hohm S. A., Lahey R., Eades W. C., Creer M. H., Nolta J. A. (2006) Blood 107, 2162–2169 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.