Abstract
The human lectin complement pathway involves circulating complexes consisting of mannose-binding lectin (MBL) or three ficolins (ficolin-1, -2, and -3) in association with three MBL/ficolin-associated serine proteases (MASP) (MASP-1, -2, and -3) and a nonenzymatic sMAP. MASP-1 and MASP-3 (MASP1 isoforms 1 and 2, respectively) are splice variants of the MASP1 gene, whereas MASP-2 and sMAP are splice variants of the MASP2 gene. We have identified a novel serum protein of 45 kDa that is associated with MBL and the ficolins. This protein is named MBL/ficolin-associated protein 1 (MAP-1 corresponding to MASP1 isoform 3). The transcript generating MAP-1 (MASP1_v3) contains exons 1–8 and a novel exon encoding an in-frame stop codon. The corresponding protein lacks the serine protease domains but contains most of the common heavy chain of MASP-1 and MASP-3. Additionally MAP-1 contains 17 unique C-terminal amino acids. By use of quantitative PCR and MAP-1-specific immunohistochemistry, we found that MAP-1 is highly expressed in myocardial and skeletal muscle tissues as well as in liver hepatocytes with a different expression profile than that observed for MASP-1 and MASP-3. MAP-1 co-precipitated from human serum with MBL, ficolin-2, and ficolin-3, and recombinant MAP-1 was able to inhibit complement C4 deposition via both the ficolin-3 and MBL pathway. In conclusion we have identified a novel 45-kDa serum protein derived from the MASP1 gene, which is highly expressed in striated muscle tissues. It is found in complex with MBL and ficolins and may function as a potent inhibitor of the complement system in vivo.
Keywords: Complement, Enzyme Inhibitors, Gene Structure, Inflammation, Innate Immunity, MASP1, MASP1 Isoform 3, Complement, Ficolin, Mannose-binding Lectin
Introduction
Activation of the complement system is accomplished via three different initiation pathways: the alternative pathway, the classical pathway, and the lectin pathway (1). In humans, four recognition molecules of the lectin pathway have been described: mannose-binding lectin (MBL),3 ficolin-1 (also called M-ficolin), ficolin-2 (also called l-ficolin), and ficolin-3 (also called H-ficolin or Hakata antigen) (2). MBL and the ficolins bind structures on different classes of microorganisms and are involved in sequestration and removal of dying host cells (2, 3). Three MBL/ficolin-associated serine proteases have been described so far (MASP-1, MASP-2, and MASP-3), as well as a protein lacking a serine protease domain named sMAP or MAp19 (4). Present consensus places MASP-2 as the main initiator of the lectin complement pathway by cleaving C4 and C2 to form the C4b2a complex leading to further downstream complement activation (5). Although the functions of the other MASPs are poorly understood, MASP-1 appears to play a role as an amplifier of complement activation (6, 7). Additionally, MASP-1 is able to cleave fibrinogen to fibrin, whereas MASP-2 is able to generate active thrombin by cleavage of prothrombin (8, 9). No conclusive biological function has yet been attributed to MASP-3 and sMAP.
MASP-1 and MASP-2 were originally named after their association with MBL (5, 10). Subsequently, MASP-3 and sMAP (also named Map19) were identified as alternative splicing variants of the MASP1 and MASP2 genes, respectively. However, the names of the MASPs are confusing because they do not correspond to the actual gene name. According to the nomenclature suggested by the Human Genome Organisation (HUGO), the proteins should have been named MASP1 isoform 1 (MASP-1), MASP1 isoform 2 (MASP-3), MASP2 isoform 1 (MASP-2), and MASP2 isoform 2 (sMAP or Map19). These names relate to the splice variants MASP1_v1, MASP1_v2, MASP2_v1, and MASP2_v2, respectively. However, the terms MASP-1, MASP-2, MASP-3, and sMAP are commonly used in the literature.
The MASP1 gene is located on chromosome 3q27-q28. MASP-1 (MASP1 isoform 1) and MASP-3 (MASP1 isoform 2) contain an identical heavy chain except for 15 C-terminal residues. The heavy chain is comprised of two C1r/C1s, urchin-EGF, bone morphogenetic protein (CUB) domains separated by an EGF domain and followed by two complement control protein domains (CCPs). The light chain contains the serine protease domain, which is different for MASP-1 and MASP-3 (11). MASP-1 is primarily expressed in the liver, whereas a more broad tissue expression pattern is observed for MASP-3 (12). The MASP2 gene is located on chromosome 1p36-p36.2. MASP-2 (MASP2 isoform 1) has a similar modular structure as MASP-1 and MASP-3 containing a heavy chain of two CUB domains, an EGF domain, and two CCP domains. The light chain is a MASP-2 unique serine protease domain, whereas sMAP (MASP2 isoform 2) is a truncated form lacking the serine protease domain and a major part of the heavy chain. Expression of the two MASP2 isoforms is predominantly localized to the liver.
In this report we describe the identification of a novel differential spliced variant (MASP1_v3) of the MASP1 gene that is found as a serum protein with a molecular mass of 45 kDa. This protein lacks the second CCP domain and the entire serine protease domain but contains 17 unique C-terminal amino acid residues. The protein is found in association with MBL and ficolins and down-regulates activation of complement via the lectin pathway. We propose to use the term MBL/ficolin-associated protein 1 (MAP-1) in addition to MASP1 isoform 3. This will indicate that the protein is not a serine protease, is derived from the MASP1 gene, and will not be mistaken for MASP-3.
EXPERIMENTAL PROCEDURES
Alignment of the MASP1 Gene and Its Splice Variants
The National Centre for Biotechnology Information Entrez data bases were searched for alternative transcript sequences of MASP1. Three splice variants were identified with the accession numbers NM001879, NM139125, and NM001031849 corresponding to MASP1_v1, MASP1_v2, and MASP1_v3, respectively. The sequences were aligned using BioEdit Software (v7.0.9.0).
Real Time Quantitative PCR Analysis
Commercially available normalized human tissue cDNA panels (Clontech) were investigated for MASP1_v1, MASP1_v2, and MASP1_v3 expression by real time quantitative PCR (qPCR) analysis on a Stratagene Mx2005 platform. TaqMan probes were labeled with 5′-FAM reporter dye and 3′-TAMRA quencher dye (DNA Technology, Risskov, Denmark). The reactions were performed with 1× TaqMan Universal PCR Master Mix, 0.4 μm each primer, and 0.25 μm TaqMan probe. The cycling parameters were 2 min at 50 °C, 10 min at 95 °C, and 45 cycles of 15 s at 95 °C and 1 min at 60 °C. All of the samples were measured in duplicate in three individual experiments on different plates using 0.7 ng of cDNA in a total volume of 10 μl. The amplifications were normalized to BCR, GAPDH, and β-actin. The data were corrected for differences in amplification efficiencies. The relative gene expression was calculated with reference to MASP1_v3 expression in heart (100 × 2 ^ (ΔCt(MASP1_v3heart) − ΔCt(x)), where x is any of the other tissues).
DNA Sequencing of MASP1_v3 Exon 9 in 100 Healthy Individuals
Sequencing of the exon 9 of the MASP1 gene spanning from positions +44,083 to +44,431 relative to the translation ATG start site (National Centre for Biotechnology Information accession number NC_000003) was performed on genomic DNA templates from 100 healthy Caucasian individuals. The fragment was amplified by using a single primer set (Table 1), where the forward primers contained a 5′-T7 sequence (5′-ttatacgactcacta-3′). PCR was performed in 20-μl volumes containing 2 ng of liver cDNA (Clontech), 0.25 μm each primer, 2.5 mm MgCl2, 0.2 mm dNTP, 50 mm KCl, 10 mm Tris·HCl, pH 8.4, and 0.4 units of Platinum TaqDNA polymerase (Invitrogen) and run at the following cycling parameters: 2 min at 94 °C; 15 cycles of 30 s at 94 °C, 60 s at 64 °C, 60 s at 72 °C; 15 cycles of 30 s at 94 °C, 60 s at 58 °C, and 60 s at 72 °C; and 5 min at 72 °C. The products were sequenced in the forward direction using the ABI BigDye cycle sequencing terminator kit (Applied Biosystems) according to the manufacturer's protocol using 5′-biotinylated T7 sequence primers. The sequence reactions were purified on a PyroMark Vacuum Prep Work station (Biotage) using streptavidin beads (GenoVision) and analyzed on an ABI Prism 3100 Genetic Analyzer (Applied Biosystems).
TABLE 1.
Oligonucleotides utilized for PCR amplification
| cDNA | Forward | Probe | Reverse | |
|---|---|---|---|---|
| Oligonucleotide (5′ → 3′) used for PCR amplification of human cDNA | MASP1_v1 | GCACCCAGAGCCACAGTG | GCCTTCCAGTGTGTGGGC | |
| MASP1_v2 | GCACCCAGAGCCACAGTG | GCCTTCCAGAGTGTGGTCA | ||
| MASP1_v3 | GCACCCAGAGCCACAGTG | CGATCTGGAGAGCGAACTC | ||
| Oligonucleotides (5′ → 3′) used for qPCR analysis of human cDNA | MASP1_v1 | CAAGATGCTCAACAATAACACAGGT | TATATACCTGTTCTGCCCAAGGAGTCTGGATGA | GCTGCCCATTCAGGTGTGAC |
| MASP1_v2 | CAAGATGCTCAACAATAACACAGGT | TATATACCTGTTCTGCCCAAGGAGTCTGGATGA | GGAAGAGGCCAGGCTCAGC | |
| MASP1_v3 | AGGCTACAAAGTGCTGAAGGAT | TCTGAAGGATGGGACGTGGAGTAAC | CACTCTGTCACTTGCTCTGAC | |
| BCR | CCTTCGACGTCAATAACAAGGAT | TCCATCTCGCTCATCATCACCGACA | CCTGCGATGGCGTTCAC | |
| β-Actina | ||||
| GAPDHb | ||||
| Oligonucleotides (5′ → 3′) used for sequencing of human genomic DNA | MASP1_v3 | CTGTTCTTCACACTGGCTG | CTGCTGAGATCATGTTGTTC |
a TaqMan Gene Expression Assays (Applied Biosystems, ACTB_Hs_00242273_A1).
b TaqMan Gene Expression Assays (Applied Biosystems, GAPDH_Hs_99999905_A1).
Recombinant Proteins
Expression of rMASP-1, rMASP-3, rMAP-1 (MASP1 isoforms 1, 2 and 3, respectively), rMASP-2 (MASP2 isoform 1), rMBL, and rficolin-3 in CHO cells was carried out as described elsewhere (13, 14).
Generation of Monoclonal and Polyclonal Antibodies
BALB/c×NMRI mice and rabbits were immunized subcutaneously three times with 25 μg of a MAP-1-specific peptide (CKKNEIDLESELKSEQVTE) coupled to diphtheria toxoid adsorbed to Al(OH)3 and mixed in 1:1 ratio with Freund's incomplete adjuvant. The peptide contains the 17 C-terminal amino acids and two additional natural N-terminal residues, lysine and cysteine (the latter was used for coupling). The coupling was conducted using (N-e-maleimidocaproyloxy)succinimide ester coupling reagents (Pierce) according to the manufacturer's recommendations. The fusion and selection of the antibodies was done essentially as described previously (15). Selected monoclonal antibodies were purified from culture supernatant by protein A affinity chromatography using the Äkta fast protein liquid chromatography system according to the manufacturer's instructions (GE Healthcare). Monoclonal antibody (mAb) 20C4 was used for immunoprecipitation, immunoaffinity purification, and immunoblotting. mAb 12B11 was used for the immunohistochemistry analysis. Rabbit polyclonal antibodies to MAP-1 were affinity-purified using a column coupled with human serum albumin to which the 17-residue MAP-1-specific peptide had been conjugated.
ELISA Specificity
ELISA was used to validate the specificity of the MAP-1 antibodies. The rMASP-1, rMASP-3, and rMAP-1 was immobilized to Maxisorp ELISA plates (Nunc) in 2-fold serial dilutions starting at 5 μg/ml. After washing/blocking in PBS, 0.05% Tween, an mAb to MAP-1 (mAb 20C4) or an antibody that reacts with the common heavy chain of MASP-1 and -3 (mAb 8B3) was applied at 5 μg/ml. Secondary detection was performed with HRP rabbit anti-mouse IgG (PO260; Dako) diluted 1:1000 in PBS, 0.05% Tween 20.
SDS-PAGE and Immunoblotting
Electrophoresis was performed on 10% or 4–12% (w/v) Bis-Tris polyacrylamide gels with discontinuous buffers using the NuPAGE® system (Invitrogen) as recommended by the manufacturer. Immunoblotting was performed using polyvinylidene difluoride membranes (PVDF-HyBond; GE Healthcare), 2 μg/ml of primary mAbs, and secondary visualization by HRP-conjugated streptavidin (P0397; Dako) diluted 1:1500 or HRP rabbit anti-mouse IgG (PO260; Dako) diluted 1:1000 in PBS, 0.05% Tween 20. The membranes were developed with 3-amino-9-ethylcarbazole (Sigma-Aldrich) (0.04% in acetone) and 0.015% H2O2 in 50 mm sodium acetate buffer, pH 5.
Immunoprecipitation
Immunoprecipitation of MAP-1 from serum was performed with the MAP-1-specific mAb 20C4 or mAb 8B3 (a monoclonal antibody reacting against the common heavy chain of MASP-1 and MASP-3) as a positive control. Additionally an IgG mAb antibody (IgG1κ) with no known specificity was applied as a negative control. A total of 10 μg of mAb 20C4, 8B3, or IgG1κ was allowed to bind to sheep anti-mouse IgG Dynabeads (M-280, catalog number 112.02D; Dynal/Invitrogen). After a washing step the beads were applied to a pool of normal human serum (diluted 1:1 in Tris-buffered saline) and incubated end over end for 1 h at 4 °C. After the final washing steps and magnetic separation the beads were boiled in SDS loading buffer and subjected to SDS-PAGE and immunoblotting probed with biotinylated antibodies to MAP-1, MBL, or ficolin-3.
The same precipitation procedure as described above was performed with mAbs to MBL (Hyb 131-11; Bioporto), ficolin-2 (FCN219) (16), and ficolin-3 (FCN334) (17). To compensate for differences in serum concentrations of MBL and ficolin-2 and -3, we precipitated from 1 ml, 300 μl, and 100 μl of serum, respectively. The samples were analyzed by SDS-PAGE and immunoblotting and probed with a biotinylated antibody to MAP-1.
Immunohistochemistry
CHO cells expressing rMAP-1 were grown in culture flasks in RPMI 1640 with 10% fetal calf serum. The cells were harvested at 80–90% confluence and fixed for 24 h in 4% formaldehyde-PBS and subsequently embedded in paraffin. Six different human liver tissues and samples from two different myocardial tissues, two skeletal muscle tissues, and two samples obtained from human aorta were also fixed and paraffin-embedded as described above. Sections of 5-μm slices were obtained with a Leitz Wetzlar microtome and placed on glass slides and stored at 4 °C until assayed. Pretreatments and analysis were performed as described previously (18). Primary antibodies against MAP-1 were the mAb 12B11 or the affinity-purified polyclonal antibody MAP-1, both diluted to 5 μg/ml. Isotype antibody controls were applied to the tissues at the same concentration. Secondary antibody was EnVisionTM antibody (HRP anti-mouse or HRP anti-rabbit; Dako). Analysis of the staining patterns was conducted under a Leica DMLB2 microscope.
Complement Activation Assay
The influence of MAP-1 on the MBL and ficolin-3-mediated complement factor C4 deposition was assessed essentially as described previously (19, 20). Briefly, mannan (MBL ligand) (Sigma-Aldrich M7504) or acetylated bovine serum albumin (AcBSA) (ficolin-3 ligand) was immobilized to Maxisorp ELISA plates (Nunc) at 10 μg/ml. After washing, rMBL or rficolin-3 (0.4 μg/ml) was added and incubated for 1.5 h. The rMAP-1 or rMASP-2 was applied for 1 h in 2-fold serial dilutions in the first dimension followed by incubation for 45 min at 37 °C with serial dilutions of serum deficient of MBL (21) or ficolin-3 (20) in the second dimension. The C4 deposition was measured using a polyclonal antibody to C4c (Q0369, Dako).
In addition we assessed the displacement of MASP-2 with MAP-1 using a pure system. The rMASP-2 was preincubated for 45 min at 20 °C in serial dilutions in the first dimension on an rMBL/mannan matrix as described above followed by incubation with dilutions of rMAP-1 in the second dimension for 45 min at 20 °C. Purified C4 (Quidel) was added at a concentration of 1 μg/ml and incubated for 45 min at 37 °C. Detection was conducted as described above.
RESULTS
Alignment of the Transcript Variants of MASP1
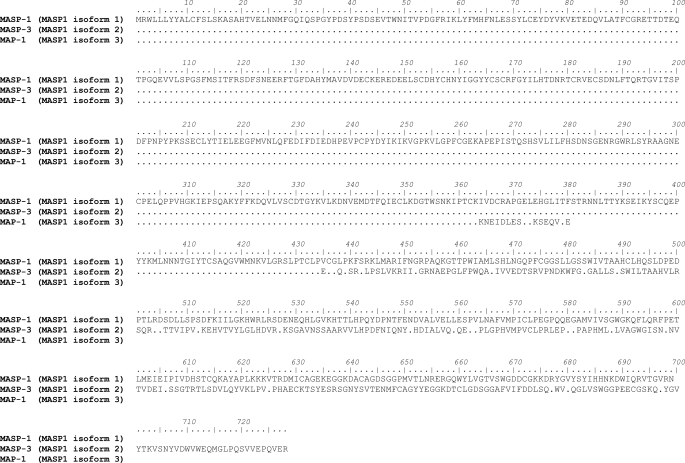
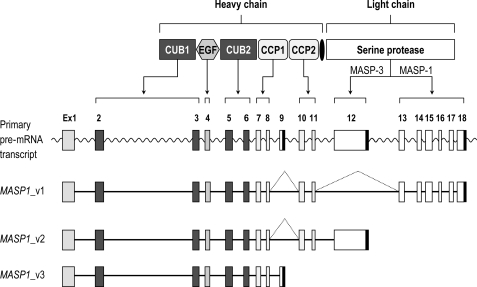
We searched the National Centre for Biotechnology Information Entrez data base for alternative transcript sequences of MASP1 and identified a novel mRNA sequence (MASP1_v3). The corresponding protein “MASP-1 isoform 3” was named MAP-1 for MBL/ficolin-associated protein 1. Alignment analysis of the nucleotide sequences of the different MASP1 transcript variants (Figs. 1 and 2) demonstrated that MASP1_v3 only shares exons 1–8 with the other two splice variants of MASP1. These exons encode most of the common heavy chain. However, because of an extra exon (exon 9) containing a stop codon, this molecule does not have a serine protease domain (Fig. 2). The corresponding MAP-1 protein sequence encodes 380 amino acids, whereas MASP-1 and MASP-3 contains 699 and 728 amino acids, respectively (Fig. 1). By alignment analysis of the deduced amino acid sequences, we found that MAP-1 lacks the second CCP domain and the entire serine protease domain but contains 17 unique C-terminal amino acid residues (Fig. 1). We calculated the theoretical molecular mass based on the deduced amino acid sequence to be 43.6 kDa with predicted signal peptide and 41.4 kDa without predicted signal peptide.
FIGURE 1.
Alignment of the isoforms of MASP1. The deduced amino acid sequences of MASP-1 (MASP1 isoform 1), MASP-3 (MASP1 isoform 2), and MAP-1 (MASP1 isoform 3) were aligned using the BioEdit Software. The dots represent identical amino acids. MASP-1 and MASP-3 contain different C-terminal serine protease domains, whereas MAP-1 does not contain a serine protease domain. Instead the protein contains 17 unique amino acids at the C-terminal end.
FIGURE 2.
Alternative splicing of the MASP1 gene. MASP1_v1 is generated by splicing out exons 9 and 12, both of which contain a stop codon sequence (marked with black boxes). The MASP1_v1 sequence contains a stop codon in exon 18. The MASP1_v2 is generated by splicing out exon 9 and terminated by the stop codon in exon 12. The MASP1_v3 is generated if exon 9 is not spliced out. The MAP-1 protein contains the exons encoding the two CUB domains, the EGF domain, and the first CCP domain together with the unique exon 9 domain.
Tissue Distribution of Human MASP1_v1, MASP1_v2, and MASP1_v3 mRNA Using Real Time Quantitative PCR Analysis
The expression of MASP1_v1, MASP1_v2, and MASP1_v3 transcripts in tissues were investigated by end point PCR analysis on cDNA panels from Clontech (data not shown). High mRNA levels of all three transcript variants were detected in the liver. Furthermore, MASP1_v3 was strongly expressed in heart tissue, and a lower mRNA level of MASP1_v3 was detected in skeletal muscle, brain, colon, prostate, and small intestine. No detectable mRNA levels were detected in bone marrow, cecum, duodenum, esophagus, jejunum, kidney, lung, lymph, ovary, pancreas, placenta, spleen, stomach, testis, thymus, or tonsil.
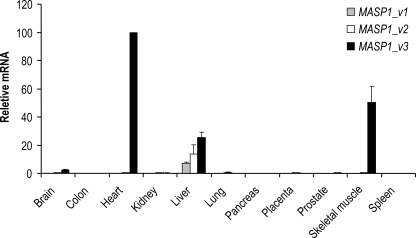
Based on the initial end point PCR analysis, tissues with MASP1_ v1, MASP1_v2, or MASP1_v3 expression were selected for further investigations using a new independent cDNA library. For quantification we used real time qPCR analysis. Confirming the previous observations, high levels of MASP1_v1 and MASP1_v2 mRNA mainly were found in liver tissue (Fig. 3), but a minor extra-hepatic expression of MASP1_v2 was also detected in all the investigated tissues, corresponding to ∼1–5% of that observed in the liver (data not shown). The mRNA concentration of MASP1_v3 was higher in liver tissue compared with MASP1_v1 and MASP1_v2 (Fig. 3). However, of particular interest was the finding that the MASP1_v3 mRNA level was expressed approximately two times higher in skeletal muscle and approximately four times higher in the heart tissue compared with the relative mRNA concentration in the liver.
FIGURE 3.
Quantitative tissue distribution of MASP1_v1, MASP1_v2, and MASP1_v3. The tissue distributions of the MASP1_v1, MASP1_v2, and MASP1_v3 transcripts were investigated using real time qPCR. A very high expression of MASP1_v3 was detected in the heart and skeletal muscle tissues. The MASP1_v3 expression was higher in liver compared with MASP1_v1 and MASP1_v2. The transcription was normalized to the mean of BCR, GAPDH, and β-actin housekeeping genes. The relative expression of the splice variants of MASP1 was calculated with the MASP1_v3 expression in the heart as reference (index, 100). The data were corrected for differences in amplification efficiencies. The qPCR system was performed in duplicate.
DNA Sequencing of MASP1 Exon 9 in 100 Individuals
To investigate a possible genetic variation of the specific exon of the MASP1_v3 transcript, we sequenced the MASP1_exon 9 in 100 healthy Caucasian individuals. No genetic variations were observed in this exon (data not shown).
Antibody Specificities
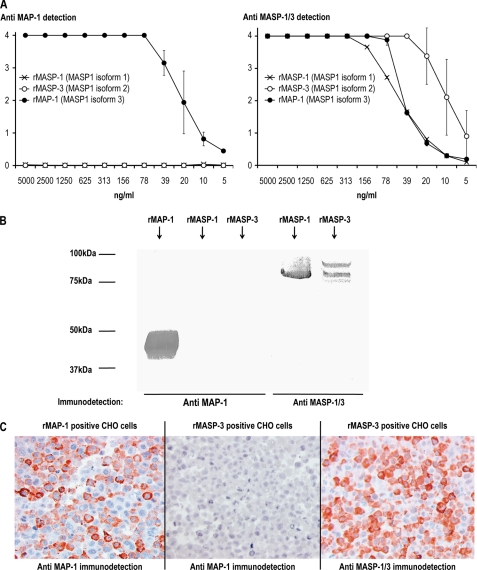
We used ELISA, immunoblotting, and immunohistochemistry to validate the specificities of the antibodies used. Fig. 4A (left side) shows the reaction pattern of anti-MAP-1 mAb 20C4 to rMASP-1, rMASP-3, and rMAP-1 immobilized to an ELISA plate in serial dilutions. No cross-reactivity to rMASP-1 or rMASP-3 was observed. An antibody to the common heavy chain of MASP-1 and MASP-3 was used as a positive control (Fig. 4A, right side). Additionally no cross-reactivity of mAb 20C4 to rMASP-1 and rMASP-3 was observed in SDS-PAGE/immunoblotting (Fig. 4B). CHO cells expressing either rMASP-3 or rMAP-1 were fixed in formaldehyde, embedded in paraffin, and used to screen for specificity in immunohistochemistry. The anti-MAP-1 antibody showed a positive staining of the MAP-1-positive cells, and no cross-reactivity to the MASP-3 cells was evident (Fig. 4C). The anti-MASP-1/3 mAb 8B3 served as a control for the immunohistochemical staining of the MASP-3 positive cells.
FIGURE 4.
Antibody specificities. A, rMASP-1, rMASP-3, and rMAP-1 (MASP1 isoforms 1, 2, and 3, respectively) were immobilized in 2-fold serial dilutions on Maxisorp plates at a starting concentration of 5 μg/ml. Immunodetection was performed with anti-MAP-1 (mAb 20C4; left side) and with anti-MASP-1/3 (mAb 8B3; right side). Immunodetection values are given as A490–650 nm. The error bars indicate two times the standard deviation of double determinations. B, SDS-PAGE/electroblotting of rMASP-1, rMASP-3, and rMAP-1. Immunodetection was performed with anti-MAP-1 (mAb 20C4; left side) and anti-MASP-1/3 (mAb 8B3; right side). C, immunohistochemical analysis of formaldehyde fixed CHO cells expressing rMASP-3 or rMAP-1. Immunodetection was performed with anti-MAP-1 (mAb 12B11; left and middle section) and with anti-MASP-1/3 (mAb 8B3; right section).
Tissue Localization of MAP-1
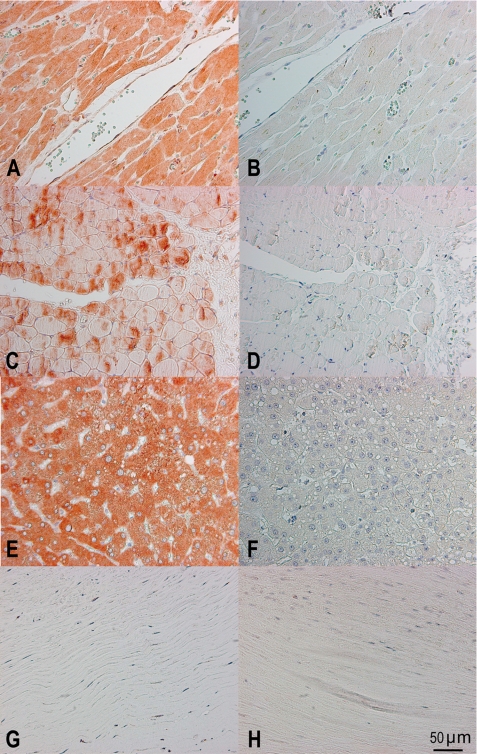
Immunohistochemistry was used to confirm the qPCR analysis and to identify specific cells and tissue areas positive for MAP-1. We analyzed samples from different individuals (two myocardial samples, two skeletal muscle samples, six liver samples, and two aortic samples). Strong staining with the anti-MAP-1 monoclonal antibody was observed in myocardium/cardiac muscle fibers, skeletal muscle fibers, and liver hepatocytes, but no staining was seen in aortic tissue (Fig. 5, left panels). This staining pattern was further validated using polyclonal anti-MAP-1 antibodies (data not shown). Difference in the inter-individual staining intensity of the tissues was observed (data not shown). No staining was evident when the tissues were analyzed with isotype control antibodies (Fig. 5, right panels).
FIGURE 5.
Immunohistochemical analysis of MAP-1 (MASP1 isoform 3) tissue localization. The left panels show staining with the specific mAb 12B11 to MAP-1. The right panels show the isotype control staining with a nonrelated IgG1k mAb. A and B, myocardium; C and D, skeletal muscle; E and F, liver tissue; G and H, aortic tissue. The bottom right corner bar indicates 50 μm on all slides.
MAP-1 Co-precipitates with Ficolin-2, Ficolin-3, and MBL
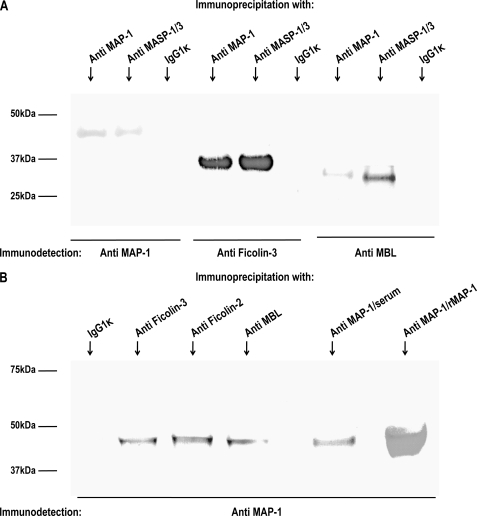
To investigate a possible association between MAP-1 and MBL and ficolin-3, we precipitated serum complexes using both an antibody to MAP-1 (mAb 20C4) and an antibody against the common heavy chain of MASP-1 and MASP-3 (mAb 8B3). An IgG1κ isotype antibody served as a negative control. The precipitates were subsequently analyzed by reduced SDS-PAGE, and immunoblotting was detected with biotinylated antibodies to MAP-1, MBL, and ficolin-3, respectively. We observed pronounced ficolin-3 co-precipitation bands, but weaker bands were also seen with MBL (Fig. 6A). The samples were not probed with antibodies against ficolin-2 because they were not applicable for reduced immunoblotting. We then reversed the immunoprecipitation using mAbs against MBL, ficolin-2, and ficolin-3 to precipitate serum complexes from 1 ml, 300 μl, and 100 μl of serum, respectively. The different serum volumes were chosen to adjust for differences in the serum concentration of MBL (2 μg/ml), ficolin-2 (5 μg/ml), and ficolin-3 (20 μg/ml), respectively. The samples were subsequently analyzed by immunoblotting probed with a biotinylated antibody to MAP-1. Distinct MAP-1 bands were observed in the precipitates from ficolin-2 and ficolin-3, and a slightly weaker band was apparent in the MBL precipitate (Fig. 6B, left side). Immunoprecipitated serum MAP-1 and rMAP-1 served as a positive control (Fig. 6B, right side). No MAP-1 band was evident when the IgG1κ isotype antibody was used as precipitation mAb.
FIGURE 6.
Immunoprecipitation of MAP-1 (MASP1 isoform 3) and MASP-1/3 (MASP-1 isoform 1 and 2) serum complexes. A, MAP-1 and MASP-1 and -3 were immunoprecipitated from serum using anti-MAP-1 (mAb 20C4) and anti-MASP-1/3 (mAb 8B3). Serum immunoprecipitation with an IgG1κ isotype antibody served as a negative control. Reduced samples were applied to SDS-PAGE, and immunoblotting was detected with biotinylated mAbs to ficolin-3 (FCN313), MBL (Hyb 131-11), or MAP-1 (mAb 20C4). B, immunoprecipitation with mAbs to MBL (Hyb 131-11), ficolin-2 (FCN219), and ficolin-3 (FCN334) from 1 ml, 300 μl, and 100 μl of serum, respectively (left side). Positive controls were MAP-1 precipitated from 100 μl serum and rMAP-1 from 1 ml of culture supernatant using mAb 20C4 (right side). Immunoprecipitation from 1 ml of serum with an IgG1κ isotype antibody served as negative control. The samples were analyzed by immunoblotting probed with a biotinylated antibody to MAP-1.
MAP-1 Inhibits Complement Activity of the Lectin Pathway
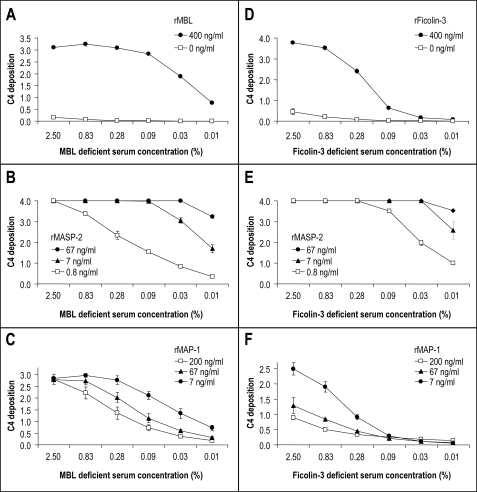
The rMBL (13) and rficolin-3 (14) were used to reconstitute the complement activity in serum deficient in MBL (21) or ficolin-3 (20), respectively. Mannan and AcBSA served as ligands for MBL and ficolin-3. Both rMBL and rficolin-3 were able to initiate C4 deposition in the MBL- and ficolin-3-deficient sera, respectively (Fig. 7, A and D). Preincubation of rMBL or rficolin-3 with serial dilutions of rMASP-2 resulted in a strong dose-dependent enhancement of the C4 deposition via both the MBL and ficolin-3 activation pathways (Fig. 7, B and E), whereas preincubation of rMBL or rficolin-3 with rMAP-1 resulted in a pronounced dose-dependent inhibition of the C4 deposition via both pathways (Fig. 7, C and F).
FIGURE 7.
Influence of rMASP-2 (MASP2 isoform 1) and rMAP-1 (MASP1 isoform 3) on the MBL- and ficolin-3-mediated complement C4 deposition. Polystyrene plates were coated with either mannan followed by 400 ng/ml rMBL or AcBSA followed by 400 ng/ml rficolin-3. MBL- or ficolin-3-deficient serum was added in dilutions from 0.01 to 2.5%. C4 depositions were measured using a polyclonal antibody to C4 and are given as A490–650 nm values. The error bars indicate two times the standard deviation of double determinations. Approximated concentrations of rMBL, rficolin-3, rMAP-1, and rMASP-2 are given in the figure labels. A–C show MBL-dependent C4 deposition on mannan, whereas D–F show ficolin-3-dependent C4 deposition on AcBSA. A, C4 deposition on a mannan-coated surface incubating rMBL at 400 ng/ml before addition of MBL-deficient serum in different dilutions. The control was without the addition of rMBL. B, dose-dependent enhancing effect of rMASP-2 on the rMBL-mediated C4 deposition. A total of 400 ng/ml of rMBL was preincubated with serial dilutions of rMASP-2 prior to the application of MBL-deficient serum as described under “Experimental Procedures.” C, dose-dependent inhibitory effect of rMAP-1 on the rMBL-mediated C4 deposition. A total of 400 ng/ml of rMBL was preincubated with serial dilutions of rMAP-1 prior to the application of MBL-deficient serum as described under “Experimental Procedures.” D, C4 deposition on an AcBSA-coated surface incubating rficolin-3 at 400 ng/ml before the addition of ficolin-3-deficient serum in different dilutions. The control was without addition of rficolin-3. E, dose-dependent enhancing effect of rMASP-2 on the rficolin-3-mediated C4 deposition. A total of 400 ng/ml of rficolin-3 was preincubated with serial dilutions of rMASP-2 prior to the application of ficolin-3-deficient serum as described under “Experimental Procedures.” F, dose-dependent inhibitory effect of rMAP-1 on the rficolin-3-mediated C4 deposition. A total of 400 ng/ml of rficolin-3 was preincubated with serial dilutions of rMAP-1 prior to the application of ficolin-3-deficient serum as described under “Experimental Procedures.”
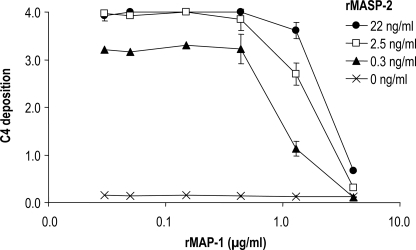
In addition we addressed a possible displacement of MASP-2 with MAP-1 using a system of pure components comprising only of rMBL, rMASP-2, rMAP-1, and purified C4. The rMASP-2 was preincubated with mannan-rMBL complexes in serial dilutions. Thereafter, rMAP-1 was added in varying concentrations followed by the addition of purified C4. The application of rMAP-1 to the system clearly reduced the levels of C4 deposition (Fig. 8).
FIGURE 8.
Influence of rMASP-2 (MASP2 isoform 1) and rMAP-1 (MASP1 isoform 3) on the complement C4 deposition in a pure system. A total of 400 ng/ml of rMBL was applied to a mannan surface followed by incubation for 45 min with serial dilutions of rMASP-2 in the first dimension. Afterward the serial dilutions of rMAP-1 were applied in the second dimension followed by application of purified C4 at 1 μg/ml. The C4 depositions (given as A490–650 nm values) were measured with a polyclonal antibody to C4. The error bars indicate two times the standard deviation of double determinations. Approximated concentrations of rMAP-1 and rMASP-2 are given in the figure labels.
DISCUSSION
We discovered an unidentified protein band with an apparent molecular mass of 45 kDa when ficolins or MBL were precipitated from human serum and subsequently subjected to immunoblotting probed with antibodies against the common heavy chain of the MASP-1 and MASP-3 (MASP1 isoforms 1 and 2, respectively) (data not shown). This prompted us to search the data bases for alternative transcript sequences of the MASP1 gene. A novel MASP1-derived transcript was found containing exons 1–8 that encodes most of the common heavy chain shared with MASP1_v1 and MASP1_v2. This transcript did not contain exons 10–18 encoding the second CCP domain and both serine protease domains of MASP1 and MASP3 (Fig. 2). However, the transcript contained an until now unidentified exon (exon 9) encoding 17 unique C-terminal amino acid residues and an in-frame a stop codon. The transcript was named MASP1_v3, and the corresponding protein was MAP-1 (MASP1 isoform 3).
Performing end point PCR analysis on cDNA libraries, we found that the MASP1_v3 transcript was expressed in different tissues including liver, brain, colon, prostate, heart, and skeletal muscles. This was different from the expression of MASP1_v1 and MASP1_v2, which were mainly expressed in the liver. Based on the initial results, tissues with a positive signal of MASP1_v1, MASP1_v2, or MASP1_v3 transcripts were selected for further quantitative expression analysis on a new cDNA library. mRNA levels of MASP1_v1 and MASP1_v2 were mainly found in liver tissue. Nevertheless, minor extra-hepatic expression of MASP1_v2 was also detected in all of the investigated tissues, corresponding to approximately to 1–5% of that observed in the liver. The MASP1_v3 mRNA level was higher in liver tissue compared with MASP1_v1 and MASP1_v2. Surprisingly the highest concentration of MASP1_v3 mRNA was found in heart tissue followed by skeletal muscle tissue, indicating that striated muscle tissue is a major source of MASP1_v3. We have previously observed some differences in the expression pattern between MASP1_v1 and MASP1_v2, suggesting that a differential genetic regulation of these two splicing forms may exist (12). Alternative splicing is widely accepted as an important mechanism for generating genetic diversity and regulation of gene expression (22). Alternative promoter usage has also been correlated with downstream alternative splicing events, differential expression levels, and different tissue specificity even in the absence of protein isoforms (23). Alternative pre- mRNA splicing and the differential inclusion or exclusion of portions of a nascent transcript into the final protein-coding mRNA is widely recognized to be a ubiquitous mechanism for controlling protein expression (22). Differences in the activities and/or specificities of splicing factors and regulators of transcription thus may well explain the different mRNA levels and tissue distribution of MASP1_v3 compared with the MASP1_v1 and MASP1_v2 variants. MASP1_v3 transcription could also be initiated by a separate MASP1_v3 promoter in analogy with what has been observed for the MBL2 gene encoding the MBL protein sequence, which is initiated by alternative promoters (12, 24). However, none of these theories are mutually exclusive. Future studies will hopefully reveal the molecular origin behind the different expression pattern of the different MASP1 transcripts.
To characterize MAP-1, we generated both monoclonal and mono-specific polyclonal antibodies raised against a peptide comprising the 17 unique C-terminal amino acids of MAP-1. When using MAP-1 specific antibodies in a panel of paraffin-embedded tissues of myocardial, skeletal muscle, liver, and aortic tissue, we could clearly demonstrate that MAP-1 is ubiquitously present in myocardial myocyte fibrils, whereas a more restricted patchy staining pattern was observed in skeletal muscle fibrils. The latter could indicate that MAP-1 is induced by muscle contractions. By contrast no MAP-1 staining could be observed in aortic tissue, indicating that this isoform is expressed in striated muscle tissue and not in smooth muscle tissue, which are indeed fundamentally different in terms of structure, function, excitation-contraction coupling, and mechanisms of contraction.
We have shown that MAP-1 is present in serum as a 45-kDa protein in complex with MBL, ficolin-2, and ficolin-3 based on different immunoprecipitation experiments, which were visualized by SDS-PAGE and immunoblotting. These experiments clearly demonstrate that MAP-1 is a novel serum protein associated with the lectin pathway initiator molecules. Recently, crystal structures of the interaction between MASP-1 and MASP-3 with MBL and the ficolins revealed that the CUB1-EGF-CUB2 domains are responsible for the binding of MASP-1 and MASP-3 to MBL and ficolins (25). Therefore it is reasonable to assume that these domains are also responsible for the interactions between MAP-1 and MBL or ficolins. Furthermore, the main initiator of the lectin complement pathway, MASP-2, has similar CUB1-EGF-CUB2 domains that also interact with MBL and ficolins (26, 27). The serine protease domain of MASP-2 is responsible for the cleavage of C4 and C2, leading to further downstream complement activation (5). Thus, we hypothesized that MAP-1 could have a potentially regulatory role in the lectin complement pathway. When complexes of rMAP1 with rMBL and rficolin-3 were incubated with MBL- or ficolin-3-deficient serum, respectively, it was clear that rMAP-1 exerts a strong dose-dependent inhibitory effect on the complement C4 deposition, whereas recombinant rMASP-2 augmented this deposition. To further investigate whether rMAP-1 could displace rMASP-2, we used a serum-free system employing recombinant MBL, MASP-2, MAP-1, and purified C4. The rMAP-1 was able to almost completely inhibit the C4 deposition despite the fact that it was added in sequence after MASP-2, thus indicating that it could displace rMASP-2 from rMBL. This is in agreement with results demonstrating that the same amino acid residues in MBL are essential not only for MASP-1 and MASP-3 binding but also MASP-2 binding (28).
Taken together these results indicate that MAP-1 functions as a potent systemic regulator of the complement system in vivo together with C1 inhibitor and C4-binding protein. The presence of MAP-1 in striated muscle tissues may also indicate that MAP-1 could function as a local inhibitor of complement activation. Because of the tissue expression pattern, we cannot exclude the possibility that MAP-1 may have other functions outside the complement system yet to be clarified.
In conclusion we describe the identification of a novel differential spliced gene product of the MASP1 gene named MAP-1 (or MASP1 isoform 3), which is found as a serum protein with a molecular mass of 45 kDa. MAP-1 lacks the second CCP domain and the entire serine protease domain but contains 17 unique C-terminal amino acid residues. MAP-1 is found in association with MBL and ficolins and is highly expressed in myocardial and skeletal muscle tissues and also in liver hepatocytes. MAP-1 inhibited complement deposition by MBL and ficolin-3 and appears to be a novel regulator of complement activation via the lectin pathway.
Acknowledgments
We thank Vibeke Witved and Anette Kliem for technical assistance.
This work was supported by Rigshospitalet, The Capital Region of Denmark, The Novo Nordisk Research Foundation, The Lundbeck Foundation, The John and Birthe Meyer Foundation, and The Carlsberg Foundation.
- MBL
- mannose-binding lectin
- MASP
- MBL/ficolin-associated serine protease
- CCP
- complement control protein
- CUB
- C1r/C1s, Urchin-EGF, bone morphogenetic protein
- EGF
- epidermal growth factor
- sMAP/Map19
- small MBL-associated protein/19-kDa MBL-associated protein
- MAP-1
- MBL/ficolin-associated protein (MASP1 isoform 3)
- BCR
- breakpoint cluster region
- GAPDH
- glyceraldehyde-3-phosphate dehydrogenase
- mAb
- monoclonal antibody
- qPCR
- quantitative PCR
- CHO
- Chinese hamster ovary
- ELISA
- enzyme-linked immunosorbent assay
- PBS
- phosphate-buffered saline
- HRP
- horseradish peroxidase
- AcBSA
- acetylated bovine serum albumin.
REFERENCES
- 1.Walport M. J. (2001) N. Engl. J. Med. 344, 1058–1066 [DOI] [PubMed] [Google Scholar]
- 2.Endo Y., Matsushita M., Fujita T. (2007) Immunobiology 212, 371–379 [DOI] [PubMed] [Google Scholar]
- 3.Turner M. W. (2003) Mol. Immunol. 40, 423–429 [DOI] [PubMed] [Google Scholar]
- 4.Fujita T. (2002) Nat. Rev. Immunol. 2, 346–353 [DOI] [PubMed] [Google Scholar]
- 5.Thiel S., Vorup-Jensen T., Stover C. M., Schwaeble W., Laursen S. B., Poulsen K., Willis A. C., Eggleton P., Hansen S., Holmskov U., Reid K. B., Jensenius J. C. (1997) Nature 386, 506–510 [DOI] [PubMed] [Google Scholar]
- 6.Chen C. B., Wallis R. (2004) J. Biol. Chem. 279, 26058–26065 [DOI] [PubMed] [Google Scholar]
- 7.Takahashi M., Iwaki D., Kanno K., Ishida Y., Xiong J., Matsushita M., Endo Y., Miura S., Ishii N., Sugamura K., Fujita T. (2008) J. Immunol. 180, 6132–6138 [DOI] [PubMed] [Google Scholar]
- 8.Krarup A., Gulla K. C., Gál P., Hajela K., Sim R. B. (2008) Biochim. Biophys. Acta 1784, 1294–1300 [DOI] [PubMed] [Google Scholar]
- 9.Krarup A., Wallis R., Presanis J. S., Gál P., Sim R. B. (2007) PLoS. ONE 2, e623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Matsushita M., Fujita T. (1992) J. Exp. Med. 176, 1497–1502 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Takahashi M., Mori S., Shigeta S., Fujita T. (2007) Adv. Exp. Med. Biol. 598, 93–104 [DOI] [PubMed] [Google Scholar]
- 12.Seyfarth J., Garred P., Madsen H. O. (2006) Mol. Immunol. 43, 962–971 [DOI] [PubMed] [Google Scholar]
- 13.Larsen F., Madsen H. O., Sim R. B., Koch C., Garred P. (2004) J. Biol. Chem. 279, 21302–21311 [DOI] [PubMed] [Google Scholar]
- 14.Hummelshoj T., Fog L. M., Madsen H. O., Sim R. B., Garred P. (2008) Mol. Immunol. 45, 1623–1632 [DOI] [PubMed] [Google Scholar]
- 15.Skjoedt M. O., Palarasah Y., Rasmussen K., Vitved L., Salomonsen J., Kliem A., Hansen S., Koch C., Skjodt K. (2010) Dev. Comp. Immunol. 34, 59–68 [DOI] [PubMed] [Google Scholar]
- 16.Hummelshoj T., Thielens N. M., Madsen H. O., Arlaud G. J., Sim R. B., Garred P. (2007) Mol. Immunol. 44, 401–411 [DOI] [PubMed] [Google Scholar]
- 17.Munthe-Fog L., Hummelshøj T., Ma Y. J., Hansen B. E., Koch C., Madsen H. O., Skjødt K., Garred P. (2008) Mol. Immunol. 45, 2660–2666 [DOI] [PubMed] [Google Scholar]
- 18.Madsen J., Kliem A., Tornoe I., Skjodt K., Koch C., Holmskov U. (2000) J. Immunol. 164, 5866–5870 [DOI] [PubMed] [Google Scholar]
- 19.Garred P., Larsen F., Seyfarth J., Fujita R., Madsen H. O. (2006) Genes Immun. 7, 85–94 [DOI] [PubMed] [Google Scholar]
- 20.Munthe-Fog L., Hummelshøj T., Honoré C., Madsen H. O., Permin H., Garred P. (2009) N. Engl. J. Med. 360, 2637–2644 [DOI] [PubMed] [Google Scholar]
- 21.Garred P., Larsen F., Madsen H. O., Koch C. (2003) Mol. Immunol. 40, 73–84 [DOI] [PubMed] [Google Scholar]
- 22.House A. E., Lynch K. W. (2008) J. Biol. Chem. 283, 1217–1221 [DOI] [PubMed] [Google Scholar]
- 23.Landry J. R., Mager D. L., Wilhelm B. T. (2003) Trends Genet. 19, 640–648 [DOI] [PubMed] [Google Scholar]
- 24.Naito H., Ikeda A., Hasegawa K., Oka S., Uemura K., Kawasaki N., Kawasaki T. (1999) J. Biochem. 126, 1004–1012 [DOI] [PubMed] [Google Scholar]
- 25.Teillet F., Gaboriaud C., Lacroix M., Martin L., Arlaud G. J., Thielens N. M. (2008) J. Biol. Chem. 283, 25715–25724 [DOI] [PubMed] [Google Scholar]
- 26.Thielens N. M., Cseh S., Thiel S., Vorup-Jensen T., Rossi V., Jensenius J. C., Arlaud G. J. (2001) J. Immunol. 166, 5068–5077 [DOI] [PubMed] [Google Scholar]
- 27.Girija U. V., Dodds A. W., Roscher S., Reid K. B., Wallis R. (2007) J. Immunol. 179, 455–462 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Teillet F., Lacroix M., Thiel S., Weilguny D., Agger T., Arlaud G. J., Thielens N. M. (2007) J. Immunol. 178, 5710–5716 [DOI] [PubMed] [Google Scholar]