Abstract
Expression of the cytokine interleukin-13 (IL13) is critical for Th2 immune responses and Th2-mediated allergic diseases. Activation of human IL13 expression involves chromatin remodeling and formation of multiple DNase I-hypersensitive sites throughout the locus. Among these, HS4 is detected in the distal IL13 promoter in both naive and polarized CD4+ T cells. We show herein that HS4 acts as a position-independent, orientation-dependent positive regulator of IL13 proximal promoter activity in transiently transfected, activated human CD4+ Jurkat T cells and primary murine Th2 cells. The 3′-half of HS4 (HS4-3′) was responsible for IL13 up-regulation and bound nuclear factor (NF) 90 and NF45, as demonstrated by DNA affinity chromatography coupled with tandem mass spectrometry, chromatin immunoprecipitation, and gel shift analysis. Notably, the CTGTT NF45/NF90-binding motif within HS4-3′ was critical for HS4-dependent up-regulation of IL13 expression. Moreover, transfection of HS4-IL13 reporter vectors into primary, in vitro differentiated Th2 cells from wild-type, NF45+/−, or NF90+/− mice showed that HS4 activity was exquisitely dependent on the levels of endogenous NF45 (and to a lesser degree NF90), because HS4-dependent IL13 expression was virtually abrogated in NF45+/− cells and reduced in NF90+/− cells. Collectively, our results identify NF45 and NF90 as novel regulators of HS4-dependent human IL13 transcription in response to T cell activation.
Keywords: DNA/Transcription, Gene/Regulation, Immunology, Methods/Mass Spectrometry, Transcription, Transcription/Promoter, Transcription/Regulation
Introduction
The T cell-derived cytokine interleukin (IL)2-13 plays a pivotal effector role in T helper type 2 (Th2) immune responses to extracellular parasites (1). When dysregulated by genetic and/or environmental factors, IL13 expression is essential for the pathogenesis of allergic diseases (2). Indeed, experimental animal models have shown that IL-13 is necessary and sufficient to induce all the cardinal features of allergic lung inflammation, including airway hyper-responsiveness, eosinophilia, goblet cell metaplasia and mucus hyper-secretion, epithelial cell damage, and fibrosis (3–5). IL13 expression and IL-13-dependent events are also amplified in human allergy (6, 7), and high IL-13 production in early life is strongly associated with the subsequent development of allergic sensitization (8–10).
The IL13 gene lies within the Th2 cytokine locus on human chromosome 5q31, which also includes IL4 and IL5. Th2 cytokine gene expression has been shown to be tightly coordinated and fine-tuned by multiple local and distant cis-regulatory elements that are located throughout the Th2 locus and are marked by developmentally conserved DNase I-hypersensitive (HS) sites (11). HS sites typically reflect the DNA binding activity of sequence-specific trans-acting factors that induce destabilization or displacement of local nucleosomes (12) and mark the location of enhancers, silencers, or locus control regions (13). In the murine Th2 locus, nuclease HS regions appear to work cooperatively (14, 15) and engage in local and long range intra-chromosomal interactions that are essential for concerted Th2 cytokine expression (16).
We recently characterized the dynamic modifications in DNase I hypersensitivity and epigenetic marks that occur at the human IL13 locus during the differentiation of naive CD4+ Th cells into a polarized IL-13/IL-4 secreting Th2 phenotype (17). Our study demonstrated that distinct regions of the IL13 locus exhibit distinct patterns of chromatin accessibility at defined stages of the Th cell differentiation process. In naive T cells, chromatin at the proximal promoter, the transcription unit, and the IL13/IL4 intergenic region was in an inaccessible state, marked by the absence of HS sites and by extensive CpG hypermethylation. During Th2 differentiation, these regions underwent profound remodeling, revealed by the appearance of numerous HS sites that co-localized with DNA hypomethylation. In contrast, the distal IL13 promoter contained two closely spaced, novel HS sites, HS4 and HS5, which were detectable in unstimulated naive CD4+ T cells and persisted throughout Th cell differentiation (17). Detection of constitutive HS sites in naive CD4+ T cells was intriguing because these cells rapidly express substantial amounts of IL-13 upon T cell receptor cross-linking (18). Early accessibility of the distal promoter suggested occupancy of HS4 and HS5 by constitutive transcription factors might poise the locus for rapid IL13 expression upon T cell activation and/or differentiation.
Defining properties of the HS5 region were characterized in previous work (19). The work presented herein was designed to investigate whether HS4 marks the location of an IL13 cis-regulatory element and to identify nucleoproteins that regulate IL13 transcription by interacting with the HS4 region. We show that HS4 does indeed act as a novel positive regulator of human IL13 promoter activity in response to T cell activation. Nuclear factor (NF) 90 and NF45 played an important role in HS4-dependent up-regulation of IL13 expression.
EXPERIMENTAL PROCEDURES
Mice
C57BL/6 wild-type (WT) mice obtained from The Jackson Laboratory and NF45+/− 3 and NF90+/− (20) mice on a C57BL/6 background were maintained under specific pathogen-free conditions. All experiments were performed according to institutional and federal guidelines.
T Cell Culture, Isolation, and Th2 Differentiation
Jurkat T cells (ATCC clone E6-1) were cultured in RPMI 1640 medium supplemented with fetal bovine serum (10%), penicillin (100 units/ml), streptomycin (100 μg/ml), and l-glutamine (2 mm). To generate murine Th2 cells, CD4+ T cells were isolated from splenocyte suspensions using the CD4+ T cell isolation kit (Miltenyi) as recommended by the manufacturer. Cells (∼5 × 106) were resuspended in Dulbecco's modified Eagle's medium supplemented with fetal calf serum (10%), HEPES (10 mm), 2-mercaptoethanol (0.1 mm), penicillin (100 units/ml), streptomycin (100 μg/ml) and l-glutamine (2 mm) and stimulated for 3 days with plate-bound anti-CD3 mAb (clone 145-2C11, 1 μg/ml) and anti-CD28 mAb (clone 37.51, 1 μg/ml) in the presence of IL-4 (1000 units/ml) and neutralizing antibodies against interferon-γ (clone R4-6A2, 5 μg/ml) and IL-12 (clone C17.8, 3 μg/ml) (21). After expansion in the presence of IL-4, neutralizing antibodies, and IL-2 (20 units/ml) for 4 days, cells were re-stimulated with plate-bound anti-CD3 and anti-CD28 mAbs and nucleofected at days 9 and 10. The efficiency of Th2 cell polarization was assessed by intracellular cytokine staining (17). Levels of NF45 and NF90 RNA were reduced by ≈50% in NF45+/− and NF90+/− Th2 cells as measured by real time PCR using predeveloped QuantiTect primer assays for murine NF45/ILF2, NF90/ILF3, and GapdH (Qiagen) with SYBR Green detection on an ABI Prism 7900 sequence detection system (data not shown).
Reporter Constructs
HS6/Luc (22) consisted of a 363-bp region encompassing the human IL13 proximal promoter (−369 to −6, relative to the IL13 ATG, GenBankTM accession number L42080) cloned into pGL3 Basic (Promega). PCR amplification of HS4 (−1650 to −1435) was performed with primers 5′ATACTCGTCGACATAAGGGGCGTTGACTCAC and 5′TTGATGTCGACTCTGACTCCCAGAAGTCTG or HS4-3′ (−1577 to −1435) with primers 5′ATACTCGGTACCATCACGGAGACCCTGTGGGAGAT and 5′TTGATGTCGACTCTGACTCCCAGAAGTCTG, and cloning of these regions into the SalI restriction site of HS6/Luc generated HS4-HS6/Luc and HS4-3′-HS6/Luc, respectively. The −1650IL13p/Luc, −1577IL13p/Luc, and −1446IL13p/Luc constructs were generated by PCR amplification using distinct forward primers (5′ATACTCGGTACCATAAGGGGCGTTGACTCAC, 5′ATACTCGGTACCATCACGGAGACCCTGTGGGAGAT, and 5′ATACTCGGTACCTGGGAGTCAGAGCCAGCGCT) and a single reverse primer (5′TTGATGCTAGCCAGTGCCAACAGGAGAGGATT). Each of these regions was then cloned into the KpnI and NheI restriction sites of pGL3 Basic. The −1577mutIL13p/Luc construct, in which the CTGTT motif within HS4-3′ was mutated by transversion of nucleotides −1529 to −1521, was generated by ligating two PCR fragments (corresponding to nucleotides −1577 to −1521 and −1527 to −6) to each other using a newly created BsmFI restriction site. The ligated product was then cloned into the SacI and NheI restriction sites of pGL3 Basic. HS4-3′mut-HS6/Luc was generated applying the same cloning strategy followed to create HS4-3′-HS6/Luc, with the exception that −1577mutIL13p/Luc was used as a PCR template. Construct sequence fidelity and HS4 orientation were assessed by sequencing.
Transient Transfections
Endotoxin-free plasmids (5 μg) were electroporated into Jurkat T cells (5 × 106) in log phase of growth using the BTX ECM830 square wave electroporator (1 pulse, 250 V, 50 ms). Cells were co-transfected with pRLTK (25 ng, Promega) to normalize for transfection efficiency. After electroporation, cells were incubated in the presence or absence of phorbol 12-myristate 13-acetate (PMA) (20 ng/ml, Sigma) and ionomycin (1 μm, Sigma). Anti-CD3/anti-CD28 mAb-activated murine Th2 cells (1 × 106) were nucleofected with endotoxin-free reporter constructs (1 μg) and pRL-TK (50 ng) using the Amaxa Nucleofector and the mouse T cell Nucleofector kit (Amaxa).
Firefly and Renilla luciferase activities in cell lysates were assessed 16 h after electroporation or nucleofection using the Dual LuciferaseTM assay system (Promega). Total protein concentrations in cell lysates were determined using the BCA assay (Pierce). Results were expressed as relative luciferase activity (RLA) units, i.e. firefly luciferase activity corrected by transfection efficiency and protein content detected in each sample or as fold induction values, i.e. ratio of RLA units measured in stimulated over unstimulated samples.
Nucleoprotein Extraction
Nuclear extracts from Jurkat T cells in log phase of growth were prepared in 2.5-liter batches. Cells were collected by centrifugation and washed twice with phosphate-buffered saline. Cell pellets were resuspended (1:5 v/v) in buffer A (3 mm MgCl2, 10 mm NaCl, 10 mm Tris, pH 7.5, 0.1 mm EGTA, 0.5 mm DTT, 1 μg/ml aprotinin, 2 μm leupeptin, 1 mm phenylmethylsulfonyl fluoride) and incubated on ice for 10 min. Nonidet P-40 (0.5%) was added, and nuclei were pelleted by centrifugation. Nuclear pellets were resuspended (1:2.5 v/v) in buffer C (1.5 mm MgCl2, 20 mm HEPES, pH 7.0, 420 mm NaCl, 25% glycerol, 0.2 mm EDTA, 0.5 mm DTT, 1 μg/ml aprotinin, 2 μm leupeptin, 0.5 mm phenylmethylsulfonyl fluoride) and incubated on ice for 30 min. After centrifugation, supernatants were collected, snap-frozen in liquid nitrogen, and stored at −80 °C.
DNA Affinity Chromatography
Nuclear extracts (5 mg) were preincubated on ice for 30 min with 5 μg of the biotinylated oligonucleotides NcoI-HS4-3′-(−1527/−1496) or NcoI-control. The latter corresponds to an unrelated region that failed to bind any of the NcoI-HS4-3′-(−1527/−1496)-binding proteins in EMSA. DNA-protein complexes were then bound to streptavidin-coated magnetic beads (500 μl, Miltenyi) for 30 min. After incubation, the mixture was loaded onto a 5-ml column (Miltenyi) attached to a magnet (Miltenyi). Magnetic beads were washed four times with wash buffer (1.5 mm MgCl2, 20 mm HEPES, 100 mm NaCl, and 0.5 mm phenylmethylsulfonyl fluoride). The column was then removed from the magnet and bead-DNA-protein complexes were eluted with wash buffer supplemented with 8.5 mm MgCl2 and 1 mm DTT. NcoI (10 units) was added to the sample, and digestion was performed for 1 h at 37 °C. After digestion, the sample was loaded again onto the original column and attached to a magnet. The bypass (i.e. DNA-protein complexes) was then collected. Eluted DNA-protein complexes from six columns were combined and concentrated by overnight precipitation with ice-cold acetone at −20 °C.
Concentrated protein samples were either trypsin-digested in solution or separated by SDS-PAGE prior to peptide digestion following standard protocols. Gels were silver-stained, and bands resulting from separation of NcoI-HS4-3′-(−1527/−1496) affinity column eluates were excised. This procedure could not be performed for NcoI-control affinity column eluates because they had low protein content.
Mass Spectrometry
Tandem mass spectrometry coupled to liquid chromatography (LC-MS/MS) analyses of digested protein gel bands were carried out using a linear quadrupole ion trap ThermoFinnigan LTQ mass spectrometer equipped with a Michrom Paradigm MS4 HPLC with a reverse phase column, a SpectraSystems AS3000 autosampler, and a nanoelectrospray source. Dependent data scanning was performed by Xcalibur version 1.4 software (23) with a default charge of 2, an isolation width of 1.5 atomic mass units, an activation amplitude of 35%, activation time of 30 ms, and a minimal signal of 100 ion counts (arbitrary units based on mass spectrometer used). Globally dependent data settings were as follows: reject mass width of 1.5 atomic mass units, dynamic exclusion enabled, exclusion mass width of 1.5 atomic mass units, repeat count of 1, repeat duration of 1 min, and exclusion duration of 5 min. Scan event series included one full scan with mass range 350–2000 Da, followed by six dependent MS/MS scans of the most intense ion. Tandem MS spectra of peptides were analyzed with TurboSEQUESTTM version 3.1 (24), and the peak list (dta files) for the search was generated by Bioworks 3.1. Parent peptide mass error tolerance was set at 1.5 atomic mass units, and fragment ion mass tolerance was set at 0.5 atomic mass units during the search. The criteria that were used for a preliminary positive peptide identification are the same as described previously, namely peptide precursor ions with a +1 charge having an Xcorr >1.8, +2 Xcorr > 2.5 and +3 Xcorr > 3.5. A dCn score >0.08 and a fragment ion ratio of experimental/theoretical >50% were also used as filtering criteria for reliable matched peptide identification (25, 26). All matched peptides were confirmed by visual examination of the spectra. All spectra were searched against a human-rat-mouse data base created from the latest version of the nonredundant protein data base downloaded March 16, 2004, from NCBI. At the time of the search, this custom protein data base from NCBI contained 168,153 entries. The results were also validated using XTandem (27) and Scaffold 2 (28, 29). Results correspond to proteins/peptides identified with a probability over 95%. For tandem mass spectrometry coupled to two-dimensional liquid chromatography (LC-LC-MS/MS or Multidimensional Protein Identification Technology (MudPIT)) analyses of digested eluates, a microbore HPLC system (Paradigm MS4, Michrom) was used with two separate strong cation exchange and reverse phase columns. Peptide elution with a 12-step gradient protocol was performed directly into a custom-built nanoelectrospray ionization source of a ThermoFinnigan LCQ-Deca XP Plus ion trap mass spectrometer. Dependent data scanning was performed as described above, with the exception that a minimal signal of 10,000 was used. Tandem MS spectra of peptides were also analyzed as described above.
Chromatin Immunoprecipitation (ChIP)
Jurkat T cells in log phase of growth were incubated in the presence or absence of PMA (20 ng/ml) and ionomycin (2 μm) for 4 h. Formaldehyde cross-linking was then performed essentially as described previously (20, 30) but was stopped by the addition of 125 mm glycine. Cells were then washed twice with phosphate-buffered saline. Chromatin fragmentation and precipitation were performed as described previously (22). Briefly, cross-linked chromatin was sonicated five times for 10 s each time and diluted 10-fold in ChIP dilution buffer (Upstate Biotechnology, Inc.) supplemented with protease inhibitors. After preclearing the sample with a salmon sperm DNA/protein A-agarose slurry (Upstate Biotechnology, Inc.), chromatin was immunoprecipitated with a mouse anti-NF90 mAb (DRBP76, 10 μg; BD Biosciences) or a rabbit anti-NF45 antibody (10 μg, Aviva). Immunoprecipitated complexes were captured, cross-links were reversed, and DNA was recovered by phenol/chloroform extraction and ethanol precipitation. Real time PCR was performed using the QuantiTect SYBR Green PCR kit (Qiagen) on an ABI Prism 7900 sequence detection system. PCR primers (5′TTAGGAAACAGGCCCGTAGA and 5′CACAAGGGTGCTTGTGAC or 5′TTCTCAACCTCAGCACTGGTGACA and 5′GACTTTGCTGTTTGCTGTCAGGCT) were designed to amplify a 244-bp region of the human distal IL13 promoter (nucleotides −1531 to −1287) and a 249-bp region of the myc1 locus, respectively (20, 30). PCR amplifications were performed in triplicate with the following cycling conditions: 15 min at 95 °C followed by 40 cycles of 15 s at 90 °C, 30 s at 59 °C, and 30 s at 72 °C. Serial dilutions of input DNA were used to generate a standard curve for each experiment. Copy number values were calculated based on standard curves, and results were expressed as the ratio between immunoprecipitated IL13 promoter targets and immunoprecipitated myc1 targets.
EMSA
Jurkat T cells in log phase of growth were incubated in the presence or absence of PMA (20 ng/ml) and ionomycin (2 μm) for 4 h. Nuclei were prepared as described in the nucleoprotein extraction section. Nuclear proteins were then extracted as described previously (30, 31). Briefly, nuclear proteins were purified after incubation with 0.3 m ammonium sulfate for 30 min at 4 °C, and the supernatants were precipitated with 0.2 g/ml ammonium sulfate. Protein pellets were collected, resuspended in buffer C25 (25 mm HEPES, 25 mm KCl, 0.1 mm EDTA, 1 mm DTT, 10% glycerol), and dialyzed against three changes of buffer C25. For probe and competitor preparation, single-stranded complementary oligonucleotides were annealed and PAGE-purified. Double-stranded oligonucleotides were then end-labeled with [γ-32P]ATP using T4 polynucleotide kinase.
Nuclear extracts (15 μg) were incubated with 32P-labeled oligonucleotide probes in binding buffer (100 mm Tris-Cl, 10 mm EDTA, 10 mm DTT, 5 mm MgCl2, 8 mm Na2HPO4, pH 7.5, 0.6 mm NaN3, 400 ng/μl bovine serum albumin, 160 mm NaCl, 30% glycerol) and poly(dI-dC) (50 ng/μl) for 30 min at 4 °C. Oligonucleotide competitors (90- or 30-fold molar excess) were preincubated with nuclear extracts for 30 min at 4 °C before addition of radiolabeled probe. DNA-protein complexes were separated on 5% (w/v) nondenaturing polyacrylamide gels in Tris/borate/EDTA (0.5 times). Electrophoresis was performed at 20 mA for 5 h at 4 °C, and gels were dried prior to autoradiography.
Western Blot Analysis
Jurkat T cells were incubated in the presence or absence of PMA (20 ng/ml) and ionomycin (1 μm) for 4 h. After stimulation, cells were incubated with agitation for 20 min in lysis buffer (50 mm Tris-Cl, 150 mm NaCl, 1 mm EDTA, 1% Triton X-100, 1 mm DTT, 1× protease inhibitor mixture (Roche Applied Science), 5 mm β-glycerophosphate, 1 mm NaF, 1 mm NaV, 1 mm benzamidine). Lysates were clarified by centrifugation at 12,000 rpm for 10 min at 4 °C. Supernatants were then collected, snap-frozen in liquid nitrogen, and stored at −80 °C. For Western blot analyses, whole cell lysates were electrophoretically separated using 10% SDS-polyacrylamide gels and transferred to nitrocellulose membranes (0.45-μm pore size). Membranes were then probed with anti-NF45 (Aviva) or anti-NF90 antibodies (BD Biosciences), washed, and incubated with horseradish peroxidase-labeled secondary antibodies (Abcam). Protein bands were visualized by chemiluminescence (ECL, Abcam). Membranes were stripped, exposed again to ensure no residual signal was detectable, and incubated with an anti-phosphoserine antibody (Abcam) or with a mixture of anti-phospho-mitogen-activated protein kinase (MAPK)/ cyclin-dependent kinase (CDK)/AKT/ataxia telangiectasia mutated (ATM)/ATM and Rad3-related (ATR) protein substrate antibodies (Cell Signaling). Protein bands were detected as described above.
RESULTS
HS4 Marks the Location of a Novel IL13 cis-Regulatory Element
Our previous high resolution analysis of DNA methylation at the IL13 locus (17) showed that the CpG dinucleotides at positions −1650 and −1435 in the distal IL13 promoter (numbering relative to the IL13 ATG, GenBankTM accession number NC_000005) represent the boundaries of a region that is DNase I-hypersensitive and hypomethylated in naive as well as differentiated Th1 and Th2 cells. This region was named HS4 (Fig. 1A).
FIGURE 1.
HS4 acts as an IL13 cis-regulatory element. A, schematic representation of DNase I HS sites (arrows) mapped throughout the IL13 locus in human primary CD4+ T cells (17). Exons I–IV are depicted as black boxes. B and C, Jurkat T cells were transiently co-transfected with IL13 luciferase (Luc) reporter constructs and pRLTK and harvested after 16 h of culture in the presence or absence of PMA (20 ng/ml) and ionomycin (1 μm). Results are expressed as RLA fold-induction (mean ± S.E.) measured in four (B) or seven (C) independent experiments. Statistical significance was calculated using the Wilcoxon two-sample test. n.s., not significant.
To begin assessing whether HS4 marks the location of a novel cis-acting element involved in the regulation of IL13 expression, and to dissect the molecular mechanisms underlying its function, we took a reductionist approach. The region encompassing HS4 was cloned in both orientations downstream of a luciferase reporter gene in a construct driven by a highly active 369-bp fragment encompassing HS6, which corresponds to the proximal IL13 promoter (17, 22). These vectors were transiently transfected into human CD4+ Jurkat T cells, which are well suited for these studies because they exhibit DNase I hypersensitivity at HS44 and up-regulate IL13 mRNA levels ≈100-fold upon activation (22). Luciferase activity was measured after 16 h of culture in the presence or absence of activating stimuli (PMA, 20 ng/ml, and ionomycin, 1 μm) that recapitulate T cell receptor-mediated signaling. Fig. 1B shows that T cell activation resulted in brisk up-regulation of proximal IL13 promoter activity, which was further significantly increased by HS4 when the region was cloned in the genomic, but not the reverse, orientation. Complementary experiments compared the activity of IL13 promoter reporter vectors that did (−1650IL13p/Luc) or did not (−1446IL13p/Luc) carry HS4 at its genomic location. Fig. 1C shows that deletion of the entire HS4 region (−1650 to −1446) significantly decreased IL13 promoter activity. However, the latter was fully preserved when only the 5′ end of HS4 (−1650 to −1577) was deleted, indicating that the IL13-enhancing properties of HS4 map to the 3′ end of the element (HS4-3′, −1577 to −1446). In all experiments, HS4 had negligible effects on IL13 promoter-driven transcription in unstimulated cells (data not shown). Collectively, these results indicate that the 3′ end of HS4 acts as a position-independent, orientation-dependent cis-regulatory element that increases IL13 promoter activity in stimulated CD4+ Jurkat T cells.
Identification of NF45 and NF90 as HS4-3′-binding Proteins
To begin exploring the molecular mechanisms underlying HS4-3′-mediated enhancement of IL13 transcription, we examined DNA/protein interactions occurring at this region in resting and activated CD4+ Jurkat T cells. EMSA analysis detected distinct complexes specifically binding to HS4-3′(data not shown), but traditional approaches (oligonucleotide competition and/or antibody supershifting guided by a reliable binding motif-predicting algorithm (32)) failed to identify the constituent proteins. Therefore, we adopted a different hypothesis-generating strategy that sought to identify HS4-3′-binding nucleoproteins by combining DNA affinity chromatography with tandem mass spectrometry. For this analysis to achieve high resolution, HS4-3′ was divided into three shorter fragments (Fig. 2A). Here, we present the results obtained for HS4-3′-(−1527/−1496), the central portion of the element.
FIGURE 2.
Isolation and identification of HS4-3′-interacting proteins. A, location and sequence of biotinylated oligonucleotides used as baits in DNA affinity chromatography. The NcoI restriction site is underlined. B, biochemical approach to the isolation and identification of HS4-3′(−1527/−1496)-binding proteins. C, total numbers of protein species recovered from DNA affinity chromatography and identified by tandem mass spectrometry approaches. D, NF45 and NF90 peptides identified by tandem mass spectrometry. aa, amino acids.
Our strategy involved coupling a biotinylated HS4-3′-(−1527/−1496) oligonucleotide, or a biotinylated unrelated negative control, to magnetic beads coated with streptavidin and using the latter as baits to isolate the relevant DNA-binding proteins from Jurkat T cell nuclear extracts. Because uncoupled magnetic beads were found to support some degree of nonspecific protein binding, we incorporated an NcoI restriction site at the 5′ end of the oligonucleotide baits (Fig. 2A). NcoI cleavage after incubation of the nuclear extracts with the magnetic beads, but before mass spectrometry analysis, maximized the recovery of DNA sequence-specific proteins.
Fig. 2B highlights the main steps in the protein isolation and identification process. Jurkat T cell nuclear proteins were incubated with NcoI-HS4-3′-(−1527/−1496) or NcoI-control oligonucleotides. The mixture was then loaded onto streptavidin-coated beads, and DNA-protein complexes were recovered from the affinity columns after digestion with NcoI. The identity of proteins bound to the HS4-3′-(−1527/−1496) oligonucleotide was established by two complementary proteomics approaches, one gel-based (SDS-PAGE followed by LC-MS/MS) and the other gel-free (MudPIT) (33). In contrast, MudPIT but not SDS-PAGE/LC-MS/MS was performed on proteins recovered from oligonucleotide control affinity columns because of low protein content. Proteins recovered from HS4-3′-(−1527/−1496) but not oligonucleotide control beads were considered to be DNA sequence-specific (Fig. 2B).
Fig. 2, C and D, and Table 1 show the results of this series of experiments. Overall, gel-based and gel-free strategies identified similar numbers of HS4-3′-(−1527/−1496)-binding proteins (Fig. 2C), 18 of which were detected by both approaches. Among these, seven proteins bound HS4-3′-(−1527/−1496) specifically (Table 1). HS4-3′-(−1527/−1496)-specific proteins identified only by SDS-PAGE/LC-MS/MS or MudPIT are listed in supplemental Tables 1 and 2. Among the HS4-3′-(−1527/−1496)-specific proteins listed in Table 1, NF45 and NF90 became the focus of subsequent work. Their identification as bona fide HS4-3′-(−1527/−1496)-binding proteins was supported not only by the recovery of the appropriate peptides (Fig. 2D) but also by several other independent findings: the detection of identical peptides when SDS-PAGE/LC-MS/MS was used to analyze Jurkat cell-derived and recombinant NF45 and NF90 (data not shown); the consistent recovery of both proteins, in line with their reported ability to bind DNA as heterodimers (31, 34, 35), and the presence in HS4-3′-(−1527/−1496) of a single CTGTT motif (−1525/−1521). Mutation of a region that includes this motif disrupted binding of the NF90-containing complex to the 3′ end of the antigen receptor-response element in the IL2 proximal promoter (IL2pARRE) (30).
TABLE 1.
HS4–3′-(−1527/−1496)-specific binding proteins identified by SDS-PAGE-LC-MS/MS and MudPIT
| Accession no.a | Protein name | Molecular mass | Total no. of unique peptides | Protein coverage |
|---|---|---|---|---|
| Dab | % | |||
| NP_006588.1 | Heat shock 70-kDa protein 8 | 70899.8 | 15 | 33 |
| XP_343269.1 | Siah-binding protein 1; FBP interacting repressor | 60249.0 | 7 | 22 |
| Q92979 | Probable ribosome biogenesis protein | 26720.3 | 6 | 36 |
| NP_005745.1 | GTPase-activating protein  SH3 domain-binding protein 1 SH3 domain-binding protein 1 |
52162.8 | 5 | 19 |
| Q12905 | Nuclear factor 45 | 44698.2 | 4 | 17 |
| NP_006550.1 | KH domain containing, RNA binding, signal transduction- associated 1 | 48228.2 | 3 | 17 |
| Q12906 | Nuclear factor 90 | 95384.9 | 2 | 5 |
a Accession numbers are from the NCBI Protein Database.
b Scaffold protein and peptide identification probability was >95%.
NF90 and NF45 Bind HS4 in Vivo
To validate the identification of NF45 and NF90 as HS4-3′-binding proteins, we next used ChIP to assess whether these proteins interact with HS4 in vivo. Cross-linked chromatin was isolated from Jurkat T cells (unstimulated or PMA/ionomycin-stimulated for 4 h) and immunoprecipitated with anti-NF90 or anti-NF45 antibodies. Real time PCR was then performed to test for enrichment of a 244-bp region spanning HS4-3′ (nucleotides −1531 to −1287). A negative control was provided by the amplification of a 249-bp region within the myc1 locus that fails to bind NF90 (30). Results were expressed as HS4-3′ relative copy number, i.e. as the ratio between immunoprecipitated IL13 promoter copies and immunoprecipitated myc1 copies. Fig. 3 shows that under basal conditions the HS4-3′-containing region was modestly enriched in chromatin samples immunoprecipitated with anti-NF90 or anti-NF45 antibodies, but T cell activation markedly increased the recruitment of endogenous NF90 and NF45 to HS4-3′. Because chromatin sonication for ChIP typically yields DNA fragments that are somewhat heterogeneous in size, ChIP analysis cannot formally prove that the docking site for NF45-NF90 complexes is located within, or is limited to, the HS4-3′ region. However, these results are fully consistent with the possibility that NF90 and NF45 interact with HS4-3′, and more generally they implicate these proteins in the regulation of endogenous IL13 in human CD4+ Jurkat T cells.
FIGURE 3.
NF90 and NF45 bind HS4 in vivo. Chromatin from Jurkat T cells, resting or activated with PMA (20 ng/ml) and ionomycin (2 μm) for 4 h, was cross-linked, sonicated, and immunoprecipitated with anti-NF90 or anti-NF45 antibodies. Target enrichment was assessed by real time PCR with primers that amplify a 244-bp region spanning HS4-3′ (nucleotides −1531 to −1287) or a 249-bp negative control region within the myc1 locus. A standard curve for each experiment was generated with serial dilutions of input DNA. Results are expressed as the mean ± S.E. of the ratio between the number of HS4 and myc1 target copies immunoprecipitated by anti-NF90 or anti-NF45 antibodies in three independent experiments. P/I, PMA + ionomycin.
Mapping of the Interactions between NF45/NF90 and HS4-3′-(−1527/−1496)
Because the NF45-NF90 complex is thought to interact with the CTGTT motif in the IL2 promoter ARRE (30) (sequence underlined in Fig. 4A), EMSA analysis was next performed to assess whether the CTGTT motif within HS4-3′-(−1527/−1496) supports binding of a similar complex. Nuclear extracts from Jurkat T cells cultured for 4 h in the presence or absence of PMA (20 ng/ml) and ionomycin (2 μm) were incubated with 32P-labeled IL2pARRE or HS4-3′-(−1527/−1496) oligonucleotide probes, with or without unlabeled oligonucleotide competitors (Fig. 4A). Fig. 4B shows that, consistent with our previous findings (30, 31), the IL2pARRE probe formed a complex (IL2p) (lanes 1, 5, and 11), which became slightly more intense in response to T cell stimulation (lane 2). This complex bound the IL2pARRE specifically because it was competed by unlabeled IL2pARRE (Fig. 4B, lane 6) but not by an oligonucleotide (mutIL2pARRE (31)) in which the element was mutated (lane 7). Similarly, the HS4-3′-(−1527/−1496) probe formed a specific complex (Fig. 4B, IL13p, lanes 3, 8, and 14), which also became slightly more intense in extracts from activated T cells (lane 4), and was selectively competed by unlabeled HS4-3′-(−1527/−1496) (lane 15), but not by mutHS4-3′-(−1527/−1496), an oligonucleotide in which the CTGTT motif had been transverted (lane 16). Of note, the IL13p and IL2p complexes had virtually identical mobility (Fig. 4B, lanes 1–4). Moreover, IL2pARRE (Fig. 4B, lane 9) but not mutIL2pARRE (lane 10) inhibited complex IL13p formation. Conversely, HS4-3′-(−1527/−1496) (Fig. 4B, lane 12) but not mutHS4-3′-(−1527/−1496) (lane 13) inhibited IL2p formation. These data show that the CTGTT motifs within HS4-3′-(−1527/−1496) and the IL2pARRE interact with similar if not identical protein complexes. In view of our previous demonstration that the CTGTT motif in the IL2pARRE binds NF45 and NF90 (30), these results suggest these proteins also bind HS4-3′-(−1527/−1496). Unfortunately, identification of NF45 and/or NF90 as constituents of the IL13p or IL2pARRE complexes could not be confirmed by antibody supershifting experiments, most likely because of low affinity interactions under EMSA conditions.
FIGURE 4.
DNA/protein interactions at HS4-3′-(−1527/−1496). A, sequence of oligonucleotides used as EMSA probes and competitors. Underlined is the reported (IL2pARRE) or predicted (HS4-3′-(−1527/−1496)) recognition motif for NF45/NF90. B, EMSA analysis was performed using nuclear proteins extracted from Jurkat T cells after a 4-h incubation with or without PMA (20 ng/ml) and ionomycin (2 μm). Competitors were added at 90-fold (lanes 6 and 7 and lanes 9 and 10) or 30-fold (lanes 12 and 13 and lanes 15 and 16) molar excess and are noted above the relevant lanes. The figure shows three independent gels (gel 1, lanes 1–4; gel 2,: lanes 5–10; and gel 3, lanes 11–16). Lanes that were not contiguous in the original gels and have been juxtaposed in the figure are separated by black lines. C, Jurkat T cells were incubated in the presence or absence of PMA (20 ng/ml) and ionomycin (1 μm) for 4 h. Levels of NF45 and NF45 serine phosphorylation were assessed by Western blot (WB) analysis of whole cell lysates (WCL) representing 800,000, 400,000, and 200,000 cell equivalents, using an anti-NF45 (upper panel) or an anti-phosphoserine (lower panel) antibody. P/I, PMA + ionomycin.
Activation-dependent Phosphorylation of NF45
Our ChIP and EMSA experiments (Fig. 3 and Fig. 4B) suggested recruitment of the NF45-NF90 complex to the distal IL13 promoter was increased by T cell activation. To begin understanding the mechanisms underlying this finding, we next assessed whether PMA/ionomycin treatment of Jurkat T cells under our experimental conditions increased NF45 and/or NF90 protein levels and/or resulted in post-translational modifications of these proteins. We focused particularly on phosphorylation, a modification previously reported for both NF45 and NF90 in T cells (36–38). As shown in Fig. 4C, Western blot analysis of lysates from Jurkat T cells incubated with or without PMA and ionomycin for 4 h revealed that activation did not affect NF45 protein levels but resulted in increased serine phosphorylation of the NF45 protein band. In contrast, NF90 levels and phosphorylation status appeared comparable under unstimulated and stimulated conditions (data not shown). These data raise the possibility that T cell activation-dependent NF45 phosphorylation may contribute to increased recruitment of the NF45-NF90 complex to the distal IL13 promoter.
HS4-dependent IL13 Up-regulation Requires the CTGTT Motif Located within HS4-3′
To further investigate the extent to which HS4-3′ contributes to IL13 up-regulation driven by full-length HS4, and to define the role of the CTGTT motif in HS4-3′ activity, the region encompassing HS4-3′ (−1577 to −1435) was cloned into HS6/Luc downstream of the luciferase reporter gene. The activity of this construct (HS4-3′-HS6/Luc) was compared with the activity of HS4-HS6/Luc. For these experiments, reporter constructs were transfected into in vitro differentiated primary murine CD4+ Th2 cells. Th2 cells are programmed for high rate IL13 expression, and as shown by our previous work (19), the murine Th2 transcriptional machinery allows optimal expression of human IL13 reporter vectors. In agreement with the results obtained in human CD4+ Jurkat T cells, HS4 strongly (>2-fold) enhanced IL13 promoter activity in murine Th2 cells (Fig. 5). Notably, the IL13 up-regulating activity of HS4 appeared to reside completely within HS4-3′, because no significant difference in luciferase expression was detected between full-length HS4 and HS4-3′.
FIGURE 5.
CTGTT motif within HS4-3′ is critical for HS4-mediated IL13 up-regulation. CD4+ T cells were isolated from WT C57BL/6 mice (n = 2) and in vitro differentiated under Th2 skewing conditions for 7 days. Th2 polarization was assessed by intracellular staining for IL-4 and IL-13. After 2–3 days in the presence of plate-bound anti-CD3 and anti-CD28 mAbs, Th2 cells were nucleofected with the IL13 promoter reporter constructs HS6/Luc, HS4-HS6/Luc, HS4-3′-HS6/Luc, and HS4-3′mut-HS6/Luc. Results are expressed as the mean ± S.E. of RLA measured in seven independent experiments. Statistical significance of individual comparisons was assessed using the Wilcoxon two-sample test. n.s., not significant; Luc, luciferase.
The role played by the NF45/NF90-binding CTGTT motif within HS4-3′ was directly assessed by introducing into HS4-3′-HS6/Luc the mutation that disrupted IL13p complex formation in EMSA (Fig. 4). Remarkably, mutation of the CTGTT motif abrogated the IL13 enhancing effects of HS4-3′ (Fig. 5, hatched bar), demonstrating DNA/protein interactions mediated by this motif are critical for HS4-3′-dependent IL13 up-regulation.
HS4-dependent IL13 Up-regulation Is Exquisitely Dependent on the Endogenous Levels of NF45 and NF90
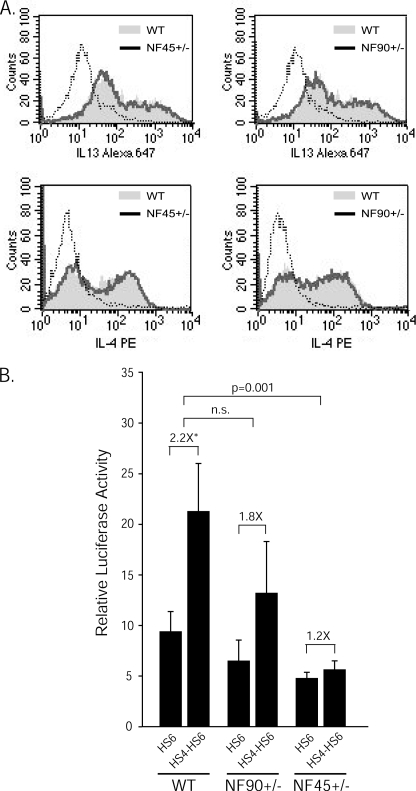
To explore more directly the functional role of NF45 and NF90 in IL13 regulation, we tested the activity of the HS4-HS6/Luc reporter construct in primary T cells from NF90- and NF45-deficient mice. Deletion of both copies of NF90 or NF45 results in perinatal lethality due to respiratory failure (20) or embryonic lethality,3 respectively. On the other hand, adult NF90+/− and NF45+/− mice are indistinguishable from WT littermates in terms of size, activity, and longevity (20).3 These mice therefore provide a unique model to assess whether HS4-dependent up-regulation of IL13 reporter activity depends on the levels of NF45 and/or NF90 in the endogenous nuclear environment.
For these experiments, primary CD4+ T cells were isolated from WT, NF45+/−, or NF90+/− mice and differentiated in vitro under Th2-polarizing conditions. Intracellular staining for IL-13 and IL-4 revealed that CD4+ T cells from NF45+/− and NF90+/− mice became Th2-polarized as efficiently as WT cells upon incubation with the appropriate differentiating stimuli (Fig. 6A). After 2–3 days in the presence of anti-CD3 and anti-CD28 mAbs, WT, NF45+/−, and NF90+/− Th2 cells were nucleofected with HS4/HS6 IL13 reporter vectors. Fig. 6B shows that HS4 strongly enhanced IL13 promoter activity in WT primary murine Th2 cells, thus confirming the results obtained in human Jurkat T cells. However, virtually no HS4-dependent up-regulation of IL13 expression was detected in Th2 cells from NF45+/− mice. A more subtle decrease in HS4 enhancing activity was observed in NF90+/− Th2 cells. Comparable results were obtained when murine Th2 cells were nucleofected with the −1577IL13p/Luc reporter construct (data not shown). These results demonstrate that HS4-dependent IL13 enhancement is exquisitely dependent on the endogenous levels of NF45, and to a lesser extent of NF90, suggesting that NF45 and NF90 act as positive regulators of IL13 expression and confirming the important role these factors play in HS4-mediated IL13 regulation.
FIGURE 6.
HS4-dependent IL13 up-regulation depends on the levels of endogenous NF45 and NF90. A, CD4+ T cells were isolated from WT (n = 3), NF45+/− (n = 4), or NF90+/− (n = 4) C57BL/6 mice and in vitro differentiated into Th2 cells for 1 week. Th2 polarization was assessed by intracellular staining for IL-4 and IL-13. PE, phycoerythrin. B, Th2 cells were re-stimulated with plate-bound anti-CD3 and anti-CD28 mAbs for 2–3 days and nucleofected with HS6/Luc or HS4-HS6/Luc. Results are expressed as the mean ± S.E. of RLA measured in nine independent experiments. Statistical significance was calculated using the Wilcoxon two-sample test (*, p ≤ 0.04). n.s., not significant.
DISCUSSION
Activation of gene expression upon engagement of cell stimulatory pathways relies on a complex interplay between cis-regulatory elements, sequence-specific trans-activating factors, and co-activators. IL13 expression in activated human CD4+ T cells is no exception. Several cis-regulatory elements have been mapped in the murine IL13 locus (reviewed in Ref. 11), and a number of putative ones have been identified in the human locus by us (17) and others (39). Here, we characterize HS4 as a novel element that enhances IL13 promoter activity in stimulated T cells. Interestingly, HS4 has regulatory properties, yet a multispecies sequence alignment (40) showed the HS4 region, and specifically the CTGTT motif critical for its positive regulatory effects, is highly conserved in Hominoids and Old World monkeys but not in New World monkeys, prosimians, or rodents (Fig. 7). Our results might appear to be at odds with the basic tenets of comparative genomics, according to which functional sequences are typically marked by strong evolutionary conservation (41, 42). On the other hand, recent experiments in mouse cells containing a copy of a human chromosome in addition to the complete mouse genome clearly showed that nonconserved sequences can be not only functional but responsible for important species-specific differences in gene expression patterns (43). Our data are consistent with these findings.
FIGURE 7.
HS4 is a recently arisen regulatory element. The ClustalW program (40) generated a multiple sequence alignment for the HS4 region in humans, 11 primate species from distinct clades, and mice. Hominoid, Old World monkey (OWM), and New World monkey (NWM) IL13 promoter sequences were previously generated in our laboratory (19). Human, mouse, and prosimian (PS) IL13 promoter sequences were obtained from GenBankTM accession number NC_000005, GenBankTM accession number NC_000077, and Ensembl GeneScaffold_442, respectively. Dashes and asterisks mark conserved positions and gaps, respectively. The CTGTT motif is bracketed. Numbering is relative to the IL13 ATG.
Lack of HS4 conservation in mice also likely explains why endogenous IL-13 production was fully preserved in NF45+/− and NF90+/− murine Th2 cells, even though NF45- and NF90-deficient nuclear environments failed to support the expression of the human IL13 reporter construct. These data suggest that, in contrast to the human IL13 promoter, the murine IL13 promoter may not be NF45/NF90-regulated. It is tempting to speculate that the emergence of an NF45/NF90-dependent regulatory element may have endowed the human IL13 locus with properties that adaptively fine-tune the critical roles played by IL13 in immunity (1) and reproduction (44).
The IL13 proximal promoter strongly activates IL13 expression in response to T cell receptor-derived signals by relying on nuclear factor of activated T cells, AP-2, and GATA-3 (45–48). The IL13 promoter enhancing activity of HS4 mapped to the 3′-half of the element and involved previously undescribed interactions with NF45 and NF90. The identification of these proteins as IL13 regulators was supported by independent but complementary lines of evidence as follows: the results of mass spectrometry analysis of proteins recovered from HS4-3′-based affinity chromatography, which were confirmed by ChIP; the existence of an NF45/NF90-binding CTGTT motif (30) within HS4-3′; and functional data from NF45- or NF90-deficient mouse models. NF90 and NF45 are DZF (C2H2 zinc finger) motif-containing proteins that bind DNA as heterodimers (31, 34) and are subunits of a supra-molecular complex involved in the regulation of multiple targets. Most relevant to our work, NF45-NF90 complexes dock onto the proximal IL2 promoter at the ARRE, a cis-regulatory element involved in IL2 transcriptional activation in response to T cell receptor signaling (20, 30, 31, 49, 50). The trans-activation domains of NF90 and NF45 have been proposed to interact with RNA polymerase II at the IL2 promoter, thereby leading to transcription (20). Indeed, antisera against NF90 or NF45 decreased basal and inducible in vitro IL2 transcription (31). Furthermore, stable overexpression of NF90 and NF45 resulted in a large increase of IL2 ARRE luciferase activity, whereas NF90 deficiency disrupted inducible IL2 transcription and was accompanied by decreased binding of RNA polymerase II at the IL2 promoter in activated murine T cells (20, 50). It is possible that similar mechanisms contribute to the positive role played by NF45 and NF90 in IL13 regulation, whereas no evidence so far supports functional or physical interactions between NF45/NF90 and the transcription factors (nuclear factor of activated T cells, AP-2, and GATA-3), which regulate IL13 proximal promoter activity.
A comparison of the interactions between NF45/NF90 and the IL13 and IL2 promoters provides insights into the role of promoter context in fine-tuning distinct functional outcomes instigated by binding of the same nucleoprotein complex. The interaction of NF45/NF90 with the AT-rich purine box in the IL2ARRE was shown to involve the specific and dynamic binding of that element by Ku70 and Ku80, and a model was proposed according to which T cell activation-induced conformational changes in the ARRE would lead to decreased Ku70 binding and, concomitantly, to increased binding of Ku80 and NF90 (30). In vivo data supported this model. In contrast, we could not find conclusive evidence for an interaction between Ku70 or Ku80 and the IL13 promoter, even though ChIP analysis showed that recruitment of NF45 and NF90 to the promoter was enhanced by T cell activation. It is noteworthy that, according to our mutational analysis, NF45/NF90 binding to the IL13 regulatory region involved a CTGTT motif fully conserved in the IL2 promoter. However, the IL2 promoter also contains a stretch of five adenines that are located immediately upstream of the CTGTT motif and are not found in IL13. We hypothesize that the lack of these adenines might create unfavorable conditions for binding of Ku proteins, whose predilection for AT-rich targets is well known (51). The inability of Ku proteins to interact with the IL13 promoter in turn suggests that the strong T cell activation-dependent enhancement in NF45/NF90 recruitment to this region may involve other proteins (perhaps including some of those identified by our mass spectrometry analysis) and/or require activation-induced post-translational modifications of NF45 and/or NF90 themselves. Our finding that cell activation increases NF45 serine phosphorylation is consistent with this possibility. The kinase(s) responsible for NF45 phosphorylation in this model remain to be defined. We propose low level binding of NF45/NF90 to HS4 in resting T cells may be compatible with, or sufficient for, maintaining the region in a constitutively accessible state, which is reflected by hypersensitivity to DNase I digestion. Enhanced recruitment of NF45/NF90 to HS4 in response to T cell activation may contribute to rapid IL13 up-regulation by facilitating proficient interactions between HS4 and the IL13 proximal promoter.
Our finding that HS4-mediated IL13 up-regulation was abrogated in NF45+/− cells and reduced in NF90+/− cells is both novel and intriguing, because NF45 and NF90 mRNA levels in these cells were reduced by only ≈50%. These data therefore suggest that HS4 function is exquisitely dependent on the endogenous levels of these proteins. It remains to be established whether such dependence reflects stoichiometric constraints imposed by NF45/NF90 molecular and/or functional interactions, or rather results from the existence of these proteins in concentrations that are limiting relative to the multiple tasks they are expected to perform.
The ability of NF90 to bind double-stranded RNA and regulate IL2 and MKP-1 mRNA stability in T cells and epithelial cells, respectively (20, 52), raises the possibility that the NF45-NF90 complex may also contribute to the regulation of IL13 expression at the post-transcriptional level. Like other cytokines, IL13 mRNA contains several AU-rich elements in its 3′-untranslated region (53), and proteins binding to these elements are known to control mRNA stability by acting in combination with NF90 (52). Further research is required to clarify the functional interplay between the promoter- and 3′-untranslated region-mediated influences of NF45 and NF90 on human IL13 expression.
Supplementary Material
Acknowledgments
Mass spectral proteomic analyses were performed by the Arizona Proteomics Consortium, which is supported by National Institutes of Health Grant ES06694 from NIEHS and Grant CA023074 from NCI and The BIO5 Institute.
This work was supported, in whole or in part, by National Institutes of Health Grant HL66391 (to D. V.) from NHLBI.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Tables 1 and 2.
P. N. Kao, unpublished results.
R. Webster and D. Vercelli, unpublished data.
- IL
- interleukin
- Th2
- T helper type 2
- ARRE
- antigen receptor-response element
- ChIP
- chromatin immunoprecipitation
- EMSA
- electromobility shift assay
- HS
- hypersensitive
- NF
- nuclear factor
- RLA
- relative luciferase activity
- WT
- wild type
- PMA
- phorbol 12-myristate 13-acetate
- mAb
- monoclonal antibody
- DTT
- dithiothreitol
- HPLC
- high pressure liquid chromatography
- LC-MS/MS
- liquid chromatography-tandem mass spectrometry
- MudPIT
- multidimensional protein identification technology.
REFERENCES
- 1.Finkelman F. D., Shea-Donohue T., Morris S. C., Gildea L., Strait R., Madden K. B., Schopf L., Urban J. F., Jr. (2004) Immunol. Rev. 201, 139–155 [DOI] [PubMed] [Google Scholar]
- 2.Vercelli D. (2008) Nat. Rev. Immunol. 8, 169–182 [DOI] [PubMed] [Google Scholar]
- 3.Wills-Karp M., Luyimbazi J., Xu X., Schofield B., Neben T. Y., Karp C. L., Donaldson D. D. (1998) Science 282, 2258–2261 [DOI] [PubMed] [Google Scholar]
- 4.Grünig G., Warnock M., Wakil A. E., Venkayya R., Brombacher F., Rennick D. M., Sheppard D., Mohrs M., Donaldson D. D., Locksley R. M., Corry D. B. (1998) Science 282, 2261–2263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Walter D. M., McIntire J. J., Berry G., McKenzie A. N., Donaldson D. D., DeKruyff R. H., Umetsu D. T. (2001) J. Immunol. 167, 4668–4675 [DOI] [PubMed] [Google Scholar]
- 6.Ghaffar O., Laberge S., Jacobson M. R., Lowhagen O., Rak S., Durham S. R., Hamid Q. (1997) Am. J. Respir. Cell Mol. Biol. 17, 17–24 [DOI] [PubMed] [Google Scholar]
- 7.Lordan J. L., Bucchieri F., Richter A., Konstantinidis A., Holloway J. W., Thornber M., Puddicombe S. M., Buchanan D., Wilson S. J., Djukanoviæ R., Holgate S. T., Davies D. E. (2002) J. Immunol. 169, 407–414 [DOI] [PubMed] [Google Scholar]
- 8.Ohshima Y., Yasutomi M., Omata N., Yamada A., Fujisawa K., Kasuga K., Hiraoka M., Mayumi M. (2002) Pediatr. Res. 51, 195–200 [DOI] [PubMed] [Google Scholar]
- 9.Lange J., Ngoumou G., Berkenheide S., Moseler M., Mattes J., Kuehr J., Kopp M. V. (2003) Clin. Exp. Allergy 33, 1537–1543 [DOI] [PubMed] [Google Scholar]
- 10.Neaville W. A., Tisler C., Bhattacharya A., Anklam K., Gilbertson-White S., Hamilton R., Adler K., Dasilva D. F., Roberg K. A., Carlson-Dakes K. T., Anderson E., Yoshihara D., Gangnon R., Mikus L. D., Rosenthal L. A., Gern J. E., Lemanske R. F., Jr. (2003) J. Allergy Clin. Immunol. 112, 740–746 [DOI] [PubMed] [Google Scholar]
- 11.Ansel K. M., Djuretic I., Tanasa B., Rao A. (2006) Annu. Rev. Immunol. 24, 607–656 [DOI] [PubMed] [Google Scholar]
- 12.Boyes J., Felsenfeld G. (1996) EMBO J. 15, 2496–2507 [PMC free article] [PubMed] [Google Scholar]
- 13.Wilson C. B., Rowell E., Sekimata M. (2009) Nat. Rev. Immunol. 9, 91–105 [DOI] [PubMed] [Google Scholar]
- 14.Lee G. R., Fields P. E., Griffin T. J., Flavell R. A. (2003) Immunity 19, 145–153 [DOI] [PubMed] [Google Scholar]
- 15.Lee G. R., Spilianakis C. G., Flavell R. A. (2005) Nat. Immunol. 6, 42–48 [DOI] [PubMed] [Google Scholar]
- 16.Spilianakis C. G., Flavell R. A. (2004) Nat. Immunol. 5, 1017–1027 [DOI] [PubMed] [Google Scholar]
- 17.Webster R. B., Rodriguez Y., Klimecki W. T., Vercelli D. (2007) J. Biol. Chem. 282, 700–709 [DOI] [PubMed] [Google Scholar]
- 18.Ribeiro-do-Couto L. M., Boeije L. C., Kroon J. S., Hooibrink B., Breur-Vriesendorp B. S., Aarden L. A., Boog C. J. (2001) Eur. J. Immunol. 31, 3394–3402 [DOI] [PubMed] [Google Scholar]
- 19.Cameron L., Webster R. B., Strempel J. M., Kiesler P., Kabesch M., Ramachandran H., Yu L., Stern D. A., Graves P. E., Lohman I. C., Wright A. L., Halonen M., Klimecki W. T., Vercelli D. (2006) J. Immunol. 177, 8633–8642 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Shi L., Godfrey W. R., Lin J., Zhao G., Kao P. N. (2007) J. Exp. Med. 204, 971–977 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Avni O., Lee D., Macian F., Szabo S. J., Glimcher L. H., Rao A. (2002) Nat. Immunol. 3, 643–651 [DOI] [PubMed] [Google Scholar]
- 22.Strempel J. M., Vercelli D. (2007) J. Biol. Chem. 282, 3738–3746 [DOI] [PubMed] [Google Scholar]
- 23.Andon N. L., Hollingworth S., Koller A., Greenland A. J., Yates J. R., 3rd, Haynes P. A. (2002) Proteomics 2, 1156–1168 [DOI] [PubMed] [Google Scholar]
- 24.Yates J. R., 3rd, Eng J. K., McCormack A. L., Schieltz D. (1995) Anal. Chem. 67, 1426–1436 [DOI] [PubMed] [Google Scholar]
- 25.Cooper B., Eckert D., Andon N. L., Yates J. R., Haynes P. A. (2003) J. Am. Soc. Mass Spectrom. 14, 736–741 [DOI] [PubMed] [Google Scholar]
- 26.Qian W. J., Liu T., Monroe M. E., Strittmatter E. F., Jacobs J. M., Kangas L. J., Petritis K., Camp D. G., 2nd, Smith R. D. (2005) J. Proteome Res. 4, 53–62 [DOI] [PubMed] [Google Scholar]
- 27.Craig R., Beavis R. C. (2004) Bioinformatics 20, 1466–1467 [DOI] [PubMed] [Google Scholar]
- 28.Keller A., Nesvizhskii A. I., Kolker E., Aebersold R. (2002) Anal. Chem. 74, 5383–5392 [DOI] [PubMed] [Google Scholar]
- 29.Nesvizhskii A. I., Keller A., Kolker E., Aebersold R. (2003) Anal. Chem. 75, 4646–4658 [DOI] [PubMed] [Google Scholar]
- 30.Shi L., Qiu D., Zhao G., Corthesy B., Lees-Miller S., Reeves W. H., Kao P. N. (2007) Nucleic Acids Res. 35, 2302–2310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Corthésy B., Kao P. N. (1994) J. Biol. Chem. 269, 20682–20690 [PubMed] [Google Scholar]
- 32.Cartharius K., Frech K., Grote K., Klocke B., Haltmeier M., Klingenhoff A., Frisch M., Bayerlein M., Werner T. (2005) Bioinformatics 21, 2933–2942 [DOI] [PubMed] [Google Scholar]
- 33.Link A. J., Eng J., Schieltz D. M., Carmack E., Mize G. J., Morris D. R., Garvik B. M., Yates J. R., 3rd (1999) Nat. Biotechnol. 17, 676–682 [DOI] [PubMed] [Google Scholar]
- 34.Reichman T. W., Muñiz L. C., Mathews M. B. (2002) Mol. Cell. Biol. 22, 343–356 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Guan D., Altan-Bonnet N., Parrott A. M., Arrigo C. J., Li Q., Khaleduzzaman M., Li H., Lee C. G., Pe'ery T., Mathews M. B. (2008) Mol. Cell. Biol. 28, 4629–4641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tao W. A., Wollscheid B., O'Brien R., Eng J. K., Li X. J., Bodenmiller B., Watts J. D., Hood L., Aebersold R. (2005) Nat. Methods 2, 591–598 [DOI] [PubMed] [Google Scholar]
- 37.Matsuoka S., Ballif B. A., Smogorzewska A., McDonald E. R., 3rd, Hurov K. E., Luo J., Bakalarski C. E., Zhao Z., Solimini N., Lerenthal Y., Shiloh Y., Gygi S. P., Elledge S. J. (2007) Science 316, 1160–1166 [DOI] [PubMed] [Google Scholar]
- 38.Pei Y., Zhu P., Dang Y., Wu J., Yang X., Wan B., Liu J. O., Yi Q., Yu L. (2008) J. Immunol. 180, 222–229 [DOI] [PubMed] [Google Scholar]
- 39.Santangelo S., Cousins D. J., Winkelmann N. E., Staynov D. Z. (2002) J. Immunol. 169, 1893–1903 [DOI] [PubMed] [Google Scholar]
- 40.Larkin M. A., Blackshields G., Brown N. P., Chenna R., McGettigan P. A., McWilliam H., Valentin F., Wallace I. M., Wilm A., Lopez R., Thompson J. D., Gibson T. J., Higgins D. G. (2007) Bioinformatics 23, 2947–2948 [DOI] [PubMed] [Google Scholar]
- 41.Waterston R. H., Lindblad-Toh K., Birney E., Rogers J., Abril J. F., Agarwal P., Agarwala R., Ainscough R., Alexandersson M., An P., Antonarakis S. E., Attwood J., Baertsch R., Bailey J., Barlow K., Beck S., Berry E., Birren B., Bloom T., Bork P., Botcherby M., Bray N., Brent M. R., Brown D. G., Brown S. D., Bult C., Burton J., Butler J., Campbell R. D., Carninci P., Cawley S., Chiaromonte F., Chinwalla A. T., Church D. M., Clamp M., Clee C., Collins F. S., Cook L. L., Copley R. R., Coulson A., Couronne O., Cuff J., Curwen V., Cutts T., Daly M., David R., Davies J., Delehaunty K. D., Deri J., Dermitzakis E. T., Dewey C., Dickens N. J., Diekhans M., Dodge S., Dubchak I., Dunn D. M., Eddy S. R., Elnitski L., Emes R. D., Eswara P., Eyras E., Felsenfeld A., Fewell G. A., Flicek P., Foley K., Frankel W. N., Fulton L. A., Fulton R. S., Furey T. S., Gage D., Gibbs R. A., Glusman G., Gnerre S., Goldman N., Goodstadt L., Grafham D., Graves T. A., Green E. D., Gregory S., Guigó R., Guyer M., Hardison R. C., Haussler D., Hayashizaki Y., Hillier L. W., Hinrichs A., Hlavina W., Holzer T., Hsu F., Hua A., Hubbard T., Hunt A., Jackson I., Jaffe D. B., Johnson L. S., Jones M., Jones T. A., Joy A., Kamal M., Karlsson E. K., Karolchik D., Kasprzyk A., Kawai J., Keibler E., Kells C., Kent W. J., Kirby A., Kolbe D. L., Korf I., Kucherlapati R. S., Kulbokas E. J., Kulp D., Landers T., Leger J. P., Leonard S., Letunic I., Levine R., Li J., Li M., Lloyd C., Lucas S., Ma B., Maglott D. R., Mardis E. R., Matthews L., Mauceli E., Mayer J. H., McCarthy M., McCombie W. R., McLaren S., McLay K., McPherson J. D., Meldrim J., Meredith B., Mesirov J. P., Miller W., Miner T. L., Mongin E., Montgomery K. T., Morgan M., Mott R., Mullikin J. C., Muzny D. M., Nash W. E., Nelson J. O., Nhan M. N., Nicol R., Ning Z., Nusbaum C., O'Connor M. J., Okazaki Y., Oliver K., Overton-Larty E., Pachter L., Parra G., Pepin K. H., Peterson J., Pevzner P., Plumb R., Pohl C. S., Poliakov A., Ponce T. C., Ponting C. P., Potter S., Quail M., Reymond A., Roe B. A., Roskin K. M., Rubin E. M., Rust A. G., Santos R., Sapojnikov V., Schultz B., Schultz J., Schwartz M. S., Schwartz S., Scott C., Seaman S., Searle S., Sharpe T., Sheridan A., Shownkeen R., Sims S., Singer J. B., Slater G., Smit A., Smith D. R., Spencer B., Stabenau A., Stange-Thomann N., Sugnet C., Suyama M., Tesler G., Thompson J., Torrents D., Trevaskis E., Tromp J., Ucla C., Ureta-Vidal A., Vinson J. P., Von Niederhausern A. C., Wade C. M., Wall M., Weber R. J., Weiss R. B., Wendl M. C., West A. P., Wetterstrand K., Wheeler R., Whelan S., Wierzbowski J., Willey D., Williams S., Wilson R. K., Winter E., Worley K. C., Wyman D., Yang S., Yang S. P., Zdobnov E. M., Zody M. C., Lander E. S. (2002) Nature 420, 520–56212466850 [Google Scholar]
- 42.Xie X., Lu J., Kulbokas E. J., Golub T. R., Mootha V., Lindblad-Toh K., Lander E. S., Kellis M. (2005) Nature 434, 338–345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wilson M. D., Barbosa-Morais N. L., Schmidt D., Conboy C. M., Vanes L., Tybulewicz V. L., Fisher E. M., Tavaré S., Odom D. T. (2008) Science 322, 434–438 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Khaled W. T., Read E. K., Nicholson S. E., Baxter F. O., Brennan A. J., Came P. J., Sprigg N., McKenzie A. N., Watson C. J. (2007) Development 134, 2739–2750 [DOI] [PubMed] [Google Scholar]
- 45.Dolganov G., Bort S., Lovett M., Burr J., Schubert L., Short D., McGurn M., Gibson C., Lewis D. B. (1996) Blood 87, 3316–3326 [PubMed] [Google Scholar]
- 46.Keen J. C., Cianferoni A., Florio G., Guo J., Chen R., Roman J., Wills-Karp M., Casolaro V., Georas S. N. (2006) Immunology 117, 29–37 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kishikawa H., Sun J., Choi A., Miaw S. C., Ho I. C. (2001) J. Immunol. 167, 4414–4420 [DOI] [PubMed] [Google Scholar]
- 48.Lavenu-Bombled C., Trainor C. D., Makeh I., Romeo P. H., Max-Audit I. (2002) J. Biol. Chem. 277, 18313–18321 [DOI] [PubMed] [Google Scholar]
- 49.Kao P. N., Chen L., Brock G., Ng J., Kenny J., Smith A. J., Corthésy B. (1994) J. Biol. Chem. 269, 20691–20699 [PubMed] [Google Scholar]
- 50.Zhao G., Shi L., Qiu D., Hu H., Kao P. N. (2005) Exp. Cell Res. 305, 312–323 [DOI] [PubMed] [Google Scholar]
- 51.Novac O., Matheos D., Araujo F. D., Price G. B., Zannis-Hadjopoulos M. (2001) Mol. Biol. Cell 12, 3386–3401 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kuwano Y., Kim H. H., Abdelmohsen K., Pullmann R., Jr., Martindale J. L., Yang X., Gorospe M. (2008) Mol. Cell. Biol. 28, 4562–4575 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Casolaro V., Fang X., Tancowny B., Fan J., Wu F., Srikantan S., Asaki S. Y., De Fanis U., Huang S. K., Gorospe M., Atasoy U. X., Stellato C. (2008) J. Allergy Clin. Immunol. 121, 853–859 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.