Abstract
Dark operative protochlorophyllide oxidoreductase (DPOR) catalyzes the light-independent two-electron reduction of protochlorophyllide a to form chlorophyllide a, the last common precursor of chlorophyll a and bacteriochlorophyll a biosynthesis. During ATP-dependent DPOR catalysis the homodimeric ChlL2 subunit carrying a [4Fe-4S] cluster transfers electrons to the corresponding heterotetrameric catalytic subunit (ChlN/ChlB)2, which also possesses a redox active [4Fe-4S] cluster. To investigate the transient interaction of both subcomplexes and the resulting electron transfer reactions, the ternary DPOR enzyme holocomplex comprising subunits ChlN, ChlB, and ChlL from the cyanobacterium Prochlorococcus marinus was trapped as an octameric (ChlN/ChlB)2(ChlL2)2 complex after incubation with the nonhydrolyzable ATP analogs adenosine 5′-(γ-thio)triphosphate, adenosine 5′-(β,γ-imido)triphosphate, or MgADP in combination with AlF4−. Additionally, a mutant ChlL2 protein, with a deleted Leu153 in the switch II region also allowed for the formation of a stable octameric complex. Furthermore, efficient complex formation required the presence of protochlorophyllide. Electron paramagnetic resonance spectroscopy of ternary DPOR complexes revealed a reduced [4Fe-4S] cluster located on ChlL2, indicating that complete ATP hydrolysis is a prerequisite for intersubunit electron transfer. Circular dichroism spectroscopic experiments indicated nucleotide-dependent conformational changes for ChlL2 after ATP binding. A nucleotide-dependent switch mechanism triggering ternary complex formation and electron transfer was concluded. From these results a detailed redox cycle for DPOR catalysis was deduced.
Keywords: ATPases, Electron Paramagnetic Resonance (EPR), Electron Transfer, Enzyme Catalysis, Iron-Sulfur Protein, Nitrogenase, Photosynthesis, Photosynthetic Pigments, Protein-Protein Interactions, Bacteriochlorophyll
Introduction
Reduction of protochlorophyllide a (Pchlide)2 to chlorophyllide a (Chlide) is a central step in the biosynthesis of chlorophyll and bacteriochlorophyll (1). Two evolutionarily unrelated enzymes are capable of catalyzing the stereospecific two-electron reduction of the C17–C18 double bond of Pchlide (Fig. 1A) (2–4). Monomeric, light-dependent Pchlide oxidoreductase (POR; NADPH Pchlide oxidoreductase, EC 1.3.1.33) drives the NADPH-dependent reduction (5–7) of Pchlide bound in the active site via absorption of light energy. This light dependence of POR catalysis prevents angiosperms from synthesizing chlorophyll in the dark (8, 9). Anoxygenic-photosynthetic bacteria make use of a different ATP-dependent Pchlide reducing system, which is termed the light-independent, dark operative Pchlide oxidoreductase (DPOR), whereas gymnosperms, mosses, ferns, algae, and cyanobacteria utilize both POR and DPOR (3). In chlorophyll-synthesizing organisms DPOR is encoded by the chlN, chlB, and chlL genes (10–12). The corresponding genes for bacteriochlorophyll-synthesizing organisms have been termed bchN, bchB, and bchL (10, 13). DPOR subunits ChlN, ChlB, and ChlL share significant amino acid sequence homologies with nitrogenase subunits NifD, NifK, and NifH, respectively (12, 14).
FIGURE 1.
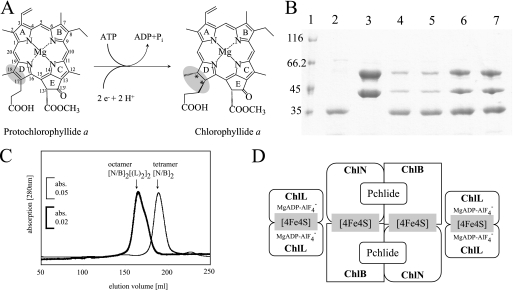
Reduction of Pchlide a by DPOR, analyses of ternary DPOR by SDS-PAGE and gel permeation chromatography, and a schematic model of the octameric DPOR complex. A, ring D is stereospecifically reduced in the presence of an electron donor and ATP. Rings A–E and the individual carbon atoms are labeled according to IUPAC nomenclature (50). B, SDS-PAGE analyses of purified recombinant ChlL, ChlN/ChlB, and ternary DPOR complexes containing ChlN, ChlB, and ChlL. Lane 1, molecular mass marker, relative molecular masses (×1000) are indicated; lane 2, affinity chromatographically purified ChlL recovered from glutathione-agarose after PreScission protease cleavage; lane 3, pure ChlN and ChlB after PreScission protease cleavage of the glutathione-agarose bound GST tag; lane 4, ternary DPOR complex in the presence of ATPγS; lane 5, ternary DPOR complex in the presence of AMP-PNP; lane 6, ternary DPOR complex in the presence of MgADP·AlF4−; lane 7, ternary DPOR complex formed with the mutant enzyme ChlLΔLeu153 in the absence of nucleotide cofactors. Each complex was formed in the presence of Pchlide. C, preparative gel permeation chromatography of (ChlN/ChlB)2 (thin line) and ternary DPOR complex (bold line). 0.08 μmol of purified ternary DPOR and 0.26 μmol of pure (ChlN/ChlB)2 after protease cleavage were analyzed on a Superdex 200 HR 26/60 column under anaerobic conditions (95% N2, 5% H2, <1 ppm O2) using a flow rate of 1 ml/min and monitoring the absorbance at 280 nm. D, octameric model of a DPOR complex showing two ChlL2 dimers carrying ATP analogs coupled to the (ChlN/ChlB)2 heterotetramer in the Pchlide-bound state.
In earlier studies it was shown that the dimeric subcomplex ChlL2 carries a redox active intersubunit [4Fe-4S] center symmetrically coordinated by residues Cys65 and Cys158 (Prochlorococcus marinus numbering). In vivo this [4Fe-4S] cluster is reduced by a ferredoxin. Subunit ChlL contains a highly conserved ATP cofactor binding motif called P-loop (35YGKGGIGK42, P. marinus numbering). Hydrolysis of two ATP molecules by dimeric subunit ChlL2 facilitates electron transfer to a second [4Fe-4S] cluster located on the heterotetrameric complex (ChlN/ChlB)2. Transient interaction of ChlL2 and (ChlN/ChlB)2 with involvement of surface-exposed residues Leu70, Val107, and Lys109 of ChlN, Gly66 and Gln101 of ChlB, and Tyr127 of ChlL was proposed (15). This initial step of DPOR catalysis was shown to resemble the corresponding electron transfer reactions in nitrogenase catalysis. In contrast, the second [4Fe-4S] cluster located on (ChlN/ChlB)2 has no direct equivalent in nitrogenase, which carries the [8Fe-7S] P-cluster and the [1Mo-7Fe-9S-1X-1homocitrate] (Fe-Moco) metallo center instead (16, 17).
Substrate recognition by the DPOR subcomplex (ChlN/ChlB)2 tolerated minor modifications of the ring substituents on rings A, B, C, and E of Pchlide (18), whereas DPOR did not utilize substrate analogs with modifications at the catalytic target on ring D. Based on these results it was proposed that the active site of DPOR covers large parts of the macrocyclic substrate and is located in close proximity to the redox active [4Fe-4S] cluster.
The individual electron transfer processes of DPOR are dependent on the transient interaction of ChlL2 and (ChlN/ChlB)2. For the related nitrogenase system the dynamic protein-protein interaction of NifH2 and (NifD/NifK)2 was extensively characterized by using nucleotide cofactor analogs and site-directed mutagenesis (16). In this context MgADP in combination with the inorganic compound AlF4− was used as an analog of the transition state of ATP γ-phosphate hydrolysis (19). This compound enabled efficient trapping of the ternary NifH2(NifD/NifK)2 complex, which was suitable for structural analysis. The structure of the inactive transition state complex revealed a minimal distance of less than 17.5 Å between the [4Fe-4S] cluster of NifH2 and the P-cluster of (NifD/NifK)2 indicating a protein conformation in the state of electron transfer (16, 20–22). In addition, deletion of Leu127 (Azotobacter vinelandii numbering) in the switch II sequence motif of NifH also enabled trapping of the ternary nitrogenase complex. In the related crystal structure the mutant NifH2 protein adopts a nucleotide-bound conformation even in the absence of ATP (23, 24).
The transient protein-protein interaction of ChlL2 and (ChlN/ChlB)2 is essential for the ATP-dependent electron transfer processes catalyzed by DPOR. Here we describe the trapping of ternary P. marinus ChlL2(ChlN/ChlB)2 complexes and their biochemical and spectroscopic characterization. Based on these experiments we describe the ATP-dependent triggering of the electron transfer processes during DPOR catalysis.
EXPERIMENTAL PROCEDURES
Heterologous Production and Purification of P. marinus DPOR
P. marinus DPOR ChlL2 and (ChlN/ChlB)2 subcomplexes were recombinantly produced and purified under anaerobic conditions (oxygen partial pressure <1 ppm) in an anaerobic chamber (Coy Laboratories) as described earlier (18). The employed lysis buffer contained 100 mm HEPES-NaOH, pH 7.5, 150 mm NaCl, and 10 mm MgCl2. N-terminal GST tags fused to ChlN and ChlL were used for affinity chromatographic purification of both complexes using Protino® glutathione-agarose (Macherey-Nagel). PreScissionTM protease (GE Healthcare) treatment was employed to specifically elute ChlL2 and (ChlN/ChlB)2 from the resin, respectively (18).
Site-directed Mutagenesis of ChlL
Three nucleotides of the plasmid pGEX-chlL (18) were deleted using the QuikChangeTM site-directed mutagenesis kit (Stratagene). The oligonucleotides GATGTAGTTATTTTTGATGTCGGTGATGTTGTATGTGGTGGA and TCCACCACATACAACATCACCGACATCAAAAATAACTACATC were employed to delete the codon UAA responsible for Leu153. Mutant protein ChlLΔLeu153 was produced and purified as described for the wild-type protein.
Determination of Protein Concentration
The BCA (bicinchonic acid) protein assay kit (Pierce) was used according to the manufacturer's instructions with bovine serum albumin as a standard.
N-terminal Amino Acid Sequence Determination
Automated Edman degradation was used to confirm the identity of purified proteins. Edman degradation was also employed for the subsequent quantification of the individual subunits in elution fractions.
DPOR Enzyme Assay
The standard DPOR activity assay containing 13 μm Pchlide, 2 mm dithionite, as an artificial electron donor, 2 mm ATP, and an ATP regenerating system was performed and analyzed as described earlier in the presence of 100 pmol of purified (ChlN/ChlB)2 and 200 pmol of ChlL2 (125 μl final volume) (15, 18).
Analysis of Ternary DPOR Protein-Protein Interaction
9 mg (76 nmol) of GST-tagged subunit ChlL2 was immobilized on 3 ml of Protino glutathione-agarose in a 10-ml gravity-flow column as described (18). This column was washed with 30 ml of lysis buffer, then the affinity resin was incubated for 60 min with an excess of purified (ChlN/ChlB)2 complex (25 mg; 120 nmol) in lysis buffer containing 30 μm Pchlide and 10 mm AMP-PNP or, alternatively, 10 mm ATPγS (Sigma). After extensive washing with 30 ml of lysis buffer containing the respective ATP analogs at a concentration of 10 mm, subunit ChlL2 was liberated from the GST tag bound to the resin via PreScission protease treatment (5 units, 18 h at 18 °C) (15). Eluate fractions were analyzed by SDS-PAGE for the co-elution of DPOR subunit ChlN with ChlB due to ternary complex formation. Identical experiments were performed employing MgADP·AlF4− instead of nonhydrolysable ATP analogs. For this purpose MgADP·AlF4− formation was initiated by addition of 10 mm MgADP and 5–50 mm NaF to a solution of lysis buffer containing 0.01–2 mm AlCl3. An alternative approach for stabilization of the DPOR protein-protein interaction was based on the mutant protein ChlLΔLeu153. ChlLΔLeu153 was purified via Protino glutathione-agarose chromatography and complex formation of this protein with (ChlN/ChlB)2 was analogously studied in the absence of ATP or nucleotide analogs. To analyze the influence of substrate binding on ternary complex formation all experiments were also performed in the absence of Pchlide.
Optimization of Conditions for Formation of the Ternary MgADP·AlF4− Complex
To identify parameters relevant for quantitative DPOR·MgADP·AlF4− complex formation, preformed complexes were subsequently analyzed in the standard DPOR assay. To determine the minimal NaF requirement 200 pmol of purified ChlL2 and 100 pmol of purified (ChlN/ChlB)2 were mixed with 10 mm MgADP and 500 μm AlCl3 in the presence of varying concentrations of NaF (10 μm to 50 mm) in a final volume of 100 μl of lysis buffer. The analogous complex formation was also studied in the presence of 13 μm Pchlide. After a 60-min incubation the individual experiments were assayed for Pchlide reduction activity by adding 2 mm dithionite, 2 mm ATP, an ATP regenerating system and additionally 13 μm Pchlide. Identical experiments were also used to determine the time-dependent complex formation induced by MgADP·AlF4−. Therefore, 20 individual assays in the absence and presence of 13 μm Pchlide were incubated for 1–70 min with 500 μm AlCl3, 25 mm NaF, and 10 mm MgADP. Subsequently, DPOR activity was analyzed. To determine a potential inhibitory effect of the individual components required for MgADP·AlF4− complex formation control reactions were performed in which only 100 mm NaF, 25 mm AlCl3 or 10 mm MgADP as well as combinations of 100 mm NaF and 10 mm MgADP or 25 mm AlCl3 and 10 mm MgADP were added.
Preparative Purification of the Ternary DPOR Complex
For preparative complex formation 30 mg of GST-ChlL2 (253 nmol) were bound to 10 ml of Protino glutathione-agarose in a 25-ml gravity flow column. After washing with 100 ml of lysis buffer, 15 ml of purified (ChlN/ChlB)2 (75 mg; 355 nmol) in lysis buffer containing 30 μm Pchlide, 500 μm AlCl3, 25 mm NaF, and 10 mm MgADP were applied to the column and incubated for 60 min at 18 °C. After a subsequent washing step with 100 ml of lysis buffer containing 10 mm MgADP, 500 μm AlCl3, and 25 mm NaF the ”trapped“ ternary DPOR complex was eluted via PreScission protease cleavage (20 units, 18 h, 18 °C).
Native Molecular Mass Determination
Preparative gel permeation chromatography of 5 ml of purified (ChlN/ChlB)2 (60 mg, 284 nmol) or the ternary complex formed in the presence of Pchlide (27 mg, 80 nmol) were performed, respectively, in an anaerobic chamber using a Superdex 200 HR 26/60 column (GE Healthcare) equilibrated with lysis buffer. For chromatography of ternary DPOR complexes the lysis buffer additionally contained 25 mm NaF and 250 μm AlCl3. The column was calibrated with protein standards (Molecular Weight Marker Kit MW-GF 1000, Sigma) at a flow rate of 1.5 ml min−1. The eluted proteins were detected at 280 nm and collected in 1.5-ml aliquots. Fractions containing the ternary DPOR complex were identified by SDS-PAGE analyses.
DPOR Activity in the Presence of ATP Analogs
To analyze the residual enzymatic activity of ternary DPOR, 100 μl (∼50 pmol) of the trapped complexes eluted in the presence of 10 mm AMP-PNP, 10 mm ATPγS, MgADP·AlF4− or formed with mutant protein ChlLΔLeu153 were analyzed in the standard DPOR assay after elution from Protino glutathione-agarose by PreScission protease treatment.
ATPase Activity of DPOR
To analyze the ATPase activity of DPOR a colorimetric assay system was employed that allowed to monitor the increase of the free phosphate concentration during DPOR catalysis in the absence of an ATP regenerating system. The commercial test system (ATPase Assay System, Innova Biosciences) makes use of a purified ATP reagent devoid of free phosphate usually resulting from spontaneous hydrolysis of ATP. To determine enzymatically formed phosphate the standard DPOR reduction assay was modified. Assays were performed in 96-well plates at 25 °C under anaerobic conditions. Each 190-μl assay contained 100 pmol of (ChlN/ChlB)2, 200 pmol of ChlL2, 2 mm dithionite, and 13 μm Pchlide in lysis buffer. In parallel, assays were performed in the absence of Pchlide. Control assays lacking subcomplexes of ChlL2 or (ChlN/ChlB)2, control reactions for which the DPOR subunits had been previously inactivated by oxygen exposure or controls in the absence of any protein components containing sole lysis buffer, were carried out to determine spontaneous ATP hydrolysis under conditions of the assay. Assays were initiated by addition of 10 μl of 10 mm ATP reagent. To quantify free phosphate the individual reactions were stopped by addition of 50 μl of the supplied Pi Color Lock Gold Mix in 5 m HCl after 2, 3, 4, 5, 10, 15, 20, 25, 30, 40, and 50 min. Free phosphate indicated by a dark green malachite complex was spectroscopically analyzed using a Fusion microplate reader (PerkinElmer Life Sciences) at 600 nm. For the quantification of phosphate concentrations in the assay mixture a phosphate standard was used to calibrate the system according to the manufacturer's instructions. To evaluate the background effect of Pchlide absorption at 600 nm, the pigment (13 μm Pchlide) or alternatively complete Pchlide reduction assays were incubated in 200 μl of lysis buffer and supplemented with 5 m HCl after 5, 10, and 30 min.
HPLC Analyses of Nucleotides Bound to ChlL2
Two mg of GST-ChlL2 (17 nmol) were bound to 1 ml of Protino glutathione-agarose and incubated in the presence of 2 mm MgATP, 2 mm MgADP or absence of nucleotides in lysis buffer for 30 min. These columns were extensively washed with 12 ml of lysis buffer. Subsequently, proteins were eluted with 2 ml of lysis buffer containing 25 mm glutathione and analyzed by HPLC for bound nucleotide cofactors according to Ref. 25. A HPLC system equipped with a multiwavelength detector (MD 2015plus, Jasco) was used. For this purpose 40-μl samples were applied to a reverse phase C18 HPLC column (hypersil 5 μm, 250 × 4.6 mm) and run with 1 ml min−1 at 5.8 megapascals at 30 °C in 100 mm potassium phosphate, pH 6.5, 10 mm tetrabutylammonium bromide, 7.5% (v/v) acetonitrile. Nucleotide absorption was recorded at 250 nm. MgATP and MgADP standards at a concentration of 100 μm each were run under identical conditions.
UV-visible Light Absorption Spectroscopy
UV-visible light spectra of purified DPOR proteins were recorded using a V-550 spectrometer (Jasco) in combination with sealed quartz cuvettes. To quantify pigments UV-visible spectra were measured in 80% acetone employing extinction coefficients of ϵ626 = 30.4 mm−1 cm−1 for Pchlide (26) and ϵ665 = 74.9 mm−1 cm−1 for Chlide (27).
Circular Dichroism Spectra of ChlL
CD spectra of the purified DPOR subcomplex ChlL2 at a concentration of 5.5 mg ml−1 (84 μm) in low salt lysis buffer (10 mm HEPES/NaOH, pH 7.5, 15 mm NaCl, and 10 mm MgCl2) were recorded using an anaerobic quartz cuvette (10-mm light path) with a J-810 spectrometer (Jasco) and a thermostated cell holder. Each spectrum was the result of seven successive spectra; each normalized against the low salt lysis buffer. Spectra were recorded at 25 °C under anaerobic conditions. Analyzed samples were pure ChlL2, ChlL2 with MgATP, and ChlL2 with MgADP. Nucleotides were used at a final concentration of 2 mm in protein samples and reference buffer.
Preparation of EPR Samples
EPR sample preparation was carried out in an anaerobic chamber. Purified ternary P. marinus DPOR was concentrated to 19.5 mg ml−1 (58 μm), using a stirred Amicon ultrafiltration cell (Millipore) equipped with 50,000 Da compounds excluding ultrafiltration membrane. Purified subcomplexes ChlL2, ChlL2 in the presence of 2 mm MgADP and ChlL2 in the presence of 2 mm MgATP, were concentrated to 100 μm (6.5 mg ml−1). 20 mm sodium dithionite was added to 100 μl of protein solution and incubated for up to 2 h. For control experiments samples containing solely ChlL2 (100 μm) and (ChlN/ChlB)2 (55 μm) were analogously reduced. Proteins were finally transferred to quartz EPR tubes with a 4-mm outer diameter and frozen in liquid nitrogen.
EPR Spectroscopy
9.5 GHz X-Band EPR spectra were recorded on a Bruker ESP300E spectrometer equipped with a rectangular microwave cavity in the TE102 mode. For low temperature measurements the samples were placed in an Oxford ESR 900 helium flow cryostat with an Oxford ITC502 temperature controller. The microwave frequency was detected with an EIP frequency counter (Microwave Inc.). The magnetic field was calibrated using a Li/LiF standard with a known g-value of 2.002293 ± 0.000002 (28). Baseline corrections were performed by subtracting a background spectrum, obtained under the same experimental conditions from a sample containing lysis buffer with 10 mm ADP, 25 mm NaF, and 500 μm AlCl3, or 2 mm ATP or 2 mm ADP, respectively. Simulations of the experimental EPR spectra have been carried out with the MATLAB toolbox EasySpin (version 3.1.0) (29).
RESULTS AND DISCUSSION
The subunit and iron-sulfur cluster composition and substrate recognition of DPOR were recently characterized (13, 15, 18). However, information regarding ATP-driven subunit interactions and the related electron transfer were still missing. This study focused on the dynamic protein-protein interaction of subunits (ChlN/ChlB)2 and ChlL2 and the related redox processes.
ATPase Activity of DPOR
The ATP-dependent DPOR system uses a ferredoxin as a natural electron donor (13, 30), whereas for in vitro experiments dithionite as an artificial chemical electron donor was used (13). Direct treatment of the catalytic (ChlN/ChlB)2 complex with the reductant dithionite did not result in substrate reduction. However, in the presence of ChlL2 and ATP DPOR catalysis followed a Michaelis-Menten type kinetic with respect to the co-substrates dithionite and ATP (13). Hence, ChlL2 containing two ATP binding sites is the functional electron donor leading to a catalytically active (ChlN/ChlB)2 protein. Taking into account the thermodyamic properties of Pchlide reduction the consumption of 4 mol of ATP/mol of reduced Pchlide was proposed. This amount of ATP correlates with the dimeric composition of ChlL2 containing two ATP binding sites and a single [FeS] cluster.
To understand the catalytic mechanism of DPOR, it is essential to elucidate the ATPase properties of DPOR. Here, we analyzed the ATPase activity of DPOR in the presence or absence of the substrate Pchlide. The obtained results were relevant for the elucidation of the mechanistic order of DPOR catalysis.
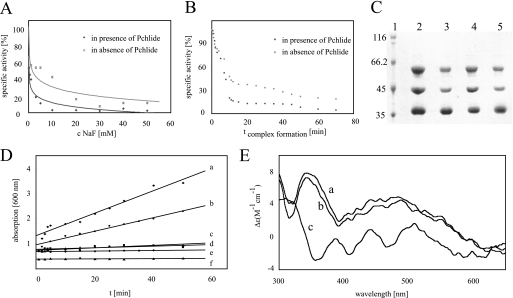
With the employed test system free phosphate liberated during DPOR catalysis was detected as a dark green metallophosphate complex. The ATPase assay system did not contain an ATP regenerating system and employed a specifically purified ATP reagent devoid of free phosphate resulting in increased sensitivity of the system. A linear increase in the free phosphate concentration over a range of 50 min was observed (Fig. 2D, line a). Control reactions devoid of (ChlN/ChlB)2, devoid of ChlL2, or assays with the addition of either oxygen inactivated DPOR or solely lysis buffer exhibited a linear background of less than 5% (Fig. 2D, lines c, d, e, and f). This background was ascribed to the fact that Pchlide and Chlide possess a spectral absorption at 600 nm. Taking into account this unspecific background a specific activity for the ATP hydrolysis of DPOR of 11 pmol min−1 mg−1 was observed (Fig. 2D, line a). Correlating this experimental value to Pchlide reduction, a ratio of ∼14 mol of ATP hydrolyzed per mol of Chlide formed was deduced. Analogously, for the related nitrogenase system an in vitro consumption of up to 36 ATP per reduced N2 versus an apparent minimal theoretical consumption of 16 ATP was observed (31–33). According to this, the minimal ATP consumption of DPOR catalysis might be in the range of 4 mol of ATP hydrolyzed per mol of Chlide formation. However, the employed conditions of the in vitro assay are not sufficient to reflect the optimal in vivo situation.
FIGURE 2.
Formation of ternary DPOR complexes is supported by Pchlide, ATPase activity of DPOR catalysis, and circular dichroism spectra of ChlL2. A, DPOR complex formation for 90 min with 10 mm MgADP, 500 μm AlCl3, and increasing concentrations of NaF ranging from 10 μm to 50 mm in the presence (dots) and absence (crosses) of Pchlide. Subsequent DPOR assays indicate the loss of DPOR activity. B, time dependence of DPOR complex formation. DPOR complex formation with MgADP·AlF4− in the presence (dark gray) and absence (light gray) of Pchlide for 1–70 min. Subsequent Pchlide reduction assays indicate the formation of inactive DPOR complexes. C, Pchlide-dependent DPOR complex formation. SDS-PAGE analyses of ternary DPOR complexes containing MgADP·AlF4− (lanes 2 and 3) and ChlLΔLeu153 (lanes 4 and 5), respectively. Lane 1, molecular mass marker, relative molecular masses (×1000) are indicated; lanes 2 and 4, complex formation in the presence of Pchlide; lanes 3 and 5, complex formation in the absence of Pchlide. D, ATPase activity of DPOR was analyzed over 50 min after addition of 500 μm ATP. Absorption at 600 nm indicates liberated phosphate as a green metallocomplex. Analyzed samples contained: a, (ChlN/ChlB)2 and ChlL2 in the presence of Pchlide; b, (ChlN/ChlB)2 and ChlL2 without Pchlide; c, sole ChlL2; d, sole (ChlN/ChlB)2; e, (ChlN/ChlB)2 and ChlL2 after oxygen exposure; f, lysis buffer. E, circular dichroism spectra of ChlL2 in the presence and absence of nucleotides. Spectrum a, ChlL2 (85 μm) + 2 mm MgATP; spectrum b, ChlL2 (85 μm) + 2 mm MgADP; spectrum c, sole ChlL2 (85 μm).
Interestingly, DPOR assays that were incubated in the absence of the substrate Pchlide also revealed significant ATPase activity and resulted in a linear increase in the free phosphate concentration over a range of 50 min (Fig. 2D, line b). Approximately 60% ATPase activity compared with the substrate containing assay was observed. This clearly indicated that DPOR is able to efficiently hydrolyze ATP in the absence of its natural substrate Pchlide. This nonproductive ATPase activity might not be relevant under in vivo conditions. Nevertheless, this in vitro observation is of interest for the understanding of the dynamic protein-protein interaction of (ChlN/ChlB)2 and ChlL2 responsible for DPOR electron transfer. Experiments in which ChlL2 was not completed by addition of subcomplex (ChlN/ChlB)2 did not result in detectable ATPase activity (Fig. 2D, line c).
From these results we conclude that the docking of ChlL2 to (ChlN/ChlB)2 is a prerequisite for the observed ATPase activity. Obviously, this protein-protein interaction is important for the ATP hydrolyzing activity of ChlL2. Additionally, this dynamic subunit interplay is also strongly influenced by the presence of the substrate Pchlide. In previous studies the high affinity of the substrate Pchlide for the (ChlN/ChlB)2 complex was demonstrated (18, 34). Because maximum DPOR ATPase activity was dependent on the presence of Pchlide and subcomplex (ChlN/ChlB)2, we postulate that Pchlide binding is the initial step of DPOR catalysis. Subsequently, the substrate-bound complex (ChlN/ChlB)2 efficiently stimulates the ATPase activity of ChlL2, which is a prerequisite for substrate reduction. Moreover, tight binding of the substrate is also beneficial in vivo to overcome the highly phototoxic nature of this hydrophobic molecule. The harmful photodynamic properties of Pchlide have only recently been demonstrated (35).
Ternary DPOR Protein-Protein Interaction
Affinity purification of the GST-ChlN subunit allows for the co-purification of stoichiometric amounts of ChlB (Fig. 1B, lane 3). However, all previous attempts to purify the ternary ChlL2(ChlN/ChlB)2 complex failed (13, 18). Therefore, a transient interaction responsible for the electron transfer of subunit ChlL2 to subcomplex (ChlN/ChlB)2 was proposed (15). In the present study three ATP analogs were employed to stabilize the observed transient protein-protein interaction and to make the ternary DPOR complex accessible to biochemical characterization. The non-hydrolyzable ATP analogs ATPγS, AMP-PNP, and MgADP in combination with the inorganic compound AlF4− were used. These individual compounds are mimicking different stages of the ATP hydrolysis process. GST-tagged ChlL2 was immobilized on a Protino glutathione-agarose column and subsequently incubated with an excess of the purified (ChlN/ChlB)2 subcomplex containing 30 μm Pchlide in the presence of the various ATP analogs at a concentration of 10 mm, respectively. After extensive washing of the column the potential ternary complexes were liberated from the affinity matrix by PreScission protease cleavage of GST-ChlL. Eluate fractions were subsequently analyzed by SDS-PAGE for the co-elution of DPOR subunits ChlN and ChlB due to ternary complex formation. In Fig. 1B, lanes 4–6, the SDS-PAGE shows the “bait” protein ChlL with a relative molecular weight of 35,000 (calculated molecular mass of 32,395) eluted in the presence of DPOR subunits ChlN with a relative molecular weight of 45,000 (calculated molecular mass of 46,199) and ChlB with a relative molecular weight of 60,000 (calculated molecular mass of 58,729). The identity of the proteins was confirmed by Edman degradation of the corresponding peptides. All experiments performed in the presence of ATP analogs resulted in formation of ternary DPOR complexes. In each case an almost identical ratio of ChlN/ChlB was observed as judged by SDS-PAGE analysis (Fig. 1B, lanes 4–6). In contrast, the ratio of components ChlN/ChlB versus ChlL varied significantly. The highest amount of ChlN/ChlB was observed in the presence of MgADP·AlF4− (Fig. 1B, lane 6), whereas in the presence of AMP-PNP or AMPγS only lower amounts of ChlN/ChlB were co-purified (Fig. 1B, lanes 4 and 5). To identify the stoichiometry of subunits ChlN, ChlB, and ChlL in the ternary DPOR, isolated complexes were analyzed by Edman degradation. A molar ratio of 1.1/1/2.2 for subunits ChlN/ChlB/ChlL incubated in the presence of MgADP·AlF4− and Pchlide was observed. Additionally, this ternary DPOR complex (0.08 μmol) was analyzed by size exclusion chromatography under anaerobic conditions. A relative native molecular weight of ∼360,000 (elution volume 166 ml) was obtained (Fig. 1C). These results are indicative of an octameric DPOR complex (ChlN/ChlB)2(ChlL2)2 with a calculated molecular mass of 339,439 Da. Elution fractions containing the octameric complex were bright green and exhibited an absorption maximum at 626 nm after extraction with 80% acetone. A molar ratio of 1.9 mol of Pchlide/mol of (ChlN/ChlB)2(ChlL2)2 was determined indicating that two molecules of Pchlide are part of the ternary DPOR complex. This result is consistent with the proposal of two active sites per (ChlN/ChlB)2 (13, 18). Fig. 1D shows a schematic representation of two ChlL2 dimers individually docked onto the Pchlide carrying the heterotetrameric (ChlN/ChlB)2 complex to form the proposed octameric protein complex. The slight shoulder in the elution diagram (elution volume 176 ml) might be indicative of minor amounts of hexameric (ChlN/ChlB)2ChlL2 complexes (Fig. 1C). Clear differences were also observed for experiments in which DPOR subunits ChlL2 and (ChlN/ChlB)2 were incubated in the absence of Pchlide. These experiments resulted in significantly reduced amounts of the co-purified (ChlN/ChlB)2 component (Fig. 2C, compare lanes 2 and 3). This result demonstrated that Pchlide binding to (ChlN/ChlB)2 probably influences ternary DPOR complex formation.
Ternary MgADP·AlF4−·DPOR Complexes Are Catalytically Inactive
The catalytic properties of the ternary MgADP·AlF4−·DPOR complex were analyzed using the standard DPOR assay. When MgADP·AlF4−-dependent complex formation was induced for 60 min the subsequent DPOR assays did not result in detectable DPOR activity, even in the presence of an increased ATP concentration up to 20 mm. Control experiments assured that 100 mm NaF, 25 mm AlCl3, and 10 mm MgADP as well as combinations of NaF and MgADP, or AlCl3 and MgADP had no inhibitory effects on DPOR catalysis. The MgADP·AlF4− analog represents the transition state of ATP hydrolysis, which has been shown to bind to the nucleotide binding site of various enzymes (16, 36–41). Because our experiments in the presence of high ATP concentrations failed to restore any DPOR activity, we concluded that MgADP·AlF4− was tightly bound to the nucleotide binding site of ChlL2 and was not displaceable by ATP. This was the prerequisite for the efficient trapping of the ternary DPOR complex.
Ternary ATPγS·DPOR and AMP-PNP·DPOR Complexes Are Catalytically Active
The ternary ATPγS·DPOR complex (100 pmol) or the ternary AMP-PNP·DPOR complex (100 pmol) were analyzed in the standard DPOR assay in the presence of 1 mm ATP. Only slightly reduced specific Chlide formation activities of 870 and 900 pmol min−1 mg−1 were obtained, compared with the specific activity of 910 pmol min−1 mg−1 for the DPOR enzyme in the absence of ATP analogs (18). Because both nonhydrolyzable ATP analogs have been shown to bind to the ATP binding sites of various ATP-dependent enzymes (42–45) we concluded that ATPγS or AMP-PNP were binding to the ATP binding site of ChlL2, however, with a much lower affinity compared with MgADP·AlF4−. Our findings support that ATP efficiently competes against these two ATP analogs for the ATP binding sites of ChlL2 under conditions of the standard DPOR assay resulting in the observed Pchlide reduction. Because MgADP·AlF4− enables the formation of significantly more stable ternary complexes all further analyses of the ATP-dependent protein-protein interaction of DPOR were performed using MgADP·AlF4− as an ATP analog.
Ternary MgADP·AlF4−·DPOR Complex Formation Is Enhanced in the Presence of Pchlide
Our initial experiments suggested that DPOR complex formation induced by MgADP·AlF4− is more efficient in the presence of Pchlide (Fig. 2C, compare lanes 2 and 3). Two alternative experimental approaches to study the influence of Pchlide on DPOR complex formation were employed.
Ternary complex formation experiments were initiated for 60 min by using 10 mm MgADP, 500 μm AlCl3 and varying concentrations of NaF (10 μm to 50 mm) (19). All experiments were performed either in the absence or presence of 13 μm Pchlide. In the subsequent DPOR assay an increasing loss of DPOR activity was observed correlating with increasing NaF concentrations (Fig. 2A). Interestingly, for all employed NaF concentrations a significantly higher decline of DPOR activity was obtained in the presence of Pchlide. For example, in the presence of 10 mm NaF, Pchlide-induced complex formation resulted in a residual activity of 5%, whereas in the absence of the substrate a relative activity of 50% was observed.
Alternatively, the influence of Pchlide on the time dependence of MgADP·AlF4−·DPOR complex formation was determined. For our initial inhibition experiments an extensive incubation time of 60 min for the formation of the ternary complex was used. To determine the time-dependent inhibitory complex formation a full time course was analyzed. Complex formation was initiated with 500 μm AlCl3, 25 mm NaF, and 10 mm MgADP in the absence or presence of 13 μm Pchlide for 1–70 min. Subsequently, DPOR activity was determined. As shown in Fig. 2B the time-dependent activity loss of the MgADP·AlF4−·DPOR complex was a slow process that was again more effective in the presence of the substrate. After a 10-min incubation with Pchlide DPOR preserved 15% activity, whereas in the absence of the substrate DPOR still showed a relative activity of 45%. From these results we concluded that substrate binding to (ChlN/ChlB)2 clearly influenced formation of the ternary MgADP·AlF4−·DPOR. In the presence of Pchlide, inactive ternary complexes were formed significantly faster at lower concentrations of AlF4−.
In the current study three independent methods demonstrated the impact of Pchlide binding on ternary DPOR complex formation. These results are consistent with recent studies that report a co-purification of (ChlN/ChlB)2 with the substrate Pchlide (18, 34). These findings imply a catalytic mechanism where substrate binding precedes the transient interaction of (ChlN/ChlB)2 and ChlL2.
EPR Analyses of [4Fe-4S] Clusters in the Ternary DPOR Complex
Because EPR analyses require milligram amounts of DPOR complexes, preparative upscaling was required as outlined under “Experimental Procedures.” An overall recovery of 80 mg (0.234 μmol) of the ternary DPOR complex at a concentration of 19.5 mg ml−1 (58 μm) was achieved.
After dithionite reduction these samples were subjected to EPR spectroscopy. In parallel highly purified protein samples of ChlL2 (100 μm) and (ChlN/ChlB)2 (60 μm) were analyzed analogously.
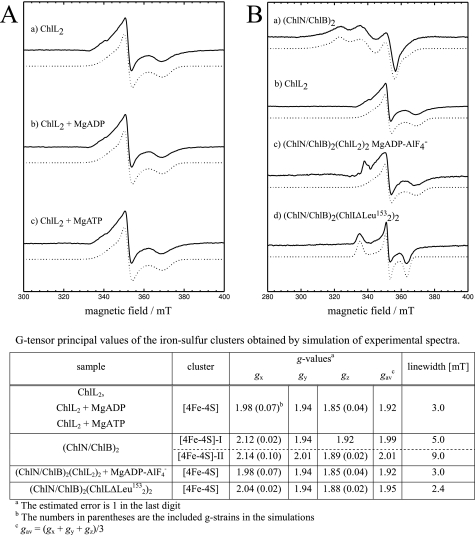
The ternary MgADP·AlF4− complex of DPOR revealed a typical EPR spectrum of a reduced [4Fe-4S]1+ cluster (Fig. 3B, spectrum c). The obtained g-values correlated well with data obtained for (ChlL)2 alone (Fig. 3B, b, and table). In an earlier investigation the EPR spectrum of the [4Fe-4S] clusters located on (ChlN/ChlB)2 was characterized as a superposition of a signal for two [4Fe-4S] clusters with different g-values of gx = 2.115, gy = 1.935, and gz = 1.917 for [4Fe-4S]-I and gx = 2.14, gy = 2.008, and gz = 1.89 for [4Fe-4S]-II (Fig. 3B, a, and table) (18). These characteristic EPR spectroscopic data allow for the clear discrimination of the [4Fe-4S] signals of ChlL2 and (ChlN/ChlB)2. From the obtained EPR data of the MgADP·AlF4−·DPOR complex we concluded that the artificial electron donor dithionite solely reduced the [4Fe-4S] cluster of ChlL2. Under native turnover conditions this reduced cluster was then able to transfer a single electron onto Pchlide via the [4Fe-4S] cluster located on (ChlN/ChlB)2. However, no evidence for a paramagnetic (ChlN/ChlB)2 [4Fe-4S]1+ cluster or a partially reduced Pchlide substrate was obtained. The employed reductant dithionite does not have the required redox potential to reduce [4Fe-4S] clusters to the all-ferrous [FeS]0 state as shown for the homologous nitrogenase system (46). The additional EPR feature at ∼335 millitesla most probably originates from traces of impurities in the sample. These data indicated that in the trapped ternary MgADP·AlF4−·DPOR complex electron flow was blocked in the state of the [4Fe-4S]1+ cluster of ChlL2. This catalytically inactive MgADP·AlF4−·DPOR complex mimics a situation immediately before the electron transfer. By analogy to various other dynamic switch proteins AlF4− can be considered to represent the trigonal bipyramidal geometry of the γ-phosphate in the transition state undergoing nucleophilic attack by a water molecule (20, 40). However, in the MgADP·AlF4− complex of DPOR the catalytic cycle was trapped before electron transfer occurred. We concluded that the entire ATP hydrolysis process is necessary for the electron transfer process in the ternary complex. Furthermore, this hydrolysis might also trigger the dissociation of this complex.
FIGURE 3.
EPR spectra of ChlL2 and ternary DPOR complexes. EPR spectra of ChlL2, (ChlN/ChlB)2, and ternary DPOR complexes after reduction with dithionite (solid), in combination with the corresponding simulations (dotted). A, ChlL2 recorded in the native state (a) and in the presence of MgADP (b) and MgATP (c) measured at 10 K. B, comparison of spectra of the (ChlN/ChlB)2 subcomplex (a) and ChlL2 (b) with the ternary octameric (ChlN/ChlB)2(ChlL2)2 complexes formed with MgADP·AlF4− (c) or mutant ChlLΔLeu153 (d). T = 10 K (a–c), T = 20 K (d). The spectrum for (ChlN/ChlB)2 exhibited a superposition of two non-identical [4Fe-4S] clusters. Experimental conditions: microwave frequency 9.5 GHz, 1 millitesla (mT) modulation amplitude, 12.5 kHz modulation frequency, 2.5 milliwatt microwave power. The table lists g-values and line widths determined from the simulations of the experimental EPR spectra.
Nucleotide-dependent Alteration of the ChIL2 Conformation
For the required changes in the dynamic protein-protein interaction of DPOR subunits the affinity of ChlL2 for (ChlN/ChlB)2 must be efficiently controlled. For this purpose in nucleotide-dependent switch proteins minor conformational changes are induced by ATP binding and hydrolysis. Such alterations have been shown for the related nitrogenase protein NifH2 by various three-dimensional structural analyses (16) as well as by CD spectroscopy in the 350–600 nm range (24, 47).
To study potential nucleotide-dependent conformational alterations, highly concentrated ChlL2 protein samples (84 μm) were analyzed by CD spectroscopy. Nucleotide binding in samples of ChlL2 was demonstrated using standard HPLC analyses (data not shown). ChlL2 revealed a significantly different spectrum in the presence of 2 mm MgATP and 2 mm MgADP when compared with the nucleotide-free sample (Fig. 2E). The spectral shape of ChlL2 in the presence of nucleotides correlates well to CD spectra obtained in a related study for NifH2 incubated with MgADP also showing positive deflections for the spectra visible at 340 and 480 nm. In contrast, ChlL2 in the absence of nucleotides and ChlL2 in the presence of ATP exhibited substantially different spectra when compared with nucleotide-free NifH2 or NifH2 in the presence of MgATP (24, 47, 48). From these experiments we concluded that ChlL2 is a nucleotide-dependent switch protein triggering an ATP-dependent conformational change that subsequently results in ternary complex formation. Our experiments also point toward minor differences for the nucleotide-dependent conformational changes of ChlL2 and NifH2.
For the related nitrogenase system it was demonstrated that such dynamic processes also have influence on the direct environment of the [4Fe-4S] cluster of NifH2 (16, 47, 49) resulting in a more negative redox potential for this cluster (16, 48). It was observed that upon nucleotide binding the geometry of the [4Fe-4S] cluster of NifH2 was slightly altered (16). In an EPR analysis a shift of spectral shape and g-values of NifH2 was observed in the presence of MgATP, whereas MgADP had no influence on the EPR spectra (48).
For ChlL2 of DPOR we demonstrated that the EPR spectra of ChlL2 (100 μm) and ChlL2 in the presence of 2 mm MgADP or MgATP exhibited no differences in the spectral shape and g-values (Fig. 3A, a–c, and table). From these observations we concluded that the nucleotide-dependent conformational changes of ChlL2 do not significantly change the periphery of the [4Fe-4S] cluster of ChlL2. Here the DPOR system clearly differs from the related nitrogenase system.
Leucine153 Is Involved in the ATP-dependent Switch Mechanism of ChlL2
In nitrogenase, binding of NifH2 to (NifD/NifK)2 is dependent on ATP-induced conformational changes. The “switch II” motif, DVLGDVVCGG of NifH2, is crucial for ATP binding and subsequent conformational rearrangements. This sequence motif couples ATP binding at the P-loop region to the [4Fe-4S] cluster inter alia by minor repositioning of cluster ligand Cys132 (A. vinelandii numbering). Deleting Leu127 of the switch II motif traps NifH2 in a conformation preventing hydrolysis of ATP thus inactivating nitrogenase (16). This trapped NifH2 variant was shown to form a stable ternary complex with (NifD/NifK)2 allowing simple co-purification (23). Leu127 of NifH corresponds to Leu153 of ChlL, conserved in all ChlL amino acid sequences.
Mutating chlL to produce the protein variant ChlLΔLeu153 resulted in a completely inactivated DPOR enzyme. Furthermore, this mutant DPOR enzyme did not show any ATPase activity (data not shown). When ChlLΔLeu153 was incubated with (ChlN/ChlB)2 in the presence of Pchlide, ternary complex formation was observed with a stoichiometry of 1.1 mol of ChlN/1 mol of ChlB/2.2 mol of ChlL (Figs. 1B, lane 7, and 2C, lane 4). When complex formation was analyzed in the absence of Pchlide, clearly reduced amounts of (ChlN/ChlB)2 were detected in the SDS-PAGE analysis (Fig. 2C, lane 5). This substrate-dependent complex formation is in accordance with the results for the ternary complex formation in the presence of MgADP·AlF4−. However, for ChlLΔLeu153 the presence of nucleotides or nucleotide analogs was not required. From these results we concluded that this mutant ChlL2 protein adopts a protein conformation with a high affinity for (ChlN/ChlB)2 analogously as described for the MgADP·AlF4−·DPOR transition state complex.
EPR spectroscopic analysis of ChlLΔLeu153 in complex with (ChlN/ChlB)2 after dithionite reduction revealed a rhombic EPR signal with similar gy and gz values as obtained for the ternary MgADP·AlF4−·DPOR complex and ChlL2 alone, respectively (Fig. 3B, d, and table). The gx value was found slightly higher (Fig. 3, table). These data indicated that the [4Fe-4S] cluster of ChlL2ΔLeu153 was solely reduced under the employed conditions. In addition, the local geometric structure of the cluster of ChlL2ΔLeu153 in the (ChlN/ChlB)2/ChlL2ΔLeu153 complex might be slightly different compared with the wild-type protein complex. This might be caused by deletion of Leu153 in close proximity to the cluster or by a slightly different conformation of the complex. The smaller line width of this cluster compared with the wild-type ChlL2 protein was probably based on a lower g-strain, which might be due to a better defined and more rigid local structure. This nucleotide-free ternary complex did not facilitate electron transfer onto (ChlN/ChlB)2. We concluded that the catalytic cycle is trapped prior to electron transfer. This is in accordance with a model in which the electron transfer to (ChlN/ChlB)2 is coupled to the ATP hydrolysis step. From the results of the spectroscopic investigation of the ternary MgADP·AlF4−·DPOR complex and the ternary ChlLΔLeu153 complexes we concluded that ChlL2 is a nucleotide-dependent switch protein.
Proposed Catalytic Redox Cycle of DPOR
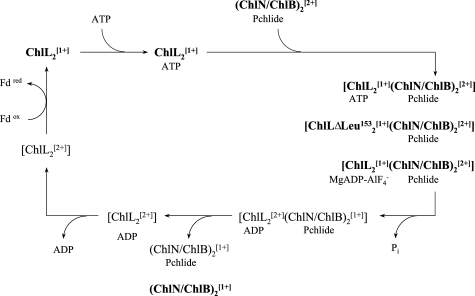
In the present study various states of the DPOR catalytic cycle have been characterized with biochemical and biophysical methods. In our proposed redox cycle (Fig. 4) these individual states are highlighted (bold).
FIGURE 4.
Proposed redox cycle of DPOR catalysis. Schematic model indicating electron transfer processes and dynamic subunit interaction during ATP-driven DPOR catalysis. Five intermediates were confirmed by EPR spectroscopy (bold), the redox state is marked by the superscript 1+ for reduced and superscript 2+ for oxidized [4Fe-4S] clusters. According to this redox cycle two consecutive single electron reductions of (ChlN/ChlB)2 are required to provide the two electrons necessary for Pchlide reduction.
Our experiments demonstrated that the single electron reduction of ChlL2 is enabled in the absence of (ChlN/ChlB)2 (18). Therefore, we conclude that during the initial step of DPOR catalysis the natural electron donor, which is a ferredoxin (13), transfers an electron onto the [4Fe-4S] cluster of the dimeric subunit ChlL2. Because the presence of ATP is not a prerequisite for effective dithionite reduction of ChlL2 we further propose that reduction of the [4Fe-4S] cluster precedes nucleotide binding. Subsequently, binding of ATP molecules results in slight conformational alterations as indicated by the CD spectroscopy data. However, these structural changes do not influence the [4Fe-4S]1+ cluster geometry of ChlL2. Binding of the substrate Pchlide to subunit (ChlN/ChlB)2 is then required for transient ternary complex formation. ATP hydrolysis by the ChlL2 subunits then facilitates the tight protein-protein interaction of ChlL2 and (ChlN/ChlB)2 and the simultaneous electron transfer process from the [4Fe-4S] cluster of ChlL2 to the [4Fe-4S] cluster of (ChlN/ChlB)2. In the present study this transient complex has been characterized with two independent techniques. The octameric (ChlL2[1+])2(ChlN/ChlB)2[2+]MgADP·AlF4− complex and the octameric (ChlL2ΔLeu153)2[1+](ChlN/ChlB)2[2+] complex represent the situation where the reduced component ChlL2 is close to the electron transfer on (ChlN/ChlB)2. EPR spectroscopic data clearly indicated that the transition state of ATP hydrolysis is not accompanied by a reduced [4Fe-4S] cluster in (ChlN/ChlB)2 or a partially reduced substrate. Therefore, complete ATP hydrolysis is needed to obtain a [4Fe-4S]1+ cluster on (ChlN/ChlB)2, which then has the ability to transfer a single electron onto Pchlide. The presence of ADP then leads to the dissociation of the ternary complex that results in the liberation of ChlL2 in the [4Fe-4S]2+ state and (ChlN/ChlB)2 in the [4Fe-4S]1+ state. The dynamic switch protein ChlL2 has to trigger two rounds of this redox catalytic cycle to supply the two electrons necessary for the reduction of the substrate. These individual electron transfer processes are completed by the stereospecific addition of two protons on the C17 and C18 carbons of Pchlide in the active site of the DPOR enzyme. Either protonated amino acids or precisely orientated water molecules might be the source for those two protons.
Acknowledgments
We thank Carl E. Bauer for providing Rhodobacter capsulatus strain ZY-5. We thank Sebastian Bruchmann and Phillip Ringel for their remarkable help during DPOR complex formation experiments. Furthermore, we thank Rita Getzlaff for N-terminal protein sequencing.
This work was supported by the Deutsche Forschungsgemeinschaft.
- Pchlide a
- protochlorophyllide a
- Chlide a
- chlorophyllide a
- EPR
- electron paramagnetic resonance
- GHz
- gigahertz
- GST
- glutathione S-transferase
- ATPγS
- adenosine 5′-(γ-thio)triphosphate
- AMP-PNP
- adenosine 5′-(β,γ-imido)triphosphate
- HPLC
- high pressure liquid chromatography
- POR
- protochlorophyllide oxidoreductase
- DPOR
- dark operative protochlorophyllide oxidoreductase.
REFERENCES
- 1.Beale S. I. (1999) Photosynth. Res. 60, 43–73 [Google Scholar]
- 2.Apel K. (2001) in Regulation of Photosynthesis (Aro E.-M., Anderson B. eds) pp. 235–252, Kluwer Akademic Publishers, Dordrecht [Google Scholar]
- 3.Fujita Y. (1996) Plant Cell Physiol. 37, 411–421 [DOI] [PubMed] [Google Scholar]
- 4.Schoefs B. (2001) Photosynth. Res. 70, 257–271 [DOI] [PubMed] [Google Scholar]
- 5.Belyaeva O. B., Griffiths W. T., Kovalev J. V., Timofeev K. N., Litvin F. F. (2001) Biochemistry 66, 173–177 [DOI] [PubMed] [Google Scholar]
- 6.Heyes D. J., Hunter C. N., van Stokkum I. H., van Grondelle R., Groot M. L. (2003) Nat. Struct. Biol. 10, 491–492 [DOI] [PubMed] [Google Scholar]
- 7.Heyes D. J., Ruban A. V., Wilks H. M., Hunter C. N. (2002) Proc. Natl. Acad. Sci. U.S.A. 99, 11145–11150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rüdiger W. (2003) in Porphyrin Handbook, Chlorophylls and Bilins: Biosynthesis, Synthesis, and Degradation (Kadish K. M., Smith K. M., Guilard R. eds) pp. 71–108, Academic Press, New York [Google Scholar]
- 9.Masuda T., Takamiya K. (2004) Photosynth. Res. 81, 1–29 [DOI] [PubMed] [Google Scholar]
- 10.Suzuki J. Y., Bollivar D. W., Bauer C. E. (1997) Annu. Rev. Genet. 31, 61–89 [DOI] [PubMed] [Google Scholar]
- 11.Bollivar D. W., Suzuki J. Y., Beatty J. T., Dobrowolski J. M., Bauer C. E. (1994) J. Mol. Biol. 237, 622–640 [DOI] [PubMed] [Google Scholar]
- 12.Burke D. H., Hearst J. E., Sidow A. (1993) Proc. Natl. Acad. Sci. U.S.A. 90, 7134–7138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bröcker M. J., Virus S., Ganskow S., Heatcote P., Heinz D. W., Schubert W. D., Jahn D., Moser J. (2008) J. Biol. Chem. 283, 10559–10567 [DOI] [PubMed] [Google Scholar]
- 14.Fujita Y., Matsumoto H., Takahashi Y., Matsubara H. (1993) Plant Cell Physiol. 34, 305–314 [PubMed] [Google Scholar]
- 15.Wätzlich D., Bröcker M. J., Uliczka F., Ribbe M., Virus S., Jahn D., Moser J. (2009) J. Biol. Chem. 284, 15530–15540 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Igarashi R. Y., Seefeldt L. C. (2003) Crit. Rev. Biochem. Mol. Biol. 38, 351–384 [DOI] [PubMed] [Google Scholar]
- 17.Howard J. B., Rees D. C. (1994) Annu. Rev. Biochem. 63, 235–264 [DOI] [PubMed] [Google Scholar]
- 18.Bröcker M. J., Wätzlich D., Uliczka F., Virus S., Saggu M., Lendzian F., Scheer H., Rüdiger W., Moser J., Jahn D. (2008) J. Biol. Chem. 283, 29873–29881 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bigay J., Deterre P., Pfister C., Chabre M. (1987) EMBO J. 6, 2907–2913 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Howard J. B., Rees D. C. (2006) Proc. Natl. Acad. Sci. U.S.A. 103, 17088–17093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Schindelin H., Kisker C., Schlessman J. L., Howard J. B., Rees D. C. (1997) Nature 387, 370–376 [DOI] [PubMed] [Google Scholar]
- 22.Duyvis M. G., Wassink H., Haaker H. (1998) Biochemistry 37, 17345–17354 [DOI] [PubMed] [Google Scholar]
- 23.Chiu H., Peters J. W., Lanzilotta W. N., Ryle M. J., Seefeldt L. C., Howard J. B., Rees D. C. (2001) Biochemistry 40, 641–650 [DOI] [PubMed] [Google Scholar]
- 24.Ryle M. J., Seefeldt L. C. (1996) Biochemistry 35, 4766–4775 [DOI] [PubMed] [Google Scholar]
- 25.Eberth A., Ahmadian M. R. (2009) in Current Protocols in Cell Biology (Bonifacio J. S., Dasso M., Harford J. B., Pippincott-Schwartz J., Yamada K. M. eds) Vol. 43, pp. 14.9.1–14.9.25, John Wiley and Sono, Inc., New York [Google Scholar]
- 26.Fujita Y., Bauer C. E. (2000) J. Biol. Chem. 275, 23583–23588 [DOI] [PubMed] [Google Scholar]
- 27.McFeeters R. F., Chichester C. O., Whitaker J. R. (1971) Plant Physiol. 47, 609–618 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Stesmans A., van Gorp G. (1989) Rev. Sci. Instrum. 60, 2949–2952 [Google Scholar]
- 29.Stoll S., Schweiger A. (2006) J. Magn. Reson. 178, 42–55 [DOI] [PubMed] [Google Scholar]
- 30.Eisen J. A., Nelson K. E., Paulsen I. T., Heidelberg J. F., Wu M., Dodson R. J., Deboy R., Gwinn M. L., Nelson W. C., Haft D. H., Hickey E. K., Peterson J. D., Durkin A. S., Kolonay J. L., Yang F., Holt I., Umayam L. A., Mason T., Brenner M., Shea T. P., Parksey D., Nierman W. C., Feldblyum T. V., Hansen C. L., Craven M. B., Radune D., Vamathevan J., Khouri H., White O., Gruber T. M., Ketchum K. A., Venter J. C., Tettelin H., Bryant D. A., Fraser C. M. (2002) Proc. Natl. Acad. Sci. U.S.A. 99, 9509–9514 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rainbird R. M., Hitz W. D., Hardy R. W. (1984) Plant Physiol. 75, 49–53 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rees D. C., Howard J. B. (2000) Curr. Opin. Chem. Biol. 4, 559–566 [DOI] [PubMed] [Google Scholar]
- 33.Erickson J. A., Nyborg A. C., Johnson J. L., Truscott S. M., Gunn A., Nordmeyer F. R., Watt G. D. (1999) Biochemistry 38, 14279–14285 [DOI] [PubMed] [Google Scholar]
- 34.Nomata J., Ogawa T., Kitashima M., Inoue K., Fujita Y. (2008) FEBS Lett. 582, 1346–1350 [DOI] [PubMed] [Google Scholar]
- 35.Walther J., Bröcker M. J., Wätzlich D., Nimtz M., Rohde M., Jahn D., Moser J. (2009) FEMS Microbiol. Lett. 290, 156–163 [DOI] [PubMed] [Google Scholar]
- 36.Chaney M., Grande R., Wigneshweraraj S. R., Cannon W., Casaz P., Gallegos M. T., Schumacher J., Jones S., Elderkin S., Dago A. E., Morett E., Buck M. (2001) Genes Dev. 15, 2282–2294 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Leslie A. G., Abrahams J. P., Braig K., Lutter R., Menz R. I., Orriss G. L., van Raaij M. J., Walker J. E. (1999) Biochem. Soc. Trans. 27, 37–42 [DOI] [PubMed] [Google Scholar]
- 38.Maruta S., Henry G. D., Sykes B. D., Ikebe M. (1993) J. Biol. Chem. 268, 7093–7100 [PubMed] [Google Scholar]
- 39.Danko S., Yamasaki K., Daiho T., Suzuki H. (2004) J. Biol. Chem. 279, 14991–14998 [DOI] [PubMed] [Google Scholar]
- 40.Duyvis M. G., Wassink H., Haaker H. (1996) FEBS Lett. 380, 233–236 [DOI] [PubMed] [Google Scholar]
- 41.Renner K. A., Howard J. B. (1996) Biochemistry 35, 5353–5358 [DOI] [PubMed] [Google Scholar]
- 42.Boll M., Fuchs G., Lowe D. J. (2001) Biochemistry 40, 7612–7620 [DOI] [PubMed] [Google Scholar]
- 43.Horio M., Gottesman M. M., Pastan I. (1988) Proc. Natl. Acad. Sci. U.S.A. 85, 3580–3584 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hegner M., Smith S. B., Bustamante C. (1999) Proc. Natl. Acad. Sci. U.S.A. 96, 10109–10114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kelman Z., Lee J. K., Hurwitz J. (1999) Proc. Natl. Acad. Sci. U.S.A. 96, 14783–14788 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Guo M., Sulc F., Ribbe M. W., Farmer P. J., Burgess B. K. (2002) J. Am. Chem. Soc. 124, 12100–12101 [DOI] [PubMed] [Google Scholar]
- 47.Ryle M. J., Lanzilotta W. N., Seefeldt L. C., Scarrow R. C., Jensen G. M. (1996) J. Biol. Chem. 271, 1551–1557 [DOI] [PubMed] [Google Scholar]
- 48.Ryle M. J., Lanzilotta W. N., Seefeldt L. C. (1996) Biochemistry 35, 9424–9434 [DOI] [PubMed] [Google Scholar]
- 49.Lanzilotta W. N., Holz R. C., Seefeldt L. C. (1995) Biochemistry 34, 15646–15653 [DOI] [PubMed] [Google Scholar]
- 50.Moss G. P. (1987) Pure Appl. Chem. 59, 779–832 [Google Scholar]