Abstract
Sirtuins catalyze NAD+-dependent protein deacetylation and are critical regulators of transcription, apoptosis, metabolism, and aging. There are seven human sirtuins (SIRT1–7), and SIRT1 has been implicated as a key mediator of the pathways downstream of calorie restriction that have been shown to delay the onset and reduce the incidence of age-related diseases such as type 2 diabetes. Increasing SIRT1 activity, either by transgenic overexpression of the Sirt1 gene in mice or by pharmacological activation by small molecule activators resveratrol and SRT1720, has shown beneficial effects in rodent models of type 2 diabetes, indicating that SIRT1 may represent an attractive therapeutic target. Herein, we have assessed purported SIRT1 activators by employing biochemical assays utilizing native substrates, including a p53-derived peptide substrate lacking a fluorophore as well as the purified native full-length protein substrates p53 and acetyl-CoA synthetase1. SRT1720, its structurally related compounds SRT2183 and SRT1460, and resveratrol do not lead to apparent activation of SIRT1 with native peptide or full-length protein substrates, whereas they do activate SIRT1 with peptide substrate containing a covalently attached fluorophore. Employing NMR, surface plasmon resonance, and isothermal calorimetry techniques, we provide evidence that these compounds directly interact with fluorophore-containing peptide substrates. Furthermore, we demonstrate that SRT1720 neither lowers plasma glucose nor improves mitochondrial capacity in mice fed a high fat diet. SRT1720, SRT2183, SRT1460, and resveratrol exhibit multiple off-target activities against receptors, enzymes, transporters, and ion channels. Taken together, we conclude that SRT1720, SRT2183, SRT1460, and resveratrol are not direct activators of SIRT1.
Keywords: Aging, Allosteric Regulation, Biophysics, Enzyme Kinetics, Enzymes, SIRT1, Enzyme Activation, Sirtuins
Introduction
Silent information regulator 2 (Sir2)3 enzymes (or sirtuins) have recently emerged as central players in the regulation of critical metabolic pathways such as insulin secretion and lipid mobilization (1–3). The seven members of the mammalian sirtuin family, SIRT1–7, each with distinct cellular localizations, act on their respective targets to effect a wide range of biological processes (4). The sirtuins regulate their targets by modulating the activity of their partner proteins through reversible deacetylation (5). The most widely studied sirtuin, SIRT1, is homologous to yeast Sir2, which was initially discovered as maintaining the acetylation state of histones in heterochromatin, thereby controlling gene expression. Since then, the discovery of numerous and diverse sirtuin substrates has implicated the sirtuins in metabolism, apoptosis, transcription, and cell survival (2, 6–13). The role of the Sir2 enzymes in mediating lifespan extension in yeast (14), worms (10), and flies (9) has led to dramatic interest in this class of enzymes as a potential means of extending longevity. Sir2 deacetylases catalyze the NAD+-dependent deacetylation of specific ϵ-amino-acetylated lysine residues from its protein substrates to form nicotinamide, the deacetylated product, and a unique metabolite, 2′-O-acetyl-ADP-ribose. The biochemical and kinetic mechanism of Sir2 deacetylases has been extensively studied (15–18).
In vivo, SIRT1 has been shown to be activated by fasting and caloric restriction (19, 20). Caloric restriction extends lifespan and produces a metabolic profile desirable for treating diseases of aging such as type 2 diabetes (21–23). Pharmacological activation of SIRT1 by the small molecule activator resveratrol has been reported to improve insulin sensitivity, increase mitochondrial content, and prolong survival of mice fed a high fat, high calorie diet (24–26). Furthermore, SIRT1 activation by transgenic overexpression of the Sirt1 gene in mice has shown similar phenotypes (27), indicating that SIRT1 may represent an attractive therapeutic target for the treatment of type 2 diabetes.
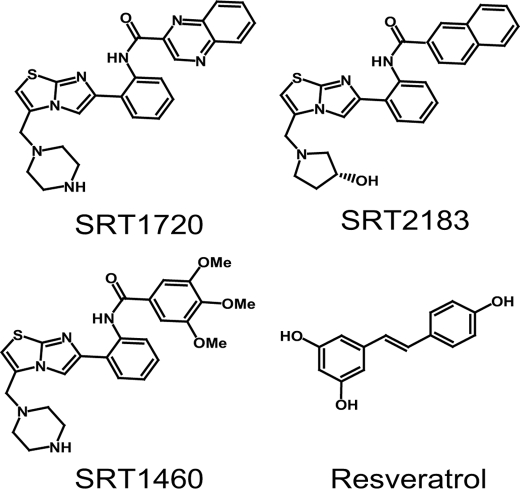
SRT1720, SRT2183, and SRT1460 were recently described by Sirtris Pharmaceuticals as SIRT1 activators (26) (Fig. 1). They are structurally unrelated to resveratrol and were reported to activate SIRT1 with potencies 1,000-fold greater than resveratrol. These compounds were identified via a high throughput fluorescence polarization assay followed by potency optimization for in vitro enzyme activity using high throughput mass spectrometry (MS). They were shown to exhibit EC1.5 (the compound concentration required to increase enzyme activity by 50%) of 0.16–2.9 μm, with 296–781% activation (when compared with 201% for resveratrol). As with other plant polyphenols such as resveratrol, the mechanism of SIRT1 activation by the Sirtris series was shown to occur through increased binding affinity by decreasing the Km of SIRT1 for acetylated peptide substrate without affecting the Km for NAD+ or the Vmax. SRT1720, the most potent activator of the series, was reported to improve glucose homeostasis, increase insulin sensitivity, and increase mitochondrial function in rodent models of type 2 diabetes (26).
FIGURE 1.
Structures of putative SIRT1 activators, SRT1720, SRT2183, SRT1460, and resveratrol.
The SIRT1 activation by the aforementioned Sirtris series was determined only with a non-native fluorophore-containing p53-derived peptide substrate in both fluorescence polarization and MS assays. Recently, SIRT1 activation by resveratrol was shown to be completely dependent on the presence of a covalently attached fluorophore in the fluorescent peptide substrate (28, 29). These results suggested that the resveratrol-mediated in vivo effects may occur through a different molecular mechanism independent of direct SIRT1 activation and raised questions about the Sirtris series of compounds and their ability to activate human SIRT1 in vitro. In the present study, to avoid any potential artifacts associated with fluorescently labeled non-native substrates, we have investigated the ability of the Sirtris series and resveratrol in activating human SIRT1 using a native peptide substrate by direct detection and quantification methods such as HPLC. Furthermore, we tested the hypothesis that the fluorophore may mimic a hydrophobic residue/pocket on the native full-length protein substrates, and therefore, the full-length substrates will behave differently from a peptide substrate. We report the first assessment of these putative SIRT1 activators on two native full-length substrates, p53 and acetyl-CoA synthetase1 (AceCS1). Herein, we report that neither the Sirtris series nor resveratrol activate SIRT1 in these native systems.
Through biophysical studies, we provide additional evidence that the Sirtris compounds interact directly with fluorophore-containing peptide substrates with micromolar binding affinities even in the absence of SIRT1. In contrast to the previous report (26), we demonstrate that SRT1720 neither lowers plasma glucose nor improves mitochondrial capacity in mice fed a high fat diet. We further demonstrate that the Sirtris series and resveratrol are highly promiscuous as they interact with multiple unrelated targets including receptors, enzymes, ion channels, and transporters. Taken together, these data suggest that SRT1720, SRT2183, SRT1460, and resveratrol are not direct SIRT1 activators.
EXPERIMENTAL PROCEDURES
Materials
SRT1720, SRT2183, and SRT1460 were synthesized according to published procedures (26). EX-527 (6-chloro-2,3,4,9-tetrahydro-1-H-carbazole-1-carboxamide) and resveratrol were purchased from Tocris Biosciences and Sigma, respectively. All compounds were stored as dry powders at room temperature in a nitrogen box and dissolved in DMSO to prepare concentrated stock solutions. Tris acetate and Triton X-100 were obtained from Research Organics and Calbiochem, respectively. HPLC solvents were from J.T. Baker. The p53-derived peptides, TAMRA-p53 peptide and native p53 peptide, were custom-synthesized by CPC Scientific, whereas 2,3-TAMRA was purchased from Molecular Probes. The amino acid sequence of all peptides used in the present study is summarized in supplemental Table S1. All reagents used were the highest quality commercially available. Acetyl-CoA synthetase1 (AceCS1) and acetylated acetyl-CoA synthetase1 (Ac-AeCS1) were provided by Dr. John Denu (University of Wisconsin, Madison, WI). All other reagents were purchased from Sigma unless otherwise noted. An Agilent 1100 HPLC system was used in all HPLC experiments, and data were processed by the Agilent ChemStation software.
SIRT1 Expression and Purification
The Escherichia coli codon-optimized cDNA for human SIRT1 was custom-synthesized at DNA2.0 (Menlo Park, CA) and cloned into a modified pET vector by using standard molecular biology techniques to generate a full-length human SIRT1 construct (amino acids 2–747) with an N-terminal His6 tag followed by a thrombin cleavage site. SIRT1 expression was accomplished in BL21gold (DE3) cells with induction at an A600 nm of 0.9 with a final concentration of isopropyl-1-thio-β-d-galactopyranoside at 50 μm for 16–20 h at 15 °C. SIRT1 was purified using a Ni2+ chelating column followed by size exclusion and anion exchange chromatography as described previously (26). This procedure typically generated SIRT1 with ∼90% purity based on SDS-PAGE visualized by Coomassie Blue staining. Typically 3–5 mg of purified SIRT1 was obtained per liter of culture.
Preparation of Native Full-length Ac-Lys382-p53
The full-length p53 cDNA was cloned into the baculovirus vector pFastBac (Invitrogen) with a His6 tag at the N terminus, and p53 was expressed in Sf9 cells as described (30). Sf9 cells expressing p53 from 2 liters of culture were lysed using a microfluidizer, and the resulting lysate was centrifuged at 100,000 × g for 40 min. The supernatant containing the soluble p53 fraction was purified by nickel-nitrilotriacetic acid-Sepharose FF column (GE Healthcare) followed by a Q Sepharose FF column (GE Healthcare). Purified p53 was acetylated by p300 as described (31). The acetylation reaction mixture (32 ml) contained 2.5 mg of purified p53, 38 μg of p300 acetylase (FLAG-tagged recombinant enzyme, Active Motif, Carlsbad, CA), 50 mm Tris-HCl, pH 8.0, 50 μm acetyl-CoA, 150 mm KCl, 1 mm DTT, 0.1 mm EDTA, and 10% glycerol. The reaction was incubated at 22 °C for 16 h. At the end of the reaction, p300 was removed by passing through an anti-FLAG M2 agarose column (Sigma-Aldrich), and the acetylated p53 (Ac-Lys382-p53) was dialyzed against a buffer containing 50 mm Tris-HCl, pH 8.0, 150 mm KCl, 10% glycerol, 0.5 mm EDTA, and 1 mm DTT. The final Ac-Lys382-p53 was confirmed to contain ∼30–70% acetylation at the Lys-382 residue by tryptic digest followed by LC-MS analysis. The purified and acetylated p53 (Ac-Lys382-p53) protein had purity greater than 98% based on SDS-PAGE visualized by Coomassie Blue staining. Typically 6 mg of purified Ac-Lys382-p53 was obtained per liter of culture.
SIRT1 Assay with TAMRA-p53 Peptide Substrate Monitored by HPLC
The SIRT1 reaction (100 μl) was carried out with 1 μm TAMRA-p53 peptide ((biotin-PEG3)-Ser-Thr-Ser-Ser-His-Ser-(Ac-Lys)-Nle-Ser-Thr-Glu-Gly-Cys(tetramethylrhodamine-5-maleimide)-Glu-Glu-NH2) (Nle stands for norleucine) (TAMRA Peptide 1 in supplemental Table S1) under the following standard SIRT1 reaction conditions: 50 mm Tris acetate, pH 7.4, 150 mm NaCl, 150 μm NAD+, 1 mm DTT, 0.01% Triton X-100, and 2% DMSO. Reactions were carried out with 5 nm SIRT1 at room temperature for 30 min, yielding ∼5% conversion of TAMRA-p53 peptide to product, allowing for the accurate measurement of ∼20-fold activation. For EC1.5 determination, resveratrol concentrations were varied at 0.01–600 μm, SRT2183 and SRT1460 were at 0.003–200 μm, and SRT1720 was at 0.005–200 μm. The reaction was quenched with trifluoroacetic acid to a final concentration of 2%, centrifuged at 16,000 × g for 10 min, and then transferred to glass HPLC vials.
The HPLC separation of the acetylated TAMRA-p53 peptide substrate and deacetylated peptide product peaks was achieved using a Vydac C18, 5 μm, 50 × 4.6-mm (218TP5405) column with accompanying guard. Mobile phase A was 100% H2O + 0.1% trifluoroacetic acid, whereas mobile phase B was 100% CH3CN + 0.1% trifluoroacetic acid. The following gradient program was used: 0–35% over 15 min followed by rapid ramp up to 100% B to wash and then 100% A to re-equilibrate the column. Flow rate was kept constant at 1.0 ml/min, column temperature was set at 25 °C, and UV detection was set at 220 nm. The retention times of the deacetylated peptide and acetylated TAMRA-p53 peptide were ∼9.1 and ∼9.3 min, respectively.
The percentage of conversion was determined by dividing the peak area of the product over the sum of the peak areas of both substrate and product peptides. This value was periodically reevaluated by comparing with a deacetylated peptide standard curve. The average rate of conversion of SIRT1 samples run in the absence of compound was compared with those in the presence of compound to establish the percentage of activation value. GraphPad Prism was used to analyze the data and calculate EC1.5 values.
SIRT1 Assay with Native p53 Peptide Monitored by HPLC
The SIRT1 reaction was carried out under the SIRT1 standard reaction conditions as described above with 1 μm native p53 peptide (Ac-Lys-Lys-Gly-Gln-Ser-Thr-Ser-Arg-His-Lys-(Ac-Lys)-Leu-Met-Phe-Lys-Thr-Glu-Gly-NH2) (Native Peptide 3 in supplemental Table S1) and 2.5 nm SIRT1 at room temperature for 30 min, yielding ∼10% conversion of substrate to product, allowing for the accurate measurement of ∼10-fold activation. Reactions were quenched and processed the same as with the TAMRA-p53 peptide. The separation of acetylated peptide substrate and deacetylated peptide product peaks was achieved using the same column, buffers, and parameters described with the TAMRA-p53 peptide, except that the gradient was altered to: 0–20% over 15 min followed by rapid ramp up to 100% B to wash and then 100% A to re-equilibrate the column. The retention times of the deacetylated and acetylated peptides were ∼10.8 and 11.7 min, respectively. Calculation of the percentage of activation was carried out as described with TAMRA-p53 peptide. The Km values for native-p53 peptide and NAD+ were obtained by fitting the plots of rate versus substrate concentration to the Michaelis-Menten equation using KaleidaGraph. To test the effect of TAMRA on SIRT1 activity by putative activators, the same SIRT1 reactions were run with or without 1, 10, and 100 μm 2,3-TAMRA, each in the presence and absence of compounds.
SIRT1 Assay with Native Full-length Ac-Lys382-p53 Substrate Monitored by ELISA and Western Blot Analysis
The SIRT1 assay mixture (50 μl) contained 150 mm Tris acetate, pH 7.4, 150 mm NaCl, 150 μm NAD+, 1 mm DTT, 0.014% Triton X-100, 3% DMSO, 100 nm Ac-Lys382-p53, and 10 nm SIRT1. The reactions were carried out in 96-well round bottom polypropylene plates (Corning, Lowell, MA). Putative activators were preincubated with SIRT1 at room temperature for 15 min, and the reactions were initiated by NAD+. The reaction mixtures were incubated at room temperature for 1 h, and the reactions were stopped by the addition of EX-527 to a final concentration of 100 μm. SIRT1 activity was measured by quantifying the levels of Ac-p53 by ELISA. The ELISA was carried out in a black MaxiSorp 96-well plate (Nunc, Rochester, NY) precoated with 0.2 μg of mouse anti-p53 antibody (Abcam, Cambridge, MA) by overnight incubation at 4 °C. Nonspecific protein binding was then blocked by block Solution (StartingBlock T20 (PBS), Thermo Scientific) for 30 min. Typically, the SIRT1 reaction mixtures were diluted 10-fold in a buffer containing 50 mm Tris, pH 8.0, 150 mm NaCl, 1% Nonidet P-40, 0.5% sodium deoxycholate, 0.1% SDS, and 100 μm EX-527. Aliquots (5 μl) of diluted SIRT1 mixture were placed in each well containing 95 μl of the above buffer in the precoated plates and incubated for 2 h at 4 °C. After washing the plates with wash buffer (PBST 0.1% Tween 20, Sigma-Aldrich), the plates were incubated with 100 μl of either 1:500 acetyl-Lys382-p53 antibody (Cell Signaling, Beverly, MA) or 1:1000 rabbit anti-p53 (FL-393) antibody (Santa Cruz Biotechnology, Santa Cruz, CA.) in block solution for 1 h at room temperature. After an additional washing with wash buffer, the plates were incubated with 100 μl of 1:10000 goat anti-rabbit IgG-horseradish peroxidase conjugate (Jackson ImmunoResearch Laboratories, West Grove, PA) in block solution for 1 h at room temperature. After a final washing with wash buffer, 100 μl of SuperSignal ELISA Pico chemiluminescent substrate (Thermo Scientific) was added to the plate for development. The plates were read using a Victor2 1420 multilabel plate reader (PerkinElmer Life Sciences) under luminescence setting. The standard curves for ELISA were linear at 1–30 ng of Ac-p53 and total p53.
Western blot analyses were conducted using the Odyssey infrared imaging system (LI-COR Biosciences, Lincoln, NE) with Ac-Lys382-p53, and total p53 was detected on the same blot by using 1:1000 mouse anti-p53 antibody (Abcam, Cambridge, MA) and 1:1000 rabbit anti-acetyl-Lys382-p53 antibody (Santa Cruz Biotechnology). The secondary antibodies used were 1:5000 IR700 goat anti-mouse and 1:5000 IR800 goat anti-rabbit (LI-COR Biosciences). SIRT1 reactions were performed and quenched as described in the ELISA procedure, and 5 μl of the reaction mixture was loaded in each well of SDS-PAGE. Quantification of total p53 levels were used to normalize the reactions for SIRT1 activity. The Km measurements of SIRT1 for p53 were performed using Western and ELISA assays by varying concentrations of p53 from 50 to 850 nm (maximal concentration available).
SIRT1 Assay with Ac-AceCS1 Substrate Monitored by HPLC
In the first step of the coupled enzyme reaction, the SIRT1 reaction was carried out using standard reaction conditions as described above with 150 nm acetylated acetyl-CoA synthetase 1 (Ac-AceCS1) and 10 nm SIRT1 at room temperature for 30 min, yielding ∼10% conversion of substrate to active AceCS1 product. This allowed for the accurate measurement of ∼10-fold activation. Reactions were quenched by nicotinamide to a final concentration of 5 mm. In the second step of the coupled reaction, saturating concentrations of AceCS1 substrates (2.5 mm ATP, 2 mm NaOAc, and 2 mm coenzyme A) were added. The AceCS1 reactions were incubated for 60 min, quenched by trifluoroacetic acid to a final concentration of 2%, centrifuged at 16,000 × g for 10 min, and then transferred to vials for HPLC analysis. Separation of AMP from ATP and other substrates and products was achieved using a SUPELCOSIL LC-18-T (Sigma, 3 μm, 150 × 4.6 mm) column with accompanying guard. Mobile phase A was 100 mm NH4OAc, pH 5.0, whereas mobile phase B was 80% A + 20% methanol. The following gradient program was used: 0% B for 10 min, 0–100% for 5 min followed by wash, and then 100% A to re-equilibrate the column. Flow rate was kept constant at 1.0 ml/min, column temperature was kept at 25 °C, and UV detection was set at 260 nm. The retention times of ATP and AMP were ∼4 and ∼8 min, respectively. Chromatograms were analyzed, and the peak area was converted to molar concentration of AMP using an AMP standard curve and then converted to the total amount of active enzyme AceCS1 product turned over by SIRT1 using the standard curves mentioned above. The average rate of conversion of SIRT1 samples run in the absence of compound was compared with those in the presence of compound to establish the percentage of activation values. The final concentrations of putative activators tested were 1, 3, 10, and 30 μm. EX-527 was tested at 100 μm. For the measurement of the Km value for Ac-AceCS1 with SIRT1, the SIRT1 reactions were performed by varying concentrations of Ac-AceCS1 at 50–750 nm (maximal concentration available).
NMR Binding Studies of SRT1460 to p53-derived Peptide Substrates
NMR chemical shift perturbation of the peptide substrates was used to monitor the molecular interaction of SRT1460 with the p53-derived peptide substrates in the absence of SIRT1 enzyme. NMR binding experiments were carried out with peptide substrates, which included the TAMRA-p53 peptide (TAMRA Peptide 1 in supplemental Table S1) and an additional pair of peptides with identical amino acid sequence and differing only in the TAMRA group (Ac-Glu-Glu-Lys-Gly-Gln-Ser-Thr-Ser-Ser-His-Ser-(Ac-Lys)-Nle-Ser-Thr-Glu-Gly-Lys(TAMRA)-Glu-Glu-NH2) and (Ac-Glu-Glu-Lys-Gly-Gln-Ser-Thr-Ser-Ser-His-Ser-(Ac-Lys)-Nle-Ser-Thr-Glu-Gly-Lys-Glu-Glu-NH2) (TAMRA Peptide 2 and Native Peptide 2 in supplemental Table S1). 1H NMR spectra were recorded on a 600-MHz Bruker DRX spectrometer equipped with a TXI cryoprobe at 25 °C. Data sets were the average of 128 scans. All the NMR spectra were collected in the presence of 10 μm peptide substrates, 50 μm SRT1460, 200 μm NAD+, and 1% DMSO-d6 in 25 mm Tris-d11, pH 8.0, 137 mm NaCl, 2.7 mm KCl, 1 mm MgCl2, and 99.9% D2O.
Surface Plasmon Resonance (SPR) Binding Studies of SRT1720 and SRT1460 to p53-derived Peptide Substrates
SPR experiments were performed in a Biacore 3000 instrument (GE Healthcare). Two p53-derived peptides used in this study have identical amino acid sequence and differ only in the TAMRA group, TAMRA Peptide 1 (supplemental Table S1) and ((biotin-PEG3)-Ser-The-Ser-Ser-His-Ser-(Ac-Lys)-Nle-Ser-The-Glu-Gly-Cys-Glu-Glu-NH2) (Native Peptide 1 in supplemental Table S1). The peptides were captured onto a neutravidin surface prepared through standard amine coupling to a CM5 sensor (GE Healthcare) at levels of 220 response units for the native peptide and 240 response units for the TAMRA peptide. SRT1720 or SRT1460 was injected over the surfaces from 3 to 50 μm. All data were collected in a buffer containing 50 mm Tris-HCl, pH 8.0, 137 mm NaCl, 3.7 mm KCl, 1 mm MgCl2, 2 mm DTT, 0.05% P20, and 1% DMSO. Data were processed in Scrubber 2 (BioLogic Software) to zero, x-align, and double reference the sensorgrams. Equilibrium binding parameters were obtained from global fits of the data to a saturation binding model in Scrubber (BioLogic Software).
Isothermal Titration Calorimetry (ITC) Binding Studies of SRT1460 to SIRT1 Peptide Substrate Complex
ITC experiments were performed in a running buffer containing 50 mm Tris-HCl, pH 8.0, 137 mm NaCl, 3.7 mm KCl, 1 mm MgCl2, 2 mm tris(2-carboxyethyl)phosphine, and 5% glycerol. The SIRT1 concentration was 41 μm in the cell. The TAMRA-p53 peptide or native p53 peptide (TAMRA Peptide 1 and Native Peptide 3 in supplemental Table S1) was dissolved in running buffer and added to SIRT1 to a final concentration of 1 mm. SRT1460 was dissolved in running buffer at 0.84 mm and used as the ligand. All titrations were performed in a Microcal VP-ITC (GE Healthcare) at 25 °C. The data were fit to a simple 1:1 interaction model in Origin ITC.
In Vivo Study
ob/ob mice (7–8 weeks old) were sourced from Charles River Laboratories and group housed four per cage on a 12:12 light:dark cycle with free access to food and water. Mice were acclimated for 1 week on a high fat diet (HFD) (60% calories from fat, RD12492, Research Diets) or chow (Purina 5015) for 1 week prior to the study. After acclimation, mice were randomized based on glucose and body weight to one of four groups, chow vehicle control, HFD vehicle control, HFD + SRT1720 30 mg/kg, and HFD + SRT1720 100 mg/kg. Mice were dosed once daily by oral gavage for 18 days with compound or vehicle (2% hydroxypropyl methylcellulose + 0.2% dioctylsulfosuccinate sodium salt), and daily body weight and food consumption were recorded. Blood glucose was monitored on days 0, 4, and 10 by tail snip and glucometer (LifeScan OneTouch Ultra glucose meter), whereas on days 1, 7, and 14, plasma glucose, triglycerides, cholesterol, and free fatty acids were analyzed on a Roche Applied Science Hitachi 912 chemistry analyzer. Plasma insulin was measure via ELISA (mouse ultrasensitive insulin kit, Alpco). Plasma exposure of compound was determined via pharmacokinetic assessment on a group of satellite mice (n = six per dose level) at 1, 2, 4, 7 and 24 h after dose on days 1 and 12 to determine steady state plasma concentrations. On day 13, food was removed at 4 p.m., and mice were fasted 16 h overnight before bleeding via retroorbital sinus at t = −60, 0, 15, 30, 60, and 120 min after glucose load (1 g/kg, oral administration (p.o.)). Mice were re-fed after the oral glucose tolerance test and allowed to recover, whereas dosing continued until the necropsy was performed 2 h after dose on day 18.
RESULTS
SRT1720, SRT2183, SRT1460, and Resveratrol Lead to Apparent Activation of SIRT1 with the Fluorogenic TAMRA-p53 Peptide Substrate
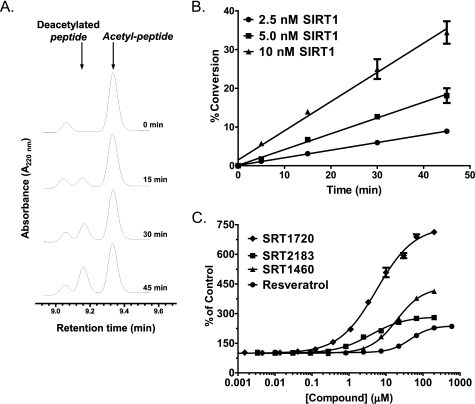
To confirm activation of SIRT1 deacetylating activity by putative SIRT1 activators reported by Milne et al. (26), we have established an HPLC method that separates deacetylated TAMRA-p53 peptide product from the acetylated substrate. This method allows an assessment of SIRT1 activity by directly measuring the product peptide and therefore avoids potential artifacts associated with indirect detection such as in the fluorescence assays. Fig. 2A shows an increase of the deacetylated peptide with concomitant decrease of the acetylated TAMRA-p53 peptide as the SIRT1 reaction progresses. Fig. 2B shows that the SIRT1 time course remains linear for up to 45 min at 2.5–10 nm SIRT1 with up to 35% conversion of the acetylated TAMRA-p53 peptide substrate. For the assessment of potential SIRT1 activation by putative activators, reaction conditions were chosen where substrate conversion was ∼5–10%, which allowed for the measurement of a maximal SIRT1 activation window of 10–20-fold. The SIRT1 assay was carried out under the conditions similar to those reported (26) for direct comparison of activation, and the results obtained with putative SIRT1 activators are shown in Fig. 2C. The maximal activation percentages of SRT1720, SRT2183, SRT1460, and resveratrol were 741, 285, 434, and 239%, respectively, with the concentration of compound required to increase enzyme activity by 50% (EC1.5 values) ranging from 0.32 μm for SRT1720 to 31.6 μm for resveratrol. These values were remarkably similar to those reported using the MS assay (26) despite some differences in the peptide substrates and SIRT1 reaction conditions used (TAMRA Peptide 1 and pH 7.4 in the present study versus TAMRA Peptide 2 and pH 8.0 in the reported study). These results confirmed that the Sirtris series and resveratrol indeed activate SIRT1 deacetylating activity with the TAMRA-p53 peptide substrate.
FIGURE 2.
Effects of SRT1720, SRT2183, SRT1460, and resveratrol on SIRT1 deacetylating activity with TAMRA-p53 peptide substrate (TAMRA Peptide 1 in supplemental Table S1) determined by HPLC. A, time course of SIRT1 reaction showing an increase in the deacetylated peptide product and a decrease in the acetylated peptide substrate. SIRT1 reactions were carried out with 10 nm SIRT1 under the conditions described under “Experimental Procedures.” B, time course of SIRT1 reactions at 2.5, 5, and 10 nm SIRT1 is shown as the percentage of conversion of TAMRA-p53 peptide substrate versus time. C, dose-response curves of SIRT1 activation by the Sirtris series and resveratrol. Error bars in B and C indicate S.D.
SRT1720, SRT2183, SRT1460, and Resveratrol Do Not Activate SIRT1 with Native p53 Peptide Substrate
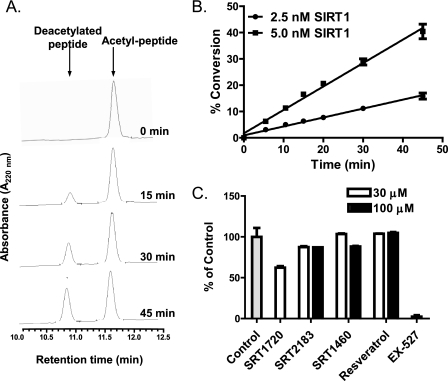
We next assessed SIRT1 activation by the same set of compounds using a native p53 peptide substrate containing the 18 amino acids flanking the native acetylation site (Lys-382) but lacking the TAMRA fluorophore (Native Peptide 3 in supplemental Table S1). To establish the assay conditions to assess the activation with a maximum activation window, we first determined the Km value for the p53 peptide to be 4.5 ± 0.17 μm by the HPLC method as described under “Experimental Procedures.” The SIRT1 Km for NAD+ was similarly determined by HPLC and was found to be 94 ± 5 μm. The Michaelis-Menten plots for the measurement of Km values for both native p53 peptide and NAD+ are shown in supplemental Fig. S1. The measurement of the SIRT1 Km for the 18-mer peptide was important in the design of our assay. The mechanism of activation of the SIRT1 modulators has been shown to occur by decreasing the Km of SIRT1 for the acetylated peptide substrate without affecting the Vmax (26). Thus, to ensure that compound activation was not overlooked, the final assay concentration of the peptide substrate was carried out at 1 μm, well below the measured Km, which allowed for a maximum assay window for assessing activation (32, 33). As shown in Fig. 3, A and B, a linear time course using a separation of acetylated peptide substrate and deacetylated peptide product by HPLC was established as discussed in the previous section.
FIGURE 3.
Effects of SRT1720, SRT2183, SRT1460, and resveratrol on SIRT1 deacetylating activity with native p53 peptide substrate (Native Peptide 3 in supplemental Table S1) determined by HPLC. A, time course of SIRT1 reaction showing an increase in the deacetylated peptide product and a decrease in the acetylated peptide substrate. SIRT1 reactions were carried out with 5 nm SIRT1 under the conditions described under “Experimental Procedures.” B, time course of SIRT1 reactions shown as the percentage of conversion of native peptide substrate versus time. C, effects of the indicated compounds on SIRT1 activity. Error bars in B and C indicate S.D.
The Sirtris series and resveratrol were assessed at 30 and 100 μm. The SIRT1 inhibitor EX-527 (34) was also evaluated. As shown in Fig. 3C, EX-527 at 100 μm completely inhibited the SIRT1 reaction as expected. The putative SIRT1 activators showed no activation of SIRT1. For SRT2183, SRT1460, and resveratrol, the relative activity of SIRT1 activity remained within ∼15% of those in the DMSO control reactions. SRT1720, which was tested only at 30 μm, exhibited ∼40% inhibition of SIRT1 activity. These results raised the possibility that the SIRT1 activation observed with the fluorophore-containing peptide was dependent on the presence of the TAMRA fluorophore. To investigate this, we repeated these experiments with the native p53 peptide in the presence or absence of varying concentrations of TAMRA fluorophore at up to 100 μm but did not observe any activation (data not shown). This result is in agreement with previous reports, which postulated that the SIRT1 activation by resveratrol requires the covalent attachment of the fluorophore to the p53 peptide (28, 29). It has been proposed that resveratrol binding to SIRT1 induces a conformational change that promotes tighter binding of the fluorophore, thus effectively reducing the Km of the peptide substrate (28).
SRT1720, SRT2183, SRT1460, and Resveratrol Have Little or No Effect on SIRT1 Deacetylating Activity with Native Full-length Substrates
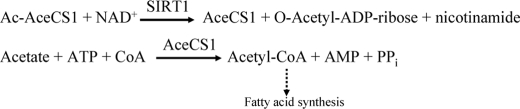
Previously, Borra et al. (28) proposed that SIRT1 activation by resveratrol with a coumarin-labeled peptide is the result of a resveratrol-induced conformational change near the coumarin binding site in SIRT1. The conformational change creates a binding pocket for the coumarin group resulting in enhanced binding of the coumarin-labeled peptide (28). Taking this hypothesis into account and considering the above data demonstrating that activation of SIRT1 was detected only in the presence of TAMRA-containing peptide, we have further hypothesized that the fluorophore may mimic a hydrophobic residue or pocket found on native, full-length acetylated substrate that promotes higher affinity for SIRT1 upon binding of putative activators. All previous studies on the assessment of SIRT1 activators were performed with p53-derived peptides of various lengths. Small peptide versus full-length protein substrates for enzymes could potentially have different binding pockets, and enzyme inhibitors are known to have distinct potency for peptide versus full-length protein substrates. Therefore, we next assessed potential activation of SIRT1 using native full-length substrates. For this purpose, we chose two full-length acetylated substrates, Ac-p53 and Ac-AceCS1. The p53 protein substrate was chosen because it is the most extensively studied SIRT1 substrate, and all previous studies on SIRT1 activators were performed using p53-derived peptides. Ac-AceCS1, recently shown to be a physiological substrate for SIRT1, is an ideal substrate for biochemical assays due to its single site acetylation (35). Studies by Starai et al. (2) and Hallows et al. (35) have demonstrated that the regulation of the AceCS activity by the Sir2 homolog initially shown in Salmonella enterica is also conserved in mammals. It was shown that mammalian AceCSs are regulated by reversible acetylation and that sirtuins activate AceCSs by deacetylation. SIRT1 was the only member of seven human Sir2 homologues capable of deacetylating AceCS1 in cellular coexpression studies. In this mechanism, AceCS1 acetylated at Lys-661 is enzymatically inactive, but upon its deacetylation by SIRT1, the enzyme becomes active, converting acetate, ATP, and coenzyme A into AMP, PPi, and acetyl-CoA, a key metabolite that is fed into fatty acid biosynthesis (35) (Scheme 1). The role of SIRT1 in this pathway has further demonstrated the role of SIRT1 in the regulation of acetate metabolism (36).
SCHEME 1.
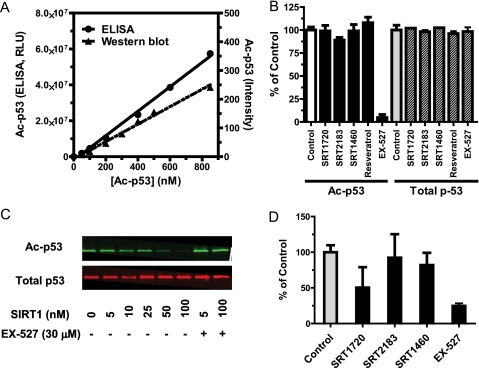
To assess p53 as a substrate, SIRT1 reactions were carried out with purified full-length Ac-Lys382-p53 (SDS-PAGE shown in supplemental Fig. S2), and SIRT1 activity was determined by ELISA that quantifies the levels of Ac-Lys382-p53 and total p53 (both acetylated and deacetylated p53) as described under “Experimental Procedures.” As with the peptide substrates, initial experiments were focused on the measurement of the Km value for Ac-Lys382-p53 to ensure that all assays were performed under conditions that would allow for an optimal assay window for observing SIRT1 activation. As shown in Fig. 4A, the Km value for Ac-Lys382-p53 was estimated to be greater than 850 nm as the SIRT1 activity with increasing concentrations of Ac-Lys382-p53 was linear up through the maximal concentration of Ac-Lys382-p53 available for testing. To our knowledge, this is the first reported estimation of the Km value of the native full-length substrate Ac-Lys382-p53 for SIRT1. The Sirtris series and resveratrol were assessed at 30 μm with the Ac-Lys382-p53 substrate concentration at 100 nm, well below the estimated Km value, and the SIRT1 reaction time course was linear under the assay conditions (data not shown). As shown in Fig. 4B, none of the putative activators exhibited SIRT1 activation as the SIRT1 activity remained within ∼10% of the control for all four compounds. As expected, the SIRT1 inhibitor EX-527 at 30 μm showed a near complete inhibition of SIRT1 activity. Total p53 levels remained the same in the presence of either putative activators or inhibitor EX-527 when compared with the DMSO control, confirming the reliability of the assay by ELISA (Fig. 4B).
FIGURE 4.
Effects of SRT1720, SRT2183, SRT1460, and resveratrol on SIRT1 deacetylating activity with purified native full-length Ac-p53 substrate. After the SIRT1 reactions, the remaining Ac-p53 and total p53 were quantified by ELISA or Western blot analysis as described under “Experimental Procedures.” A, SIRT1 activity is linear with increasing concentrations of Ac-p53 at up to 850 nm, demonstrating that the Km value for Ac-p53 is greater than 850 nm. RLU, relative luminescent units. B, effects of the indicated compounds at 30 μm on SIRT1 activity. The levels of Ac-p53 and total p53 were quantified by ELISA. C, deacetylation of Ac-p53 increases with increasing concentrations of SIRT1, whereas the total p53 quantity is unaffected. D, effects of the indicated compounds at 30 μm on SIRT1 activity with Ac-p53 substrate by Western blot. The levels of Ac-p53 were corrected for total p53 loaded onto each lane. Error bars in B and D indicate S.D.
For more direct visualization of substrates in the assay, we also utilized Western blot analysis using two separate antibodies, one specific for total p53 and another specific for Ac-Lys382-p53. A decrease in the levels of Ac-Lys382-p53 was observed at increasing concentrations of SIRT1 (Fig. 4C), and the reaction was linear for 60 min at 5–25 nm SIRT1 (data not shown). In the presence of the SIRT1 inhibitor EX-527 at 30 μm, the levels of Ac-Lys382-p53 were the same as those in the absence of SIRT1 (Fig. 4C). Total p53 levels remained the same at all concentrations of SIRT1 either in the presence or in the absence of EX-527 (Fig. 4C). Next we confirmed that the Km value for Ac-Lys382-p53, which yielded a linear dose-response curve up to 850 nm Ac-Lys382-p53 (Fig. 4A), is in complete agreement with the data obtained by ELISA. The Sirtris series and resveratrol were assessed at 30 μm, again exhibiting no activation, whereas EX-527 showed SIRT1 inhibition as expected (Fig. 4D). Subsequent to the detection of Ac-Lys382-p53, analysis of the same blot for total p53 confirmed that the same amount of total p53 was present in each sample analyzed and was used to normalize the results. These results were consistent with those generated by ELISA, and no activation of SIRT1 by the Sirtris compounds was observed. As with the native peptide, we investigated whether activation might be observed in the presence of added fluorescent TAMRA fluorophore at up to 100 μm, and similarly, no activation was observed (data not shown).
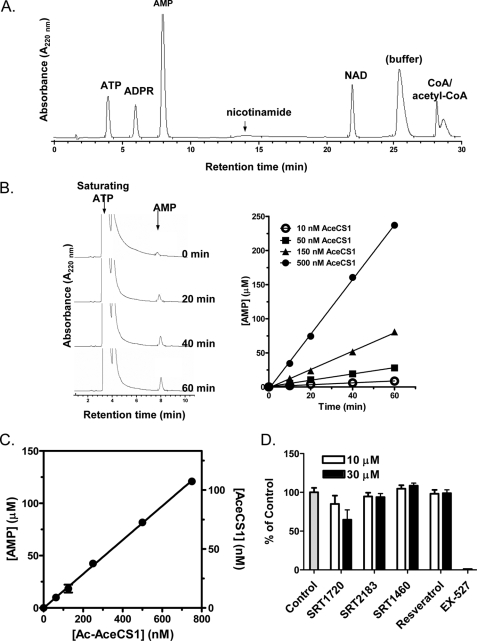
For the assessment of SIRT1 putative activators with the more metabolically relevant native substrate Ac-AceCS1, we took advantage of the knowledge that Ac-AceCS1 and AceCS1 are enzymatically inactive and active, respectively, as shown previously (35). We developed a discontinuous coupled enzyme assay in which SIRT1 reactions were carried out first with its functionally inactive substrate Ac-Lys661-AceCS1. The reaction is quenched, either with nicotinamide or with EX-527, and then the substrates (acetate, CoA, and ATP) for active AceCS1 (the SIRT1 product) are then added to saturating conditions relative to the previously reported Km values (37, 38). This allowed the measurement of AceCS1 activity under its saturating conditions. In the second step, AceCS1 reactions are quenched, and the AMP product was separated and quantified by HPLC using the conditions described under “Experimental Procedures.” As shown in Fig. 5A, the HPLC conditions established separated AMP from all components of the SIRT1 and AceCS1 reactions to allow accurate quantification of AMP. The amount of quantified AMP was used for the calculation of the precise amount of active AceCS1 product converted by SIRT1 using a calibrated standard curve. For the standard curve generation, the time courses for active AceCS1 at 10–500 nm were generated. As shown in Fig. 5B, AceCS1 reactions were linear for up to 60 min with 10–500 nm AceCS1.
FIGURE 5.
SIRT1 reaction with full-length Ac-AceCS1 substrate. SIRT1 reactions were monitored by measuring AceCS1 activity as described under “Experimental Procedures.” A, separation of the components for the SIRT1 and AceCS1 reactions (1 nmol of ATP, ADPR, AMP, nicotinamide, NAD, CoA, and acetyl-CoA each) by HPLC. Low extinction coefficient of nicotinamide results in lower relative peak area, although quantification of nicotinamide was accomplished at higher concentrations with high reproducibility. B, time course of AceCS1 reactions carried out under the conditions described under “Experimental Procedures” at AceCS1 concentrations of 10–500 nm. The HPLC chromatogram is shown for 150 nm AceCS1. C, SIRT1 activity is linear with increasing concentrations of Ac-AceCS1 at up to 750 nm, demonstrating that the Km value for Ac-AceCS1 is greater than 750 nm. D, effects of SRT1720, SRT2183, SRT1460, and resveratrol on SIRT1 activity with Ac-AceCS1 substrate. Error bars in C and D indicate S.D.
As the Km value of SIRT1 for Ac-AceCS1 has not been previously reported, we varied the concentration of Ac-AceCS1 through the highest concentration possible in our assay. The observed linear increase in the rate of AMP formation indicated that the Km of SIRT for Ac-AceCS1 was greater than 750 nm (Fig. 5C).
We next investigated the effect of the Sirtris series and resveratrol on the modulation of SIRT1 activity with the Ac-AceCS1 at 150 nm, well below the Km value. Although EX-527 ablated SIRT1 activity, all four compounds had little or no effect on the SIRT1 activity at 1, 3, 10, and 30 μm (data shown for 10 and 30 μm). As with experiments on the native peptide (Fig. 3), SIRT1 activity in the presence of 30 μm SRT1720 was inhibited by ∼35% (Fig. 5D).
NMR and SPR Binding Studies Demonstrate That SRT1460 or SRT1720 Interacts with TAMRA Peptide, whereas It Has Little or No Interaction with Native Peptide
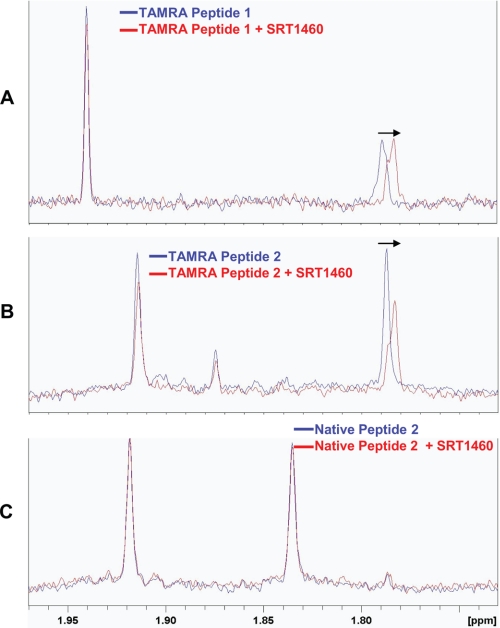
We next assessed whether the SIRT1 activation, observed with the Sirtris series only in the presence of TAMRA-containing peptide substrate, is due to a direct interaction of the compounds with the TAMRA-containing substrate. Perturbation of the NMR chemical shift is a commonly used method to detect intermolecular binding interactions, which can alter the electronic environment of an NMR-observable nuclei. In this study, the proton chemical shift perturbation of the lysine acetyl (CH3) group in the peptide substrates was evaluated as a probe to monitor the molecular interaction of a representative Sirtris compound, SRT1460, with the p53 peptide substrates in the absence of the SIRT1 enzyme. The lysine acetyl (CH3) resonance at 1.78 ppm of the TAMRA-p53 peptide (TAMRA Peptide 1 in supplemental Table S1), which is the peptide substrate used in our biochemical assay discussed above, showed a clear upfield chemical shift upon the addition of SRT1460 (Fig. 6A), which could indicate a direct interaction of SRT1460 with the peptide. To determine the requirement of the TAMRA group for an intermolecular interaction between the peptide substrate and SRT1460, a pair of p53-derived peptides with identical amino acid sequences and differing only in the TAMRA group was assessed (TAMRA Peptide 2 and Native Peptide 2 in supplemental Table S1). These peptide substrates were chosen for this study because the TAMRA Peptide 2 is the identical peptide used in the fluorescence polarization and MS assays by Sirtris (26). As in the case with the TAMRA Peptide 1, the TAMRA Peptide 2 also showed a similar upfield shift of the acetyl (CH3) resonance at 1.78 ppm upon the addition of SRT1460 as shown in Fig. 6B. However, the Native Peptide 2 (supplemental Table S1) does not show any shift of its acetyl (CH3) resonance at 1.83 ppm upon the addition of SRT1460 (Fig. 6C). Because the two peptides differ only in the TAMRA group, the observed upfield shift in the acetyl (CH3) resonance (Fig. 6B) is likely to arise from a direct binding interaction of SRT1460 with the TAMRA-containing peptides. An attempt to verify these results with SRT1720 was made; however, limited solubility of SRT1720 prohibited these studies under NMR experimental conditions.
FIGURE 6.
NMR chemical shift perturbation studies of SRT1460 with p53-derived peptide substrates in the absence of SIRT1. A, 1H NMR spectrum of 10 μm of the TAMRA Peptide 1 (supplemental Table S1) in the presence (red) or absence (blue) of 50 μm SRT1460. B, 1H NMR spectrum of 10 μm of TAMRA Peptide 2 in the presence (red) or absence (blue) of 50 μm SRT1460. C, 1H NMR spectrum of 10 μm of the Native Peptide 2 in the presence (red) or absence (blue) of 50 μm SRT1460. Arrows indicate the upfield shift of the acetyl (CH3) signal at 1.78 ppm in A and B (blue) upon the addition of 50 μm SRT1460 (red), whereas the acetyl (CH3) signal at 1.83 ppm in C showed no shift (blue and red) upon the addition of 50 μm SRT1460. The amino acid sequence of the TAMRA Peptide 2 and the Native Peptide 2 are identical and differ only in the TAMRA group (supplemental Table S1).
To further validate the interaction between TAMRA-containing peptide substrates and SRT1460 by NMR, SPR methods were developed to evaluate potential binding between ligand and peptide substrates in the absence of the SIRT1 enzyme. Supplemental Fig. S3, A and B, show the SPR response data for a series of SRT1720 injections from 3 to 50 μm over the TAMRA Peptide 1 and the Native Peptide 1 that have identical amino acid sequences and differ only in the TAMRA group (supplemental Table S1). Concentration-dependent binding was observed only with the TAMRA-containing peptide but not with the native peptide, which showed only the baseline responses. Due to its weak binding, a full saturation curve could not be generated, and therefore, the KD value for SRT1720 to the binding of the TAMRA Peptide 2 could not be accurately estimated. A similar interaction with the TAMRA Peptide 2 was also observed for SRT1460 in SPR, but the binding affinity was much weaker (data not shown). Despite the lack of saturation in the SPR binding experiments, these data do confirm a clear dose-dependent interaction of SRT1720 with the TAMRA peptide in agreement with those observed by NMR for SRT1460.
ITC Studies Demonstrate That SRT1460 Binds to the SIRT1-TAMRA Peptide Substrate Complex but Not to the SIRT1-Native Peptide Substrate Complex
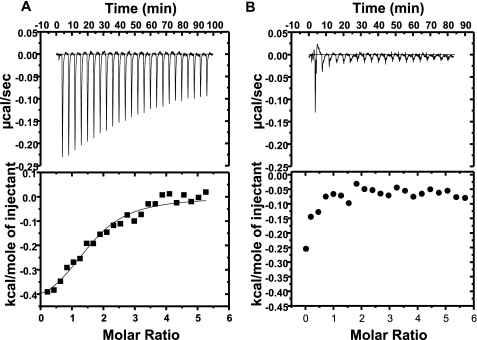
Finally, ITC was used to define the binding energetics of compounds to the SIRT1 peptide substrate complex under the conditions previously described (26). Fig. 7 shows representative binding isotherms for SRT1460 injected into the SIRT1 peptide substrate complex. Examination of the titrations reveals that SRT1460 demonstrates a clear binding isotherm with the SIRT1-TAMRA-p53 peptide complex (Fig. 7A), but no isotherm was observed with the SIRT1-native p53 peptide complex (Fig. 7B). The KD value for SRT1460 was found to be 13 μm, similar to the 16.2 μm previously reported (26). However, the enthalpy observed with this titration is significantly reduced when compared with that reported previously (26). In these experiments, the binding affinity is principally entropically driven with an enthalpy of only −0.992 kcal/mol when compared with the reported value of −6.1 kcal/mol. This difference could be due to a shorter construct of SIRT1 used in the previous study (versus full-length SIRT1 in the present study) as well as a slightly different TAMRA peptide substrate sequence (TAMRA Peptide 1 in the present study versus TAMRA Peptide 2 in the previous study (26)). However, the requirement of a TAMRA group to generate a binding isotherm to the SIRT1 peptide substrate complex is completely consistent with the direct interactions between SRT1460 and TAMRA-containing peptides observed in NMR and SPR.
FIGURE 7.
ITC titrations of SIRT1 peptide substrate complexes with SRT1460. Top panels, binding isotherm for titrations of SRT1460 to the SIRT1-TAMRA Peptide 1 complex (A) and the SIRT1-Native Peptide 3 complex (B). Bottom panels, integrated fit with a one-site binding model. The amino acid sequence of the TAMRA Peptide 1 and the Native Peptide 3 are shown in supplemental Table S1.
SRT1720 Neither Lowers Plasma Glucose nor Improves Mitochondrial Capacity in High Fat-fed ob/ob Mice
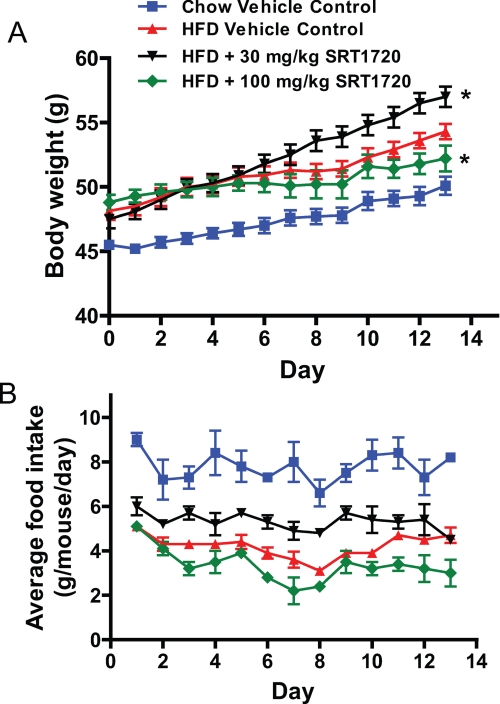
Despite a lack of evidence for the Sirtris series of compounds as direct SIRT1 activators, we investigated whether the in vivo efficacy demonstrated by SRT1720 in several rodent models of diabetes (26) could be validated and attributed to indirect activation of SIRT1. We therefore attempted to reproduce the in vivo efficacy for SRT1720 in mouse models of type 2 diabetes previously shown (26). ob/ob mice (7–8 weeks old) were placed on an HFD and dosed with SRT1720 at 30 and 100 mg/kg, oral administration (p.o.), once daily for 18 days. Body weight and food intake were monitored daily, and fed glucose levels were measured every 3–4 days. It became clear early on in the study that the mice were not tolerating the 100 mg/kg SRT1720 dose resulting in the death of three out of eight mice at days 7, 10, and 13, respectively. The remaining mice in this group showed reduced food intake and body weight gain (Fig. 8, A and B), and thus, the data obtained at the end of 18 days could not be easily interpreted. The 30 mg/kg dose group tolerated the dosing schedule but showed increased food intake and weight gain when compared with HFD controls as shown in Fig. 8, A and B. These data did not agree with the reported data where no change in body weight was reported for the ob/ob mice study on an HFD dosed with 100 mg/kg daily dosing of SRT1720 for 1 week (26). (Food intake was not assessed in the reported study.) Furthermore, the 30 mg/kg group had no effect on any glucose parameters (fed, fasting, or area under the curve during an oral glucose tolerance test), although reductions in fasting insulin were observed. To ensure that the lack of efficacy at 30 mg/kg was not due to lower exposure in our study, we measured the exposure from plasma and determined the average steady state Cmax and area under the curve values of 1.3 μm and 8110 ng/h/ml, respectively, for the 30 mg/kg dosing group, which were similar to those previously reported for the 100 mg/kg dose group (26). Reported changes in citrate synthase activity in the diet-induced obese mice study treated with SRT1720 (26) prompted us to look for evidence of changes in mitochondrial biogenesis in these chronically dosed ob/ob mice at 30 mg/kg. Cytochrome c oxidase activity and citrate synthase activity were measured in multiple tissues, but the data did not show any evidence to suggest enhanced mitochondrial function. Overall, our present study showed that the 30 mg/kg dose had no effect on glycemic control despite exposures that matched those previously reported (26) but instead showed increased food intake and weight gain when compared with HFD controls. The high dose of 100 mg/kg was not well tolerated and resulted in reduced weight gain, reduced food intake, and death.
FIGURE 8.
Effect of chronic dosing of SRT1720 on (A) body weight and (B) food intake in ob/ob mice fed a high fat diet. ob/ob mice were placed on a HFD (60% calories from fat) for 1 week before being dosed with SRT1720 at the indicated doses once daily by oral gavage. Food intake per cage and individual body weights were recorded daily over 13 days of treatment. The graph represents mean ± S.E. for all groups (n = 8 for chow and HFD control groups and SRT1720 at 30 mg/kg groups; SRT1720 at 100 mg/kg group had n = 8 on day 1, which had dropped to n = 5 by day 13). *, p < 0.05 when compared with HFD control by two-way ANOVA and Student's t test.
SRT1720, SRT2183, SRT1460, and Resveratrol Interact with Multiple Unrelated Targets
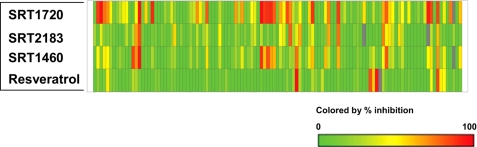
Selectivity assessment is critical to determine whether or not any compound can serve as a potential pharmacological tool or drug. Based on our data above demonstrating that the Sirtris series and resveratrol do not directly interact with SIRT1, we have assessed the in vitro selectivity of these compounds at 10 μm against a broad panel of over 100 targets including receptors, enzymes, ion channels, and transporters (supplemental Table S2). The results shown in Fig. 9 and supplemental Table S3 illustrate that these compounds are highly promiscuous. SRT1720, SRT2183, and SRT1460 at 10 μm exhibited >50% inhibition of 38, 14, and 20 targets, respectively, out of over 100 targets assessed. Resveratrol appeared less promiscuous but still showed >50% inhibition of seven targets at 10 μm (supplemental Table S3). It is not surprising that the Sirtris series have similar profiles as they are structurally related. The same 22 targets were inhibited by two out of the three Sirtris compounds by >50%. All three Sirtris compounds exhibited >50% inhibition of four targets (adenosine A3 receptor, urotensin receptor (GPR14), phosphodiesterase 2, and norepinephrine transporter). Only one target, norepinephrine transporter, was inhibited by all three Sirtris compounds and resveratrol by >50%, demonstrating that the Sirtris series and resveratrol have different target selectivity profiles. (supplemental Table S3). These data demonstrate that neither the Sirtris series nor resveratrol would serve as useful pharmacological tools due to their highly promiscuous profiles.
FIGURE 9.
Selectivity profiles of SRT1720, SRT2183, SRT1460, and resveratrol. All four compounds were tested at 10 μm against over 100 targets listed in supplemental Table 1. A range of 0–100% inhibition is shown in a colored scale.
DISCUSSION
In this report, we have focused on assessment of the purported SIRT1 activators SRT1720, SRT2183, SRT1460, and resveratrol by employing SIRT1 assays that allow direct quantification and detection to avoid any potential artifacts frequently associated with indirect detection methods such as in a fluorescence assay. To achieve a maximum activation assay window, we determined the Km values for each assay condition and carefully established all in vitro SIRT1 assays at peptide/protein substrate concentrations well below the Km values (32, 33). Furthermore, putative activators were assessed under the SIRT1 assay conditions where controls (−activator) have only 5–10% of substrate conversion to product, which allowed a large measurable 10–20-fold activation assay window. We have confirmed that the Sirtris compounds and resveratrol activate SIRT1 deacetylating activity with TAMRA peptide substrate by direct quantification of deacetylated product by HPLC. By similar direct quantification of deacetylated peptide product converted from the native p53 peptide by SIRT1, we demonstrated that the SIRT1 activation by these compounds requires the presence of the fluorophore covalently attached to the peptide substrate as no activation was observed with native peptide lacking the fluorophore or with native peptide in the presence of TAMRA. Borra et al. (28) previously proposed that SIRT1 activation by resveratrol with a coumarin-labeled peptide is the result of a resveratrol-induced conformational change near the coumarin binding site in SIRT1, which creates a binding pocket for the coumarin group resulting in enhanced binding of the coumarin-labeled peptide. Based on this proposal and our present results showing that the SIRT1 activation is observed only in the presence of the fluorophore-containing peptide substrate, we further hypothesized that the fluorophore may mimic a hydrophobic residue/pocket found on native, full-length substrate that promotes higher affinity for SIRT1 upon binding to activators. Toward this goal, we assessed two native full-length substrates, p53 and AceCS1. We have developed highly sensitive quantification/detection methods such as ELISA/Western blot and HPLC, which enabled the use of full-length Ac-p53 and Ac-AceCS1 substrates, respectively. To our knowledge, this is the first report of the assessment of SIRT1 activators in SIRT1 enzyme assays using native full-length substrates. Our results show that the Sirtris series of compounds and resveratrol have little or no effect on SIRT1 activity even with these two full-length protein substrates. To further assess a potential involvement of the fluorophore in the interaction with compounds, we have utilized NMR and SPR binding interaction studies. These studies demonstrated that SRT1460 and SRT1720 can interact directly with the TAMRA peptide substrates with micromolar affinities even in the absence of SIRT1. Likewise in the presence of SIRT1, the ITC study demonstrated that the binding of SRT1460 is observed only in the presence of the fluorophore-containing peptide substrate.
We assessed in vivo efficacy of SRT1720 in high fat-fed ob/ob mice. In contrast to the data reported (26), we did not observe a decrease in plasma glucose at the dose where the plasma SRT1720 exposure is comparable with that reported. Instead, we observed increased food intake and weight gain in mice treated with SRT1720 when compared with HFD controls. Finally, we have assessed a broad selectivity of these activators against over 100 targets and showed that these compounds are highly promiscuous as they interact with multiple unrelated targets. While this manuscript was in preparation, Beher et al. (39) also described that resveratrol does not activate SIRT1 activity with native p53 peptide substrate, which is in complete agreement with our results and those previously published (28, 29).
In summary, our detailed assessment of the Sirtris series and resveratrol involving several biochemical assays with native substrates and biophysical studies employing NMR, SPR, and ITC demonstrated that these compounds are not direct SIRT1 activators. We also demonstrated that SRT1720 does not show beneficial effects in a rodent diabetes model, which is in contrast to that previously reported (26). The broad selectivity assessment against over 100 targets including receptors, enzymes, ion channels, and transporters show that the Sirtris series and resveratrol are highly promiscuous and would not serve as useful pharmacological tools for studying SIRT1 pathways. In the literature, resveratrol has been widely referred to as a SIRT1 activator (for selected recent references, see Refs. 40–44) and routinely used to activate SIRT1 in various cellular assays, with only a few questioning the original study that reported its ability to activate SIRT1 in an artificial substrate-based fluorescent assay (28, 29, 39, 45). Likewise, the Sirtris compounds have been referred to as SIRT1 activators in recent publications (46–48). Our present data are significant for the field as we provided strong evidence that neither the Sirtris series nor resveratrol are direct SIRT1 activators.
Acknowledgments
We thank Stephen Orena for assistance with ELISA; Kieran Geoghegan, Lise Hoth, and Tom McLellan for LC-MS analysis; Michael Norcia, Roger J. Schilling, Nicole Rauccio, Payal Rana, and Carol Petras for development of alternative biochemical assays; Steve Orena and Delin Ren for cell-based assays; Yanfang Liu, Benjamin Maciejewski, Levenia Baker, and Anne Brodeur for assistance with in vivo study design and execution; and John Denu (University of Wisconsin) for providing Ac-AceCS1, and AceCS1.
This article was selected as a Paper of the Week.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1–S3 and Tables S1–S3.
- Sir2
- silent information regulator 2
- AceCS
- acetyl-CoA synthetase
- HFD
- high fat diet
- Nle
- norleucine
- EX-527
- 6-chloro-2,3,4,9-tetrahydro-1-H-carbazole-1-carboxamide
- TAMRA
- 6-carboxytetramethylrhodamine
- DTT
- dithiothreitol
- ELISA
- enzyme-linked immunosorbent assay
- ITC
- isothermal titration calorimetry
- PEG
- polyethylene glycol
- SPR
- surface plasmon resonance
- MS
- mass spectrometry
- LC-MS
- liquid chromatography-mass spectrometry.
REFERENCES
- 1.Moynihan K. A., Grimm A. A., Plueger M. M., Bernal-Mizrachi E., Ford E., Cras-Méneur C., Permutt M. A., Imai S. (2005) Cell Metab. 2, 105–117 [DOI] [PubMed] [Google Scholar]
- 2.Starai V. J., Celic I., Cole R. N., Boeke J. D., Escalante-Semerena J. C. (2002) Science 298, 2390–2392 [DOI] [PubMed] [Google Scholar]
- 3.Liang F., Kume S., Koya D. (2009) Nat. Rev. Endocrinol. 5, 367–373 [DOI] [PubMed] [Google Scholar]
- 4.Smith B. C., Hallows W. C., Denu J. M. (2008) Chem. Biol. 15, 1002–1013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sauve A. A., Wolberger C., Schramm V. L., Boeke J. D. (2006) Annu. Rev. Biochem. 75, 435–465 [DOI] [PubMed] [Google Scholar]
- 6.Rusche L. N., Kirchmaier A. L., Rine J. (2003) Annu. Rev. Biochem. 72, 481–516 [DOI] [PubMed] [Google Scholar]
- 7.Gasser S. M., Cockell M. M. (2001) Gene 279, 1–16 [DOI] [PubMed] [Google Scholar]
- 8.Dryden S. C., Nahhas F. A., Nowak J. E., Goustin A. S., Tainsky M. A. (2003) Mol. Cell Biol. 23, 3173–3185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rogina B., Helfand S. L. (2004) Proc. Natl. Acad. Sci. U.S.A. 101, 15998–16003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tissenbaum H. A., Guarente L. (2001) Nature 410, 227–230 [DOI] [PubMed] [Google Scholar]
- 11.Vaziri H., Dessain S. K., Ng Eaton E., Imai S. I., Frye R. A., Pandita T. K., Guarente L., Weinberg R. A. (2001) Cell 107, 149–159 [DOI] [PubMed] [Google Scholar]
- 12.Luo J., Nikolaev A. Y., Imai S., Chen D., Su F., Shiloh A., Guarente L., Gu W. (2001) Cell 107, 137–148 [DOI] [PubMed] [Google Scholar]
- 13.Langley E., Pearson M., Faretta M., Bauer U. M., Frye R. A., Minucci S., Pelicci P. G., Kouzarides T. (2002) EMBO J. 21, 2383–2396 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kaeberlein M., McVey M., Guarente L. (1999) Genes Dev. 13, 2570–2580 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Borra M. T., Langer M. R., Slama J. T., Denu J. M. (2004) Biochemistry 43, 9877–9887 [DOI] [PubMed] [Google Scholar]
- 16.Jackson M. D., Schmidt M. T., Oppenheimer N. J., Denu J. M. (2003) J. Biol. Chem. 278, 50985–50998 [DOI] [PubMed] [Google Scholar]
- 17.Sauve A. A., Schramm V. L. (2004) Curr. Med. Chem. 11, 807–826 [DOI] [PubMed] [Google Scholar]
- 18.Smith B. C., Denu J. M. (2007) J. Am. Chem. Soc. 129, 5802–5803 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cohen H. Y., Miller C., Bitterman K. J., Wall N. R., Hekking B., Kessler B., Howitz K. T., Gorospe M., de Cabo R., Sinclair D. A. (2004) Science 305, 390–392 [DOI] [PubMed] [Google Scholar]
- 20.Heilbronn L. K., Civitarese A. E., Bogacka I., Smith S. R., Hulver M., Ravussin E. (2005) Obes. Res. 13, 574–581 [DOI] [PubMed] [Google Scholar]
- 21.Facchini F. S., Hua N., Abbasi F., Reaven G. M. (2001) J. Clin. Endocrinol. Metab. 86, 3574–3578 [DOI] [PubMed] [Google Scholar]
- 22.Civitarese A. E., Carling S., Heilbronn L. K., Hulver M. H., Ukropcova B., Deutsch W. A., Smith S. R., Ravussin E. (2007) PLoS Med. 4, e76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Barzilai N., Banerjee S., Hawkins M., Chen W., Rossetti L. (1998) J. Clin. Invest. 101, 1353–1361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Baur J. A., Pearson K. J., Price N. L., Jamieson H. A., Lerin C., Kalra A., Prabhu V. V., Allard J. S., Lopez-Lluch G., Lewis K., Pistell P. J., Poosala S., Becker K. G., Boss O., Gwinn D., Wang M., Ramaswamy S., Fishbein K. W., Spencer R. G., Lakatta E. G., Le Couteur D., Shaw R. J., Navas P., Puigserver P., Ingram D. K., de Cabo R., Sinclair D. A. (2006) Nature 444, 337–342 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lagouge M., Argmann C., Gerhart-Hines Z., Meziane H., Lerin C., Daussin F., Messadeq N., Milne J., Lambert P., Elliott P., Geny B., Laakso M., Puigserver P., Auwerx J. (2006) Cell 127, 1109–1122 [DOI] [PubMed] [Google Scholar]
- 26.Milne J. C., Lambert P. D., Schenk S., Carney D. P., Smith J. J., Gagne D. J., Jin L., Boss O., Perni R. B., Vu C. B., Bemis J. E., Xie R., Disch J. S., Ng P. Y., Nunes J. J., Lynch A. V., Yang H., Galonek H., Israelian K., Choy W., Iffland A., Lavu S., Medvedik O., Sinclair D. A., Olefsky J. M., Jirousek M. R., Elliott P. J., Westphal C. H. (2007) Nature 450, 712–716 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Banks A. S., Kon N., Knight C., Matsumoto M., Gutiérrez-Juárez R., Rossetti L., Gu W., Accili D. (2008) Cell Metab. 8, 333–341 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Borra M. T., Smith B. C., Denu J. M. (2005) J. Biol. Chem. 280, 17187–17195 [DOI] [PubMed] [Google Scholar]
- 29.Kaeberlein M., McDonagh T., Heltweg B., Hixon J., Westman E. A., Caldwell S. D., Napper A., Curtis R., DiStefano P. S., Fields S., Bedalov A., Kennedy B. K. (2005) J. Biol. Chem. 280, 17038–17045 [DOI] [PubMed] [Google Scholar]
- 30.Sun X. Z., Nguyen J., Momand J. (2003) Methods Mol. Biol. 234, 17–28 [DOI] [PubMed] [Google Scholar]
- 31.Wang Y. H., Tsay Y. G., Tan B. C., Lo W. Y., Lee S. C. (2003) J. Biol. Chem. 278, 25568–25576 [DOI] [PubMed] [Google Scholar]
- 32.Groebe D. R. (2006) Drug Discov. Today 11, 632–639 [DOI] [PubMed] [Google Scholar]
- 33.Segel I. H. (1993) in Enzyme Kinetics: Behavior and Analysis of Rapid Equilibrium and Steady-state Enzyme Systems, pp. 227–272, Wiley-Interscience, New York [Google Scholar]
- 34.Napper A. D., Hixon J., McDonagh T., Keavey K., Pons J. F., Barker J., Yau W. T., Amouzegh P., Flegg A., Hamelin E., Thomas R. J., Kates M., Jones S., Navia M. A., Saunders J. O., DiStefano P. S., Curtis R. (2005) J. Med. Chem. 48, 8045–8054 [DOI] [PubMed] [Google Scholar]
- 35.Hallows W. C., Lee S., Denu J. M. (2006) Proc. Natl. Acad. Sci. U.S.A. 103, 10230–10235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.North B. J., Sinclair D. A. (2007) Trends Biochem. Sci. 32, 1–4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ishikawa M., Fujino T., Sakashita H., Morikawa K., Yamamoto T. (1995) Tohoku J. Exp. Med. 175, 55–67 [DOI] [PubMed] [Google Scholar]
- 38.Luong A., Hannah V. C., Brown M. S., Goldstein J. L. (2000) J. Biol. Chem. 275, 26458–26466 [DOI] [PubMed] [Google Scholar]
- 39.Beher D., Wu J., Cumine S., Kim K. W., Lu S. C., Atangan L., Wang M. (2009) Chem. Biol. Drug Des. 74, 619–624 [DOI] [PubMed] [Google Scholar]
- 40.Ramadori G., Gautron L., Fujikawa T., Vianna C. R., Elmquist J. K., Coppari R. (2009) Endocrinology 150, 5326–5333 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sulaiman M., Matta M. J., Sundaresan N. R., Gupta M. P., Periasamy M., Gupta M. (2010) Am. J. Physiol. Heart Circ. Physiol., in press [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Chen L., Feng Y., Zhou Y., Zhu W., Shen X., Chen K., Jiang H., Liu D. (2010) J. Inorg. Biochem., in press [DOI] [PubMed] [Google Scholar]
- 43.Gracia-Sancho J., Villarreal G., Jr., Zhang Y., García-Cardeña G.Cardiovasc. Res. 85, 514–519 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Albani D., Polito L., Forloni G. (2010) J. Alzheimers Dis. 19, 11–26 [DOI] [PubMed] [Google Scholar]
- 45.Um J. H., Park S. J., Kang H., Yang S., Foretz M., McBurney M. W., Kim M. K., Viollet B., Chung J. H. (2010) Diabetes, in press [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Feige J. N., Lagouge M., Canto C., Strehle A., Houten S. M., Milne J. C., Lambert P. D., Mataki C., Elliott P. J., Auwerx J. (2008) Cell Metab. 8, 347–358 [DOI] [PubMed] [Google Scholar]
- 47.Yamazaki Y., Usui I., Kanatani Y., Matsuya Y., Tsuneyama K., Fujisaka S., Bukhari A., Suzuki H., Senda S., Imanishi S., Hirata K., Ishiki M., Hayashi R., Urakaze M., Nemoto H., Kobayashi M., Tobe K. (2009) Am. J. Physiol. Endocrinol. Metab. 297, E1179–E1186 [DOI] [PubMed] [Google Scholar]
- 48.Smith J. J., Kenney R. D., Gagne D. J., Frushour B. P., Ladd W., Galonek H. L., Israelian K., Song J., Razvadauskaite G., Lynch A. V., Carney D. P., Johnson R. J., Lavu S., Iffland A., Elliott P. J., Lambert P. D., Elliston K. O., Jirousek M. R., Milne J. C., Boss O. (2009) BMC Syst. Biol. 3, 31. [DOI] [PMC free article] [PubMed] [Google Scholar]