Abstract
We report that upon UV radiation insult, mammalian cells specifically down-regulate Mcm10, a protein essential for the initiation and elongation phases of DNA replication. The levels of a majority of replication factors remain unaffected under this condition, implying that Mcm10 is a key node in the regulation of the replication machinery. High doses of ionizing gamma radiation and exposure to a combination of DNA-damaging chemicals do not decrease Mcm10 protein levels, demonstrating that Mcm10 down-regulation is triggered only by UV-specific damage. The decrease of Mcm10 protein levels is not caused by transcriptional inhibition or cleavage by apoptotic enzymes, but results from degradation by the 26 S proteasome. UV-triggered degradation of Mcm10 requires its linker or C-terminal domain. In addition, Mcm10 down-regulation is not limited to cells from a particular lineage. Therefore, our study reveals a mechanism by which mammalian cells effectively inhibit the replication machinery during stress to prevent it from drifting toward a catastrophic path of genomic instability.
Keywords: DNA, DNA/Damage, DNA/Protein Interaction, DNA/Repair, DNA/Replication, Protein/Degradation
Introduction
Proliferating eukaryotic cells tightly regulate DNA replication so that it occurs only once per cell cycle and on experiencing DNA damage, this process is rapidly inhibited by the regulation of key proteins to avoid aberrant replication. DNA replication begins by binding of a six-subunit origin recognition complex to the origin of replication followed by recruitment of several additional initiation factors, including CDC6, Cdt1, and MCM, in the M and G1 phase of the cell cycle. An increase in cyclin-dependent kinase activity at the G1-S transition promotes the unwinding of DNA and the recruitment of the DNA synthesis factors, CDC45, GINS complex, RPA, and the DNA polymerase α-primase. Mcm10 is a conserved DNA replication protein with homologs from Saccharomyces cerevisiae to Homo sapiens (1, 2). In Xenopus and humans, the chromatin-bound Mcm2–7 helicase complex is essential for the loading of Mcm10, but in budding yeast, it is reported that Mcm10 is recruited to the chromosome before the Mcm2–7 helicase (3, 4). Although the step at which Mcm10 is recruited to the replication origin remains debatable and may vary between species, it has been proven that Mcm10 is essential for replication initiation and elongation steps (5–9). Therefore, it is expected that cells must tightly regulate the activity of Mcm10, and this is apparent from the natural control of Mcm10 activity; this protein undergoes inhibitory phosphorylation at the end of the S phase, where its activity needs to be curtailed, leading to its dissociation from chromatin at G2 phase and eventually degradation in mitosis (2, 10, 11).
The essential requirement of Mcm10 at multiple stages of DNA replication raises the possibility that the cell may regulate its Mcm10 activity in stressed environments to block DNA replication and cell cycle progression. Stress resulting from different DNA-damaging agents, such as UV and gamma radiation or mutagenic chemicals, manifests itself in a particular manner, which could incite a similar or specific cellular response. In response to UV irradiation, cells induce a dose-dependent regulation of transcription, and the genes that are repressed within 12–24 h include DNA polymerase-α, primase, Fen1, RFC, cyclin B1, cyclin A, Cdk7, and Cdc6 (12). The activity of replication protein will be inhibited faster by proteolysis than down-regulation of its transcription but very few proteolytic targets have been identified (13–15). This study evaluated changes in key replication factors following stress. In this attempt, we have proven that Mcm10, an essential replication factor, is down-regulated following UV stress. In the present study, we demonstrate that upon UV exposure, Mcm10 protein levels decrease, whereas gamma radiation and DNA-damaging chemicals have no effect on Mcm10 stability. The degradation of Mcm10 is mediated by the 26 S proteasome and is independent of the cell cycle phase. Because Mcm10 is essential for the initiation and elongation phases of DNA replication, this study provides a possible mechanism by which UV radiation induces the arrest of DNA replication by inhibiting a key molecule of the DNA replication machinery.
EXPERIMENTAL PROCEDURES
Cell Culture, Chemicals, Antibodies, Cell Cycle Synchronization, and Fractionation
Cell lines were maintained in Dulbecco's modified Eagle's medium or MEM supplemented with fetal bovine serum and antibiotics. Specific chemicals and antibodies used in this study are mentioned under supplemental information. U2OS cells were synchronized in late G1 phase by incubation with 0.5 mm mimosine for 24 h, released into drug-free medium, and UV-irradiated at 4-h intervals using the UV cross-linker CL-1000 from UVP. Cells were harvested 2 h after irradiation for protein and DNA analysis. Synchronization of HeLa cells at the mitotic phase was accomplished by incubation with 0.04 μg/ml nocodazole for 15 h, and samples were subsequently collected in the manner described above. Fractionation of U2OS cells was performed as described earlier (16).
RNAi Silencing and Reverse Transcriptase-PCR
Genes were silenced by transfection with 40–80 nm of specific, small inhibitory RNA (siRNA)3 duplexes on three consecutive days. The cells were harvested 24 h after the last transfection, and the levels of protein and mRNA were evaluated by immunoblotting and reverse transcriptase-PCR, respectively. For reverse transcriptase-PCR, RNA was extracted using the TRIzol method, and 0.25–1 μg of RNA was used for cDNA synthesis. The primers used for PCR are described under supplemental information.
Plasmid Construction, Transfection, Stable Cell Line, Immunoblotting, and Immunofluorescence
Full-length Mcm10 was subcloned into EcoRV and NotI sites of pCDNA3, which carries a sequence encoding the hemagglutinin tag at the N-terminal of the insert. Mcm10 cDNA was amplified by PCR, digested with NotI and HindIII and subcloned into pCTAP (which allows for tandem affinity purification with a C-terminal streptavidin-binding peptide and a calmodulin-binding peptide tag). Similarly, Mcm10 cDNA was cloned into the BamHI and NotI sites of pMX-puro-HA and BamHI and EcoRI sites of pMX-puro-3NLS-GST-HA vector, respectively. The sequences of cloning primers have been provided under supplemental information. Cells of almost equal confluency were lysed in proportionate volumes of Laemmli buffer for immunoblotting. To demonstrate equal protein loading in each lane, immunoblotting was performed and a nonspecific protein band was displayed. For expression of Mcm10, 293T cells were transfected with pCTAP-MCM10 and harvested 48 h later. 293T cells were transfected with pMX-puro-MCM10 along with helper plasmids (that express the viral VSV-G envelope protein as well as the Gag and Pol proteins) to generate viral particles. To obtain stable U2OS cell lines expressing Mcm10, U2OS cells were infected with the viral particles and selected with 1 μg/ml puromycin. To determine the levels of Mcm10 fragments, lysates of the stable cell lines were immunoblotted with an HA antibody, and equal protein loading was demonstrated in all lanes by immunoblotting with a control antibody. For immunofluorescence, cells were grown on coverslips, fixed in 4% formaldehyde, permeabilized with 0.2% Triton X-100, blocked with 10% fetal bovine serum, 0.1% Tween-20, and then stained with a specific primary antibody followed by a FITC-conjugated anti-rabbit or anti-mouse secondary antibody. Finally, the cells were observed under the microscope after mounting with vector shield mounting reagent with DAPI.
Recovery and BrdUrd Labeling
HeLa cells were exposed to 40 J/m2 UV and harvested every 4 h to assay Mcm10 levels. For BrdUrd immunofluorescence, cells were incubated with 100 μm BrdUrd over the same time period followed by fixing with 4% formaldehyde, treatment with 2 m HCl, and neutralization with 0.1 m sodium borate buffer (pH 8.5). The fixed cells were permeabilized with 0.2% Triton X-100, blocked with 3% bovine serum albumin and incubated with a FITC-conjugated BrdUrd antibody.
RESULTS
Mcm10 Is Down-regulated after UV Irradiation
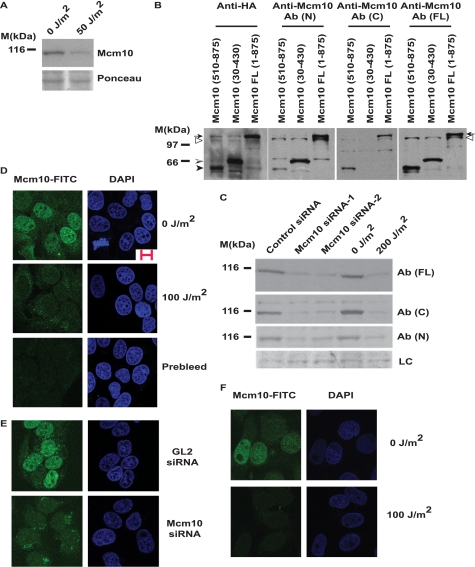
To determine the effect of UV irradiation on the replication machinery, we surveyed the levels of known replication factors post-UV exposure. The human osteosarcoma cell line, U2OS, was exposed to 50 J/m2 UV radiation, and in an immunoblot with the Mcm10 antibody, we observed that the intensity of a 110-kDa band decreased within 1 h (Fig. 1A). Mcm10 is an intensely studied protein, and therefore, we wanted to confirm that: (a) the 110-kDa band is indeed Mcm10 and not a cross-reactive band, and (b) the decrease in the band intensity is due to complete degradation and not proteolysis of a particular segment, leaving the rest of the protein unaltered. To address these points, we raised several antibodies that recognize different parts of Mcm10. HA-tagged full-length (1–875 amino acids), N-terminal (30–430 amino acids), and C-terminal (510–875 amino acids) Mcm10 were cloned into the pCTAP vector. The specific Mcm10 fragments were expressed in 293T cells and detected with an HA antibody (Fig. 1B, left panel). Ab (FL) is a polyclonal rabbit antibody raised against the full-length protein. As seen in Fig. 1B (right panel), this antibody recognized the C-terminal, N-terminal, and full-length proteins. Ab (N) is a polyclonal rabbit antibody raised against the full-length protein, but showed weak immunoreactivity against the C-terminal protein (Fig. 1B, middle-left panel). Ab (C) is a C-terminal commercial antibody from Calbiochem raised against a peptide that corresponds to exon 14 (amino acids 584–659) of Mcm10 (Fig. 1B, middle-right panel). A lower molecular mass band below the 30–430 amino acids fragment in lane 2 was recognized by all of the antibodies that have immunoreactivity toward the N-terminal of Mcm10 (anti-HA, Ab (N), Ab (FL)), but not by Ab (C). Thus, this band seems to be a degradation product that arose from the expression of the 30–430 amino acids fragment of Mcm10. Therefore, we obtained antibodies that recognized either the N-terminal, C-terminal, or full-length Mcm10 proteins.
FIGURE 1.
Mcm10 is down-regulated following UV irradiation. A, U2OS cells were exposed to 50 J/m2 UV radiation and harvested after 1 h. Mcm10 levels were analyzed. B, 293T cells were transfected with pCTAP-Mcm10-FL (1–875), -30 to 430 or -510 to 875. At 48 h after transfection, the expression of Mcm10 was analyzed. The anti-HA antibody identifies the mobility of the 510–875 amino acids (black arrowhead), 30–430 amino acids (shaded arrowhead), and full-length Mcm10 proteins (arrow) (left panel). The hollow arrowhead points to a cross-reactive band. C, down-regulation of the 110-kDa Mcm10 protein band is confirmed by detection with different antibodies and siRNA. LC refers to loading control, a nonspecific band that displays equal protein loading in different lanes. D, indirect immunofluorescence confirms the degradation of Mcm10 after UV irradiation. Unperturbed (top panel) and 100 J/m2 UV-irradiated (middle panel) U2OS cells were fixed, and immunofluorescence was performed using a rabbit anti-Mcm10 [Ab (N)] antibody. Localization of Mcm10 was displayed with an anti-rabbit FITC secondary antibody (left panel), and DNA was stained with DAPI (right panel). The prebleed of the anti-Mcm10 [Ab (N)] antibody did not show any signal (bottom panel). The scale bar is 10 microns. E, RNAi confirms the Mcm10 immunofluorescence signal. HeLa cells were transfected with GL2 or MCM10 siRNA oligos and later processed for immunofluorescence with the anti-Mcm10 [Ab (N)] antibody. F, indirect immunofluorescence with a different Mcm10 antibody, Ab (FL), confirms the degradation of Mcm10 post-UV irradiation. Unperturbed and 100 J/m2 UV-irradiated U2OS cells were fixed, and immunofluorescence was performed using the anti-Mcm10 [Ab (FL)] antibody.
When we immunoblotted the non-irradiated and UV-irradiated lysates, there was a decrease in Mcm10 signal with all three of these antibodies (Fig. 1C). This result completely ruled out the possibility that the disappearance of the Mcm10 band was due to partial proteolysis. Also, we did not observe the appearance of any other low molecular mass bands, which would indicate truncation of Mcm10. We performed RNA-mediated interference (RNAi) depletion against two segments of Mcm10; siRNA-1 targeted nucleotides 1062–1082 of the Mcm10 coding sequence, while siRNA-2 targeted nucleotides 3225–3245 of the 3′-UTR of Mcm10. Both siRNAi-1 and siRNA-2 caused a decrease in the same 110-kDa band, establishing that UV radiation triggered the down-regulation of Mcm10 (Fig. 1C). We then performed indirect immunofluorescence with the N-terminal Mcm10 antibody, Ab (N), and observed that in an asynchronous population, around 40–50% of cells retained the Mcm10 protein (Fig. 1D). To confirm that the FITC signal was from the Mcm10 protein, RNAi against MCM10 was performed in HeLa cells (Fig. 1E). There was no signal with prebleed or after Mcm10 siRNA, authenticating the Mcm10 signal. Following UV irradiation, low level of Mcm10 was observed, verifying its degradation (Fig. 1D). Immunofluorescence was also carried out with a different Mcm10 antibody, Ab (FL), which confirmed the down-regulation of Mcm10 post-UV irradiation (Fig. 1F). To exclude any artifact of visualization or nonspecific effects, we examined the localization of cyclin A under similar conditions, which did not show any change in its level upon UV treatment (supplemental Fig. S1B).
UV-triggered Mcm10 Down-regulation Is Not Due to Transcriptional Inhibition
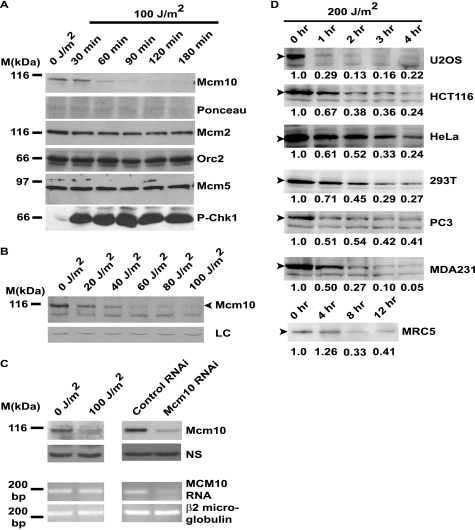
UV radiation triggers major transcriptional reprogramming, affecting many genes. To ascertain whether UV irradiation affects other replication factors, we evaluated the levels of key replication proteins including replicative helicases, Mcm2, Mcm3, Mcm4, Mcm5, Mcm6, and Mcm7 and replication initiators, Orc2, Orc3, Orc4, and Orc5 (Fig. 2A and supplemental Fig. S1A). We observed that the levels of these key replication proteins remained unchanged, and that the decrease of Mcm10 was uniquely regulated, as has been reported for Cdt1 (13). The Mcm10 decrease around 1–2 h post-UV exposure seems to be temporally separated from the instantaneous Cdt1 down-regulation, thereby displaying the redundancy of the cellular surveillance mechanisms for maintaining genomic stability (Fig. 2A). Although 40 J/m2 was sufficient to cause Mcm10 down-regulation, we used a higher UV dose in all future experiments to firmly establish the involvement of the factors mediating this pathway (Fig. 2B). Jeanette Cook and co-workers (14) have reported that after 3 h of exposure to 30 J/m2 UV radiation, Cdc6 is down-regulated by a different ubiquitin ligase, and therefore, it seems that the cell utilizes Mcm10 as an independent node for the inhibition of the replication machinery.
FIGURE 2.
UV-triggered Mcm10 down-regulation is not due to transcriptional inhibition. A, down-regulation of replication proteins following UV irradiation is specific to Mcm10. U2OS cells were exposed to 100 J/m2 UV and then harvested at the indicated time points. DNA damage was confirmed by the phosphorylation of Ser-345 of Chk1 (P-Chk1). B, dose-course experiment was performed in U2OS cells. The cells were exposed to different doses of UV radiation as indicated and then harvested after 2 h. C, mRNA level of Mcm10 is not altered within 2 h after 100 J/m2 UV irradiation. U2OS cells were UV-irradiated and harvested 2 h later. Protein (first panel from top) and mRNA levels were estimated (left panel, third panel from top). D, different cell lines were exposed to 200 J/m2 UV radiation, and the cells were harvested at the indicated time points. Numbers indicate the relative intensity of the Mcm10 band, which is denoted by the black arrowhead. LC refers to a loading control.
Yoshida and Inoue (17) have reported that the repressing transcription factor E2F4 is recruited to the TOPBP1 and MCM10 promoter after a few hours (4–24 h) of UV exposure. The authors suggested that this would lead to a decrease in Mcm10 mRNA levels, although no data demonstrating changes in the Mcm10 mRNA has been shown. To ascertain whether the decrease in the Mcm10 protein that we observed post-UV exposure is due to transcriptional inhibition, we exposed U2OS cells to UV irradiation and evaluated the Mcm10 mRNA levels after 2 h by reverse transcriptase-PCR. We found that there was no significant change in Mcm10 mRNA levels (Fig. 2C, left panel, third panel from top). On the other hand, Mcm10 mRNA levels were reduced after siRNA depletion, indicating that our PCR amplification was not at a saturating level (Fig. 2C, right panel, third panel from top). Therefore, the observed UV-mediated decrease of the Mcm10 protein is not due to transcriptional inhibition and, instead, is likely due to proteolysis. It has been previously reported that Mcm10 protein expression is not mainly regulated at the transcriptional level, but depends on degradation (10). Our initial work was largely performed with the osteosarcoma cell line, U2OS, and we wanted to ascertain that the Mcm10 decrease was a regulatory event adopted by cells from varying sources. Therefore, we compared the following cell lines: human embryonic kidney cells, 293T; prostate cancer cell line, PC3; breast cancer cell line, MDA231; human colon cancer cell line, HCT116 and cervical cancer cell line, HeLa. Upon exposure to UV, Mcm10 decreased in all the cell lines tested, although the rate of degradation varied between cell lines (Fig. 2D). We also tested the regulation of Mcm10 in a primary human fetal lung fibroblast cell line, MRC-5, and observed that Mcm10 levels decreased after UV shock. Therefore, Mcm10 down-regulation was not limited to cells from a particular lineage.
Mcm10 Down-regulation Is Specific to UV-mediated Damage and Is Not Caused by Gamma Radiation or Genotoxic Chemicals
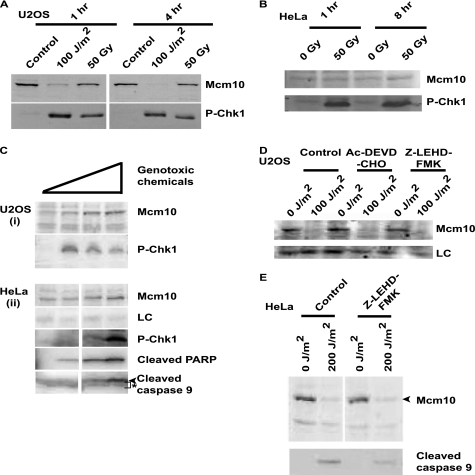
We wanted to test whether Mcm10 is down-regulated after exposure to other agents of DNA damage such as ionizing gamma radiation and DNA-damaging chemicals. Interestingly, in U2OS and HeLa cells, until 4 and 8 h (respectively) after exposure to high doses of gamma radiation (50 Gy), we did not observe any change in Mcm10 levels (Fig. 3, A and B). With this treatment, phosphorylation at serine 345 of Chk1, a standard marker for DNA damage, was observed (Fig. 3, A and B, bottom panel). We then exposed U2OS and HeLa cells to high doses of DNA-damaging chemicals, which included a combination of the following drugs: 25 milliunits/ml bleomycin, 10 μm cisplatin, 10 μm doxorubicin, and 100 μm cyclophosphamide. The cells were harvested 8 h later. To our surprise, exposure of U2OS and HeLa cells to this massive dose of genotoxic chemicals did not decrease Mcm10 protein levels (Fig. 3C, i and ii). Therefore, the down-regulation of Mcm10 is conducted by a pathway that is specifically induced by UV-mediated DNA damage. Exposure to genotoxic chemicals for 8 h led to DNA damage and activation of apoptotic machinery, as evidenced by cleavage of PARP and caspase 9 (Fig. 3C, ii). However, under this condition of activated apoptosis, Mcm10 levels did not decrease, establishing that the apoptotic machinery does not target Mcm10. Further, we determined the kinetics of the activation of apoptosis, as evaluated by the appearance of cleaved caspases 7 and 9 and the decrease of Mcm10 after UV exposure (supplemental Fig. S2A). We observed that there was a significant decrease in Mcm10 levels within 2–3 h of UV-irradiation of HeLa cells, but the activation of caspase 7 and 9 occurred only after 6 h, demonstrating that the apoptotic pathway is activated after the Mcm10 decrease and, therefore, cannot be the cause of its down-regulation (supplemental Fig. S2A). We exposed U2OS cells to UV radiation in the presence of Ac-DEVD-CHO and Z-LEHD-FMK, inhibitors of the key DNA damage-activated apoptotic enzymes, caspase 3 and caspase 9, respectively (Fig. 3, D and E). Although we observed a clear inhibition of caspase 9, the decrease of Mcm10 was unaffected, thereby verifying that Mcm10 down-regulation is not mediated by the apoptotic pathway. The above results conclusively establish that the down-regulation of Mcm10 post-UV exposure is not caused by the apoptotic machinery.
FIGURE 3.
Mcm10 down-regulation occurs specifically after UV irradiation and is not mediated by the apoptotic pathway. A and B, U2OS and HeLa cells were exposed to UV and gamma radiation as indicated and harvested after different time points. C, Mcm10 levels do not decrease after exposure to DNA-damaging chemicals. Lane 1 represents U2OS cells not exposed to genotoxic chemicals. Lane 2 represents U2OS cells treated with a combination of 1 milliunit/ml bleomycin, 2 μm cisplatin, 2 μm doxorubicin, and 10 μm cyclophosphamide. Lane 3 represents treatment with 5 milliunit/ml bleomycin, 5 μm cisplatin, 5 μm doxorubicin, and 20 μm cyclophosphamide. Lane 4 represents treatment with 25 milliunit/ml bleomycin, 10 μm cisplatin, 10 μm doxorubicin, and 100 μm cyclophosphamide. U2OS (i) and HeLa (ii) cells were incubated with the specified concentrations of these chemicals and harvested 8 h later. Cleaved PARP (ii, fourth panel) and cleaved caspase 9 (ii, bottom panel) were also activated at higher concentrations of these chemicals (the arrowhead in the bottom panel denotes the cleaved caspase 9 band and * corresponds to a cross-reactive band). D, apoptotic inhibitors do not stabilize Mcm10 after exposure to UV radiation. U2OS cells were incubated in the presence of 2 μm Ac-DEVD-CHO and Z-LEHD-FMK for 4 h, exposed to 100 J/m2 UV radiation and harvested 1 h later. E, HeLa cells were incubated in the presence of 2 μm Z-LEHD-FMK, exposed to 200 J/m2 UV radiation and harvested after 4 h. LC refers to loading control.
UV-triggered Degradation of Mcm10 Is Mediated by the 26 S Proteasome
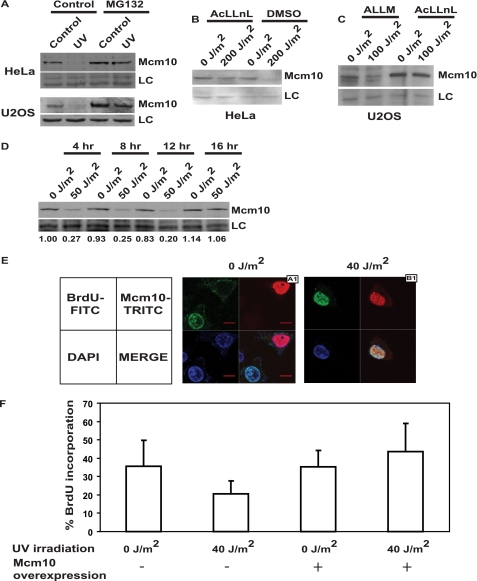
Because the apoptotic machinery was ruled out as a down-regulator of Mcm10 after UV exposure, we ascertained the involvement of other proteolytic pathways. We used other inhibitors to confirm the involvement of the proteasome; MG132 is an inhibitor of the 26 S proteasome, AcLLnL inhibits the proteasome and calpains, while ALLM inhibits calpains but not the proteasome. HeLa and U2OS cells were incubated in the presence of 50 μm MG-132, 25 μm AcLLnL, or 25 μm ALLM for 4 h followed by 200 J/m2 and 100 J/m2 UV exposure and were harvested after 4 h and 2 h, respectively. We observed that MG132 and AcLLnL treatment blocked the Mcm10 decrease, but that ALLM had no effect (Fig. 4, A, B, and C). These observations confirmed the role of proteasome-mediated proteolysis in the down-regulation of Mcm10 after UV treatment. It has been previously reported that MG132 stabilized Mcm10, whereas ALLM had no effect on Mcm10 levels (10).
FIGURE 4.
Mcm10 stabilization partially rescues the block of replication in UV checkpoint arrest. A–C, proteasome inhibitors MG132 and AcLLnL abrogated the effect of UV radiation on Mcm10, whereas the calpain-specific inhibitor ALLM had no effect. HeLa and U2OS cells were treated with different inhibitors followed by UV irradiation and analyzed for Mcm10 levels. LC refers to loading control. D, Mcm10 levels recover after 12–16 h of exposure to 50 J/m2 UV radiation. HeLa cells were UV-irradiated and then harvested at a 4-h interval until 16 h. Numbers indicate the relative intensity of the Mcm10 band. E, non-irradiated (A1) and UV-irradiated (B1) HeLa cells were visualized for DNA staining, BrdUrd incorporation, and Mcm10 overexpression by DAPI, FITC, and TRITC immunofluorescence, respectively. The arrangement of immunofluorescence images obtained from a single field is illustrated in the left panel of E. The bottom right square of each field is a merge of DAPI, FITC, and TRITC images. F, Mcm10 overexpression enhances recovery of replication following UV checkpoint arrest. The y-axis of the bar graph depicts the percentage of total cells that have incorporated BrdUrd between 16 and 24 h after UV irradiation. The average and standard deviation of the % BrdUrd incorporation values obtained from four independent experiments have been plotted.
Mcm10 Overexpression Enhances Recovery of Replication following UV Checkpoint Arrest
We observed that endogenous Mcm10 protein levels were low in UV-irradiated cells until 4–12 h after UV irradiation and begin to recover after 12–16 h, followed by a reinitiation of DNA replication around 16–24 h (Fig. 4D). We hypothesized that if Mcm10 is one of the limiting factors for replication, its overexpression during this period may enhance the recovery of DNA replication. On the basis of BrdUrd incorporation, we measured the rate of DNA replication during 16–24 h of UV irradiation in Mcm10-overexpressing cells and non-Mcm10-overexpressing cells (Fig. 4E). The average and standard deviations of four independent experiments are shown in Fig. 4F. All cells within randomly selected microscope fields were analyzed for Mcm10 overexpression and BrdUrd incorporation, a few examples of which have been displayed in supplemental Fig. S4. Following UV irradiation, BrdUrd incorporation was observed to be higher in Mcm10-overexpressing cells than in non-Mcm10-overexpressing cells. BrdUrd incorporation without UV irradiation was carried out similarly, and we observed that Mcm10 overexpression did not significantly alter the rate of DNA replication (Fig. 4F). Therefore, we conclude that Mcm10 is a limiting factor whose activity regulates DNA replication following stress. The rate of DNA replication varied in different experiments, as is depicted by the standard deviation, but we consistently observed complementation of DNA replication on exogenous Mcm10 expression after UV irradiation (Fig. 4F).
Mcm10 Is Required for Normal S Phase Progression
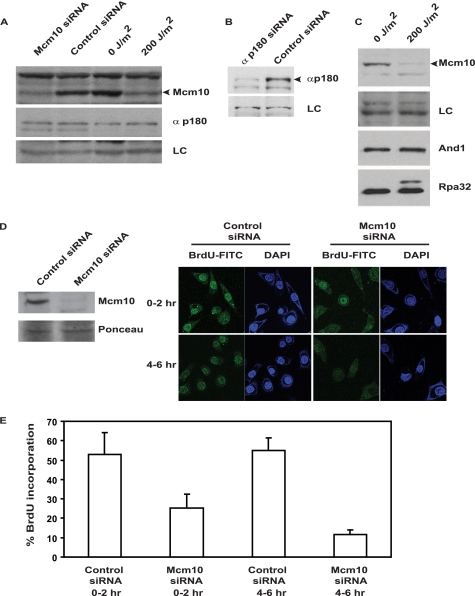
We observed that the levels of the p180 subunit of polymerase-α were not decreased after Mcm10 RNAi or UV irradiation, and therefore, the effect of Mcm10 degradation on replication elongation occurred independently of the regulation of p180 stability (Fig. 5A). Stability of And1 was also not altered following UV irradiation (Fig. 5C). To test the requirement of Mcm10 in the elongation step, we depleted Mcm10 by RNAi and then blocked the cells at the G1/S transition by treatment with hydroxyurea for 20 h (Fig. 5D). Mcm10-depleted cells were released from the G1/S block in the presence of BrdUrd. On the basis of BrdUrd incorporation, we measured the rate of DNA replication from 0–2 h and 4–6 h after release from hydroxyurea (Fig. 5, D and E). The absence of Mcm10 during S phase led to a decrease in the incorporation of BrdUrd from 53 to 25% during 0–2 h after release from hydroxyurea. Similarly, incorporation of BrdUrd decreased from 55% to 12% during 4–6 h after release from hydroxyurea, signifying that Mcm10 is required for progression through S phase. It has been proven that Mcm10 is essential for the replication initiation step because it recruits several replication factors (4, 18). Therefore, UV-triggered degradation of Mcm10 blocks not only replication initiation, but also S phase progression.
FIGURE 5.
Mcm10 is essential for normal S phase progression. A, UV irradiation and RNAi decreased the Mcm10 protein, but did not significantly alter the levels of p180 subunit of DNA polymerase-α. HeLa cells were either transfected with Mcm10 siRNA-2 or UV-irradiated as indicated and harvested after 4 h. The lysates were used to assay levels of Mcm10 and the p180 subunit of DNA polymerase-α. The black arrowhead depicts the Mcm10 band. LC refers to loading control. B, band of the p180 subunit of polymerase-α as denoted with a black arrowhead was confirmed by siRNA in HeLa cells. C, immunoblots of control and UV-irradiated HeLa cells display the level of indicated proteins. D and E, Mcm10 depletion decreases BrdUrd incorporation as cells transverse through the S phase. D, left panel displays the Mcm10 depletion whereas the right panel displays BrdUrd incorporation in control and Mcm10 siRNA cells from 0–2 h and 4–6 h after release from hydroxyurea. HeLa cells transfected with either control or Mcm10 siRNA were visualized for BrdUrd incorporation (by FITC immunofluorescence) at the indicated intervals after release from hydroxyurea. The cells were stained with DAPI to display nuclei. E, bar graph depicts the percentage of BrdUrd incorporation in control and Mcm10 siRNA cells at the indicated intervals after release from hydroxyurea. The average and standard deviation of three independent experiments have been plotted.
The C-terminal Half of Mcm10 Contains Signals for UV-triggered Degradation
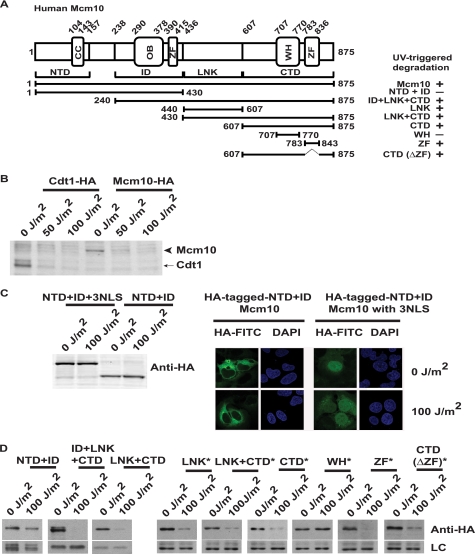
We generated stable U2OS cells expressing HA-tagged Mcm10 (utilizing the pMX-retroviral vector that is based on the moloney murine leukemia virus) to determine the segments of Mcm10 that are required for UV degradation. Full-length Mcm10 can be broadly divided into N-terminal (NTD), inner (ID), linker (LNK), and C-terminal (CTD) domains (Fig. 6A). A coiled-coil motif is present within the N terminus of Mcm10, which is required for homodimerization. The inner and C-terminal domains, which contain zinc finger (ZF) and winged helix (WH) motifs, bind to single- and double-stranded DNA and the p180 subunit of DNA polymerase-α (19, 20). We observed that exogenous Mcm10 was degraded upon 50 J/m2 UV radiation (Fig. 6B). Exogenous Mcm10 is expressed from the viral LTR promoter, and therefore, along with previous observations where we did not observe any changes in Mcm10 mRNA until 2 h after UV irradiation, this conclusively ruled out any specific contribution of the endogenous promoter in Mcm10 down-regulation (Fig. 2C). We observed that the NTD+ID fragment was not proteolyzed following UV stress, but because this fragment was cytosolic, its mislocalization could be a possible reason for its lack of degradation (Fig. 6C). Therefore, we expressed the NTD+ID fragment in fusion with a nuclear localization signal, which guided the protein to the nucleus, but this fragment was still not degraded post-UV irradiation. Therefore, the degradation signal does not lie within the N-terminal domain, which is required for dimerization. Thus, it is clear that dimerization of Mcm10 is not essential for UV-induced modification and subsequent degradation. We observed that the LNK+CTD fragment, which contains the ZF and WH motifs, was rapidly degraded following UV irradiation (Fig. 6D). We fragmented these domains into many smaller subdomains and observed that the ZF, but not the WH, motif was degraded by UV irradiation, indicating that the degradation signal lies within the ZF motif. However, the CTD domain lacking the ZF motif was also degraded, indicating that the ZF motif is sufficient, but not essential, for UV-triggered degradation (Fig. 6D). We also observed that the 178-amino acid LNK domain was degraded following UV irradiation. Therefore, we conclude that the degradation signal lies within the C-terminal half of Mcm10.
FIGURE 6.
The degradation signal lies within the C terminus of Mcm10. A, schematic representation of Mcm10 and the fragments cloned into the pMX-puro vector. OB, ZF, and WH refer to oligonucleotide binding, zinc finger, and winged helix motifs, respectively. B, exogenous Mcm10 is down-regulated after exposure to UV radiation. Stable U2OS cells expressing Cdt1-HA and Mcm10-HA were exposed to UV radiation at the indicated doses and harvested 2 h later. The black arrowhead points to Mcm10-HA, and the arrow points to Cdt1-HA. C, NTD+ID fragment is resistant to UV degradation irrespective of its cellular localization. U2OS cells expressing the NTD+ID fragment with and without a nuclear localization signal (NLS) were exposed to 100 J/m2 UV radiation and harvested 2 h later. Immunofluorescence with a HA antibody displays the localization of the NTD+ID fragment with or without a nuclear localization signal. D, Mcm10 down-regulation requires the C-terminal domain. U2OS cells expressing different Mcm10 fragments were exposed to 100 J/m2 UV radiation and harvested 2 h later. * indicates that the respective domains were cloned in fusion with the nuclear localization signal. LC refers to loading control. UV-triggered degradation of Mcm10 fragments is summarized on the right side of A.
UV-triggered Mcm10 Degradation Is Independent of DNA Replication and Occurs in All Cellular Fractions
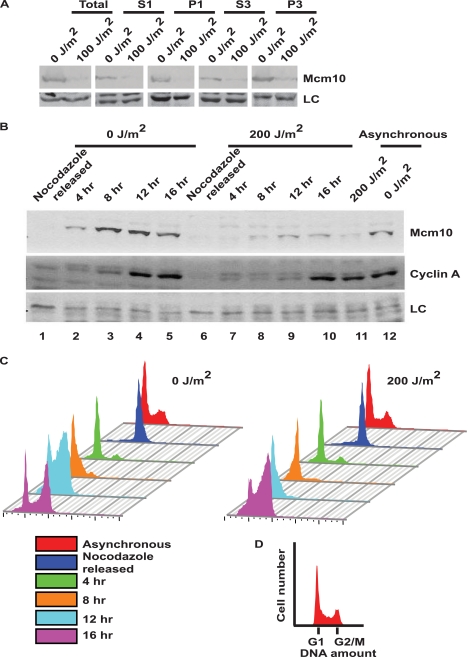
To determine if the UV-triggered pathway targets only chromatin-bound or nuclear soluble proteins, we fractionated UV-irradiated cells using the protocol of Mendez and Stillman (16). The cells were lysed in isotonic buffer to isolate the nuclear fraction (P1) from the cytosolic fraction (S1). Nuclei were then lysed in hypotonic buffer to separate the nucleoplasmic proteins (S3) from the chromatin and nuclear matrix proteins (P3). Although Mcm10 was largely present in the chromatin and nuclear matrix fraction, UV irradiation caused a Mcm10 decrease in all fractions (Fig. 7A). Therefore, the UV-triggered pathway not only targets the Mcm10 bound to chromatin, but also targets soluble protein present in nucleoplasm. Mcm10 is naturally proteolyzed in M phase, and we wanted to clearly distinguish UV-triggered degradation from cell cycle-regulated degradation (10, 11). HeLa cells were blocked with nocodazole, and cells arrested in M phase were collected by mitotic-shake off. The obtained mitotic cells were then re-plated in drug-free medium and harvested at regular intervals until 16 h. Flow cytometry measurements and levels of cyclin A demonstrate a block in M phase and subsequent progression through the cell cycle (Fig. 7, B and C). Mcm10 was absent in nocodazole-released cells, demonstrating the natural proteolysis of Mcm10 in M phase. Mcm10 levels began to increase after release and peaked around 12 h, displaying the natural cycling of Mcm10 levels (Fig. 7B). To evaluate the effect of UV radiation on the Mcm10 protein during different phases of the cell cycle, HeLa cells were UV-irradiated before harvesting at different time intervals. At 12–16 h after nocodazole release, cell cycle-regulated degradation was absent, but UV irradiation led to a Mcm10 decrease (Fig. 7B, compare lanes 4, 5, with 9, 10). This result demonstrates the UV-induced degradation under the conditions where cell cycle-regulated degradation is not observed and clearly distinguishes the two pathways of Mcm10 degradation. Similarly, we synchronized cells in late G1 phase with mimosine and released them in drug-free medium to collect cells at different phases. We observed that Mcm10 degradation occurred throughout the cell cycle (supplemental Fig. S3, A and B).
FIGURE 7.
Mcm10 down-regulation is independent of cell cycle phase. A, Mcm10 is degraded in all of the cellular fractions. The S1, P1, S3, and P3 fractions correspond to the cytoplasmic, nuclear, nuclear soluble, and chromatin-bound fractions, respectively. U2OS cells were exposed to UV radiation and harvested 2 h later for fractionation. Because of the weak signal in the S1 and S3 fractions, their exposure was almost three times longer than the other fractions in this figure. B and C, Mcm10 is down-regulated in all cell cycle phases post-UV stress, whereas its natural degradation only occurs during mitosis. B, HeLa cells were blocked at the mitotic phase using nocodazole, released into drug-free medium and were either non-irradiated (lanes 1–5) or UV-irradiated (lanes 6–10) at the indicated time points. After 2 h, the control and UV-irradiated cells were harvested and used for immunoblotting and flow cytometry. Cyclin A levels (middle panel) display progression through the cell cycle and LC refers to loading control. C, flow cytometry of propidium iodide-stained DNA of UV-irradiated and non-irradiated HeLa cells, as described in B, show a M phase block and release. The color key denotes cells obtained at different time points. D, FACS histogram shows peaks corresponding to the different phases of the cell cycle.
Cell Cycle-regulated Degradation of Mcm10 Utilizes the C-terminal Half of Mcm10
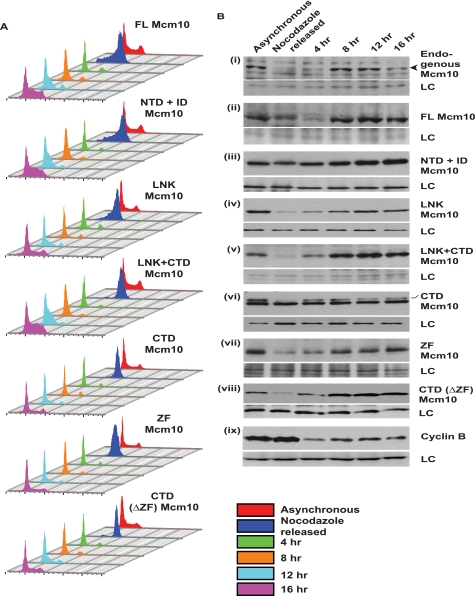
Because Mcm10 is naturally proteolyzed during M phase, we wanted to identify the domains of Mcm10 that are essential for cell cycle-regulated degradation. Stable U2OS cell lines expressing different fragments of Mcm10 were blocked with nocodazole, which arrested the cells in M phase. The obtained mitotic cells were released from the block and harvested at regular intervals to collect cells in different phases of the cell cycle (Fig. 8, A and B). The flow cytometry profile of propidium iodide-stained DNA for all stable cell lines expressing different fragments of exogenous Mcm10 demonstrates a block in M phase and subsequent progression through the cell cycle (Fig. 8A). As expected, endogenous Mcm10 levels were low in M phase and peaked around 12 h after nocodazole release (Fig. 8B, i). Full-length Mcm10 expressed from the retroviral vector showed a degradation pattern similar to that of the endogenous protein, validating our assay for evaluating the cell cycle-regulated degradation of Mcm10 (Fig. 8B, ii). The NTD+ID domain was resistant to cell cycle-regulated degradation, but the LNK and CTD domains were proteolyzed in M phase (Fig. 8B, iii–vi). The 61-amino acid ZF motif was sufficient for M phase proteolysis of Mcm10 (Fig. 8B, vii). However, the CTD domain lacking the ZF motif was also degraded in M phase, signifying that though ZF motif is sufficient, it is not essential for M phase proteolysis of Mcm10 (Fig. 8B, viii). From the above data, we conclude that the C-terminal half of Mcm10 is required for cell cycle and UV-triggered degradation of Mcm10.
FIGURE 8.
Cell cycle-regulated degradation of Mcm10 utilizes the same domains that are required for UV-triggered degradation. A and B, to identify the domains required for cell cycle-regulated degradation of Mcm10, stable cell lines of U2OS expressing different fragments of Mcm10 were arrested in M phase using nocodazole and then harvested at the indicated time points after release into drug-free medium. A shows the flow cytometry analysis of U2OS stable cell lines expressing different fragments of Mcm10, and B shows the levels of corresponding Mcm10 fragments. The color key denotes cells obtained at different time points. The endogenous Mcm10 band is denoted by the black arrowhead, whereas the CTD fragment has been marked. LC refers to loading control.
DISCUSSION
UV-triggered Targeting of Mcm10
Over the past few years, regulatory mechanisms that transduce to the replication machinery after cellular stress have been identified. The replication initiator, Cdt1, is degraded within a few minutes following UV and gamma irradiation of cells (13). A relatively slower rate of proteolysis was observed for the p12 subunit of polymerase-δ and the replication initiator, Cdc6 (14, 15). We screened a large number of known replication factors, but could not identify any other protein that is down-regulated (Fig. 2A and supplemental S1A). Inhibition of Mcm10, which naturally dissociates from chromatin at the end of S phase and is, therefore, probably involved in maintaining replication licensing, would be a fitting choice for inactivation of the replication machinery. Targeting Mcm10 inhibits replication initiation and we have demonstrated that if Mcm10 is depleted by RNA interference, it leads to a decrease in the progression through S phase. Therefore, the degradation of Mcm10 following UV irradiation would block S phase progression (Fig. 5D). An increase in BrdUrd incorporation in Mcm10-overexpressing cells signifies that Mcm10 enhances recovery of replication following UV irradiation. Therefore, apart from serving to stall replication, it also seems that a low Mcm10 level is one of the limiting factors preventing reinitiation of DNA replication.
Degradation Pathway of Mcm10
The kinetics of Mcm10 degradation raises the possibility of the involvement of the apoptotic machinery. The following evidence rules out the role of apoptosis in Mcm10 regulation. First, MG132 and AcLLnL (proteasome inhibitors), but not ALLM (inhibitor of calpains), stabilized Mcm10 following UV exposure. Second, inhibition of caspase 9 did not stabilize Mcm10. Most importantly, when we induced apoptosis by exposing cells to high levels of damaging chemicals, Mcm10 levels did not decrease. In this communication, we demonstrate that Mcm10 down-regulation post-UV exposure is blocked by MG132, a proteasome inhibitor. The identification of E3 ubiquitin ligase is being addressed separately.4 Mcm10 was proteolyzed after UV irradiation, but not after exposure to gamma radiation or DNA strand-damaging chemicals. Genotoxic agents lead to different forms of damage. Gamma radiation causes mostly double strand breaks, while UV exposure leads to the formation of cyclobutane-pyrimidine dimers and 6–4 photoproducts. The machinery that senses these forms of damage is different, although there is considerable functional overlap. It is, therefore, logical to draw the conclusion that Mcm10 down-regulation, at the least, invokes some unique UV damage-specific sensors. We determined the kinetics of Mcm10 degradation in different cell lines and noted that Mcm10 was degraded faster in U2OS cells (almost 80% degradation in 2 h), which are positive for the p53 and retinoblastoma proteins (Fig. 2D). However, the p53 and retinoblastoma protein-deficient HeLa cells also degrade Mcm10, although at a slower rate (less than 50% degradation in 2 h). The fine dissection of this pathway requires comparison between cell lines deficient in key checkpoint proteins as the knockdown of checkpoint proteins achieved by RNAi is frequently not sufficient to establish their roles.
Active replication was not essential for Mcm10 down-regulation after UV, as cells blocked in G1 and those in G2 phase also showed a decrease in Mcm10. Therefore, this pathway is not only resilient to changes in CDK activity but is also not dependent on collision or recognition of DNA lesions by active replication forks. Apart from being unaffected by DNA replication, the UV-triggered degradation of Mcm10 occurs during the phases of the cell cycle where it is presumed to be inert and inactive. Why would this mechanism target Mcm10 even after exit from S phase? It is possible that Mcm10 may have an unknown role in G2 phase or that this mechanism serves to prevent any aberrant replication that ensues after DNA damage. High levels of Mcm10 have been reported in neuroblastomas, where the MYC oncogene is believed to regulate Mcm10 expression (21). Because we are claiming that Mcm10 down-regulation is a pathway that the cell utilizes to inhibit replication under stress, then the obvious question is how the cell inhibits replication under other stresses, such as gamma radiation where Mcm10 levels remain unaffected. Apparently different targets are down-regulated. Cdt1 is known to be degraded after UV and gamma irradiation. Elucidation of specific and common targets of stress-induced pathways would help us better understand the regulation of the cell to maintain genomic stability. However, this study emphasizes that during stress, the cell inhibits the activity of replication initiator, Mcm10, to preclude DNA replication.
Supplementary Material
Acknowledgments
We thank Aparna Sankaran for the initial work on this project and subsequent follow-up experiments. We thank Nidhi Gupta for the Mcm10 antibody and the standardization of the immunofluorescence work. We acknowledge the help of Md. Muntaz Khan, Kanchan Rawat, and Rashmeet Dhindsa in the protein analysis and immunofluorescence work. We thank Dr. Takeshi Senga for critical suggestions and the plasmids pMX-GST-NLS and pMX-Cdt1, Dr. Toshio Kitamura for the pMX retroviral system, and Dr. Ramesh Juyal for the rabbit immunizations. We thank Prateek Arora for help in the flow cytometry analysis.
This work was supported by sanctioned projects from the Department of Biotechnology and Science of Technology of the Government of India and a NET fellowship (to A. S. and M. K.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1–S4 and information.
M. Kaur and S. Saxena, manuscript in preparation.
- siRNA
- small inhibitory RNA
- FITC
- fluorescein isothiocyanate
- DAPI
- 4′,6-diamidino-2-phenylindole
- HA
- hemagglutinin
- TRITC
- tetramethylrhodamine isothiocyanate
- BrdUrd
- bromodeoxyuridine
- RNAi
- RNA interference
- LNK
- linker domain
- CTD
- C-terminal domain
- ID
- inner domain
- NTD
- N-terminal domain.
REFERENCES
- 1.Solomon N. A., Wright M. B., Chang S., Buckley A. M., Dumas L. B., Gaber R. F. (1992) Yeast 8, 273–289 [DOI] [PubMed] [Google Scholar]
- 2.Izumi M., Yanagi K., Mizuno T., Yokoi M., Kawasaki Y., Moon K. Y., Hurwitz J., Yatagai F., Hanaoka F. (2000) Nucleic Acids Res. 28, 4769–4777 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Homesley L., Lei M., Kawasaki Y., Sawyer S., Christensen T., Tye B. K. (2000) Genes Dev. 14, 913–926 [PMC free article] [PubMed] [Google Scholar]
- 4.Wohlschlegel J. A., Dhar S. K., Prokhorova T. A., Dutta A., Walter J. C. (2002) Mol. Cell 9, 233–240 [DOI] [PubMed] [Google Scholar]
- 5.Merchant A. M., Kawasaki Y., Chen Y., Lei M., Tye B. K. (1997) Mol. Cell. Biol. 17, 3261–3271 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Das-Bradoo S., Ricke R. M., Bielinsky A. K. (2006) Mol. Cell. Biol. 26, 4806–4817 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ricke R. M., Bielinsky A. K. (2004) Mol. Cell 16, 173–185 [DOI] [PubMed] [Google Scholar]
- 8.Chattopadhyay S., Bielinsky A. K. (2007) Mol. Biol. Cell 18, 4085–4095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhu W., Ukomadu C., Jha S., Senga T., Dhar S. K., Wohlschlegel J. A., Nutt L. K., Kornbluth S., Dutta A. (2007) Genes Dev. 21, 2288–2299 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Izumi M., Yatagai F., Hanaoka F. (2001) J. Biol. Chem. 276, 48526–48531 [DOI] [PubMed] [Google Scholar]
- 11.Izumi M., Yatagai F., Hanaoka F. (2004) J. Biol. Chem. 279, 32569–32577 [DOI] [PubMed] [Google Scholar]
- 12.Gentile M., Latonen L., Laiho M. (2003) Nucleic. Acids Res. 31, 4779–4790 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Higa L. A., Mihaylov I. S., Banks D. P., Zheng J., Zhang H. (2003) Nat. Cell Biol. 5, 1008–1015 [DOI] [PubMed] [Google Scholar]
- 14.Hall J. R., Kow E., Nevis K. R., Lu C. K., Luce K. S., Zhong Q., Cook J. G. (2007) Mol. Biol. Cell 18, 3340–3350 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhang S., Zhou Y., Trusa S., Meng X., Lee E. Y., Lee M. Y. (2007) J. Biol. Chem. 282, 15330–15340 [DOI] [PubMed] [Google Scholar]
- 16.Méndez J., Stillman B. (2000) Mol. Cell. Biol. 20, 8602–8612 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yoshida K., Inoue I. (2004) Oncogene 23, 6250–6260 [DOI] [PubMed] [Google Scholar]
- 18.Park J. H., Bang S. W., Jeon Y., Kang S., Hwang D. S. (2008) Biochem. Biophys. Res. Commun. 365, 575–582 [DOI] [PubMed] [Google Scholar]
- 19.Robertson P. D., Warren E. M., Zhang H., Friedman D. B., Lary J. W., Cole J. L., Tutter A. V., Walter J. C., Fanning E., Eichman B. F. (2008) J. Biol. Chem. 283, 3338–3348 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Okorokov A. L., Waugh A., Hodgkinson J., Murthy A., Hong H. K., Leo E., Sherman M. B., Stoeber K., Orlova E. V., Williams G. H. (2007) EMBO Rep. 8, 925–930 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Koppen A., Ait-Aissa R., Koster J., van Sluis P. G., Ora I., Caron H. N., Volckmann R., Versteeg R., Valentijn L. J. (2007) Eur. J. Cancer 43, 2413–2422 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.