Abstract
Tricyclic antidepressants (TCAs) have been used for decades, but their orientation within and molecular interactions with their primary target is yet unsettled. The recent finding of a TCA binding site in the extracellular vestibule of the bacterial leucine transporter 11 Å above the central site has prompted debate about whether this vestibular site in the bacterial transporter is applicable to binding of antidepressants to their relevant physiological target, the human serotonin transporter (hSERT). We present an experimentally validated structural model of imipramine and analogous TCAs in the central substrate binding site of hSERT. Two possible binding modes were observed from induced fit docking calculations. We experimentally validated a single binding mode by combining mutagenesis of hSERT with uptake inhibition studies of different TCA analogs according to the paired mutation ligand analog complementation paradigm. Using this experimental method, we identify a salt bridge between the tertiary aliphatic amine and Asp98. Furthermore, the 7-position of the imipramine ring is found vicinal to Phe335, and the pocket lined by Ala173 and Thr439 is utilized by 3-substituents. These protein-ligand contact points unambiguously orient the TCA within the central binding site and reveal differences between substrate binding and inhibitor binding, giving important clues to the inhibition mechanism. Consonant with the well established competitive inhibition of uptake by TCAs, the resulting binding site for TCAs in hSERT is fully overlapping with the serotonin binding site in hSERT and dissimilar to the low affinity noncompetitive TCA site reported in the leucine transporter (LeuT).
Keywords: Computer Modeling, Drug Action, Membrane Proteins, Site-directed Mutagenesis, Seratonin Transporters, Imipramine, LeuT, Serotonin, Transporter, Tricyclic
Introduction
The serotonergic system plays an important role in many psychiatric disorders. Its role in depression is well established (1). The majority of antidepressants, including TCAs,6 cause increased synaptic serotonin (5-HT) levels via blockade of 5-HT reuptake into the presynaptic neuron (2–4) by competitive inhibition of hSERT. TCAs have been in clinical use since the 1950s, with imipramine being the first and most prominent compound (5). In severely depressed hospitalized patients, TCAs appear to be more efficacious than selective serotonin reuptake inhibitors (6). TCAs remain in widespread clinical use, especially for treatment-resistant depression (7).
hSERT belongs to the neurotransmitter sodium symporter family (2, 8). These transporters utilize the electrochemical gradient of sodium and chloride ions to accumulate 5-HT against its own gradient (9–11). No experimentally solved structures of the monoamine transporters exist, including hSERT and the dopamine and norepinephrine transporters. However, the structure of LeuT, a bacterial homolog of the neurotransmitter sodium symporters, in a substrate-occluded conformation, was reported in 2005 (12). Two sodium ions (12) and a chloride ion bind near the central substrate site (13–14) structurally and functionally coupling sodium and chloride binding to substrate binding. Recently, different transport mechanisms have been suggested (15, 16).
Subsequently, a low affinity noncompetitive binding site for TCAs in the extracellular vestibule of the LeuT 11 Å above the central binding site was identified (17, 18). The relevance of the LeuT vestibular site for TCA binding to the physiologically relevant target, hSERT, is a matter of debate. This study identifies the central binding site, not the putative vestibular site, as relevant for TCA binding to hSERT and furthermore describes and validates the orientation of TCAs within this site.
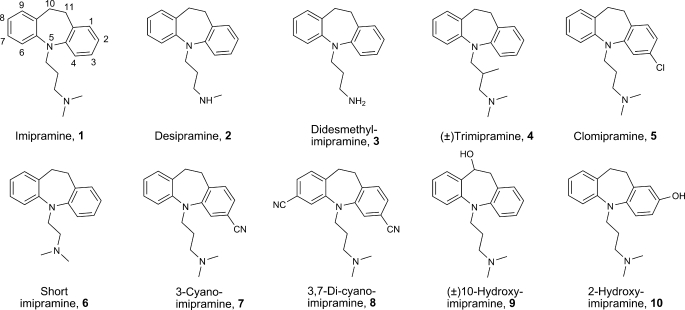
In this paper, we present induced fit docking studies of imipramine and selected analogs in the previously described homology models of hSERT (19). We present binding affinity studies of 10 imipramine analogs (Fig. 1) in wild type (WT) hSERT and 27 single point and two double mutated hSERT in a Paired Mutation Ligand Analog Complementation (PaMLAC) study. Several specific protein-ligand interaction points are identified to elucidate the orientation of imipramine within the binding site. These findings pin imipramine to the central binding site similar to the one found for 5-HT (19). Contrary to the low affinity noncompetitive TCA site in LeuT (17, 18), the central inhibitor binding site identified in this study readily accounts for the high affinity and competitive nature of TCA inhibition of hSERT.
FIGURE 1.
Chemical structures of the TCAs included in this study. The numbering of atoms in the tricyclic skeleton is indicated for imipramine.
We also present molecular dynamics (MD) simulations of imipramine bound to central and vestibular sites in hSERT along with molecular docking studies of TCAs in LeuT. These simulations are consistent with stable high affinity binding of TCA to the central binding site of hSERT as well as with unstable low affinity binding of TCA to a vestibular binding site in LeuT and in hSERT.
EXPERIMENTAL PROCEDURES
Site-directed Mutagenesis
Mutagenesis of hSERT cDNA in the pcDNA3 vector (Invitrogen) was carried out using complementary oligonucleotide primer pairs mismatched at the site of the desired point mutation in a polymerase reaction with Phusion high fidelity DNA polymerase (Finnzymes). The polymerase reaction was digested for 12 h with DpnI and used for transformation of supercompetent Solopack Gold (Stratagene) XL10 Escherichia coli according to the manufacturer's instructions. Colonies representing possible mutant clones were grown overnight at 37 °C in LB medium supplemented with 200 ng/ml ampicillin in 96-well deep well plates (Millipore) in a gyratory shaker. DNA was purified from these cultures using the Montage plasmid miniprep kit (Milipore) and subjected to sequencing on an ABI 3100 (Applied Biosystems) automated sequencer using BigDye Terminator version 3.1 (Applied Biosystems) chemistry to identify the introduced mutation. Clones carrying the desired mutation were cultured in larger volumes and subjected to midiprep plasmid purification using the Nucleobond (Macherey-Nagel) or the PureYield (Promega) plasmid midiprep kits. Full-length sequencing of the hSERT cDNA gene in the mutant midiprep DNA was carried out to verify that no unwanted mutations had been introduced.
Cell Culture
HEK-293 MSR cells (Invitrogen) were cultured as monolayer cultures in Dulbecco's modified Eagle's medium (BioWhitaker) supplemented with 10% fetal calf serum (Invitrogen), 100 units/ml penicillin, 100 μg/ml streptomycin (BioWhitaker), and 6 μg/ml Geneticin (Invitrogen) at 95% humidity and 5% p(CO2) at 37 °C. Cells were detached from the culture flasks by Versene (Invitrogen) and trypsin/EDTA (BioWhitaker) treatment for subculturing or seeding into white TC-microtiter plates (Nunc).
Uptake Assay
Transfection and measurement of [3H]5-HT (PerkinElmer Life Sciences) uptake was performed as described by Larsen et al. (20) except that HEK-293 MSR cells (Invitrogen) were used instead of COS-1 cells.
Protein Modeling
Two previously described homology models are included in this study: one based on an alignment of the hSERT and LeuT developed by us, model A (19), and one based on the comprehensive alignment of neurotransmitter sodium symporters by Beuming et al. (21), model B. The two alignments are identical around the ligand binding site and only differ slightly in the alignment of more distant helices 4, 5, 9, and 12 (19). The models were built as described by Celik et al. (19). The two sodium ions were manually included in the sites observed in the LeuT structure (12). As previously described (19), the chloride ion was manually placed in the site between Tyr121, Ser336, Asn368, and Ser372. The four residues and the chloride ion were then minimized in MacroModel 9.1 (Schrödinger LLC, 2006, New York) with the OPLS-AA force field (23) until convergence. The full protein was minimized to a root mean square (r.m.s.) deviation of 0.3 Å with the “refinement only” option in the protein preparation workflow in Impact 4.0 (Schrödinger LLC, 2006, New York).
Ligand Modeling
Imipramine and four analogs (desipramine, short imipramine, clomipramine, and 3-cyanoimipramine) were drawn in Maestro (Schrödinger LLC, 2006, New York) as charged on the alkylamine and minimized with the OPLS-AA force field as implemented in MacroModel 9.1 (Schrödinger LLC, 2006, New York) for 10,000 steps of conjugate gradient iterations or until convergence, according to default settings. The two conformations of the azepine ring system were located by a Monte Carlo conformational search in MacroModel 9.1 (Schrödinger LLC, 2006, New York); both conformations were included as input structures for the docking simulations to account for the conformational flexibility of this ring system, which is not automated in the docking program.
Docking in LeuT
Docking of imipramine and three analogs (desipramine, clomipramine, and 3-cyanoimipramine) into the extracellular vestibule of the three structures of the LeuT (Protein Data Bank codes 2A65, 2Q6H, and 2Q72) was performed with Glide 5.0 (Schrödinger, LLC, 2008, New York) (25). Clomipramine and imipramine, respectively, are co-crystallized in the two latter structures. These differ slightly in the conformation of the EL4 loop; thus, both are included. The binding site in 2Q72 and 2Q6H was defined from the co-crystallized ligand (setups I and II, respectively). This is not possible in the 2A65 structure, in which the binding site was defined from residues Arg30, Asp401, and Asp404. For comparison, a similar setup was included for 2Q6H (setups III (2Q6H) and IV (2A65), respectively). The four ligands were docked in a total of four setups (Table 1). The XP GlideScore was employed in all simulations (26).
TABLE 1.
GlideScores (kcal/mol) and RMSDs (Å) for TCAs docked in the extracellular vestibule of the LeuT
The setups are explained under “Experimental Procedures.” RMSD, r.m.s. deviation.
| Setup | I |
II |
III |
IV |
||||
|---|---|---|---|---|---|---|---|---|
| Score | RMSD | Score | RMSD | Score | RMSD | Score | RMSD | |
| kcal/mol | Å | kcal/mol | Å | kcal/mol | Å | kcal/mol | Å | |
| Imipramine | −8.4 | 1.95 | −7.6 | 0.70 | −7.8 | 1.65 | −6.4 | 0.89 |
| Desipramine | −8.4 | 1.52 | −7.9 | 1.48 | −7.4 | 0.49 | −6.7 | 1.17 |
| Clomipramine | −8.6 | 1.66 | −7.9 | 1.65 | −7.8 | 0.54 | −6.8 | 1.63 |
| 3-Cyanoimipramine | −8.2 | 1.73 | −7.2 | 1.46 | −8.1 | 1.47 | −6.3 | 1.51 |
Induced Fit Docking in hSERT
Initial docking simulations of imipramine in hSERT were performed to evaluate the docking parameters (see the supplemental material for details). Imipramine, desipramine, short imipramine, clomipramine, and 3-cyanoimipramine were docked into the hSERT binding site in models A and B employing the induced fit docking methodology from Schrödinger LLC (2008) (27). An energy window of 50 kcal/mol was used, and the number of poses to keep was set to 100. The XP GlideScore was employed in the final redocking stage. The rest of the settings were default. In both models, the binding site was defined from residues Asp98 (19, 28) and Ile172 (20, 29).
Molecular Dynamics Simulations
MD simulations were performed according to a previously described protocol (30) as described in the supplemental material.
Organic Synthesis
Novel compounds (short imipramine, didesmethylimipramine, 10-hydroxyimipramine, and 3,7-dicyanoimipramine) were synthesized as described in the supplemental material. Roche Applied Science generously donated 3-cyanoimipramine and Ciba-Geigy generously donated 2-hydroxyimipramine. The other TCAs were purchased from Sigma and used as received. See the supplemental material for information regarding organic synthesis and MD simulation protocols as well as extended tables of the inhibition experiments and docking simulations. Files of the validated models are given in the supplemental material.
RESULTS
Modeling Studies
The conformation of the TCA ring system is a key starting point for our modeling studies. Two possible conformations (see Fig. 2a) were included as initial structures for the docking studies. We first tested standard docking studies of TCAs in the extracellular vestibule of the LeuT and reproduced the binding mode observed in the crystal structures (17, 18) with high accuracy. We then proceeded with standard docking studies to search for a model of imipramine binding in the central binding site of hSERT models. These did not provide any relevant poses due to a lack of space for TCAs in the occluded binding site. Instead, induced fit docking (27) was performed, allowing imipramine to be introduced into the binding site of hSERT. Results from the modeling studies of imipramine binding in the extracellular vestibule of LeuT and the central hSERT binding site are presented below along with results from induced fit docking of four imipramine analogs to the central hSERT binding site.
FIGURE 2.
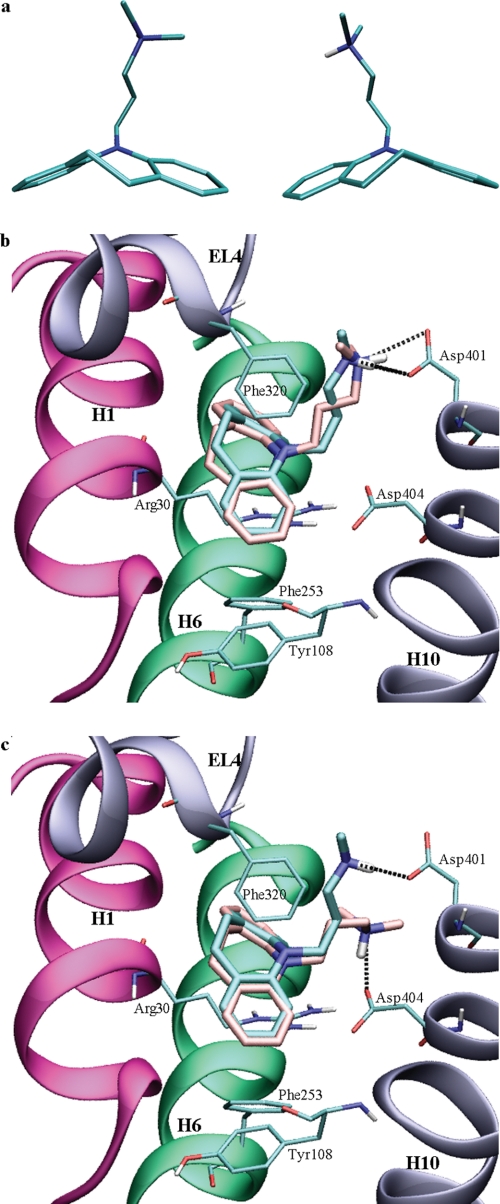
Modeling TCAs in the extracellular vestibule of the LeuT. a, two conformations of the tricyclic ring system in TCAs. Binding mode of imipramine in the extracellular vestibule of the LeuT. The co-crystallized imipramine is shown in cyan. Docked poses with the alkylamine interacting with Asp401 (b) and Asp404 (c), respectively, are shown in pink.
Docking of TCAs in the Extracellular Vestibule of the LeuT
We docked imipramine, desipramine, clomipramine, and 3- cyanoimipramine into the extracellular vestibule of three different crystal structures of the LeuT. Because no TCA is present in the original LeuT structure (12), two protocols were followed for defining the binding site in the vestibule. One protocol used the bound inhibitor in the vestibule site, whereas the other protocol used residues Arg30, Asp401, and Asp404 to define the center of the binding site. The latter protocol was employed on the original LeuT structure (12) as well as a structure with clomipramine co-crystallized (18) to be able to compare the results. We noted that imipramine, desipramine, clomipramine, and 3-cyanoimipramine are introduced readily in the extracellular vestibule of all three LeuT structures and closely resemble the binding mode observed for the TCAs in the crystal structures (17, 18) (see Fig. 2). The r.m.s. deviation for heteroatoms in the docked TCAs compared with the crystallized imipramine bound in the LeuT as well as the XP Glidescores (26) are tabulated in Table 1. The position of the ring system is stable in the docking simulations, the prime component of the r.m.s. deviation being the position of the alkylamine chain and particularly the methylated amine. In the crystal structures, the TCAs interact with Asp401 on the edge of the extracellular vestibule of the LeuT. Alternatively, the alkylamine can interact with Asp404, which is part of a stable water-mediated salt bridge in the extracellular vestibule.
Induced Fit Docking of TCAs in hSERT
Early binding experiments with tritiated imipramine are consistent with two sodium ions being necessary for TCA binding to porcine SERT (31), and we have found that coordination of two sodium ions increases protein stability of hSERT (19). A chloride ion enhances imipramine binding but is not absolutely required (32). In accordance, initial modeling studies of imipramine in two hSERT homology models (19) included two sodium ions as present in LeuT (12) but did not have the chloride ion subsequently found to be located nearby (13, 14, 30). In the present study, standard docking did not provide any poses of imipramine in the central binding site due to a lack of space to accommodate the compound (see supplemental material) since that imipramine is much larger than the cognate substrates of LeuT and hSERT. To surmount this hurdle, several different induced fit docking protocols were originally employed in an attempt to increase the size and flexibility of the binding site sufficiently to introduce imipramine, resulting in two overall binding modes of imipramine being identified (see supplemental material). Based on the preliminary data, we carried out further induced fit docking calculations in hSERT homology models, including both of the two sodium ions and the chloride ion. These induced fit docking simulations of imipramine in the hSERT models yielded 11 poses (see Table 2). In three of these poses (all from model A), imipramine is bound in the central binding site with a salt bridge to Asp98. The GlideScores of these poses are −9.0 to −10.2 kcal/mol. This binding mode is similar to the most abundantly populated binding mode from the initial studies, referred to as cluster 1, and will be further described below. Another pose from model A shows the same overall position of the ring system (GlideScore is −8.1 kcal/mol), although the fused ring system is rotated 180° compared with cluster 1. Another four poses (two in model A and two in model B) show imipramine in other orientations within the central binding site of the hSERT; three of these include a salt bridge to Asp98, whereas one pose does not include this interaction (model B). Imipramine is located in the extracellular vestibule of the hSERT models in three poses.
TABLE 2.
Induced fit docking calculations of TCAs in the hSERT
Induced fit docking calculations of TCAs in the hSERT yielded 141 different poses, clustered into two binding modes in the central binding site. Poses not belonging to these two clusters were characterized with respect to the presence of a salt bridge to Asp98. Results from model A are tabulated in the rows in normal type, and results from model B are shown in the rows in boldface type.
| Ligand | Cluster 1 | Cluster 2 | Binding sitea | Binding siteb | Vestibule | Total |
|---|---|---|---|---|---|---|
| Imipramine | 3 (1)c | 0 | 2 | 0 | 1 | 7 |
| 0 | 0 | 1 | 1 | 2 | 4 | |
| Desipramine | 2 | 2 | 1 | 0 | 16 | 21 |
| 6 (1) | 3 | 3 | 3 | 14 | 30 | |
| Short imipramine | 7 | 3 | 2 | 0 | 0 | 12 |
| 0 | 0 | 2 | 0 | 2 | 4 | |
| Clomipramine | 3 | 0 | 1 | 0 | 0 | 5 |
| 1 | 0 | 0 | 2 | 5 | 8 | |
| 3-Cyanoimipramine | 1 | 0 | 0 | 0 | 36 | 37 |
| 10 (1) | (1) | 0 | 0 | 2 | 14 | |
| Total | 33 (3) | 8 (1) | 12 | 6 | 78 | 141 |
a These poses include a salt bridge to Asp98.
b These poses show the TCA rotated in the binding site without an interaction to Asp98.
c Poses listed in parentheses reflect the possible rotation of the TCAs where the two aromatic rings have exchanged their three-dimensional positions.
Four imipramine analogs were included in the induced fit docking study: desipramine, short imipramine, clomipramine, and 3-cyanoimipramine. These four compounds resulted in 130 poses. In 29 of the poses, the tricyclic moiety is located as in cluster 1. The binding mode of desipramine and short imipramine in another eight poses is denoted cluster 2. Interactions between the alkylamine and Asp98 are found in yet another 11 poses with alternative orientations of the ring system, whereas five poses do not include the salt bridge to Asp98. The imipramine analogs are found in the extracellular vestibule of hSERT in 78 poses; these will be further discussed below. All data are tabulated in supplemental Table 2, and statistics for binding of each ligand are presented in Table 2. The computed GlideScores suggest a preference for cluster 1.
Two Putative Binding Modes for TCAs in the Central Binding Site of hSERT
Two overall binding modes were found for TCAs in the central hSERT binding site (Fig. 3). Both include salt bridges between the TCA alkylamine and Asp98 with distances of 3.6 ± 0.6 Å. In addition, hydrogen bonds from the alkylamine to Tyr95, Phe335, and Ser336 were noted in most poses. These features are similar to those of 5-HT in the hSERT (19).
FIGURE 3.
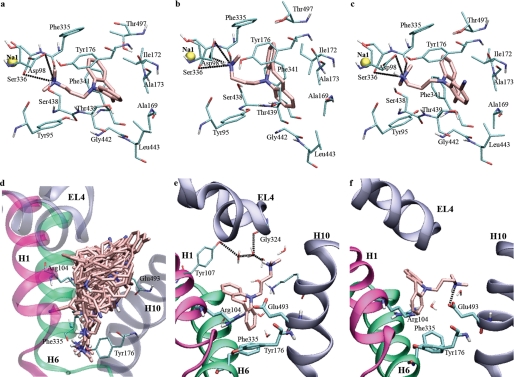
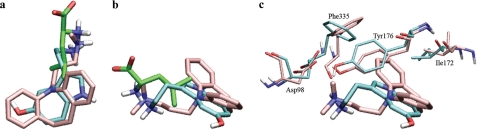
Binding modes of TCAs in the central and vestibular binding sites of hSERT. a, a representative pose of bound imipramine in the experimentally validated cluster 1. The TCA is lying in the membrane plane in the central binding site. The salt bridge to Asp98 is indicated, as are other possible hydrogen bonds to the amino group. b, a pose of bound imipramine in cluster 2. The ring system is nearly perpendicular to the membrane plane. c, binding of 3-cyanoimipramine in the central binding site; the binding mode shown here is from the experimentally validated cluster 1. The 3-cyano group is inserted between Ala173 and Thr439. d, 35 poses of TCAs in the extracellular vestibule of the hSERT, all positioned with the alkylamine pointing toward the central binding site. e, snapshot from the initial part of the MD simulation of imipramine in the extracellular vestibule of the hSERT. Water-mediated interactions are shown to the loop in the tip of EL4. f, snapshot from the final part of the simulation of imipramine in the extracellular vestibule of the hSERT. The ligand has rotated in the vestibule binding site to form a hydrogen bond to Glu493, thus disrupting the salt bridge between Arg104 and Glu493.
The binding mode of TCAs in the central binding site of the hSERT denoted cluster 2 (see Fig. 3b) was only observed for desipramine, short imipramine, and 3-cyanoimipramine. While the alkylamine is still located approximately parallel to the plane of the membrane with a salt bridge to Asp98, the ring system is rotated and nearly perpendicular to the membrane plane (Fig. 3). The position of the ring system is restricted in cluster 1, whereas it appears much more flexible in cluster 2, with translations of 1–2 Å occurring in the plane of the ring system. TCAs may form π-stacking interactions to Phe341, but despite the multiple aromatic residues in the binding site, no other possible interactions are distinctive in cluster 2.
In the binding mode denoted cluster 1 (see Fig. 3a), the two aromatic rings are located such that one ring can form orthogonal π-stabilization with Phe335 and possibly Tyr176, along with parallel displaced π-stacking with Phe341. Thr497 is also located in the vicinity of the aromatic ring and could possibly interact with a TCA substituted on the 1-position of the skeleton (for atom numbering, see Fig. 1). The other aromatic ring is located in a pocket lined by Ala169, Ala173, Tyr176, and Thr439. This pocket is an extension of the hydrophilic pocket described for the binding of the 5-HT hydroxyl-group, lined by Ala173, Ser438, and Thr439 (19). In this binding mode, the alkylamine and the center of the ring system are located almost parallel to the membrane plane within the central binding site. The curvature of the central ring forms a hydrophobic “pocket” in which the Ile172 side chain is nested. Clomipramine and 3-cyanoimipramine can have their substituent on either of the equivalent rings, leading to possible interactions with Phe335 or Ala173. The position of short imipramine is more flexible than that of imipramine in the binding site; although seven poses from model A were found in cluster 1, the shorter alkylamine of short imipramine leads to greater fluctuations in the position of the tricyclic moiety that may represent a strained and suboptimal orientation of the inhibitor.
Imipramine in the Extracellular Vestibule of the hSERT?
The induced fit docking simulations produced 78 poses with TCAs located in the extracellular vestibule of the hSERT; 30 poses were of desipramine, and 36 were of 3-cyanoimipramine. Desipramine was positioned mainly with the alkylamine pointing toward the central binding site, whereas 3-cyanoimipramine was bound either as in the LeuT structure (17, 18) or farther into the vestibule of hSERT, displacing the salt bridge between Arg104 and Glu493 and opening the aromatic lid to allow the ring system to be located between the two gates to the binding site. Twenty poses have the same overall binding mode as observed in the LeuT; some of these poses show r.m.s. deviation values as low as 1 Å for all heteroatoms. However, a majority of the poses (35 poses) with TCAs located in the extracellular vestibule displayed a binding mode with the alkylamine pointing toward the binding site. No particular location seems more favorable here. Instead, the ligands are randomly oriented and located all the way down the vestibule (Fig. 3d).
Stability of TCA Binding to hSERT
We performed MD simulations for 5 ns on two poses of imipramine in the hSERT dimer, one with imipramine bound in the occluded binding site as in cluster 1 and another that included imipramine in the extracellular vestibule of the hSERT, in a binding mode similar to the one observed for TCAs in the LeuT (17, 18). The binding of imipramine in the occluded binding site was maintained throughout the simulation and exhibited a steady binding in a stable protein.
During the MD simulation of imipramine in the extracellular vestibule, water-mediated interactions to residues in the tip of the EL4 loop were only maintained for ∼2 ns and later disrupted (Fig. 3, e and f). The disruption was caused by a translation and rotation of the inhibitor in the vestibule, which also disrupted the salt bridge between Arg104 and Glu493. This salt bridge corresponds to a stable water-mediated salt bridge between Arg30 and Asp404 in the LeuT (12, 30). An ionic hydrogen bond was later formed between the imipramine alkylamine and Glu493. The tricyclic moiety of imipramine meanwhile interacted with Trp103 and Arg104 in π-π and cation-π type interactions (see Fig. 3f) in addition to other residues. Studying the interaction energies between imipramine and each hSERT monomer indicated that the average nonbonded energy throughout the simulation was approximately −145 kcal/mol in the occluded binding site but only approximately −35 kcal/mol in the extracellular vestibule. This indicates a high degree of solvation of the inhibitor and an unstable manner of imipramine binding in the extracellular vestibule of hSERT. We speculate that the lack of stabilization of the TCA in the extracellular vestibule of the hSERT is due to the lack of an acidic residue corresponding to Asp401 in the LeuT.
Experimental Studies
HEK-293 MSR cells transiently transfected with WT and mutant hSERT were used in uptake inhibition assays with [3H]5-HT. Km values for the different mutants were determined and were used to transform IC50 to Ki values (33) for inhibitors.
Initial uptake inhibition studies with short imipramine, 3-cyanoimipramine, and clomipramine identified Asp98 and Ala173 as particularly important residues for the interaction with imipramine analogs, immediately pointing to cluster 1 from the docking studies. In turn, the orientation of the aromatic rings of imipramine in cluster 1 implicated Phe335. Fifteen mutants of these three residues (i.e. Asp98, Ala173, and Phe335) were then studied. Of these, three single mutants and one double mutant, together with three analogs of imipramine varied at the 3- and 7-positions, suggested to be vicinal to Ala173 and Phe335 in cluster 1, proved pivotal for determining the orientation of TCAs in hSERT. All results are tabulated in Table 3, and the most important findings are visualized in Fig. 4. To probe for additional possible orientations of imipramine, we produced 14 mutants at five additional positions within the binding site and included five additional imipramine analogs. Ki values for these combinations are also included in Table 3. No consistent conclusions supporting other orientations of imipramine than those predicted by IFD calculations could be derived from these data.
TABLE 3.
Mean Ki (nm) for inhibition of [3H]5-HT uptake in HEK293-MSR cells transiently transfected with hSERT WT or mutant cDNA
Data are from at least three independent experiments. 95% confidence intervals are listed in supplemental Table 3.
| Imipramine | Desipramine | Didesmethylimipramine | Short imipramine | Trimipramine | 2-Hydroxyimipramine | 10-Hydroxyimipramine | Clomipramine | 3-Cyanoimipramine | 3,7-Dicyanoimipramine | |
|---|---|---|---|---|---|---|---|---|---|---|
| hSERT | 31.8 | 1870 | 2223 | 2020 | 3530 | 83.6 | 304 | 16.41 | 5.43 | 4.57 |
| D98E | 225 | 2820 | 201 | 1581 | 8750 | 410 | 255 | 19.4 | ||
| A169I | 14.03 | 3310 | 21.0 | 50.9 | 186.2 | 18.20 | 1.079 | |||
| A173L | 4.07 | 174.2 | 18.32 | 57.5 | 152 | 2.45 | 28.2 | 7.87 | ||
| A173M | 7.31 | 48.0 | 71.9 | 25.0 | 156 | 7.69 | 37.4 | 34.9 | 18.79 | 140 |
| A173C | 12.65 | 504 | 382 | 455 | 1567 | 9.48 | 15.14 | 3.57 | ||
| A173S | 11.40 | 766 | 1374 | 1076 | 2260 | 16.37 | 13.87 | 2.19 | ||
| A173T | 15.63 | 2270 | 488 | 488 | 2820 | 8.41 | 41.9 | 2.24 | ||
| A173D | 11.83 | 807 | 509 | 242 | 13.80 | 17.38 | 3.48 | |||
| Y175F | 40.6 | 277 | 993 | 279 | 4930 | 48.98 | 22.9 | 1.875 | ||
| Y176F | 156.7 | 929 | 607 | 1091 | 9480 | 339 | 37.8 | 1.135 | ||
| F335N | 61.0 | 614 | 813 | 4320 | 6680 | 58.2 | 36.7 | 7.91 | ||
| F335H | 57.0 | 59.3 | 885 | 38.3 | 11.14 | 36.1 | ||||
| F335S | 82.8 | 1089 | 2590 | 7000 | 9930 | 97.7 | 27.2 | 6.79 | ||
| F335A | 31.3 | 1191 | 596 | 1496 | 7000 | 124.2 | 460 | 14.39 | 3.71 | 18.49 |
| F335G | 71.8 | 1259 | 2240 | 3140 | 5200 | 127.5 | 61.2 | 3.24 | ||
| F335L | 17.10 | 492 | 2450 | 3800 | 74.1 | 450 | 35.3 | 12.19 | 30.4 | |
| V343A | 75.5 | 2720 | 4620 | 8610 | 209 | 25.8 | 12.42 | |||
| V343L | 1538 | 2630 | 3310 | 6120 | 4540 | 2690 | 447 | 31.9 | ||
| V343I | 152.4 | 320 | 700 | 537 | 35.3 | 22.5 | 11.83 | |||
| T439A | 14.72 | 1611 | 697 | 1611 | 1114 | 23.4 | 7.82 | 1.114 | ||
| T439S | 15.03 | 582 | 457 | 1524 | 1556 | 22.9 | 15.38 | 0.811 | ||
| T439V | 14.96 | 832 | 525 | 1349 | 2400 | 27.2 | 14.13 | 1.706 | ||
| L443S | 24.3 | 1047 | 2310 | 5090 | 57.0 | 36.1 | 4.12 | |||
| L443T | 16.03 | 1726 | 5710 | 8070 | 50.4 | 28.8 | 5.35 | |||
| C473E | 14.35 | 83.2 | 49.3 | 8.26 | ||||||
| C473M | 34.7 | 112.8 | 62.7 | 13.58 | ||||||
| C473L | 10.23 | 129 | 31.6 | 33.3 | 6.41 | |||||
| A173M/F335L | 49.2 | 272 | 309 | 380 | 951 | 65.9 | 575 | 362 | 187.1 | 1122 |
| A173M/F335A | 9.62 | 80.4 | 318 | 530 | 54.7 | 121.3 | 50.7 |
FIGURE 4.
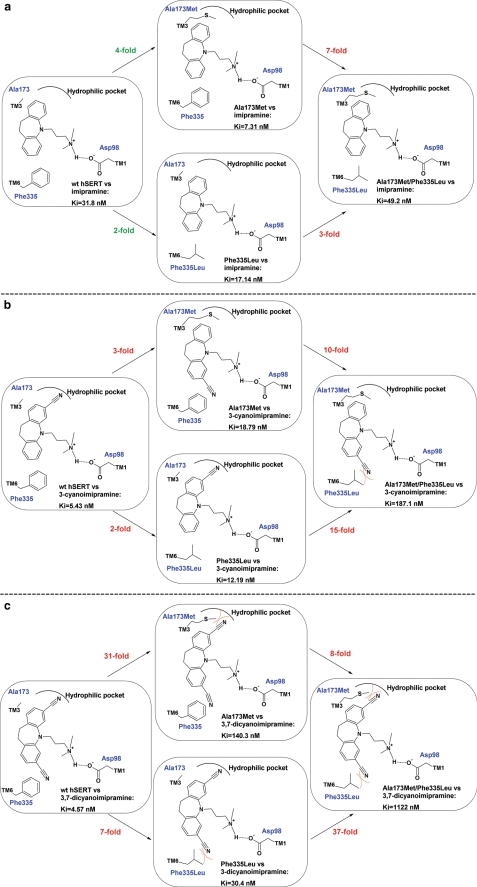
Schematic illustrations of the PaMLAC study of TCA orientation within the central binding site. Imipramine (a), 3-cyanoimipramine (b), and 3,7-dicyanoimipramine (c) inhibition of uptake by WT hSERT and three mutants, A173M, F335L, and A173M/F335L, is used to deduce the orientation of TCAs in hSERT. Decreases in inhibitory potencies are listed in red, and increases are shown in green. The salt bridge to Asp98 was established by comparing the selectivity of the WT and D98E mutant for imipramine and short imipramine (see “Ionic Interaction between TCAs and Asp98” under “Results”). Imipramine is relatively insensitive to the mutations of Ala173 and Phe335. The substituent of 3-cyanoimipramine confers increased affinity by utilizing the hydrophilic pocket lined by Ala173 and can adapt to the challenge of the A173M mutation by binding in a rotated orientation facing Phe335 instead. The double mutant precludes this adaptation and exhibits a dramatic loss of affinity for the monosubstituted TCA. 3,7-Dicyanoimipramine binds to WT hSERT with similar affinity to 3-cyanoimipramine, but the symmetric disubstitution results in a failure to adapt by rotation to the A173M mutation, resulting in a dramatic loss of affinity already for the single mutation. The protein-ligand interactions between Asp98 and the alkylamine; the hydrophilic pocket and the 3-position; and Phe335 and the 7-position are in full agreement with the binding mode found in cluster 1 and pin down the orientation of imipramine within the central binding site of hSERT.
Ionic Interaction between TCAs and Asp98
For both tryptamines (19) and N-methylated tryptamines (19, 28), it has been shown that the alkylamine interacts directly with the acidic side chain of hSERT Asp98. Because virtually all inhibitors of hSERT, including the TCAs, contain such an alkylamine, a priori it seems plausible that this functional group mimics the one in the endogenous substrate.
This hypothesis was tested in the same way as for the substrate analogs (19), namely by combining the extended side chain of D98E with short imipramine, in which the alkyl chain connecting the tricyclic moiety and the amine is shortened by one methylene unit compared with imipramine (Table 3). Short imipramine exhibited a remarkable loss of affinity compared with imipramine in WT hSERT (2020 and 31.8 nm, respectively), in agreement with both clusters proposed from the induced fit docking simulations. Evidently, the shortened alkylamine does not allow optimal orientation of the amine to the side chain of Asp98 without simultaneous movement of the ring system. To satisfy the PaMLAC paradigm, this compromised affinity must be reversed by the complementing D98E mutation. We find a slight but not statistically significant increase in D98E affinity for short imipramine relative to WT hSERT affinity for imipramine (1581 nm compared with 2020 nm). However, taken together with the affinity of D98E for imipramine, our findings show a marked, statistically significant (p < 0.0001), shift and full reversal of selectivity of short imipramine (D98E/WT Ki ratio of 0.8) compared with imipramine (D98E/WT Ki ratio of 7.1). These results are consistent with the proposed ionic interaction between the protonated alkylamine of imipramine and the acidic side chain of Asp98.
The TCA 3-Position Is Vicinal to Ala173
The binding mode of imipramine observed in cluster 1 suggests that the 3-position of imipramine is pointing toward a pocket lined by Ala169, Ala173, Tyr176, and Thr439 (Fig. 4a, left). We have shown that mutation of Ala173 to leucine and methionine extends these longer hydrophobic side chains into this hydrophilic pocket and changes its propensities to favor hydrophobic substituents and disfavor hydrophilic substituents (19). We wanted to exploit the ability of Ala173 mutations to change the properties of this part of the binding site in order to identify interacting substituents of imipramine analogs. The substitution of the 3-position with either a chlorine or a cyano group results in stronger binding of clomipramine, 3-cyanoimipramine, and 3,7-dicyanoimipramine than of imipramine to WT hSERT (Ki values of 16.41, 5.43, 4.57, and 31.8 nm, respectively) in accordance with a nonutilized hydrophilic pocket near the 3-position of imipramine (Fig. 4, a and b). Conversely, if the binding mode observed in cluster 1 is correct, we would expect A173M and A173L mutations to disfavor 3-substituted TCAs, especially partially polarized substituents, such as a cyano group (see Fig. 4b, center). Indeed, we found that the affinity of A173M and A173L is superior to the affinity of WT hSERT for all tested analogs of imipramine, including 2- and 10-substituted TCAs, whereas the 3-substituted analogs clomipramine, 3-cyanoimipramine, and 3,7-dicyanoimipramine exhibited a completely opposite pattern (Table 3). For example, the selectivity of imipramine (WT/A173M Ki ratio of 4.4), 2-hydroxyimipramine (WT/A173M Ki ratio of 10.9), and 10-hydroxyimipramine (WT/A173M Ki ratio of 8.1) was reversed for clomipramine (WT/A173M Ki ratio of 0.47), 3-cyanoimipramine (WT/A173M Ki ratio of 0.29), and 3,7-dicyanoimipramine (WT/A173M Ki ratio of 0.033). All 3-substituted imipramine analogs exhibited statistically significant reductions in selectivity for A173M compared with WT hSERT; selectivity was shifted 9-fold (p = 0.005) for clomipramine, 15-fold (p < 0.0001) for 3-cyanoimipramine, and 133-fold (p < 0.0001) for 3,7-dicyanoimipramine. Thus, mutations of Ala173 to methionine and leucine fully reversed the selectivity for the 3-substituted imipramine analogs in a manner correlating with the size, number, and hydrophilic bulk of 3- and 7-substituents, which immediately pointed to cluster 1 as the correct binding mode in terms of the location of the 3-position (Table 3 and Fig. 3).
The TCA 7-Position Is Vicinal to Phe335
The vicinity of the 3-position to Ala173 along with the rigidity of the tricyclic azepine system dictates a limited number of possible orientations of the equivalent 7-position. Induced fit docking results pointed to the location of the 7-position as being close to the side chain of Phe335 when the 3-position is near Ala173. The symmetry of imipramine means that the occurrence of a repulsion between a substituent on the 3-position of the TCA and the side chain introduced from the A173M mutation could result in preference for a fully rotated orientation of 3-substituted TCAs, placing the 3-substituent at a position normally occupied by the 7-position. Consequently, A173M mutated hSERT would still be able to bind the 3-substituted TCAs by accommodating the substituent near Phe335 (Fig. 4b, center). This reorientation may account for the fact that we observed only a modest yet statistically significant decreased affinity of 3-cyanoimipramine in A173M (Ki = 18.79 nm, p = 0.0027) and F335L (Ki = 12.19 nm, p < 0.0001) relative to WT hSERT (Ki = 5.43 nm), whereas imipramine showed a slight but statistically significant increase in affinity when introducing the A173M (Ki = 7.31 nm, p < 0.0001) mutation in hSERT compared with WT hSERT (Ki = 31.8 nm) (see Fig. 4). We observed a similar increase in the affinity of imipramine for F335L (Ki = 17.10 nm) relative to WT hSERT.
The possible rotational reorientation of 3-substituted TCAs in A173M hSERT presented us with both a challenge and an opportunity. We predicted that the permissible rotational adaptation of 3-cyanoimipramine, when going from the WT hSERT to the single mutated A173M or F335L (Fig. 4b, center), should no longer be possible for the 3,7-dicyanoimipramine because the A173M or F335L mutation would preclude a rotation of the 3-substituted imipramine analog to alleviate a sterical or polar-nonpolar clash (Fig. 4c, center). In our view, the double mutated transporter would be unable to accommodate, for example, 3-cyanoimipramine if the binding mode in cluster 1 is correct (Fig. 4b right). Likewise, the considerable 10–15-fold change in affinity for 3-cyanoimipramine, when going from A173M or F335L to A173M/F335L, would be expected to be similar or even higher for 3,7-dicyanoimipramine already when going from the WT hSERT to the single mutated A173M and F335L (Fig. 4c, center). Indeed, we noted a marked, statistically significant reduction of affinity for 3-cyanoimipramine when going from the A173M (10-fold, p = 0.0129) or F335L (15-fold, p = 0.0041) single point mutations to the A173M/F335L double mutated transporter (Fig. 4c, center versus right). In comparison, the affinity of the unsubstituted imipramine was only affected moderately when changing from A173M or F335L to the A173M/F335L double mutant of hSERT (7- and 3-fold, respectively); the affinity of imipramine did not differ significantly between WT and the A173M/F335L double mutant of hSERT (Fig. 4a). These results reflect the ability of hSERT to adapt to the repulsion between the partially polarized substituent on the 3-position and the hydrophobic side chains of either A173M or F335L by binding the ligands in the equivalent rotated orientation in accordance with the prediction from the induced fit docking with imipramine binding as in cluster 1. The fact that the A173M mutation induces the most dramatic loss of affinity (31-fold; Fig. 4c) and that the loss of affinity for 3,7-dicyanoimipramine is most dramatic when introducing the A173M mutation in the F335L (37-fold; Fig. 4c) mutated hSERT compared with introducing the F335L mutation in A173M (8-fold; Fig. 4c) mutated hSERT indicates that the interaction to the hydrophilic part of the binding site lined by Ala173 and Thr439 is the most important contributor to the increased hSERT affinity observed for clomipramine, 3-cyanoimipramine, and 3,7-dicyanoimipramine compared with imipramine.
DISCUSSION
Determining which moieties of TCAs interact with which residues in hSERT serves at least three purposes. First, the precise location of TCAs in their relevant pharmacological target can be determined and evaluated against the surprising vestibular TCA site in LeuT (17, 18). Second, reliable orientation of the TCAs within their binding site may aid the understanding of interactions between proteins and high affinity ligands in general and may specifically facilitate the development of new antidepressants or other compounds targeting monoamine transporters. Third, clues about the underlying molecular principles for perception of a ligand as a nontransportable inhibitor versus a transportable substrate can be identified and used to describe, on a molecular level, the necessary initial steps of the transport mechanism.
We have previously created homology models of hSERT (19) utilizing the structure of the LeuT as the template. By combining site-directed mutagenesis with transport inhibition studies using ligand analogs, in a paradigm we coined PaMLAC, we experimentally validated a single binding mode of 5-HT in the central binding site (19). PaMLAC studies can be based on initial knowledge or predictions about protein-ligand interactions in order to design novel ligand analogs and relevant protein mutations to study in combination. Aside from providing an extra layer of evidence, the PaMLAC paradigm also gives details about direct protein-ligand contact points unobtainable in standard mutagenesis screens and thereby novel opportunities to understand protein-ligand interactions in detail. The same methodology is here used to describe TCA binding to hSERT.
We found that mutating Asp98 to Glu, thereby lengthening the side chain by one methylene unit, reversed the selectivity of the accordingly truncated short imipramine compared with imipramine. In keeping with the findings on the alkylamine of tryptamines and N-methylated tryptamines (19, 28), our findings locate the alkylamine near Asp98 according to the PaMLAC methodology and suggest that the alkylamine is extended from the azepine heterocycle in a manner similar to that of the ethylamine of 5-HT extending from the indole heterocycle. Inherently, overlapping and similar binding modes of substrate and TCAs in the central binding site are implied.
Interactions of TCAs in the central binding site of hSERT mainly include π-stacking and other hydrophobic interactions, consistent with the hydrophobic nature of the tricyclic moiety. Consonant with its dramatic effect on TCA affinity (29), the side chain of Ile172 is nestled into the shallow hydrophobic cavity formed by the curved tricyclic system of the TCA in the main binding mode (cluster 1) observed in induced fit docking. Cluster 1 also provides an opportunity for substituted imipramine analogs to interact with Ala169, Ala173, Tyr176, Phe335, and Thr439.
We have previously shown that a hydrophilic pocket exists to accommodate the 5-hydroxy moiety of 5-HT (19). The well known increased affinity of clomipramine relative to imipramine for hSERT suggested that the substituent on the 3-position was a likely candidate for a favorable interaction with the hydrophilic pocket; this was also predicted by the binding mode denoted cluster 1 from the induced fit docking (Fig. 4). Accordingly, mutants of Ala173 (Leu, Met, and Thr), Phe335 (Leu), and Cys473 (Glu, Met, and Leu) were the only studied mutants that had statistically significant reductions in affinity for clomipramine relative to imipramine, whereas WT and other mutants exhibited equipotent or increased affinity of clomipramine relative to imipramine. It is noteworthy that both Ala173 and Cys473 line the pocket predicted by cluster 1 to be near the 3-position, whereas Phe335 is predicted by our model to be near the symmetrically equivalent 7-position.
The symmetry of imipramine implies that repulsions imposed on substituted imipramine analogs by mutated residues can be bypassed by the ligand simply binding in a rotated orientation. For monosubstituted imipramine analogs, that would mean weakened responses from mutants in terms of reduced affinity. Conversely, symmetrically disubstituted analogs of imipramine would then be sensitive to single point mutations (Fig. 4c, left versus center) to a similar degree as the monosubstituted imipramine analogs would be for the double point mutations (Fig. 4b, center versus right). Thus, the two mutations causing such changes in affinity can be viewed as being located vicinal to the two substituent positions. To test that notion, we synthesized the symmetric disubstituted 3,7-dicyanoimipramine. We studied 3-cyanoimipramine and 3,7-cyanoimipramine with imipramine as a reference in single and double mutants of Ala173 and Phe335 and observed the expected pattern; imipramine was insensitive to the mutations, 3-cyanoimipramine was most sensitive to the double mutation, and 3,7-dicyanoimipramine was sensitive for the single mutations. We therefore conclude that the side chains of Ala173 and Phe335 are vicinal to the 3- and 7-position of imipramine (Fig. 4).
From the shifts in affinity, we can also infer that the pocket lined by Ala173 in WT hSERT is the principal component of the increased affinity of the 3-substituted TCAs clomipramine and 3-cyanoimipramine. This is consistent with the binding mode observed in cluster 1. Together with the alkylamine-Asp98 interaction and the interaction of the 7-position with Phe335, these experimentally validated protein-ligand interactions effectively pin down the orientation of imipramine in the central binding site as described by cluster 1 (Fig. 3a).
The binding mode here validated for TCAs resembles the one we have earlier validated for 5-HT in the same binding site (19). 5-HT is much smaller than the TCAs, but the 5-HT indole ring is positioned corresponding to one of the TCA aromatic rings and the central azepine-ring, leaving the 5-hydroxyl group to be positioned similarly to the 3-position in the TCAs (see Fig. 5). In comparison with the high affinity TCA site in hSERT described here, the low affinity TCA site described for LeuT (17, 18) is clearly different in location.
FIGURE 5.
Comparison of leucine, 5-HT, and imipramine in the central binding site of LeuT and hSERT. Leucine (green), 5-HT (cyan), and imipramine (pink) are depicted in two views of the occluded binding site of hSERT. a, the aromatic heterocycle of 5-HT and one of the aromatic rings and the azepine ring of imipramine overlap closely; b, the amino groups from the three compounds have similar locations in the binding site. c, rotations of binding site side chains are the main differences between allowing 5-HT (cyan) and imipramine (pink) to be located in the central binding site. The binding mode shown represents cluster 1. When imipramine is bound, the side chains of Tyr176 and especially Phe335 may rotate and open the aromatic lid that is normally occluding the substrate. The second aromatic ring of imipramine is responsible for this failure to close the binding site and may prevent the initial conformational changes associated with translocation upon substrate binding, thereby locking the transporter in a partially outward facing conformation. The 5-HT hydroxyl group is positioned in the same general area as one of the aromatic rings in imipramine and may correspond to polar substituents on the TCA 3-position. Ile172, earlier found to be important for TCA affinity (29), is nestled into the hydrophobic groove formed by the curvature of the imipramine azepine system.
A major question to be answered is why substrates are transported and inhibitors, such as imipramine, are not. How do competitive inhibitors prevent hSERT from entering the cycle of conformational changes that eventually translocates the bound ligand? Tyr176 and Phe335 together form an aromatic lid, normally occluding the bound substrate from the extracellular vestibule. When imipramine is bound, Tyr176 forms a hydrogen bond to Asp98 similar to that observed in substrate binding (12, 19, 30, 34); however, the side chain dihedral angles of Tyr176 and Phe335 are slightly shifted opening the binding site (19, 30). The obstruction of lid closure by the second aromatic ring of imipramine must be deleterious for initiation of translocation. All inhibitors of hSERT have strong similarities with the substrate (i.e. they contain a protonated amine and an aromatic moiety). But the inhibitors also carry additional bulk compared with the substrate in terms of an extra aromatic ring. This unifying feature of the inhibitors indicates that the bulk of the extra ring system is key to achieving inhibition, and this notion is fully consistent with our validated model and the inhibitory mechanism based on inability to close the binding site. Moreover, binding of imipramine in a site overlapping the substrate site and stabilization of hSERT in a partially outward facing conformation is expected to result in a mode of inhibition that is competitive with transport of extracellular substrate.
It is noteworthy that LeuT Phe253, equivalent to hSERT Phe335, has been found recently to be rotated by 30° when a competitive inhibitor is bound to LeuT (16). Restricting lid closure when TCAs are bound may prevent the initial conformational changes in TM6a that would ordinarily lead to translocation of the bound ligand. Accordingly, closure of the aromatic lid when serotonin is bound must be a critical first step in movement of the rocking bundle, most prominently helices TM1 and TM6, that in a concerted manner seal off the extracellular pathway and open the intracellular pathway (15).
By induced fit docking, we have identified a diffuse site in the vestibule of hSERT similar to the TCA site in LeuT (17, 18) and suggested in the dopamine transporter for amphetamine binding (35). Binding of imipramine in this site in hSERT is unstable in MD simulations, consistent with it being a low affinity TCA site as described for LeuT (17, 18) and for the allosteric site for hSERT (36). In LeuT, the TCA binding site (17, 18) may be a consequence of the versatile modulatory substrate site (37). The apparently ubiquitous ionic interaction between the amine of the TCA and the acidic side chain of Asp401 in the vestibule of LeuT (17, 18) is not plausible in hSERT, where the charge reversal of the equivalent side chain of Lys490 is expected to result in repulsion of the protonated amine (38). Consequently, the difficulty in reconciling this observation with the fact that TCA inhibition is ∼1000-fold more potent at hSERT than at LeuT argues strongly against the notion that the vestibular TCA site in LeuT represents the primary TCA site in hSERT. Instead, this putative site could be congruent with the allosteric site (36, 39). Similarly, the dramatic changes in TCA affinity by mutation of the primary site observed in the present study (240-fold) and in the study of Henry et al. (29) (172-fold) is in sharp contrast to the very modest 2-fold changes in affinity that can be obtained by mutagenesis of the putative vestibular site (17). Furthermore, the fact that TCA binding in hSERT and human norepinephrine transporter is competitive with substrate (9, 40, 41), sodium-dependent (32, 42), and partially chloride-dependent (32, 43, 44) indicates that TCAs use the same site as the substrate, a site next to the cotransported ions in Na+/Cl−-dependent transporters (12–14, 19, 45). The overlapping of TCA and substrate sites is in accordance with findings for other competitive inhibitors as well (22, 24, 34, 35). We assert that the central TCA site modeled and biochemically validated in the present study is the site relevant for competitive high affinity inhibition of hSERT as well as Na+/Cl−-dependent transporters in general.
Supplementary Material
Acknowledgments
We thank Bente Ladegaard for skillful technical assistance and Donald Smith for commenting on the manuscript.
This work was supported by the iNANO Center, the Danish Center for Scientific Computing, the Danish National Research Foundation, the Danish Natural Science Research Council, and the Carlsberg, Novo Nordisk, and Lundbeck Foundations.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Tables 1–6 and Schemes 1–4.
- TCA
- tricyclic antidepressant
- 5-HT
- serotonin
- hSERT
- human SERT
- WT
- wild type
- PaMLAC
- paired mutation ligand analog complementation
- MD
- molecular dynamics
- r.m.s.
- root mean square.
REFERENCES
- 1.Schildkraut J. J. (1965) J. Neuropsychiatry Clin. Neurosci. 7, 524–533 [DOI] [PubMed] [Google Scholar]
- 2.Nelson N. (1998) J. Neurochem. 71, 1785–1803 [DOI] [PubMed] [Google Scholar]
- 3.Masson J., Sagné C., Hamon M., El Mestikawy S. (1999) Pharmacol. Rev. 51, 439–464 [PubMed] [Google Scholar]
- 4.Torres G. E., Gainetdinov R. R., Caron M. G. (2003) Nat. Rev. Neurosci. 4, 13–25 [DOI] [PubMed] [Google Scholar]
- 5.Kuhn R. (1958) Am. J. Psychiatry 115, 459–464 [DOI] [PubMed] [Google Scholar]
- 6.Anderson I. M. (2000) J. Affect. Disord. 58, 19–36 [DOI] [PubMed] [Google Scholar]
- 7.Bauer M., Whybrow P. C., Angst J., Versiani M., Möller H. J. (2002) World J. Biol. Psychiatry 3, 5–43 [DOI] [PubMed] [Google Scholar]
- 8.Saier M. H., Jr. (2000) Microbiology 146, 1775–1795 [DOI] [PubMed] [Google Scholar]
- 9.Rudnick G. (1977) J. Biol. Chem. 252, 2170–2174 [PubMed] [Google Scholar]
- 10.Rudnick G. (2006) J. Membr. Biol. 213, 101–110 [DOI] [PubMed] [Google Scholar]
- 11.Rudnick G. (1998) J. Bioenerg. Biomembr. 30, 173–185 [DOI] [PubMed] [Google Scholar]
- 12.Yamashita A., Singh S. K., Kawate T., Jin Y., Gouaux E. (2005) Nature 437, 215–223 [DOI] [PubMed] [Google Scholar]
- 13.Forrest L. R., Tavoulari S., Zhang Y. W., Rudnick G., Honig B. (2007) Proc. Natl. Acad. Sci. U.S.A. 104, 12761–12766 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zomot E., Bendahan A., Quick M., Zhao Y., Javitch J. A., Kanner B. I. (2007) Nature 449, 726–730 [DOI] [PubMed] [Google Scholar]
- 15.Forrest L. R., Zhang Y. W., Jacobs M. T., Gesmonde J., Xie L., Honig B. H., Rudnick G. (2008) Proc. Natl. Acad. Sci. U.S.A. 105, 10338–10343 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Singh S. K., Piscitelli C. L., Yamashita A., Gouaux E. (2008) Science 322, 1655–1661 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhou Z., Zhen J., Karpowich N. K., Goetz R. M., Law C. J., Reith M. E., Wang D. N. (2007) Science 317, 1390–1393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Singh S. K., Yamashita A., Gouaux E. (2007) Nature 448, 952–956 [DOI] [PubMed] [Google Scholar]
- 19.Celik L., Sinning S., Severinsen K., Hansen C. G., Møller M. S., Bols M., Wiborg O., Schiøtt B. (2008) J. Am. Chem. Soc. 130, 3853–3865 [DOI] [PubMed] [Google Scholar]
- 20.Larsen M. B., Elfving B., Wiborg O. (2004) J. Biol. Chem. 279, 42147–42156 [DOI] [PubMed] [Google Scholar]
- 21.Beuming T., Shi L., Javitch J. A., Weinstein H. (2006) Mol. Pharmacol. 70, 1630–1642 [DOI] [PubMed] [Google Scholar]
- 22.Andersen J., Taboureau O., Hansen K. B., Olsen L., Egebjerg J., Strømgaard K., Kristensen A. S. (2009) J. Biol. Chem. 284, 10276–10284 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jorgensen W. L., Maxwell D. S., Tirado-Rives J. (1996) J. Am. Chem. Soc. 118, 11225–11236 [Google Scholar]
- 24.Koldsø H., Severinsen K., Tran T. T., Celik L., Jensen H. H., Wiborg O., Schiøtt B., Sinning S. (2010) J. Am. Chem. Soc. 132, 1311–1322 [DOI] [PubMed] [Google Scholar]
- 25.Friesner R. A., Banks J. L., Murphy R. B., Halgren T. A., Klicic J. J., Mainz D. T., Repasky M. P., Knoll E. H., Shelley M., Perry J. K., Shaw D. E., Francis P., Shenkin P. S. (2004) J. Med. Chem. 47, 1739–1749 [DOI] [PubMed] [Google Scholar]
- 26.Friesner R. A., Murphy R. B., Repasky M. P., Frye L. L., Greenwood J. R., Halgren T. A., Sanschagrin P. C., Mainz D. T. (2006) J. Med. Chem. 49, 6177–6196 [DOI] [PubMed] [Google Scholar]
- 27.Sherman W., Day T., Jacobson M. P., Friesner R. A., Farid R. (2006) J. Med. Chem. 49, 534–553 [DOI] [PubMed] [Google Scholar]
- 28.Barker E. L., Moore K. R., Rakhshan F., Blakely R. D. (1999) J. Neurosci. 19, 4705–4717 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Henry L. K., Field J. R., Adkins E. M., Parnas M. L., Vaughan R. A., Zou M. F., Newman A. H., Blakely R. D. (2006) J. Biol. Chem. 281, 2012–2023 [DOI] [PubMed] [Google Scholar]
- 30.Celik L., Schiøtt B., Tajkhorshid E. (2008) Biophys. J. 94, 1600–1612 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Talvenheimo J., Fishkes H., Nelson P. J., Rudnick G. (1983) J. Biol. Chem. 258, 6115–6119 [PubMed] [Google Scholar]
- 32.Talvenheimo J., Nelson P. J., Rudnick G. (1979) J. Biol. Chem. 254, 4631–4635 [PubMed] [Google Scholar]
- 33.Cheng Y., Prusoff W. H. (1973) Biochem. Pharmacol. 22, 3099–3108 [DOI] [PubMed] [Google Scholar]
- 34.Beuming T., Kniazeff J., Bergmann M. L., Shi L., Gracia L., Raniszewska K., Newman A. H., Javitch J. A., Weinstein H., Gether U., Loland C. J. (2008) Nat. Neurosci. 11, 780–789 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Indarte M., Madura J. D., Surratt C. K. (2008) Proteins 70, 1033–1046 [DOI] [PubMed] [Google Scholar]
- 36.Plenge P., Mellerup E. T. (1985) Eur. J. Pharmacol. 119, 1–8 [DOI] [PubMed] [Google Scholar]
- 37.Shi L., Quick M., Zhao Y., Weinstein H., Javitch J. A. (2008) Mol. Cell 30, 667–677 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Horn A. S., Trace R. C. (1974) Br. J. Pharmacol. 51, 399–403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rudnick G. (2007) ACS Chem. Biol. 2, 606–609 [DOI] [PubMed] [Google Scholar]
- 40.Apparsundaram S., Stockdale D. J., Henningsen R. A., Milla M. E., Martin R. S. (2008) J. Pharmacol. Exp. Ther. 327, 982–990 [DOI] [PubMed] [Google Scholar]
- 41.Horn A. S., Coyle J. T., Snyder S. H. (1971) Mol. Pharmacol. 7, 66–80 [PubMed] [Google Scholar]
- 42.Lee C. M., Snyder S. H. (1981) Proc. Natl. Acad. Sci. U.S.A. 78, 5250–5254 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lee C. M., Javitch J. A., Snyder S. H. (1982) J. Neurosci. 2, 1515–1525 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lingjaerde O., Jr. (1971) Acta Physiol. Scand. 83, 257–268 [DOI] [PubMed] [Google Scholar]
- 45.Noskov S. Y., Roux B. (2008) J. Mol. Biol. 377, 804–818 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.