Abstract
Xenopus Oct25 is a POU family subclass V (POU-V) transcription factor with a distinct domain structure. To investigate the contribution of different domains to the function of Oct25, we have performed gain of function analyses. Deletions of the N- or C-terminal regions and of the Hox domain (except its nuclear localization signal) result in mutants being indistinguishable from the wild type protein in the suppression of genes promoting germ layer formation. Deletion of the complete POU domain generates a mutant that has no effect on embryogenesis. However, disruption of the α-helical structures in the POU domain, even by a single amino acid mutation, causes reversal of protein function. Overexpression of such mutants leads to dorsalization of embryos and formation of secondary axial structures. The underlying mechanism is an enhanced transcription of genes coding for antagonists of the ligands for ventralizing bone morphogenetic protein and Wnt pathways. Corresponding deletion mutants of Xenopus Oct60, Oct91, or mouse Oct4 also exhibit such a dominant-negative effect. Therefore, our results reveal that the integrity of the POU domain is crucial for the function of POU-V transcription factors in the regulation of genes that promote germ layer formation.
Keywords: Differentiation, Embryonic Stem Cell, Signal Transduction, Transcription, Xenopus, Oct4 Homologous Proteins, POU-specific Domain, Embryonic Development, Gain-of-Function
Introduction
During Xenopus embryogenesis, formation of germ layers and body plan is primarily induced by two maternal factors, VegT and β-catenin. VegT induces the nodal related genes (Xnrs) that encode ligands for the nodal signaling pathway, which is the major signal involved in mesoderm and endoderm induction (1–3). β-Catenin, in collaboration with VegT, establishes the dorsalizing signals in the Spemann organizer, and the organizer harbors quite a few secreted proteins, such as Chordin, Noggin, Cerberus, and Dkk1, that antagonize the BMP2 and the Wnt signaling pathways (4–9). To guarantee the correct formation of germ layers and body plan, these signals are finely tuned so that they can function at the right level and time and in the right locations. In Xenopus laevis, three Oct4 homologous factors, Oct60, Oct25, and Oct91, are expressed during oogenesis and early embryogenesis (10–12). We have found that Xenopus Oct factors play an important role in the regulation of the activities of VegT, β-catenin, and nodal and BMP4 signaling pathways and prevent premature and incorrect differentiation of embryonic cells to ensure correct formation of germ layers and of body axes (13–16).
Oct4 is a central player in embryonic stem (ES) cells. On the one hand, it maintains the self-renewal and pluripotency of ES cells (17–19), and on the other hand, it has the capability to introduce pluripotency into somatic cells (20–25). However, the molecular mechanisms underlying the functions of Oct4 have not been clearly understood. Because of the functional homology between mammalian Oct4 and Xenopus Oct proteins (14, 26), the analysis of these proteins might provide important insights into the molecular mechanisms by which Oct4 performs its functions.
Oct4 and its relatives are members of the POU family transcription factors of subclass V (POU-V). This protein family is characterized by a unique POU-specific domain (POU) located at the N-terminal region and a POU homeobox (Hox) domain at the C-terminal region. These two domains are joined by a variable linker region. Subclasses of this family are divided by the features of the POU and the linker sequences (27). In the classical point of view, POU factors regulate transcription of target genes via interaction between the two conserved domains, POU and Hox, and the octamer motif, ATGCAAAT (18), or certain variants (28). To achieve a higher specificity, Oct4 may form protein complexes with other transcriptional regulators. One well known example is the Oct4-Sox2 complex on the Oct4 and fgf4 promoters in ES cells (29, 30). In addition to the POU and Hox domains that are responsible for DNA binding, both the N- and C-terminal regions contain gene transactivation domains (31–33). Therefore, each region seems to play its part in the function of the Oct4 protein.
In this study, we have investigated each region of the Oct4-related protein Oct25 for its relevance in Xenopus embryogenesis. A series of deletion or point mutations were analyzed for their effects on embryonic development and gene transcription. Interestingly, disturbance of the POU domain structure but not of the Hox domain created a dominant-negative effect. Overexpression of corresponding mutants in Xenopus embryos led to a strongly dorsalized phenotype. Accordingly, the genes that promote mesoderm and endoderm germ layer differentiation and specify the dorso-ventral body axis, like Xnrs, Siamois, Chordin, Goosecoid, Dkk1, and cerberus, are strongly up-regulated. The dorsalizing effect is dependent upon functional VegT and β-catenin/TCF signaling and overrides the ventralizing activity of BMP signaling. Although DNA binding of corresponding Oct25 mutants to an Oct target sequence is abolished, they still exhibit protein/protein interactions with signal transducers and transcriptional regulators. We also demonstrate that a similar reversal of function as described for Oct25 can be obtained with the homologous proteins Oct60, Oct91, and mouse Oct4 by disturbance of the POU domain at corresponding positions. We suggest that differential binding of co-repressors and/or co-activators is responsible for the observed functional reversal.
EXPERIMENTAL PROCEDURES
Embryos and Explants
Embryos were obtained with in vitro fertilization and cultured in 0.1× MBSH (1× MBSH: 88 mm NaCl, 2.4 mm NaHCO3, 1 mm KCl, 0.82 mm MgSO4, 0.41 mm CaCl2, 0.33 mm Ca(NO3)2, 10 mm HEPES, pH 7.4). Animal cap explants were cut from uninjected or injected embryos at stage 8.5. Control and injected embryos or animal cap explants were cultured to desired stages and collected for further analyses.
Whole Mount in Situ Hybridizations
Standard procedures for whole mount in situ hybridization were used (34).
Plasmid Construction, in Vitro Transcription, and Microinjection
Plasmid construction was made by using a PCR-based strategy. All the mutants of Oct25, Oct60, Oct91, and mouse Oct4 (supplemental Fig. S1) that were used for RNA microinjection were ligated into a pCS2+ vector. GFP fusions of Oct25, Oct25ΔNLS, and Oct25ΔPOU(273–301) used for cell transfection were subcloned into a pCS2+eGFPmcs vector. The GST-tagged fusions of Oct25 and its mutants used for EMSAs were subcloned into pGEX-4T1 (Amersham Biosciences). For luciferase assays, the promoter region −257/+24 of Xnr3 (the first nucleotide of transcription start site defined as position +1) (35) was amplified from X. laevis genomic DNA and subcloned into pGL3-basic vector (Promega) to generate Xnr3Luc. All constructs were confirmed by sequencing.
Plasmids used for making antisense probes were as follows (restriction enzymes for linearizing plasmids and RNA polymerases for in vitro transcription are indicated in parentheses): pBS+Xbra (SalI/T7), pCS2+Xsox17a (ClaI/T7), Chd (EcoRI/T7), Gsc (EcoRI/T7), pGEM3-keratin (EcoRI/Sp6), pCS2+Xsox2 (EcoRI/T7), XAG2 (XhoI/T3), XMyoD (SalI/T7), NCAM (BglII/Sp6), and pBS+XHex (BamHI/T7). All plasmids used for in vitro transcription of RNAs for microinjection were cut with NotI, and RNAs were transcribed with mMessage mMachine SP6 kit (Ambion) except for pSP64T-dnXAR1 and pSP64T-BMP4, which were cut with BamHI and SalI, respectively. All in vitro transcripts were cleaned up with RNeasy kit (Qiagen).
Different doses (see below) of RNAs coding for mutants of Oct25, Oct60, Oct91, and Oct4 were injected into the equatorial region of either two dorsal blastomeres, two ventral blastomeres or all four blastomeres of four-cell stage embryos. 400 pg of VegT RNA, 100 pg of BMP4 RNA, 800 pg of dnXAR1 RNA, and 400 pg of dnTCF3 RNA were injected per embryo. 50 ng of an antisense morpholino-oligonucleotide (Gene Tools) against VegT (VegTMO, 5′-TTCCCGACAGCAGTTTCTCATTCCA-3′) were injected per embryo to knock down the VegT activity in Xenopus embryos.
DNA Transfection and Imaging
HEK293 cells (ATCC CRL 1573) were grown at 37 °C under 5% CO2 in Dulbecco's modified Eagle's medium (Invitrogen) supplemented with 10% fetal calf serum (Biochrom, Berlin, Germany). To analyze the subcellular localization of Oct25 proteins, the cells were plated on chambered cover glasses (Nunc, Rochester, NY) at a density of 80,000 cells per chamber in 2 ml of medium. After 16 h of incubation at 37 °C, cells were transfected with 200 ng of expression plasmids for enhanced GFP or Oct25-EGFP fusions using the Nanofectine transfection reagent (PAA, Pasching, Austria) according to the manufacturer's instructions. 24 h after transfection, the living cells were analyzed using an IX71 fluorescence microscope (Olympus, Hamburg, Germany) equipped with a digital camera (C4742, Hamamatsu, Hamamatsu, Japan), a 100-watt mercury lamp, and a standard fluorescein isothiocyanate (excitation, HQ470/40; emission, HQ525/50) filter set.
Quantitative RT-PCR
RNAs were extracted from embryos or animal cap explants using QiaZol (Qiagen), treated with DNase I, and purified with RNeasy kit (Qiagen). cDNAs were prepared from total RNAs using RevertAidTM first strand cDNA synthesis kit (Fermentas). The method and primers for quantitative RT-PCR (qRT-PCR) were exactly performed as described previously (14, 15).
Luciferase Assays
Each 40 pg of reporter plasmids GscLuc(−1500), Xnr1Luc(−907), Xnr3Luc, 6×DE, SiaLuc(−802), TopFlash, FopFlash, pGL3-basic, Xnr1Luc(−279), Xnr1Luc(−279-Oct), Xnr1Luc(−279-TCF), Xnr1Luc(−279-Tbox), and Xnr1Luc(−279-TCF-Tbox)) were injected alone or together with 400 pg of Oct25ΔPOU(273–301) or Oct25ΔPOU(273–281). Injected embryos were collected at gastrula stage, and luciferase assays were made according to the method described previously (15).
EMSA and GST Pulldown Assays
EMSAs and GST pulldown assays using bacterially expressed proteins were performed essentially as described previously (13, 15).
RESULTS
Nuclear Localization Signal Is Essential for the Activity of Oct25
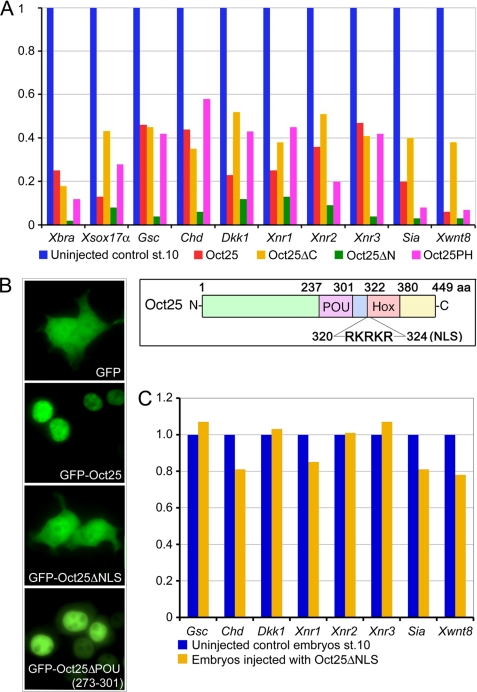
Oct25 and other Oct4 homologous proteins reveal a similar domain structure, including the diverse N- and C-terminal regions and the well conserved POU-specific (aa 237–301) and Hox-specific (aa 322–380) domains. These two domains are spaced by a linker region (aa 302–321). Moreover, a nuclear localization signal (RKRKR, aa 320–324) exists at the beginning of the Hox domain (Fig. 1 and supplemental Fig. S1). We investigated the contribution of each region to the function of Oct25 by generating deletion constructs (supplemental Fig. S1) that were overexpressed in Xenopus embryos. qRT-PCR showed that deletion mutants either lacking the C-terminal region (Oct25ΔC), N-terminal region (Oct25ΔN), or lacking both the C- and N-terminal regions (Oct25PH) inhibited mesodermal and endodermal marker gene expression (Fig. 1A), which is reminiscent of the wild type protein effect (14–16, 26). Therefore, these data suggested that Oct25 lacking these regions does not alter its function in repression of mesendoderm formation. However, when both the POU and the Hox domains, which are responsible for DNA binding, were removed from Oct25, the resulting mutant (Oct25ΔPH) did not generate any significant effect on embryogenesis after RNA injection (Table 1 and supplemental Fig. S2A).
FIGURE 1.
Analysis of the effects of the N- and C-terminal regions and the NLS of Oct25 on gene expression. A, similar to overexpression of wild type Oct25, microinjections of RNA coding for Oct25 lacking the N-terminal region (Oct25ΔN), the C-terminal region (Oct25ΔC), or both regions (Oct25PH) lead to repression of mesodermal and endodermal inducers promoting germ layer formation. For each 400 pg of Oct25, Oct25ΔN, Oct25ΔC, or Oct25PH RNA was injected into the equatorial region of all four blastomeres at the four-cell stage. The inset shows a scheme of Oct25 structural domains. B, wild type Oct25 fused to GFP (GFP-Oct25) shows an exclusively nuclear distribution. The mutant lacking the NLS (Oct25ΔNLS) fused to GFP (GFP-Oct25ΔNLS) disperses throughout the whole cell, resembling GFP protein alone, although the mutant Oct25ΔPOU(273–301) fused to GFP localizes primarily in the nuclei. C, 1 ng of RNA of the Oct25 mutant lacking the NLS (Oct25ΔNLS) was injected into embryos. No significant effect on gene expression is observed.
TABLE 1.
Numbers of embryos injected with RNAs for different mutants and the respective numbers of phenotype occurrence
| Injected RNA (dose) | Total no. of injected embryosa | Occurrences of phenotype (%)a | Phenotypeb |
|---|---|---|---|
| Oct25ΔN (400 pg) | 65 | 58 (89%) | I |
| Oct25ΔC (400 pg) | 79 | 68 (87%) | I |
| Oct25PH (400 pg) | 52 | 48 (92%) | I |
| Oct25ΔPH (1000 pg) | 92 | 83 (90%) | II |
| Oct25ΔNLS (1000 pg) | 75 | 70 (93%) | II |
| Oct25ΔHox-(330–381) (1000 pg) | 103 | 89 (86%) | I |
| Oct25ΔPOU-(237–301) (600 pg) | 87 | 80 (92%) | II |
| Oct25ΔPOU-(237–301) (1200 pg) | 95 | 82 (86%) | II |
| Oct25ΔPOU-(273–301) (1000 pg) | 117 | 99 (85%) | III |
| Oct25ΔPOU-(250–301) (1000 pg) | 69 | 54 (78%) | III |
| Oct60ΔPOU-(248–276) (1000 pg) | 71 | 56 (78%) | III |
| Oct91ΔPOU-(264–292) (1000 pg) | 83 | 73 (88%) | III |
| Oct4ΔPOU-(177–205) (1500 pg) | 96 | 61 (64%) | III |
a Combined numbers of two or three experiments are shown.
b Phenotype I means failure of blastopore formation in injected embryos during gastrulation and loss of body axis formation during tailbud stage, which is similar to the phenotype after wild type Oct25 overexpression. Phenotype II means that the injected embryos did not show any essential difference from uninjected control embryos. Phenotype III means that the injected embryos were dorsalized.
Oct4 has a nuclear localization signal (NLS) responsible for its residence in the nucleus (36). The NLS motif is also present at the beginning of the Oct25 Hox domain (Fig. 1 and supplemental Fig. S1). A mutant Oct25 without the NLS motif (Oct25ΔNLS) fused to GFP revealed a diffuse distribution throughout the cells, similar to GFP alone (Fig. 1B). In contrast, the wild type Oct25 was localized exclusively in the nuclei, suggesting that the motif is indeed required for the nuclear localization of the wild type protein. Consequently, Oct25ΔNLS had no strong effect on mesendodermal gene expression when injected into embryos (Fig. 1C). This indicates that the NLS is essential for the activity of Oct25.
Removal of aa 273–301 in the POU Domain of Oct25 Revealed a Dorso-anteriorizing Activity
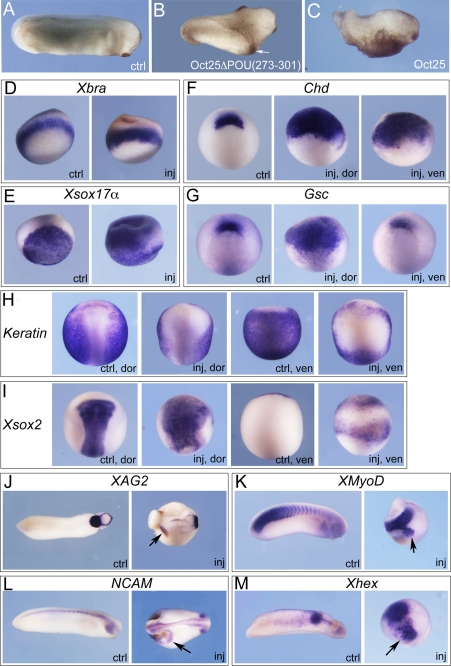
Overexpression of further deletion mutants of Oct25 were examined regarding their effects on embryogenesis. The effect of the mutant Oct25Δ(330–381) lacking the complete Hox domain but retaining the NLS was still similar to the wild type protein (Table 1; supplemental Fig. S2B). However, when the complete POU domain was missing, the mutant Oct25Δ(237–301) revealed no strong effect on embryonic development anymore (Table 1; supplemental Fig. S2C). Therefore, the POU domain is crucial for the function of Oct25. Surprisingly, when half of the POU domain from aa 273 to 301 was deleted (Oct25ΔPOU(273–301)), injection of this mutant into ventral blastomeres caused formation of prominent partial secondary axes lacking heads but including cement glands (Table 1; Fig. 2B). This effect is contrary to the overexpression of wild type protein, which caused differentiation defects and malformations of posterior structures (Fig. 2C). Moreover, deletion of this region did not alter the nuclear localization in HEK293 cells transfected with Oct25ΔPOU(273–301)-GFP plasmid (Fig. 1B). The production of partial secondary axis was inhibited when wild type Oct25 RNA was co-injected (supplemental Fig. S3A; supplemental Table S1).
FIGURE 2.
Oct25ΔPOU(273–301) mutant reveals a strong dorsalizing activity in embryos. A and B, injection of a total of 1 ng of Oct25ΔPOU(273–301) RNA into two ventral blastomeres at the four-cell stage leads to formation of partial secondary axes (indicated with arrow in B) as compared with an uninjected control (ctrl) embryo (A). C, ventral injection of 800 pg of wild type Oct25 RNA leads to suppression of posterior structures. d–M, characterization of the resulting phenotype by whole mount in situ hybridization for selected marker genes. A total of 1 ng of RNA was injected into either two dorsal blastomeres or two ventral blastomeres at the four-cell stage as indicated for phenotype analyses. D, the pan-mesodermal marker gene Xbra is not altered significantly. E, ectopic expression of the endodermal gene Xsox17α in the marginal zone and the ectoderm. F, during gastrulation, Chordin (Chd) expression is detected in an uninjected control embryo (ctrl) at the dorsal blastopore lip. In an embryo with dorsal injection of Oct25ΔPOU(273–301) RNA, the expression domain of Chd is highly expanded (inj, dor). Moreover, Chd is also ectopically induced at the ventral side upon ventral injection (inj, ven). G, at gastrula stage, Goosecoid (Gsc) is also expressed in the dorsal lip (ctrl); dorsal injection of Oct25ΔPOU(273–301) RNA up-regulates Gsc transcription significantly (inj, dor), but ventral injection does not induce ectopic expression at the ventral side (inj, ven). H, at neurula stage, expression of epidermal keratin (keratin) is detected throughout the epidermis excluding the neural fold (ctrl, dor; ctrl, ven). In contrast, it is severely reduced in both the dorsal (inj, dor) and ventral side (inj, ven) in response to injection of Oct25ΔPOU(273–301) RNA. I, neural fold marker Xsox2 is strongly increased in response to dorsal injection of the Oct25 mutant RNA (inj, dor), and ectopic expression is observed in response to ventral injection (inj, ven) compared with uninjected embryos (ctrl, dor; ctrl, ven). J, during tailbud stage, injection of Oct25ΔPOU(273–301) RNA causes ectopic expression of XAG2, a gene that marks the most anterior structure, i.e. the cement gland. K, ectopic formation of somites is observed in injected embryos, as indicated by XMyoD expression. L, ectopic formation of neural tissue as indicated by NCAM expression. M, ectopic anterior endoderm formation, as revealed by ectopic expression of Xhex in addition to its regular expression domain. Arrows indicate ectopic gene expression.
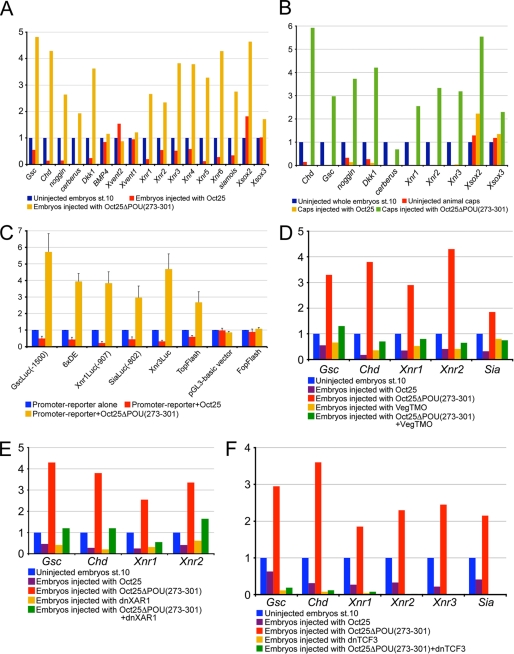
To characterize this phenotype in more detail, we performed whole mount in situ hybridization to analyze marker gene expression in Oct25ΔPOU(273–301)-injected embryos. During gastrulation, the pan-mesodermal marker Xbra was not significantly altered compared with uninjected control embryos (Fig. 2D). However, the endodermal gene, Xsox17α, revealed ectopic expression, which extended to the mesodermal and even ectodermal areas (Fig. 2E). Expression of Chordin (Chd) and Goosecoid (Gsc) was dramatically enhanced when Oct25ΔPOU(273–301) RNA was injected dorsally. Interestingly, ventral injection led to ectopic activation of Chd but not of Gsc (Fig. 2, F and G). Injection of wild type Oct25 has been shown to lead to a down-regulation of Xbra, Xsox17α, and Gsc (14, 16). During neurulation, both dorsal and ventral injections resulted in a severe reduction of epidermal keratin (keratin) expression (Fig. 2H). Xsox2, a neural plate marker gene, was significantly increased at the dorsal side and ectopically activated at the ventral side (Fig. 2I). Therefore, the injected embryos developed extra neural tissue at the cost of the epidermis. During the tailbud stage, injected embryos formed extra tissue that was derived from all three germ layers as indicated by ectopic expression of XAG2 (Fig. 2J), a gene that marks the cement gland as the most anterior structure, the somite gene XMyoD (Fig. 2K), the neural gene NCAM (Fig. 2L), and the anterior endodermal marker gene Xhex (Fig. 2m). The supplemental Table S2 lists the numbers of embryos in these whole mount experiments showing significant changes in gene expression. We further analyzed the expression of genes that are involved in germ layer formation by qRT-PCR. In whole embryos, the organizer genes that specify dorsal structures, Chd, Gsc, noggin, cerberus, Dkk1, and Xnr3, were all strongly up-regulated (Fig. 3A). Noteworthy, all these genes were significantly down-regulated by injection of wild type Oct25 RNA. The nodally related genes Xnr1, Xnr2, Xnr4, Xnr5, Xnr6, and Siamois (Sia), a target gene of maternal β-catenin pathway, as well as the neural plate genes Xsox2 and Xsox3 were also dramatically augmented. However, the genes in the BMP signaling pathway (BMP4, Xvent1, and Xvent2), which specify ventro/posterior structures, were not affected or slightly down-regulated. Next, we figured out how these genes respond to injection of Oct25ΔPOU(273–301) in animal caps. Indeed, the genes responsible for mesendoderm formation and neural induction, for instance Chd, Gsc, noggin, cerberus, Dkk1, Xnr1–3, Xsox2, and Xsox3, were also induced or enhanced in injected animal caps (Fig. 3B).
FIGURE 3.
Analysis of the Oct25ΔPOU(273–301) effect on gene transcription. A, injection of 1 ng of Oct25ΔPOU(273–301) RNA leads to an up-regulation of genes that induce mesoderm and endoderm and dorsalize body axis but does not generate an appreciable effect on the genes in the BMP pathway (BMP4, Xvent1, and Xvent2). B, injection of 1 ng of Oct25ΔPOU(273–301) RNA also increases the transcription of mesendoderm inducing genes and germ layer dorsalizing genes in animal caps. C, Oct25ΔPOU(273–301) stimulates luciferase reporter activity for the promoters of Gsc (GscLuc(−1500)), Xnr1 (Xnr1Luc(−907)), Xnr3 (Xnr3Luc), Sia (SiaLuc(−802)), the artificial promoter composed of six repeats of the distal element on Gsc promoter (6×DE), and the Wnt-responsive artificial promoter reporter TopFlash. For control, Oct25ΔPOU(273–301) does not have any strong effect on the pGL3-basic vector or on FopFlash, the negative control reporter for TopFlash. In each luciferase assay, 40 pg of reporter plasmid and 400 pg of RNA were injected. D–F, induction of genes by Oct25ΔPOU(273–301) is severely compromised after blocking the activities of VegT, activin/nodal, and Wnt/β-catenin signaling pathways. D, specific knockdown of VegT by 40 ng of an antisense morpholino (VegTMO) leads to reduced transcription of Gsc, Chd, Xnr1, Xnr2, and Sia. Injection of 1 ng of Oct25ΔPOU(273–301) RNA results in strong up-regulation of these genes. Up-regulation is lost when the RNA is co-injected with VegTMO. E, blocking of the nodal/activin signaling pathway by injection of 800 pg of a dominant-negative activin receptor I (dnXAR1) reduces the transcription of genes, like Gsc, Chd, Xnr1, and Xnr2, although injection of 1 ng of Oct25ΔPOU(273–301) RNA stimulates these genes. This stimulation is weakened by co-injection of dnXAR1 and Oct25ΔPOU(273–301) RNAs. F, when the Wnt/β-catenin signaling pathway is inhibited by injection of 400 pg of dominant-negative TCF3 (dnTCF3) RNA, transcription of Chd, Gsc, Xnrs, and Sia is dramatically decreased. Consequently, co-injection of 1 ng of Oct25ΔPOU(273–301) cannot stimulate the transcription of these genes anymore.
To test the response of gene transcription following overexpression of Oct25ΔPOU(273–301), we used promoter/ luciferase reporter assays. Promoter reporters for Xnr1 (Xnr1Luc(−907) (15)), Sia (SiaLuc(−802) (15)), Gsc (GscLuc(−1500) (37)), and Xnr3 (Xnr3Luc) were all dramatically stimulated by Oct25ΔPOU(273–301) (Fig. 3C). In addition, we also observed strong stimulation of two artificial promoters, one is composed of six catenated repeats of activin-responsive distal elements (6×DE) on the Gsc promoter (37) and the other is the Wnt-responsive reporter TopFlash (Fig. 3C). Control experiments revealed that Oct25ΔPOU(273–301) did not exert any appreciable effect on pGL3-basic vector alone or on FopFlash, the negative control reporter for TopFlash (Fig. 3C), suggesting a specific stimulation of the promoter/reporters by Oct25ΔPOU(273–301). In summary, experiments with whole embryos, animal cap explants, and luciferase reporters support the notion that the Oct25ΔPOU(273–301) mutant stimulates transcription of genes responsible for mesendoderm formation and dorsalization of embryos. Thus, Oct25 with partial removal of the POU-specific domain from aa 273 to 301 displays a dominant-negative effect.
Stimulation of Germ Layer Formation by Oct25ΔPOU(273–301) Is Dependent on the Activities of VegT, Nodal/Activin, and β-Catenin Signaling
Maternal VegT and β-catenin induce mesendoderm formation via transcriptional activation of ligands of the nodal signaling pathway and dorsalizing genes, like Sia and Xnr3. Therefore, we asked whether the induction of partial secondary axis formation in embryos by Oct25ΔPOU(273–301) relies on the activities of VegT, β-catenin, or nodal/activin signaling. We used an antisense morpholino-oligonucleotide against VegT (VegTMO) to knock down the function of VegT in embryos. First, we tested the efficiency of VegTMO. Vegetal injection of VegTMO led to severely reduced body axis, strongly pigmented belly side, and gradual death after stage 30. VegTMO-injected embryos were rescued by VegT RNA injection to form a nearly normal body plan (supplemental Fig. S4; supplemental Table S1). We then injected Oct25ΔPOU(273–301) RNA or VegTMO alone and in combination and analyzed gene expression (Fig. 3D). Again, genes like Gsc, Chd, Xnr1, Xnr2, and Sia were up-regulated by Oct25ΔPOU(273–301) alone. In contrast, injection of VegTMO resulted in a reduction of these genes. When Oct25ΔPOU(273–301) RNA was co-injected with VegTMO, we did not observe transcriptional stimulation of Gsc, Chd, Xnr1, Xnr2, and Sia. The results suggest that stimulation of these genes by Oct25ΔPOU(273–301) is dependent on VegT activity. Furthermore, we demonstrated that up-regulation was also dependent on the nodal/activin signaling pathway, because blocking with a dominant-negative activin receptor, dnXAR (38), prevented the Oct25ΔPOU(273–301) mutant from stimulating these genes to high levels anymore (Fig. 3E). Finally, when the β-catenin signaling was blocked by dominant-negative TCF3 (dnTCF3 (39)), transcription of these genes, including Xnr3, was almost completely abolished. Inhibition of β-catenin signaling also caused a failure in stimulation of these genes by co-injection of Oct25ΔPOU(273–301) RNA (Fig. 3F). This is especially obvious for the dorsalizing genes Xnr3 and Sia, which are direct targets of the Wnt pathway, but somewhat less obvious for Gsc, Chd, and Xnr1, which are also activated by the nodal/activin pathway. In summary, these experiments reveal that Oct25ΔPOU(273–301) alone was not able to stimulate transcription of genes responsible for germ layer formation without endogenous inducing activities of VegT, nodal/activin, and especially β-catenin signaling.
Oct25ΔPOU(273–301) Rescues BMP4-ventralized Embryos
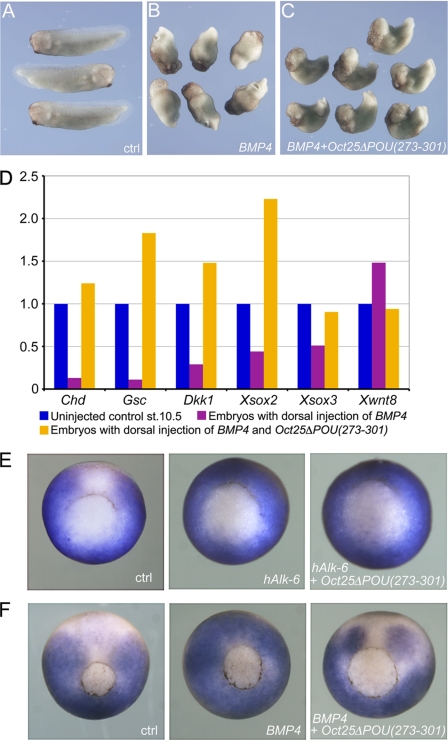
We further explored whether Oct25ΔPOU(273–301) could rescue embryos that were ventralized by the BMP signaling pathway. Dorsal injection of BMP4 RNA caused formation of “belly pieces” without dorsal structures (Fig. 4B). Co-injection of a low dose of Oct25ΔPOU(273–301) RNA (400 pg) together with BMP4 RNA already led to formation of discernible body axes (data not shown). A better rescue effect was achieved with a higher dose (1 ng) of Oct25ΔPOU(273–301) RNA. In this case, the embryos displayed clear dorsal-ventral and anterior-posterior body axes, although normal embryos did not form (Fig. 4C; supplemental Table S1). As expected, gene expression analysis after dorsal injection of BMP4 RNA showed an inhibition of dorsal genes, like Chd, Gsc, Dkk1, Xsox2, and Xsox3, whereas the ventral gene Xwnt8 was up-regulated. However, transcript levels of these genes were reversed by co-injection of Oct25ΔPOU(273–301) RNA, as shown by up-regulation of dorsal genes and down-regulation of the ventral marker gene Xwnt8 (Fig. 4D). Therefore, we conclude that Oct25ΔPOU(273–301) can rescue the ventralized phenotype caused by dorsal activation of BMP signaling and thus reveals an anti-ventralizing activity.
FIGURE 4.
Oct25ΔPOU(273–301) antagonizes BMP4 activity in embryos. A, uninjected control (ctrl) embryos at tailbud stage. B, embryos show no dorsal structures upon dorsal injection of 100 pg of BMP4 RNA. C, embryos, co-injected with 100 pg of BMP4 and 1 ng of Oct25ΔPOU(273–301) RNAs together, display clear dorsal structures. D, gene expression analysis demonstrates that dorsal injection of BMP4 RNA leads to repression of dorsal genes, like Chd, Gsc, Dkk1, Xsox2, and Xsox3, and up-regulation of the ventral gene Xwnt8. This tendency is completely reversed when Oct25ΔPOU(273–301) RNA is co-injected. E, expression of the BMP-target Xvent-2 in embryos without injection (ctrl), after animal injection of hAlk-6 mRNA (1000 pg) alone, and hAlk-6 mRNA (1000 pg) in combination with Oct25ΔPOU(273–301) (500 pg). The ectopic activation on the dorsal side is not affected by Oct25ΔPOU(273–301). F, expression of Xvent-2 in embryos without injection (ctrl), after animal injection of BMP4 mRNA (1000 pg) alone, and BMP4 mRNA (1000 pg) in combination with Oct25ΔPOU(273–301) (500 pg). The ectopic activation of Xvent-2 is inhibited by Oct25ΔPOU(273–301).
To investigate how Oct25ΔPOU(273–301) interacts with the BMP signaling pathway, we co-injected Oct25ΔPOU(273–301) RNA with constitutively active type I BMP receptor RNA, hAlk-6 (40), and analyzed the expression pattern of the BMP4 target gene Xvent2. We found that the ectopic activation of the Xvent2 gene by hALK-6 RNA injection could not be rescued by co-injection of Oct25ΔPOU(273–301) (Fig. 4E). In contrast, the effect of BMP4 RNA injection was antagonized by Oct25ΔPOU(273–301) co-injection (Fig. 4F). These experiments indicate that the BMP-antagonizing and dorsalizing effect of Oct25ΔPOU(273–301) is caused at the level of the ligand and not downstream via the receptor or components of the transduction pathway.
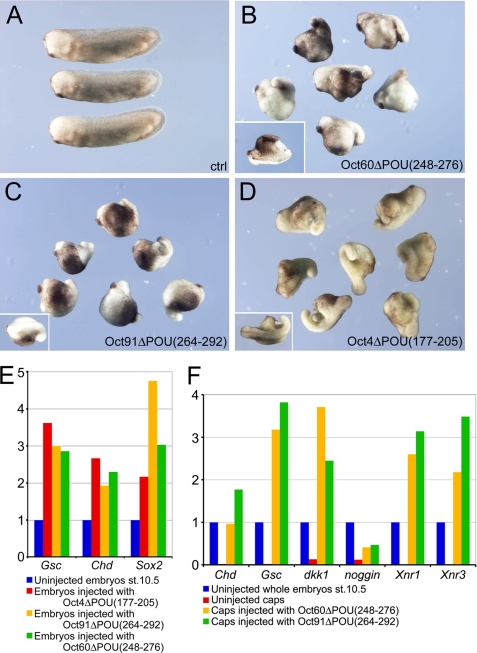
Oct60, Oct91, and Mammalian Oct4 Missing the Corresponding Regions of aa 273–301 in Oct25 Displayed Similar Dorsalizing Activities
Xenopus Oct25, Oct60, and Oct91 as well as mammalian Oct4 behave similarly in the repression of mesendoderm formation (14). We asked whether Oct60, Oct91, and mouse Oct4 with the deletion of the regions corresponding to aa 273–301 in Oct25 could also have a reversed function. For Oct60, aa 248–276 in the POU-specific domain were deleted to generate the mutant Oct60ΔPOU(248- 276); for Oct91, aa 264–292 were deleted to generate Oct91ΔPOU(264–292), and for Oct4, aa 177–205 in the POU-specific domain were removed to create Oct4ΔPOU(177–205) (supplemental Fig. S1). Ventral injections of all three mutants gave rise to dorsalized embryos as revealed by prominent protrusions from the ventro/posterior side (Fig. 5, B–D). Whole mount in situ hybridizations showed that injection of Oct60ΔPOU(248–276) RNA or Oct91ΔPOU(264–292) RNA resulted in enhanced Chd expression at the dorsal side or ectopic activation of Chd at the ventral side (supplemental Fig. S5, B and C; supplemental Table S2). qRT-PCR demonstrated that in whole embryos, Gsc, Chd and Sox2 were dramatically up-regulated in response to Oct4ΔPOU(177–205) RNA, Oct91ΔPOU(264–292) RNA, or Oct60ΔPOU(248–276) RNA injections (Fig. 5E). Moreover, we found activation of genes, like Chd, Gsc, noggin, Dkk1, Xnr1, and Xnr3, in animal caps that were injected with Oct91ΔPOU(264–292) or Oct60ΔPOU(248–276) RNA (Fig. 5F). These data indicate that deletion of the partial POU-specific domain in other members of POU-V transcription factors can also reverse their functions, as in the case of Oct25.
FIGURE 5.
Analysis of the effect after deletion of the region in Oct60, Oct91, and Oct4 corresponding to aa 273–301 in Oct25. A, uninjected control embryos at tailbud stage. B–D, ventral injections of 1 ng of Oct60ΔPOU(248–276) RNA (B), 1 ng of Oct91ΔPOU(264–292) RNA (C), and 1.5 ng of Oct4ΔPOU(177–205) RNA (D) lead to a dorsalized phenotype. The insets show embryos with partial double axis. E, qRT-PCRs reveal an up-regulation of dorsal genes in embryos injected with RNAs coding for Oct4ΔPOU(177–205), Oct91ΔPOU(264–292), or Oct60ΔPOU(248–276). F, qRT-PCRs of animal caps after injection with Oct91ΔPOU(264–292) or Oct60ΔPOU(248–276) RNAs also show stimulation of genes responsible for mesendoderm induction and body axis dorsalization.
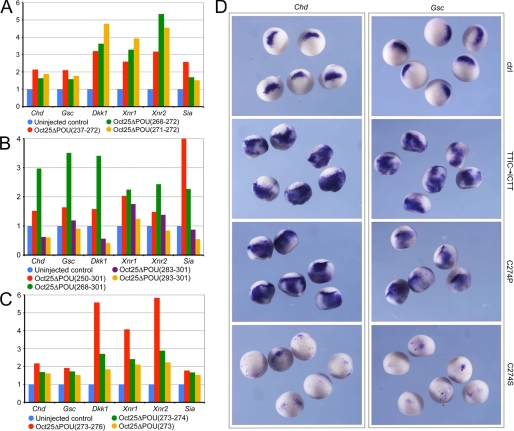
Reversal of Oct25 Function by Single Amino Acid Deletions or Mutations
As Oct25 with loss of the C-terminal half of the POU-specific domain showed a dominant-negative effect, we have asked whether the depletion of the N-terminal half in the POU domain also led to a similar effect or if we could pinpoint the amino acids that were responsible for such an effect. Actually, deletion of the N-terminal half of the POU domain from aa 237 to 272 (Oct25ΔPOU(237–272)) also caused an up-regulation of dorsalizing genes. Deletions of smaller stretches in this region from aa 268 to 272 (Oct25ΔPOU(268–272)) or even only two amino acids 271 to 272 (Oct25ΔPOU(271–272)) resulted in increased transcription of these genes (Fig. 6A). Such an effect was also observed when a longer stretch of aa 268–301 (Oct25ΔPOU(268–301)) was depleted (Fig. 6B). When most part of the POU-specific domain from aa 250 to 301 (Oct25ΔPOU(250–301)) was removed, we could still clearly observe the dorsalized phenotype in embryos upon RNA injection (Table 1; supplemental Fig. S3B) and up-regulation of the dorsalizing genes (Fig. 6B). However, when a region from aa 283 to 301 of the C-terminal end of POU domain (Oct25ΔPOU(283–301)) was missing, there was no significant dorsalizing effect anymore. If only aa 293–301 (Oct25ΔPOU(293–301)) were deleted, this mutant showed a gene repression effect resembling that of wild type Oct25 (Fig. 6B). The results suggest that the middle region of the POU domain is critical for repression of gene transcription by Oct25. As a matter of fact, deletion of aa 273–276 (Oct25ΔPOU(273–276)), 273–274 (Oct25ΔPOU(273–274)), or only a single amino acid 273 (Oct25ΔPOU(273)) indeed caused an up-regulation of dorsalizing genes (Fig. 6C).
FIGURE 6.
Mutations of single amino acids cause reversal of Oct25 function and lead to an up-regulation of dorsal mesodermal genes. A, Oct25 mutants with N-terminal deletion of the POU-specific domain (aa 237–272), a few amino acids (aa 268–272), or only two amino acids (aa 271–272) cause prominent up-regulation of gene transcription in embryos. B, Oct25 mutants with depletion of a broad region of the POU-specific domain (aa 250–301 or 268–301) also have a stimulating effect on gene transcription. When a small region close to the C terminus of the POU-specific domain (aa 283–301) is truncated, this mutant exhibits either no or only rather weak stimulating activity. Up-regulation is completely lost when aa 293–301 are deleted. C, deletion of four amino acids (aa 273–276), two amino acids (aa 273–274), or a single amino acid (aa 273) leads to a strong up-regulation of gene transcription. D, whole mount in situ hybridizations demonstrate that either the change in the order of the four amino acids TTIC to ICTT (TTIC → ICTT) or the mutation of the amino acid Cys-274 to Pro (C274P) lead to an up-regulation of the transcription of Chd and Gsc. When Cys-274 is mutated to Ser, the resulting mutant represses expression of these two genes. RNA for each mutant was injected at a total of 1 ng per embryo, except for Oct25(C274S) RNA, which was injected at a total dose of 300 pg per embryo. ctrl, control.
To gain additional support whether the sequence of the middle part of the POU domain would change the function of Oct25, we made a mutant in which the amino acids TTIC were reversed to ICTT (Oct25(TTIC → ICTT)). Moreover, another two mutants were generated, in which the amino acid cysteine 274 (Cys-274) was changed to proline (Oct25(C274P)) or serine (Oct25(C274S)), respectively, because proline is structurally different from and serine is similar to cysteine. Injection of Oct25(TTIC → ICTT) RNA and Oct25(C274P) RNA caused a significant up-regulation of Chd and Gsc expression (Fig. 6D; supplemental Table S3). However, the mutant Oct25(C274S) did not reveal such an effect but instead repressed these genes severely (Fig. 6D; supplemental Table S2). Hence, the results clearly show that disruption of the POU domain structure leads to a reversal of Oct25 function.
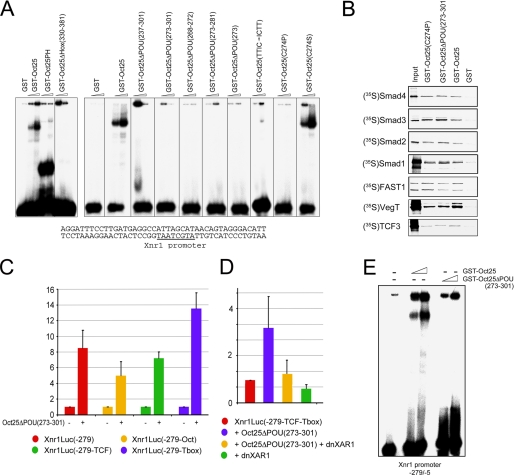
Alteration of the POU Domain Structure Leads to the Loss of DNA Binding Activity but Retains Protein Interaction Properties
As Oct25 binds DNA via its POU- and Hox-specific domains, we explored whether the mutants described above show any changes in their DNA binding activity. We made use of the Xnr1 promoter that contains an Oct25-binding motif (15) to test the DNA-binding properties of the mutants. EMSAs demonstrated again that wild type Oct25 bound to a double-stranded oligonucleotide containing the Oct25-binding motif of the Xnr1 promoter. The mutant Oct25PH containing only the POU and Hox domains showed strong affinity for this target. When the Hox domain was deleted (Oct25ΔHox(330–381)), the mutant did not bind the promoter anymore (Fig. 7A). Moreover, all the mutants in which the POU-specific domain was completely or partially deleted (Oct25ΔPOU(237–301), Oct25ΔPOU(273–301), Oct25ΔPOU(268–272), Oct25ΔPOU(273–281), and Oct25ΔPOU(273)) exhibited a loss of DNA binding activity. The mutants Oct25(TTIC → ICTT) and Oct25(C274P) were also not able to bind the DNA fragment (Fig. 7A). In contrast, the mutant Oct25(C274S) showed DNA binding activity. This situation is not unexpected, because Oct-1, a POU family protein binding to the octamer motif, has a Ser instead of Cys in the corresponding position of Cys-274 in Oct25 (supplemental Fig. S6).
FIGURE 7.
DNA binding, protein interaction, and luciferase assays using Oct25 mutants. A, EMSAs (8% PAGE) show that the GST-Oct25 fusion protein binds to a canonical octamer motif (underlined) within the Xnr1 promoter. Binding is also observed for the mutant containing only the POU and Hox domains (Oct25PH). Deletion of the Hox-specific domain results in loss of DNA binding activity. GST alone was used as control. The mutants with a truncation of either the complete POU-specific domain (aa 237–301) or different regions of the POU-specific domain lose their DNA interaction capacity. DNA binding is also lost, when the amino acids TTIC are changed to ICTT (TTIC → ICTT) or Cys-274 is mutated to Pro (C274P) but is retained in the (C274S) mutant. B, GST pulldown assays show that Oct25(C274P) and Oct25(ΔPOU(273–301) still interact with Smad4, Smad2, Smad3, Smad1, FAST1, VegT, or TCF3. C, luciferase reporter activities driven by the wild type Xnr1 promoter (Xnr1Luc(−279) or the indicated deletions (15) in the absence or presence of Oct25ΔPOU(273–301). D, luciferase reporter assays driven by the −279(−TCF-Tbox) Xnr1 promoter upon co-injection with Oct25ΔPOU(−273–301) and dnXAR1. E, EMSA of the −279/−5 Xnr1 promoter fragment (6% PAGE) reveals binding of Oct25 but no interaction with Oct25ΔPOU(−273–301) GST fusion proteins.
Oct25 represses transcription of target genes of VegT, β-catenin, or nodal signaling via formation of protein complexes with signal transducers like VegT, TCF3, FAST1, and the Smad (derived from Caenorhabditis elegans small and Drosophila MAD genes) transducers on the target gene promoters (15, 16). We therefore explored whether the mutants have lost the binding capacities to these proteins. However, in GST pulldown assays, both the mutants Oct25(C274P) and Oct25ΔPOU(273–301) displayed similar interaction activities compared with that of wild type Oct25 (Fig. 7B). As expected, the mutant Oct25ΔPOU(273–301) stimulated transcription from the wild type Xnr1Luc(−279) luciferase reporter (Fig. 7C) (15). We found that mutant reporters lacking the Oct-binding site (Xnr1Luc(−279-Oct)), the TCF/LEF-binding site (Xnr1Luc(−279-TCF)), or the Tbox-binding site (Xnr1Luc(−279-Tbox)) (15) were also strongly stimulated by co-injection of Oct25ΔPOU(273–301) RNA (Fig. 7C). This means that up-regulation of the promoter does not require each motif by itself. Also, a promoter mutant lacking both, the TCF/LEF and the Tbox sites (Xnr1Luc(−279-TCF-Tbox)), displays a strong activation by Oct25ΔPOU(273–301) (Fig. 7D). However, this stimulation is severely reduced by co-injection of dnXAR1. Therefore, we suggest that the activation of this Xnr1 promoter fragment does not only involve complex formation between dorsalizing Oct25 mutants and the transcription factors VegT and TCF but that it also involves interaction with additional transcriptional regulators, which are provided by the nodal/activin pathway. To exclude an artificial binding of the Oct25 mutant to the −279/−5 Xnr1 promoter fragment, we performed EMSAs with the wild type and the mutant protein. Although Oct25 was bound, no interaction occurred with the Oct25ΔPOU(273–301) mutant (Fig. 7E).
DISCUSSION
Here, we report a detailed study on the functional domains of the Oct4 homologous protein Oct25 in X. laevis. Although quite a few studies have been carried out on the functions of the mammalian Oct4 protein domains, we discovered a previously undefined phenomenon, i.e. when the POU-specific domain structure is disturbed, either by deletion of a few amino acids or by mutation of a single amino acid, the resulting mutated proteins exhibit dorsalizing activity in Xenopus embryos. This indicates a reversal of protein function, because the wild type protein was previously shown to repress mesodermal and endodermal germ layer formation (14–15, 26).
The Oct4 homologous proteins are composed of the N-terminal region, the POU-specific domain, the linker region, the nuclear localization signal, the Hox-specific domain, and the C-terminal region. In this study, we show that Oct25 without nuclear localization signal (Oct25ΔNLS) has apparently no effect on embryonic development and gene expression. This is not surprising, because the mutant protein distributes primarily in cytoplasm instead of the nucleus. In a study of an Oct4 mutant without NLS (Oct4ΔNLS), Oct4 and Oct4ΔNLS form a dimer and, consequently, the endogenous Oct4 protein in ES cells is retained in the cytoplasm by Oct4ΔNLS and prevents binding to target gene promoters (36). Such a mechanism may also apply to Oct25ΔNLS.
The mutants with complete removal of one or two regions or domains display two types of effects. Either the transcription of genes for mesendoderm formation is repressed or these genes are not significantly affected. Oct25 deletion mutants lacking the N-terminal region (Oct25ΔN) or the C-terminal region (Oct25ΔC) inhibit transcription of genes responsible for mesendoderm formation. We have previously shown that this effect is also caused by overexpression of wild type Oct25 protein (14–16), suggesting that the N- and C-terminal regions are functionally redundant to regulate at least a subset of target genes. A similar conclusion was reported in ES cell-based complementation assays (41). Oct4 proteins lacking either the N- or C-terminal region were able to substitute the wild type Oct4 function in rescuing the ES cell phenotype in the ZHBTc4 ES cell line. This does not automatically mean that the two deletion mutants are identical, because it seems that at least Oct4 lacking the C-terminal region can support the expression of Ebaf/Lefty1, although the mutant without the N-terminal region cannot (41). Like the Oct25ΔN and Oct25ΔC mutants, the Oct25 mutant lacking both N- and C-terminal regions (Oct25PH) also exerts an inhibitory effect on mesendodermal gene transcription. By analogy, it would be expected that the equivalent mutant of Oct4 could rescue the phenotype in ES cell complementation assays as well. However, unlike Oct4 lacking the N- or C-terminal region, Oct4 lacking both regions cannot rescue this ES cell phenotype (41). It is known that Oct4 may play dual roles for gene transcription. On the one hand, it stimulates transcription of genes supporting self-renewal and pluripotency, and on the other hand it represses genes promoting differentiation (42–43). It may be that when both transactivation domains in the N- and C-terminal region are missing, the remaining part of the protein is still able to exert its repression effect on gene transcription, but the capacity for gene activation is somehow lost.
The difference in the activities of the Oct25 mutant lacking the complete POU domain (Oct25ΔPOU(237–301)) and the mutant lacking the complete Hox domain but retaining the NLS (Oct25ΔHox(331–381)) suggests that the POU domain but not the Hox domain is a prerequisite for delivering the regulatory effect on genes examined in this study. This notion is supported by the effect of a mutant in which the C-terminal half of the POU domain (Oct25ΔPOU(273–301)) is missing. Contrary to the wild type protein that inhibits germ layer formation, Oct25ΔPOU(273–301) induces ectopic germ layer formation in embryos as shown by the presence of partial secondary axis. Additionally, a series of analyses clearly revealed enhanced or ectopic transcription of genes that induce germ layer formation, like Xnrs and Sia, which are target genes of maternal VegT and β-catenin, and the organizer genes, like Chd, noggin, cerberus, and Dkk1, which code for antagonists against BMPs and Wnts. Consequently, we were able to rescue BMP4-ventralized embryos to form dorsal structures by using the Oct25ΔPOU(273–301) mutant. Noteworthy, overexpression of the mutant in embryos did not generate any significant quantitative effect on the transcription of BMP4 and its downstream targets Xvent2 and Xvent1. This observation suggests that Oct25ΔPOU(273–301) does not achieve its dorsalizing effect via transcriptional suppression of BMP. Rather, it induces or enhances transcription of genes coding for proteins such as Xnr3, Chd, Cerberus, and Dkk1, which are antagonists of BMPs and Wnts via direct interaction. As a matter of fact, Oct25ΔPOU(273–301) RNA injection antagonized the effects of BMP4 on ectopic expression of Xvent2 but not the effects of the constitutively active BMP receptor hAlk-6. Therefore, we conclude that the underlying mechanism for the dorsalization of embryos by the mutant is due to the inhibition of ligand activities of BMP and Wnt signaling pathways. Although the wild type Oct25 inhibits the activities of VegT, β-catenin, as well as the nodal pathways, but promotes BMP4 signaling pathway (13–16), we show here that the Oct25ΔPOU(273–301) mutant has an opposite activity. These results also substantiate our previous conclusion that the endogenous Oct proteins serve as general inhibitors of differentiation signals in a way that germ layer formation can precisely occur.
The dominant-negative effect was not only observed in the mutant missing the C-terminal half of the POU domain but also in the mutant missing a stretch of amino acids located toward the N terminus of the POU domain (Oct25ΔPOU(237–272), Oct25ΔPOU(268–272), and Oct25ΔPOU(271–272)). However, complete removal of the POU domain leads to a mutant protein that has essentially lost both DNA binding activity and its capability of gene regulation. Therefore, it seems that the middle part of the POU domain is critical for gene regulation. This postulation is confirmed by the deletion of four amino acids, two amino acids, or a single amino acid, suggesting that even a minor disturbance will cause a functional reversal. As a matter of fact, a change in the order of the four amino acids TTIC to ICTT or a mutation of the amino acid cysteine at position 274 to a structurally different proline again results in a dominant-negative effect. When the same amino acid is mutated to the structurally similar amino acid serine, the function of the mutant as compared with wild type Oct25 is not significantly altered. This confirms that the configuration of the POU domain is essential for the regulation of gene transcription. Sequence comparison between Oct4 homologous proteins (supplemental Fig. S6) reveals that the POU domain is well conserved and especially that amino acids within the middle part of POU domain are identical. It is therefore rational that partial deletions of the POU domain in Xenopus Oct60 (Oct60ΔPOU(248–276)), Oct91 (Oct91ΔPOU(264–292)), or mouse Oct4 (Oct4ΔPOU(177–205)) all exhibit a similar effect to that of Oct25ΔPOU(273–301). These data also imply that Oct4ΔPOU(177–205) may exert a dominant-negative effect on gene transcription in ES cells. Therefore, it would be interesting to know whether similar mutants that reverse the function of POU-V factors do naturally occur. A search for sequence variants within an 11.3-kb region of the human OCT3/4 gene revealed a high degree of polymorphism, but no sequence variation was detected in exons 2 and 3 encoding the POU-specific domain (44). However, a novel Oct3/4 transcript retaining the complete 225-bp intron 2 sequence as a putative novel exon has recently been described (45). This insertion disturbs the coding sequence starting at the center of the POU domain. The biological significance of this novel splice variant is not yet known, but it is expressed in human ES cells and is down-regulated following the onset of differentiation.
The strong activation of the differentiation genes by Oct25ΔPOU(273–301) and Oct25(C274P) raises the question for the underlying molecular mechanisms. Formation of a partial secondary axis lacking heads but including cement gland structures is reminiscent of the blockage of the BMP4 pathway by a dominant-negative type II receptor (46). Also, suppression of epidermal keratin and expansion of the neural field as shown by Xsox2 after overexpression of Oct25ΔPOU(273–301) are characteristic of an inhibition of the BMP4 signaling pathway. In line with that, we were able to rescue the BMP4 overexpression phenotype by co-injection of Oct25ΔPOU(273–301).
How the dorsalizing mutant can stimulate transcription of genes that promote germ layer formation and dorso-anteriorization of embryos is still not clear. A canonical viewpoint for the transcriptional regulation of genes by POU family proteins is that they can interact with the enhancers via the octamer motif, with the POU domain binding the ATGC half-site and the Hox domain binding the AAAT half-site. The POU domain is composed of a cluster of four α-helices, and the Hox domain also contains three α-helices. In each domain, helices α2 and α3 form a typical helix-turn-helix motif for DNA interaction. In the POU protein Pit-1, amino acids Ser, Gln, Thr, and Arg in helix α3 of the POU domain and the amino acids Arg, Val, Cys, Asn, and Gln in helix α3 of the Hox domain directly contact DNA base pairs (47). Interestingly, these α3 helices are identical in all POU family proteins, including those in subclass V (supplemental Fig. S6). This reflects that the helix structure is crucial for DNA binding, and its destruction will essentially lead to the loss of DNA interaction. Consequently, all mutants with changes in the POU or Hox domain with the exception of C274S cannot bind the promoter via the octamer sequence anymore. However, only those mutants with alterations in the POU domain demonstrate a reversed activity in the regulation of genes involved in mesendoderm formation. Hence, the observed dominant-negative effect cannot be solely interpreted by the loss of DNA interaction. However, there might exist DNA sequences other than the canonical octamer motif for protein binding, and mutants with disruption of the POU domain might bind artificially via their Hox domain to other target sites. But when the Hox domain except for the NLS is removed from Oct25(C274P), the resulting mutant Oct25(C274P)ΔHox(330–381) still exhibits dorsalizing activity (supplemental Fig. S3C). Thus, it is excluded that disruption of the POU domain leads to artificial binding of the Hox domain and that the dorsalized phenotype is due to alternative DNA binding.
Stimulation of differentiation genes by Oct25ΔPOU(273–301) is severely compromised in response to functional knockdown of VegT or blocking of the β-catenin and nodal/activin signaling pathways. We have previously reported that Oct25 forms complexes with VegT, TCF3, FoxH1, or Smad transducers to block transcription of VegT, β-catenin, or nodal/activin target genes (15, 16). Interestingly, the dorsalizing mutants Oct25(C274P) and Oct25ΔPOU(273–301) can still interact with these proteins like the wild type Oct25. Moreover, the dorsalizing mutant Oct25ΔPOU(273–301) stimulates the Xnr1 promoter constructs in which the Oct-binding site, the TCF-binding site, or the Tbox binding site are missing. Even removal of both the TCF- and Tbox-binding sites does not abolish promoter activity. However, this promoter is also subject to regulation by the nodal/activin signaling pathway. Indeed, we could show that the stimulation obtained with the dorsalizing mutant is severely reduced by dnXAR1. We assume that the activity of the dorsalizing mutants is not due to direct interaction with DNA but must be mediated by protein/protein interactions. This assumption is further supported by previous observations that Oct25 represses gene transcription also in the absence of the octamer motif (15, 16). Co-repressors or co-activators that are differentially recruited to the regulatory complexes of Oct25 or of the dorsalizing mutants could lead to an opposite effect on the regulation of corresponding genes. It will therefore be interesting to investigate further components of the regulatory complex containing Oct25 mutants that promote dorsalization and formation of a partial secondary axis.
Supplementary Material
Acknowledgments
We thank Cornelia Donow, Nicole Heymann, Sabine Schirmer, and Xuena Zhang for skillful technical assistance. We are grateful to Drs. K. Cho, P. ten Dijke, and M. Taira for their generous gifts of plasmids.
This work was supported by Deutsche Forschungsgemeinschaft Grants SFB497/A1 (to W. K.) and SFB497/B9 (to F. O.), the European Social Fund, and by the Ministry of Science, Research and the Arts Baden-Württemberg Margarete von Wrangell scholarship (to K. B.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1–S6 and Tables S1 and S2.
- BMP
- bone morphogenetic protein
- GST
- glutathione S-transferase
- RT
- reverse transcriptase
- qRT
- quantitative RT-PCR
- NLS
- nuclear localization signal
- ES
- embryonic stem
- Hox
- homeobox
- GFP
- green fluorescent protein
- aa
- amino acid
- EMSA
- electrophoretic mobility shift assay.
REFERENCES
- 1.Clements D., Friday R. V., Woodland H. R. (1999) Development 126, 4903–4911 [DOI] [PubMed] [Google Scholar]
- 2.Hyde C. E., Old R. W. (2000) Development 127, 1221–1229 [DOI] [PubMed] [Google Scholar]
- 3.White R. J., Sun B. I., Sive H. L., Smith J. C. (2002) Development 129, 4867–4876 [DOI] [PubMed] [Google Scholar]
- 4.Behrens J., von Kries J. P., Kühl M., Bruhn L., Wedlich D., Grosschedl R., Birchmeier W. (1996) Nature 382, 638–642 [DOI] [PubMed] [Google Scholar]
- 5.Wodarz A., Nusse R. (1998) Annu. Rev. Cell Dev. Biol. 14, 59–88 [DOI] [PubMed] [Google Scholar]
- 6.Agius E., Oelgeschläger M., Wessely O., Kemp C., De Robertis E. M. (2000) Development 127, 1173–1183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Takahashi S., Yokota C., Takano K., Tanegashima K., Onuma Y., Goto J., Asashima M. (2000) Development 127, 5319–5329 [DOI] [PubMed] [Google Scholar]
- 8.Wessely O., Agius E., Oelgeschläger M., Pera E. M., De Robertis E. M. (2001) Dev. Biol. 234, 161–173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.De Robertis E. M., Larraín J., Oelgeschläger M., Wessely O. (2000) Nat. Rev. Genet. 1, 171–181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hinkley C. S., Martin J. F., Leibham D., Perry M. (1992) Mol. Cell. Biol. 12, 638–649 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Whitfield T., Heasman J., Wylie C. (1993) Dev. Biol. 155, 361–370 [DOI] [PubMed] [Google Scholar]
- 12.Whitfield T. T., Heasman J., Wylie C. C. (1995) Dev. Biol. 169, 759–769 [DOI] [PubMed] [Google Scholar]
- 13.Cao Y., Knöchel S., Donow C., Miethe J., Kaufmann E., Knöchel W. (2004) J. Biol. Chem. 279, 43735–43743 [DOI] [PubMed] [Google Scholar]
- 14.Cao Y., Siegel D., Knöchel W. (2006) Mech. Dev. 123, 614–625 [DOI] [PubMed] [Google Scholar]
- 15.Cao Y., Siegel D., Donow C., Knöchel S., Yuan L., Knöchel W. (2007) EMBO J. 26, 2942–2954 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cao Y., Siegel D., Oswald F., Knöchel W. (2008) J. Biol. Chem. 283, 34168–34177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nichols J., Zevnik B., Anastassiadis K., Niwa H., Klewe-Nebenius D., Chambers I., Schöler H., Smith A. (1998) Cell 95, 379–391 [DOI] [PubMed] [Google Scholar]
- 18.Pesce M., Schöler H. R. (2001) Stem Cells 19, 271–278 [DOI] [PubMed] [Google Scholar]
- 19.Pan G. J., Chang Z. Y., Schöler H. R., Pei D. (2002) Cell Res. 12, 321–329 [DOI] [PubMed] [Google Scholar]
- 20.Takahashi K., Yamanaka S. (2006) Cell 126, 663–676 [DOI] [PubMed] [Google Scholar]
- 21.Takahashi K., Tanabe K., Ohnuki M., Narita M., Ichisaka T., Tomoda K., Yamanaka S. (2007) Cell 131, 861–872 [DOI] [PubMed] [Google Scholar]
- 22.Wernig M., Meissner A., Foreman R., Brambrink T., Ku M., Hochedlinger K., Bernstein B. E., Jaenisch R. (2007) Nature 448, 318–324 [DOI] [PubMed] [Google Scholar]
- 23.Yu J., Vodyanik M. A., Smuga-Otto K., Antosiewicz-Bourget J., Frane J. L., Tian S., Nie J., Jonsdottir G. A., Ruotti V., Stewart R., Slukvin I. I., Thomson J. A. (2007) Science 318, 1917–1920 [DOI] [PubMed] [Google Scholar]
- 24.Park I. H., Zhao R., West J. A., Yabuuchi A., Huo H., Ince T. A., Lerou P. H., Lensch M. W., Daley G. Q. (2008) Nature 451, 141–146 [DOI] [PubMed] [Google Scholar]
- 25.Kim J. B., Sebastiano V., Wu G., Araúzo-Bravo M. J., Sasse P., Gentile L., Ko K., Ruau D., Ehrich M., van den Boom D., Meyer J., Hübner K., Bernemann C., Ortmeier C., Zenke M., Fleischmann B. K., Zaehres H., Schöler H. R. (2009) Cell 136, 411–419 [DOI] [PubMed] [Google Scholar]
- 26.Morrison G. M., Brickman J. M. (2006) Development 133, 2011–2022 [DOI] [PubMed] [Google Scholar]
- 27.Wegner M., Drolet D. W., Rosenfeld M. G. (1993) Curr. Opin. Cell Biol. 5, 488–498 [DOI] [PubMed] [Google Scholar]
- 28.Nishimoto M., Miyagi S., Katayanagi T., Tomioka M., Muramatsu M., Okuda A. (2003) Biochem. Biophys. Res. Commun. 302, 581–586 [DOI] [PubMed] [Google Scholar]
- 29.Ambrosetti D. C., Schöler H. R., Dailey L., Basilico C. (2000) J. Biol. Chem. 275, 23387–23397 [DOI] [PubMed] [Google Scholar]
- 30.Okumura-Nakanishi S., Saito M., Niwa H., Ishikawa F. (2005) J. Biol. Chem. 280, 5307–5317 [DOI] [PubMed] [Google Scholar]
- 31.Okamoto K., Okazawa H., Okuda A., Sakai M., Muramatsu M., Hamada H. (1990) Cell 60, 461–472 [DOI] [PubMed] [Google Scholar]
- 32.Imagawa M., Miyamoto A., Shirakawa M., Hamada H., Muramatsu M. (1991) Nucleic Acids Res. 19, 4503–4508 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Vigano M. A., Staudt L. M. (1996) Nucleic Acids Res. 24, 2112–2118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Harland R. M. (1991) Methods Cell Biol. 36, 685–695 [DOI] [PubMed] [Google Scholar]
- 35.McKendry R., Hsu S. C., Harland R. M., Grosschedl R. (1997) Dev. Biol. 192, 420–431 [DOI] [PubMed] [Google Scholar]
- 36.Pan G., Qin B., Liu N., Schöler H. R., Pei D. (2004) J. Biol. Chem. 279, 37013–37020 [DOI] [PubMed] [Google Scholar]
- 37.Ring C., Ogata S., Meek L., Song J., Ohta T., Miyazono K., Cho K. W. (2002) Genes Dev. 16, 820–835 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hemmati-Brivanlou A., Melton D. A. (1992) Nature 359, 609–614 [DOI] [PubMed] [Google Scholar]
- 39.Molenaar M., van de Wetering M., Oosterwegel M., Peterson-Maduro J., Godsave S., Korinek V., Roose J., Destrée O., Clevers H. (1996) Cell 86, 391–399 [DOI] [PubMed] [Google Scholar]
- 40.Wacker S. A., McNulty C. L., Durston A. J. (2004) Dev. Biol. 266, 123–137 [DOI] [PubMed] [Google Scholar]
- 41.Niwa H., Masui S., Chambers I., Smith A. G., Miyazaki J. (2002) Mol. Cell. Biol. 22, 1526–1536 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Boiani M., Schöler H. R. (2005) Nat. Rev. Mol. Cell Biol. 6, 872–884 [DOI] [PubMed] [Google Scholar]
- 43.Jaenisch R., Young R. (2008) Cell 132, 567–582 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hussain S. K., Sequerra R., Bertucci C., Hastings N. C., Rieder M., Schwartz S. M. (2008) BMC Genet. 9, 15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Atlasi Y., Mowla S. J., Ziaee S. A., Gokhale P. J., Andrews P. W. (2008) Stem Cells 26, 3068–3074 [DOI] [PubMed] [Google Scholar]
- 46.Frisch A., Wright C. V. (1998) Development 125, 431–442 [DOI] [PubMed] [Google Scholar]
- 47.Jacobson E. M., Li P., Leon-del-Rio A., Rosenfeld M. G., Aggarwal A. K. (1997) Genes Dev. 11, 198–212 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.