Abstract
We report the first crystal structure of a 1:2 hormone·receptor complex that involves prolactin (PRL) as the ligand, at 3.8-Å resolution. Stable ternary complexes were obtained by generating affinity-matured PRL variants harboring an N-terminal tail from ovine placental lactogen, a closely related PRL receptor (PRLR) ligand. This structure allows one to draw up an exhaustive inventory of the residues involved at the PRL·PRLR site 2 interface, consistent with all previously reported site-directed mutagenesis data. We propose, with this description, an interaction model involving three structural components of PRL site 2 (“three-pin plug”): the conserved glycine 129 of helix α3, the hydrogen bond network involving surrounding residues (glycine cavity), and the N terminus. The model provides a molecular basis for the properties of the different PRL analogs designed to date, including PRLR antagonists. Finally, comparison of our 1:2 PRL·PRLR2 structure with those of free PRL and its 1:1 complex indicates that the structure of PRL undergoes significant changes when binding the first, but not the second receptor. This suggests that the second PRLR moiety adapts to the 1:1 complex rather than the opposite. In conclusion, this structure will be a useful guiding tool for further investigations of the molecular mechanisms involved in PRLR dimerization and activation, as well as for the optimization of PRLR antagonists, an emerging class of compounds with high therapeutic potential against breast and prostate cancer.
Keywords: Cell Surface Receptor, Hormones, Ligand-binding Protein, Protein Structure, Receptor Structure-function, Antagonist, Cytokine Receptor, Dimerization
Introduction
Competitive prolactin (PRL)5 receptor antagonists have recently emerged as a new class of molecules with high therapeutic potential for treating unresolved PRL-sensitive pathologies (1–3). The main indications include hyperprolactinemia resulting from dopamine-resistant prolactinomas and tumors affecting peripheral PRL-target tissues such as the prostate and the breast. In the latter cases, PRL receptor (PRLR) antagonists are currently viewed as one of the most promising approaches to counteract the undesirable growth promoting actions mediated by locally produced PRL (4–7) or by constitutively active PRLR mutants (8).
PRL is a member of a long established hormone family, also including growth hormone (GH) and placental lactogen (PL), with which it shares high structural homology (9, 10). The biochemical, structural, and dynamic properties of GH have been well characterized (11–13). The determination of the x-ray structure of human GH (hGH) bound to the dimerized extracellular domain (ECD) of its receptor (13) was a crucial step toward the understanding of GHR activation mechanism (14). This structure identified two asymmetric binding sites on hGH, called sites 1 and 2, which were proposed to trigger hormone-induced sequential dimerization of the GH receptor, leading to activation (11). More recently, this model has been revisited based on the comparison of unliganded and liganded human GHR-ECD (12); it is now believed that GHR exists as an inactive dimer at the cell membrane, which is activated by hormone-induced relative rotation of subunits within a dimeric receptor (12). In addition to the characterization of the structural changes occurring in GH and GHR upon their interaction, the GH·GHR2 three-dimensional structures have also largely contributed to the development of GHR antagonists. This novel class of molecules consists of modified GH-core proteins, which harbor one steric mutation within binding site 2, namely the replacement of the helix α3 Gly with long and charged residues such as Lys or Arg. Initially introduced in the bovine hormone (G119R-bGH) (15), the Gly substitution in the human hormone was also shown to generate an antagonist (G120K/R-hGH), and the crystal structure of the hGH·hGHR2 complex revealed that the Arg/Lys could actually impede the formation of a functional receptor dimer (14). Subsequently, these studies have resulted in the approval of a GHR antagonist, Somavert® (Pegvisomant for injection), as a drug for the treatment of acromegaly (16).
In the absence of any three-dimensional structure of PRL and more importantly of PRL·PRLR complexes, we initially took advantage of the homology between PRL and GH systems to engineer the first PRLR antagonists. Based on the paradigms established from the GH·GHR interaction, our working hypothesis was that PRL also induced PRLR dimerization by interacting with two receptor molecules via two distinct binding sites. This assumption was supported by mutational studies (10, 17, 18) and by the observation that mutation of the conserved helix α3 glycine (Gly129) also generated PRLR antagonists (19, 20). However, despite obvious similarities, GH·GHR and PRL·PRLR also appeared to have their own distinctive features. For example, the G129R-hPRL antagonist was shown to exhibit significant residual agonistic properties in various PRLR-mediated bioassays (3), whereas, to the best of our knowledge, there are no similar observations for G120R-hGH in GH-responsive bioassays. In addition, although mutation of Gly120 in GHR antagonists does not affect binding affinity for GHR at the cell surface, every mutation of Gly129 tested so far in hPRL was shown to decrease 10-fold their affinity for cell-expressed PRLR, which is clearly detrimental to the efficacy of these antagonists (2, 20).
Understanding the molecular features responsible for these hormone specificities has been repeatedly hampered by the lack of a crystal structure of the 1:2 PRL·PRLR2 complex (21). Indeed, the first structural data reported for the PRL system involved PRLR-ECDs bound to non-PRL ligands: a 1:1 complex between hGH and the hPRLR (22), and a 1:2 complex between ovine PL (oPL) and the rat (r) PRLR (23). As for hPRL, its structure in solution was obtained by NMR, confirming its four anti-parallel α-helix bundle fold (24). More recently, we reported the first set of x-ray coordinates for a PRL-core hormone and determined the structure of the pure antagonist Del1–9-G129R-hPRL (20), and the structure of this compound bound to the hPRLR-ECD (1:1 complex) was then determined (25).
Although titration of hPRL by hPRL-ECD in NMR experiments suggested changes in chemical shifts at high receptor/hormone ratio (24), the lack of structural data for the 1:2 complex between PRL and its receptor has remained detrimental to our understanding of this hormonal system. For example, it has been impossible to this date to evaluate the putative structural changes occurring in the intermediate 1:1 hormone·receptor complex upon interaction with the second receptor. Also, the ligand-receptor interactions involving binding site 2, which is the targeted area for PRLR antagonists generation (3), remain structurally uncharacterized.
One of the recurrent problems encountered for the determination of the PRL·PRLR2 structure is the poor stability of this ternary complex in vitro. Although the formation of 1:2 complexes has been demonstrated by various approaches, including gel filtration, native electrophoresis, and surface plasmon resonance, interaction at binding site 2 appears to be too transient to stabilize ternary complexes (20, 24, 26). To circumvent this problem, we aimed at generating an affinity-matured hPRL variant able to form stable 1:2 complexes. Because the structure of oPL·PRLR2 suggested that the elongated N terminus of oPL could contribute to the binding of the second receptor moiety (23), we iteratively substituted the N-terminal residues of hPRL by their homologs from oPL. We here report that this strategy was successful in increasing significantly hPRL binding site 2 affinity and, thereby, stabilizing the PRL·PRLR2 ternary complex. This allowed us to solve the crystal structure of the 1:2 complex involving the most potent affinity-matured hPRL and the rPRLR-ECD.
EXPERIMENTAL PROCEDURES
Site-directed Mutagenesis
N-terminal mutations (replacements and elongations) were performed using the QuikChange II mutagenesis kit from Stratagene (La Jolla, CA). Mutations were introduced iteratively using the pT7L-hPRL expression plasmid as initial template (27). Forward and reverse (complementary) primers as well as iterative templates are shown in Table 1. After transformation, Escherichia coli BL21(DE3) colonies were analyzed for their DNA content; plasmids were sequenced to verify the presence of the expected mutations. Sequences encoding the ECD of human or rat PRLR were inserted into the pQE-70 expression plasmid (Qiagen) containing a His6 tag at the C-terminal end as described (20, 24). Subcloning constraints led to the addition of 4 amino acids just before the His6 tag (Gly211-Ser212-Arg213-Ser214 for hPRLR-ECD and Arg211-Ser212-Arg213-Ser214 for rPRLR-ECD).
TABLE 1.
Mutagenesis primers
Mutated nucleotides are underlined.
| hPRL variants | Template | Primers |
|---|---|---|
| Nter | hPRL | FOR 5′ GA GAT ATA CAT ATG GCA CAG CAT CCA CCA TAC TGT CCC GGC GGG G 3′ |
| REV 5′ C CCC GCC GGG ACA G TA TGG TGG ATG CTG TGC CAT ATG TAT ATC TC 3′ | ||
| T14P | hPRL | FOR 5′ CGA TGC CAG GTG CCT CTT CGA GAC CTG TTT GAC C 3′ |
| REV 5′ GGT CAA ACA GGT CTC GAA GAG GCA CCT GGC ATC G 3′ | ||
| PGGA | Nter | FOR 5′ CAT CCA CCA TAC TGT CGA AAC CAG CCA GCC CGA TGC CAG GTG 3′ |
| REV 5′ CAC CTG GCA TCG GGC TGG CTG GTT TCG ACA GTA TGG TGG ATG 3′ | ||
| PGGA-T14P | PGGA | FOR 5′ CGA TGC CAG GTG CCT CTT CGA GAC CTG TTT GAC C 3′ |
| REV 5′ GGT CAA ACA GGT CTC GAA GAG GCA CCT GGC ATC G 3′ | ||
| Full oPL | PGGA-T14P | FOR 5′ GT CGA AAC CAG CCA GGC AAA TGC CAG ATC CCT CTT CGA GAC C 3′ |
| REV 5′ G GTC TCG AAG AGG GAT CTG GCA TTT GCG TGG CTG GTT TCG AC 3′ |
Production of Recombinant Proteins
Recombinant hPRL (WT and variants) and PRLR ECDs were overexpressed in 0.5- to 1-liter cultures of E. coli BL21(DE3) or M15(REP4), respectively, and purified as described previously (20). Briefly, proteins were overexpressed as insoluble inclusion bodies, which were solubilized in 8 m urea (5 min at 55 °C, then 2 h at room temperature) and refolded by continuous dialysis (72 h, 4 °C) against 100 volumes of 25 mm NH4HCO3, pH 8.6. Solubilized proteins were then loaded onto a HiTrapQ anion-exchange column (Amersham Biosciences) equilibrated in 25 mm NH4HCO3, pH 8.6. Human PRL (WT and variants) and receptor ECDs were eluted along a NaCl gradient (0–500 mm), and the major peak was collected, quantified, and kept frozen until use. Purity of the various hPRL variant/receptor ECD batches was >95% as judged from SDS-PAGE analysis.
Surface Plasmon Resonance
The rat and human PRLR-ECDs were immobilized covalently onto nitrilotriacetic acid-derivatized nitrilotriacetic acid sensor chips as described previously (20). Surface densities ranging from 750 to 2300 resonance units (≈ pg.mm−2) were obtained.
Binding assays were performed at least in triplicate as previously described (20). Briefly, for site 1 characterization, the hPRL variants (concentrations ranging from 0.78 to 200 nm) were injected onto the PRLR-ECD surfaces. For sites 2+3 characterization, the PRLR-ECD surface was first saturated with each hPRL variant (350 nm). Rat or human PRLR-ECDs (concentrations ranging from 23 nm to 28.6 μm) were then injected for 8 min onto the pre-formed PRLR-ECD·PRL complexes, followed by a 5-min dissociation period. No nonspecific signal could be detected when injecting PRLR-ECD over the PRLR-ECD surface in the absence of hPRL.
The association and dissociation profiles were double-referenced using the Scrubber 2.0 software (BioLogic Software), i.e. both the signals from the reference surface (ethanolamine derivatized) and from blank experiments (using running buffer instead of protein) were subtracted. The steady-state SPR responses (Req, experimental or extrapolated) were plotted against the concentration (C) of the PRL variant (site 1) or of the PRLR-ECD (site 2) and fitted using the following equation, Req = (Rmax * C)/(Kd + C), where Kd is the equilibrium dissociation constant, and Rmax the maximal binding capacity of the surface. Kinetic parameters (kon and koff) were determined using a nonlinear least squares algorithm implemented in the BIAevaluation 4.1 software (Biacore). Further details on the equations used for data fitting are provided as supplemental information.
Analytical Ultracentrifugation
Sedimentation velocity experiments were carried out at 25 °C in an XL-I analytical ultracentrifuge (Beckman-Coulter) equipped with double UV and Rayleigh interference detection. Samples were prepared in 25 mm NH4HCO3, 150 mm NaCl, pH 8.6, and spun using an An60Ti rotor and 12-mm double-sector aluminum centerpieces. The partial specific volume of hPRLFull-oPL (0.736 ml.g−1), rPRLR-ECD (0.728 ml.g−1), and their complexes (0.732 ml.g−1) were estimated from their amino acid sequences using the software Sednterp 1.09 (available on-line from The Boston Biomedical Research Institute). The same software was used to estimate the buffer viscosity (η = 1.002 cP) and density (ρ = 1.0032 g.ml−1). hPRLFull-oPL (300 μl at 20 μm) and rPRLR-ECD (300 μl at 20 μm) were spun at 50,000 rpm, whereas the hPRLFull-oPL·rPRLR-ECD mixture (300 μl at 5 μm/15 μm) was spun at 36,000 rpm. Absorbance and interference profiles were recorded every 3 min. Sedimentation coefficient distributions c(s) were determined using the software Sedfit 11.3 (28). Theoretical sedimentation coefficients of the complexes were calculated from the Prl complex PDB file using Hydropro 7c (29) with a hydrated radius of 3.1 Å for the atomic elements.
Crystallization and X-ray Diffraction Data Collection
Initial crystallization screening was performed in 96-well sitting drop crystallization plates (Greiner Bio-One) using a Cybi-Disk robot from Cybio. Crystallization screens were set up using several commercially available high throughput crystallization screening kits (Hampton Research). Small diamond-shaped crystals appeared after 1 month at 18 °C in condition 25 of the MemFac crystallization kit. The crystallization condition was optimized manually in Linbro plates using the hanging drop method. Crystals of 40 × 40 × 80 μm3 final size were recorded at European Synchrotron Radiation Facility (ESRF, Grenoble, France) on ID14 beam line at 3.5-Å resolution. They belonged to space group P21212 with unit cell dimensions of a = 178.31 Å, b = 59.61 Å, c = 72.43 Å. Unfortunately all our efforts to refine the structure were not sufficient to reach acceptable statistics. This was probably due to the low completeness of the data set (80%), because crystals died very rapidly.
A last check of the original Greiner plates after 1 year showed large crystals in different conditions of both MemFac and Index crystallization kits. All of these were tested on the ID14-4 beam line at ESRF without addition of cryoprotectant before flash-freezing in liquid nitrogen. Half of the crystals were protein crystals, but only one did not display high mosaicity and could provide x-ray data. The crystallization reservoir was composed of 100 mm sodium citrate, pH 5.6, 100 mm lithium sulfate, 12% polyethylene glycol 4000, which corresponds to the MemFac no. 13 condition. A complete data set was recorded at 3.7-Å resolution. Despite several attempts to optimize manually the crystallization conditions, none of the newly recorded data sets diffracted better than this one. The recorded images were reduced, scaled, and merged with programs MOSFLM and SCALA (30, 31). The intensities were then converted to the structural factor amplitudes with TRUNCATE (30, 31). This new crystal belonged to space group P43212 with unit cell dimensions of a = b = 92.12 Å, c = 215.85 Å. This crystal is referred to as Prl complex in the text.
Phase Determination and Structure Refinement
The crystal structure of Prl complex was solved by molecular replacement using the program PHASER (32), with the 1:2 complex structure of oPL·rPRLR2 (Protein Data Bank code 1F6F) as search model (23). The complete complex was easily positioned in one block in the unit cell, and the position of each domain was refined by rigid body refinement using CNSv1.1 (33).
Refinement of the structure at 3.8-Å resolution was carried out by multiple cycles of manual rebuilding using the program Coot (34) and refinement using CNSv1.1, resulting in a final model with a R factor of 28.1% and an Rfree factor of 38.2%. These values all together with poor geometry quality led us to proceed to a second structure refinement procedure. The structure issued from the CNS rigid body refinement was used as the first model. This second structure refinement was performed with Phenix (35). The model was build in O (36) with the lego algorithm using structure fragments from the high resolution rebuild data base. Crystallographic refinement was accomplished using the maximum likelihood target with amplitudes. No I/sigma cutoff was applied as weak reflections with large experimental error estimates are automatically down-weighted in the likelihood-based target function. The main improvement in map quality was due to the new bulk solvent procedure that is more robust than in CNS v1.1 due to the grid search to optimize the bulk-solvent parameters ksol and Bsol. Because the number of reflections was not enough to resolve the system, atomic displacement parameters were refined by groups of five residues and periodically reset to the average value.
After several rounds of manual model building in O and refinement in Phenix, the final R and free R factors were lowered to 25.2% and 32.3%, respectively. The geometry is of good quality with 88.9% and 8.9% of the residues in the most favored and allowed regions of the Ramachandran plot, respectively. These values are in accordance with the validation criteria expected at this resolution, especially for structures with only one molecule per unit cell (37).
Most of the amino acid residues have been positioned in the electron density, nevertheless the Prl complex structure presents some missing parts, especially in the hormone (Met(−3)PRL–Gln(−1)PRL, His46PRL–Ile51PRL, Glu140PRL–Glu143PRL, Gln157PRL–Ala159PRL, and Asn198PRL–Cys199PRL), and in the second receptor (Met0PRLR2–Pro3PRLR2, Thr28PRLR2–Gly31PRLR2, Gln115PRLR2–Lys119PRLR2, Thr133PRLR2–Phe140PRLR2, and Met202PRLR2–Ser214PRLR2) (PRLR1 and PRLR2 refer to receptor molecules interacting with hPRL site 1 and site 2, respectively). Concerning PRLR1, because this molecule is involved in tight packing contacts, only the C-terminal region was not visible in the electron density (Pro203PRLR1–Ser214PRLR1).
A summary of the crystallographic data and refinement statistics is given in Table 2. An example of the map quality is presented in supplemental Fig. S1. The refined structure was validated using the program MolProbity (38). The characterization of the secondary structure elements was performed using programs Molscript (39) and Stride (40). The figure panels showing three-dimensional structures were generated using the PyMOL Molecular Graphics System.6
TABLE 2.
Summary of the crystallographic data
| Prl complex | |
|---|---|
| Data collection | |
| Beam line | Id14 ESRF |
| Space group | P43212 |
| Unit cell parameter (Å) | a = b = 92.12, c = 215.85 |
| Resolution (Å) | 3.7–107 (3.7–3.9)a |
| Total no. of reflections | 51,126 (7,639) |
| No. of unique reflections | 10,464 (1,483) |
| Completeness (%) | 99.2 (99.9) |
| Rmergeb (%) | 9.3 (17.6) |
| I/σ(I) | 15.6 (6.5) |
| Multiplicity | 4.9 (5.2) |
| Refinement | |
| Resolution (Å) | 3.80–15 |
| No. of reflections used (%) | 98.7 (9437) |
| R/Rfree (%)c | 25.2/32.3 |
| Cross-validated estimated coordinate error from sigma (Å) | 0.43 |
| No. of residues positioned/total | 569/633 |
| Mean B value (Å2) | 94.1 |
| Root mean square deviation bond lengths (Å) | 0.009 |
| Root mean square deviation bond angles (°) | 1.247 |
| Ramachandran plot (%) | |
| Most favored A/B/C | 88.9 |
| Allowed | 8.9 |
a Values in parentheses are for the highest resolution shell.
b Rmerge = ΣhΣj|〈I〉h − Ih,j|/ΣhΣjIh,j, where 〈I〉h is the mean intensity of symmetry equivalent reflections.
c Σ|Fobs − Fcalc|/ΣFobs. The formula for Rfree is the same as that for R, except that it is calculated with a portion of the structure factors that had not been used for refinement.
RESULTS AND DISCUSSION
Rationale for the Design of hPRL Variants
We focused our mutational strategy on the N-terminal tail of hPRL based on two experimental observations. First, the N terminus of oPL was shown to account for a large part of the energy of interaction with the second PRLR-ECD (23), indicating that it represents an important feature of binding site 2. Second, deletion of the nine N-terminal residues in G129R-hPRL abolished the residual agonistic activity of this partial antagonist, further highlighting the functional role of the PRL N terminus in the receptor activation process (2, 20).
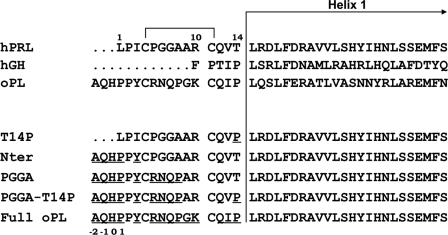
Interestingly, the N-terminal tail is the most divergent region among PRL/GH/PL hormones (10), and non-primate PLs display the longest N terminus (three additional residues compared with PRLs). Sequence comparison shows that three main features distinguish the N-terminal sequences of hPRL versus oPL (Fig. 1). The first difference is the presence of a proline right before the helix α1 of oPL (Pro14, hPRL numbering); variant hPRLT14P was generated to test its potential role. The second difference is the AQHPPY motif located before the first cysteine (Cys4PRL), which elongates the N terminus of oPL by three residues compared with hPRL. Because this AQHPPY motif was identified as an important feature of binding site 2 of oPL (23), the hPRLNter variant was generated by substituting AQHPPY for LPI. The third difference is the short stretch forming the loop constrained on its extremities by the Cys4PRL –Cys11PRL disulfide bond. With the exception of a positive charge in position 10 (ArgPRL/LysoPL), this sequence is not conserved and shifts a proline from position 5 in hPRL (PGGA) to position 8 in oPL (RNQP). Variant hPRLPGGA substitutes RNQP for PGGA in hPRLNter, and hPRLPGGA,T14P also includes the T14P replacement. Finally, the so-called hPRLFull-oPL variant was obtained by substituting the entire oPL N terminus (17 residues) for the native hPRL N-terminal sequence. With respect to hPRLPGGA,T14P, hPRLFull-oPL includes three additional replacements: Ala9 → Gly, Arg10 → Lys, and Val13 → Ile.
FIGURE 1.
N-terminal sequences of hPRL mutants analyzed in this study. Sequences of the N-terminal/helix α1 for 3 natural ligands of the PRL/GH/PL family members are shown. The native N terminus of hPRL was iteratively modified to mimic that of oPL (elongation/mutation). Numbering corresponds to hPRL sequence, numbering for elongated mutants starts from −2 as indicated. The initial methionine Met(−3), found in recombinant proteins, is not indicated.
Structural and Functional Characterization of hPRL Variants
The five hPRL variants were produced and purified with similar yields as hPRL. CD experiments showed that the secondary structure and the thermodynamic parameters for heat denaturation of the different variants were close to those of hPRL (see supplemental Table 1), indicating that the N-terminal mutations did not modify significantly the folding or stability of the hormone. N-terminal mutations also had only a minimal impact on the global affinity for membrane receptors determined by cell-based radio receptor assays, with at most a 2-fold increase in the affinity of elongated variants for both rat and human PRLR (see supplemental Table 2). Accordingly, the biological activity of hPRL variants was not significantly affected, with at most a 4-fold bioactivity decrease for variant hPRLT14P in the bioassay involving the human receptor (see supplemental Table 3).
Identification of Affinity-matured hPRL Variants and Characterization of Their Complexes
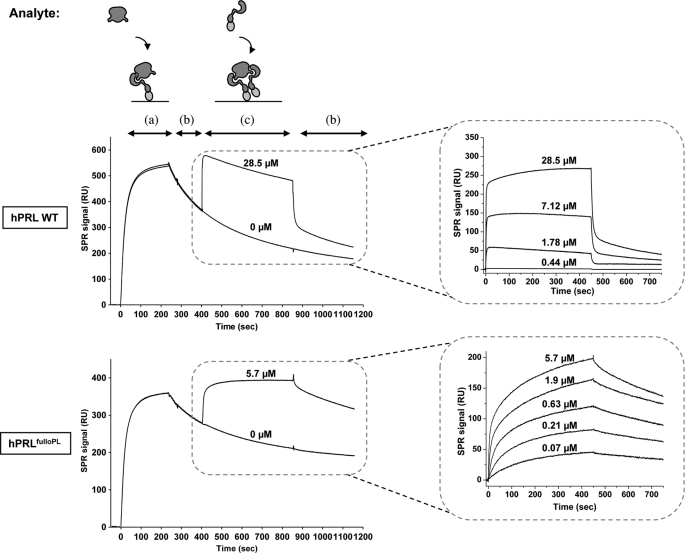
To decipher more specifically the impact of the mutations on site 1 and/or sites 2+3 binding, we used the SPR methodology that we recently set up to monitor sequentially the interaction of hPRL with two separate PRLR ECDs (20). Table 3 summarizes the equilibrium dissociation constants (Kd) of the complexes formed by the different N-terminal variants with the rat or the human PRLR-ECD. For the rPRLR-ECD, site 1 affinities of hPRLT14P, hPRLNter, and hPRLPGGA were close to that of hPRLWT, whereas those of hPRLPGGA,T14P and hPRLFull-oPL were, respectively, 3-fold and 8-fold higher, mostly due to a 5-fold decrease of the dissociation rate koff, from 10−3 s−1 for hPRLWT to 2 × 10−4 s−1 for hPRLFull-oPL (Fig. 2). As for their sites 2 plus 3 affinities, with the exception of hPRLT14P, significant increases were observed for all variants, from 3-fold for hPRLNter and hPRLPGGA to 30-fold for hPRLPGGA,T14P and hPRLFull-oPL (Table 3, Fig. 2, and supplemental Figs. S2 and S3). These very significant improvements could be attributed both to a stabilization of the site 2 interaction and to a faster formation of ECD-ECD contacts (site 3), leading to a significant increase of the half-life of the 1:2 complex formed by hPRLFull-oPL with respect to hPRLWT (see supplemental “Results”). For the human PRLR-ECD, only marginal site 1 or sites 2+3 affinity improvements were observed, 3-fold and 2-fold, respectively, for hPRLFull-oPL (Table 3).
TABLE 3.
Equilibrium dissociation constants of the complexes between the hPRL variants and PRLR-ECD (rat or human)
|
Kd |
||||
|---|---|---|---|---|
| Site 1 | Sites 2+3 | |||
| nm | ||||
| hPRL variants | Rat ECD | Human ECD | Rat ECD | Human ECD |
| WT | 10.2 ± 1.5 | 6.5 ± 1.0 | 11,000 ± 700 | 32,800 ± 1,800 |
| T14P | 12.6 ± 1.3 | ND | 26,300 ± 9,600 | NDa |
| Nter | 8.3 ± 0.4 | 5.9 ± 0.1b | 4,900 ± 1,000 | 32,800 ± 14,700 |
| PGGA | 6.2 ± 1.4 | ND | 3,700 ± 1,900 | ND |
| PGGA-T14P | 3.5 ± 0.2 | ND | 373 ± 72 | ND |
| Full-oPL | 1.2 ± 0.1 | 2.1 ± 0.6 | 312 ± 36 | 16,400 ± 2,800 |
a ND, not determined.
b From Ref. 20.
FIGURE 2.
Real-time SPR measurement of the affinity of hPRL and its affinity matured variant hPRLFulloPL toward rat PRLR-ECD. Left panels: the rat PRLR-ECD was immobilized on a nickel-nitrilotriacetic acid surface, then the hormone (350 nm) (a), buffer (dissociation phase) (b) and rPRLR-ECD (concentrations as indicated) (c) were successively injected onto the surface to measure site 1 (a) and sites 2+3 (c) interactions. The top panels correspond to hPRLWT, and the bottom panels to hPRLFulloPL. Right panels: sites 2+3 binding sensorgrams corresponding to injections of various concentrations of rPRLR-ECD onto pre-formed 1:1 ECD·PRL complexes, involving either hPRLWT (top) or hPRLFulloPL (see supplemental Fig. S2 for other hPRL variants), after subtraction of control sensorgrams corresponding to injections of buffer instead of rPRLR-ECD.
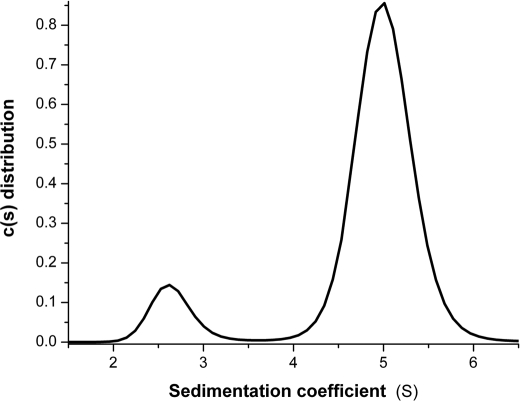
These promising results prompted us to investigate whether hPRLFull-oPL and rPRLR-ECD formed stable 1:2 complexes in solution. A mixture of hPRLFull-oPL and rPRLR-ECD (ratio 1/2) indeed appeared to elute mostly as a 1:2 complex in size-exclusion chromatography (data not shown). Analytical ultracentrifugation experiments were conducted to further characterize the hPRLFull-oPL·rPRLR-ECD complex (Fig. 3). Both hPRLFull-oPL and rPRLR-ECD sedimented as single species compatible with a monomeric form, with respective sedimentation coefficients of 2.4 ± 0.2 S and 2.5 ± 0.1 S. The hPRLFull-oPL·rPRLR-ECD mixture (1/3 ratio), was then analyzed, revealing, apart from residual free species, a larger species with a sedimentation coefficient of 5.0 ± 0.3 S (compatible with a 1:2 complex).
FIGURE 3.
Analytical ultracentrifugation analysis of the hPRLFull-oPL·rPRLR-ECD assembly in solution. Continuous sedimentation coefficient distribution analysis of a hPRLFulloPL·rPRLR-ECD mixture (1/3 molar ratio). Sedimentation coefficients are expressed in Svedbergs, where 1S = 10−13 s.
Global Description of the Prl Complex Structure and Comparison with Other Hormone·Receptor 1:2 Complexes
Having shown that hPRLFull-oPL and rPRLR·ECD could indeed form relatively stable 1:2 complexes, co-crystallization trials were carried out leading to the resolution of the 1:2 hPRLFull-oPL/(r(PRLR1·PRLR2)-ECD) complex structure referred to as Prl complex in the text.
The hydrodynamic characteristics of the Prl complex were calculated from the crystal structure, giving a hydrodynamic radius of 3.8 nm and a sedimentation coefficient of 4.99 S, which is in excellent agreement with the analytical ultracentrifugation experimental value (5.0 ± 0.3 S). The dimensions and shape of the complex are therefore consistent with that of the complex in solution.
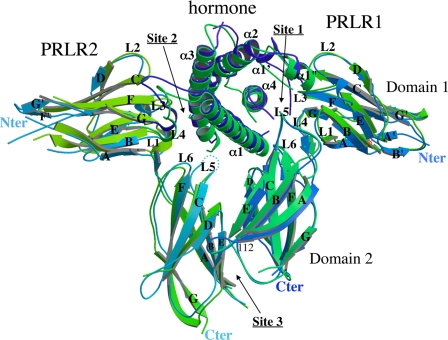
As expected, the structure is globally similar to the 1:2 x-ray structures available for the other members of the hormone/receptor family reported in the PDB (1HWG: hGH·hGHR2; 3HHR: hGH·hGHR2; 1F6F: oPL·rPRLR2; and 1KF9: hGH· hGHR2). The hormone consists of four core α-helices and two long overhand loops, which correspond to the classic long-chain cytokine fold. The two receptors are each divided in two fibronectin type-III domains termed D1 and D2. The D1 (0–98) and D2 (104–205) domains are mainly composed of seven β-strands that form a sandwich of two anti-parallel β-sheets (ABE and DCFG, see Fig. 4). In D1 domains, most of the strands are shorter than in D2, or split in two strands, but the structure is stabilized by the presence of the two disulfide bonds (Cys13–Cys23 and Cys52–Cys63). In the D1 domain of PRLR1, the N terminus (Met0PRLR1–Lys6PRLR1) seems to be highly flexible. In the 1:2 oPL·rPRLR2 structure complex, it is bent at residue Lys6PRLR1, just at the end of strand A′, and thus oriented toward loop 1PRL. In the Prl complex structure it forms an additional β-strand A′ (Pro4PRLR1–Glu8PRLR1) (Fig. 4) aligned with strand A, which occupies a space between the two small strands B′ and G′ pushing them apart but without forming a β-sheet. This particular N terminus conformation is also observed in the 1:1 hPRL·hPRLR complex (3D45)(25).
FIGURE 4.
Secondary structure assignment of the 1:2 Prl complex (hPRLFull-oPL·rPRLR2). The Prl complex (blue) is superimposed on the 1:2 oPL·rPRLR2 complex (green) (1F6F (23)) for comparison.
The only few interactions between domains D1 and D2 involve the interdomain β-strand with loops L1 and L6 forming a hinge around which the two domains can rotate. In PRLR1, this hinge is bordered on one side by interactions between Arg13PRLR1 and both Glu102PRLR1 and Tyr190PRLR1 (just above the conserved WSXWS195 sequence) and on the other side by contacts between Trp139PRLR1 and both Trp72PRLR1 and Lys17PRLR1. The three latter residues are all part of binding site 1. In PRLR2, only some of these interactions are conserved. Arg13PRLR2 is slightly far-off but Glu102PRLR2 and Tyr190PRLR2 are still at van der Waals distances. On the other side of the hinge, as loop L5 is not seen in the electronic density, the contacts are completely different. They are replaced by van der Waals' contacts between Tyr99PRLR2, which is part of binding site 2, with His188PRLR2. On both receptors, the movement between D1 and D2 seems to be governed by residues already involved in hormone-receptor interactions. Consequently, the difference in sequence between the two hormones hPRL and oPL at residues involved in the two binding sites is presumably sufficient to apply a difference in the orientation of the receptors domains. This is in accordance with a conformational change following hormone binding.
The hormone interacts with the two receptors in an asymmetric manner. Binding site 1 is a flat surface of ∼1180 Å2, which is slightly larger than in the oPL·PRLR2 complex (∼1000 Å2), but smaller than in hGH·hGHR2 complex (∼1300 Å2). It is closely similar to that in the recently determined structure of the 1:1 hPRL·hPRLR complex (25), although 7 of 28 residues involved in the binding interface differ between human and rat PRLR (supplemental Fig. S4). Only three substitutions at positions 46, 73, and 74 appear to lead to local modifications of the interaction network sufficient to explain the shift of region Thr52PRL–Asn56PRL. This region, which is part of loop 1PRL (Tyr44–Asn56), was shown for hGH to adopt a defined structure only when the receptor is bound (13). Only Thr52PRL–Asn56PRL is visible in the Prl complex electron density. It adopts a different conformation in the complex than in free PRL, but without forming well defined secondary structure elements.
Finally, the Prl complex globally shows the same receptor-receptor interface (also called stem-stem) as described in the 1:2 complex structure involving rPRLR with oPL (1F6F). It is mainly governed by van der Waals' contacts. Nevertheless, due to tighter packing in this region, an additional loop has been built in Prl complex (Glu112PRLR1–Lys120PRLR1) (Fig. 4). Functional analysis of this region is ongoing based on this structural data.
Binding Site 2
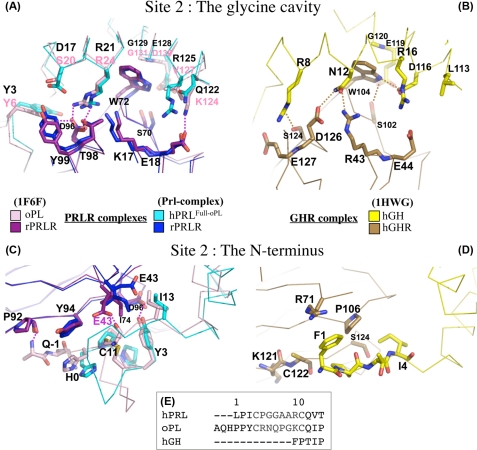
Our SPR experiments showed that binding of PRLR to hPRLFull-oPL site 2 was significantly less tight than at binding site 1. Accordingly, the surface of the PRL·PRLR2 interface was ∼700 Å2, close to that reported in 1:2 complexes involving the two other ligands (oPL and hGH). In our Prl complex structure, binding site 2 can be divided into two structural regions, which share residue Asp96PRL (Fig. 5, A and C): the glycine cavity and the N terminus.
FIGURE 5.
Important binding site 2 contact residues in the complexed hPRL, oPL, and hGH hormones. A and B correspond to the glycine binding pocket. C and D correspond to the hormone N terminus binding interface. In A and C, the superimposition of hPRLFull-oPL·rPRLR2 and oPL·rPRLR2 complex structures are represented. In B and D the hGH·hGHR2 complex is shown. E, alignment of the N-terminal sequences of the three hormones. Color codes are as indicated.
The Glycine Cavity
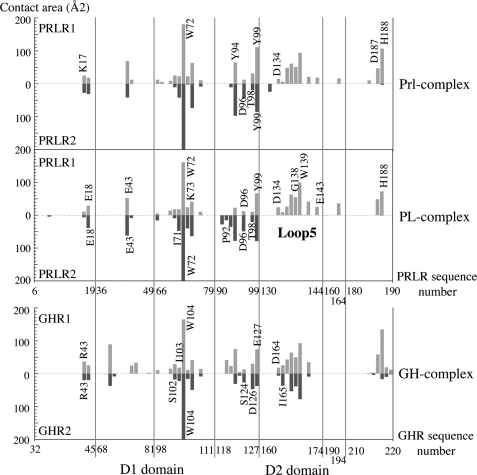
The cavity is made of two individual components: Gly129PRL itself, which forms the bottom of the pocket, and large surrounding amino acid residues, which form the walls. Upon binding, this cavity is filled with Trp72PRLR2 (equivalent to Trp104GHR2). This structural feature is a hallmark of the PRL/GH binding site 2 (Fig. 5, A and B), and its functional importance is highlighted by the fact that all substitutions of Gly129PRL and its GH and oPL homologs prevent receptor docking, leading to receptor antagonists (3, 14, 20). The walls of the cavity involve residues of α-helices 1 and 3 that interact with each other and/or with residues of PRLR2 to form a hydrogen bond network that locks the interaction. The comparison of the three complexes indicates that most of the interacting residues are topologically equivalent, although not strictly sequence conserved (Figs. 5A and 6).
FIGURE 6.
Comparison of receptor residues involved in binding sites 1 and 2 in the three 1:2 complexes of the family. Buried receptor residues in PRL (Prl-complex), PL (PDB code 1F6F), and GH (PDB code 1HWG) complexes are presented according to the sequence alignment. Large portions of the sequences are omitted for clarity.
Despite the fact that oPL·PRLR2 and PRL·PRLR2 complexes are more similar to each other than to the GH·GHR2 complex with respect to primary and tertiary structures, the hydrogen bond network is maintained in the three 1:2 complexes. This network cannot be described at the atomic level in the Prl complex due to the low resolution of the structure. However, it allows the determination that Asp17PRL and Arg21PRL are in close proximity with Asp96PRLR2 and Thr98PRLR2, and thus might be involved in tight interactions. These interactions are conserved in the GH complex, between Arg8GH and Ser124GHR2, Asn12GH, and Asp126GHR2. In the oPL complex, Ser20oPL and Arg24oPL are not in contact with each other, but the interaction between Arg24oPL and both Asp96PRLR2 and Thr98PRLR2 are maintained. This side of the cavity wall is thus made of four residues involved in conserved interactions in the three complexes although residue pairs are different. There is one additional interaction in the Prl complex between Asp17PRL and Tyr99PRLR2, which is observed neither in the oPL complex (due to the shorter side chain of Ser20oPL) nor in the GH complex (due to steric hindrance between Arg8GHand Glu127GHR2).
The other side of the wall is less conserved. In the oPL complex there is a hydrogen bond between Lys124oPL and Glu18PRLR that could also exist in the Prl complex between Arg125PRL and Glu18PRLR as the two residues are in a pre-disposed orientation. But in the GH complex, the equivalent hormone residue (Asp116GH) interacts with Trp104GHR2 instead of Glu44GHR2. A direct interaction with Trp72PRLR2 is also present in the oPL complex, but it involves the carbonyl of Val127PL. Additional interactions strengthening site 2 were also noticed in the GH complex only. These involve a hydrogen bond between Asn12GH and Arg43GHR2 and between Glu119GH and Ser102GHR2 (beyond the central tryptophan residue), a stacking contact between Arg16GH and Trp169GHR2 (Loop L5), and a van der Waals' interaction between Tyr103GH (loop between α-helices 2 and 3) and Ile165GHR2 (loop L5) (not shown on Fig. 5B). Due to the absence of electronic density at the level of loop L5 in PRLR2, and to the very different structure of the loop between α-helices 2 and 3 in the hormones, these interactions were not observed in the Prl complex. This may partly account for the markedly lower affinity of the second receptor monitored by SPR in the hPRL·rPRLR2 complex (11 μm) compared with the hGH·hGHR2 complex (3.8 nm) (41).
In summary, hydrogen bonds surround the tryptophan in both hGH·hGHR2 and oPL·rPRLR2 complexes. In the Prl complex, the equivalent residues are in close proximity compatible with tight interactions on one side of the tryptophan, and a little more spaced out on the other side even if their respective positions could predispose them for interacting.
The N Terminus
Comparison with the oPL·rPRLR2 Complex
The second structural region of binding site 2 involves the N terminus of the hormone (Fig. 5, C and D). Because the affinity-matured hPRLFull-oPL that we crystallized harbored the oPL N terminus, one would have expected the PRL and oPL complexes to exhibit the same interactions at this level, which is not exactly the case (Fig. 5C). There are three hydrogen bonds in the oPL·rPRLR2 complex, between Tyr3PRL and Asp96PRLR2, between Pro92PRLR2 carbonyl and Ala1oPL main chain, and between Cys14PL (equivalent to Cys11PRL) and Glu43PRLR2. The latter cannot occur in the Prl complex due to the very different position of Glu43PRLR2. Furthermore no density could be observed at the level of the N-terminal Ala of hPRLFull-oPL. Consequently, only Tyr3PRL is close enough to interact with the receptor (residue Asp96PRLR2) like in the oPL complex.
Comparison with the hGH·GHR2 Complex
The N terminus of hGH is also involved in the binding to GHR2, although only hydrophobic interactions appear to be involved (Fig. 5D). Pro2GH is at van der Waals' distance from Pro106GHR2, as are Phe1GH from Arg71GHR2, and Cys122GHR2 and Ile4GH from Ser124GHR2. The latter interaction is conserved in the two structures involving PRLR (I1e3PRL–Asp96PRLR2). It is noteworthy that, although the N terminus of hPRL (+9 residues) and oPL (+12) is much longer than that of hGH (Fig. 5E), most of the extra residues are involved in the small N-terminal loop constrained on each extremity by the disulfide bond between Cys4 and Cys11. Because this loop is oriented away from the receptor (Fig. 5C), the part of the N terminus really available for interaction with the receptor is of similar length in hGH and hPRL, and elongated by only three residues in oPL and in the affinity-matured mutant hPRLFull-oPL. Clearly, the substitution of the natural hPRL N terminus by that of oPL stabilized the 1:2 hPRLFull-oPL·rPRLR2 complex by increasing both site 1 and site 2 affinities (Table 3). The addition of a single hydrogen bond is likely responsible for the stabilization of PRLR2 docking. However, although all hPRL mutants but hPRLT14P harbored Gln(−1) and Tyr3 in their sequences, significant increase of site 2 affinity was observed only for hPRLPGGA-T14P and hPRLFull-oPL. This suggests that the entire N-terminal sequence of oPL is required for the N terminus to adopt a suitable folding for the stabilization of the 1:2 complex. Determining the crystal structure of wild-type hPRL complexed to a PRLR-ECD dimer would help in understanding the actual network of interactions generated by the native N terminus hPRL sequence.
The Three-pin Plug Interaction Hypothesis
As described above, three features can be distinguished within the two regions that constitute the binding site 2: the helix α3 glycine 129, the hydrogen bond network involving surrounding residues (glycine cavity) and the N terminus. We attempted to rank the importance of these three components in light of the available mutational data. Clearly, the glycine residue is the major feature of hPRL binding site 2, as highlighted by the fact that any mutation of Gly129 tested to date generated antagonists (20). In contrast, mutations of the two other components failed to generate antagonists, and at best weakened agonistic properties. Truncation of up to the 13 N-terminal residues of hPRL or elongation of three residues (hPRLNter) affected only very marginally its affinity for the full-length transmembrane PRLR or the hPRLR-ECD, as well as its bioactivity in PRLR-mediated cell assays (20, 42). With respect to the hydrogen bond network, we focused on Arg21, which is involved in the only hydrogen bond that was strictly conserved among the three hormone families. We generated R21A and R21W mutants, both of which led to a significant decrease of agonism (10-fold or more) in the Ba/F-hPRLR and HL-5 cell-based bioassays (data not shown), confirming the functional importance of the hydrogen bond between Arg21PRL and Thr98PRLR2.
Despite the fact that mutation G129R induced at least a 150-fold decrease in site 2 binding affinity (that was actually undetectable by SPR (20)), this sole mutation was not sufficient to obtain a pure antagonist (19, 43). This suggests that when the glycine pocket is hindered, one or both other regions can ensure a limited level of receptor triggering. The involvement of the N terminus was experimentally demonstrated, because its deletion (Del1–9-G129R-hPRL) knocked down residual agonism (2), whereas insertion of the AQHPPY motif (G129R-hPRLNter) boosted the residual agonistic activity to twice the level of G129R-hPRL (20). This could suggest the N terminus provides enough residual interaction energy to allow a limited recruitment of the second PRLR, or to induce a limited reorientation of the latter within a pre-formed hPRLR dimer. To test the effect of the hydrogen bond network in residual agonism, we combined R21A and G129R mutations. The double mutant displayed similar residual agonism, but slightly improved antagonism compared with G129R-hPRL (>2 fold, data not shown), which partly agrees with our hypothesis. However, the effect of this double mutation is difficult to interpret precisely, because the replacing residue (Arg129) may partly compensate the loss of Arg21 by establishing hydrogen bonds, which could attenuate the intrinsic effect of mutation R21A alone.
In summary, we can propose that two of the three components of site 2 are needed to achieve detectable agonism, whereas mutation of Gly129PRL is mandatory to generate antagonists. Mutation of the hydrogen network appeared to be more effective in agonists, while the importance of the N terminus was only apparent in antagonists.
Finally, as already observed for the oPL complex, it is noteworthy that the regions of the PRLR domain D1 involved in interactions with PRL binding sites 1 and 2 are very similar, although individual binding residues are not strictly identical (Fig. 6). In the GH complex, this symmetry is extended to the receptor domain D2. This could not be assessed in both oPL and PRL complexes due to the absence of loop L5PRLR2 in the electron density. There are much less mutational data available to be correlated with our structural observations for the receptor than for the ligand. In addition, the residues that were identified as important for hormone binding could not be assigned to a particular binding site, because functional assays were performed using cell-based bioassays (dimerized membrane receptor) and not SPR (monomeric ECD). Mutation of any of the four cysteines of D1 (44), Arg13PRLR, Glu18PRLR, and Phe64PRLR (45) were shown to decrease the affinity for PRL by 300-fold or more. It is likely that the loss of binding observed for the cysteine mutants results from structural alterations of D1, because these cysteines are involved in intramolecular disulfide bonds. This is probably also the case for the mutation of Phe64PRLR, because this residue is clearly outside both binding sites, but is within the core of D1 and in contact with Phe20PRLR, which is close to the two disulfide bonds. Arg13PRLR participates in maintaining the hinge through interactions involving Tyr190PRLR (see above), therefore its mutation could also alter the global folding of the extracellular domain. Finally, Glu18PRLR is the only residue of this short list to be involved in direct interactions with both binding site 1 and site 2 of PRL. In oPL, this residue is also involved in the hydrogen bond network with the hormone residues surrounding the glycine pocket (the second site 2 component that we identified).
Comparison of Free and Bound Hormone Structures
From the numerous hGH structures that already exist for the free hormone (PDB codes 1HUW and 1HGU), 1:1 (PDB codes 1A22, 1HWH, and 1BP3), and 1:2 complexes (PDB codes 1HWG, 1KF9, and 3HHR), it has been proposed (46) that GH structure is modified upon binding to GHR1, leading to a conformation that allows recruitment of GHR2. Our newly determined 1:2 structure provides the clues to address this issue for the PRL·PRLR2 complex. To that end, we superimposed free and bound hormones, choosing helix α2 as a superimposition region, because it is the only α-helix not involved in receptor contacts.
Concerning the GH family, as already mentioned, the structures of hGH in the 1:1 and 1:2 complexes are accurately superimposed (data not shown). In contrast, free hGH shows clear differences in the bending of the three α-helices involved in GHR binding. A clear inclination is observed for the upper part of α1, correlated with the same movement of the lower part of α4 despite the absence of direct interaction of those regions with the receptors. This movement seems to be induced by the displacement of the small helix α1′, due to receptor binding at site 1. On the opposite side of the hormone, there is a clear correlated bending of helices α1 and α3 in the regions involved in binding site 2.
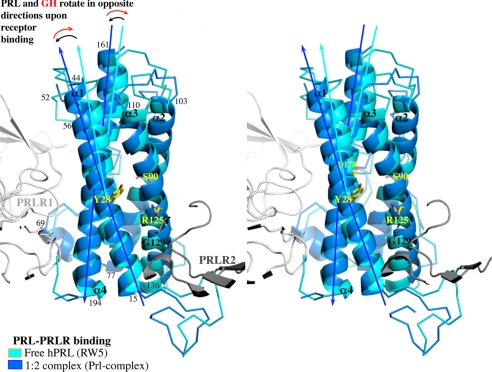
Most of these observations can be transposed to the PRL family. Despite the low resolution of our Prl complex structure, the electron density is clear enough to allow a precise backbone comparison. The structures of PRL in the 1:1 and 1:2 complexes are perfectly superimposed, except for the 18 N-terminal residues, as expected. On the contrary, only helix α2 can be superimposed when comparing free and complexed PRL. The difference in the bending of the three other α-helices is much more pronounced than in the GH family. Most interestingly, the three α-helices bend in opposite ways in PRL and GH upon binding (Fig. 7).
FIGURE 7.
Comparison of secondary structure elements between free and receptor bound PRL hormones. Stereo view of the superimposition of free hPRL (PDB code 1RW5, cyan) with bound hPRL from Prl complex (blue). Residues indicated in yellow correspond to the hinge of each α-helix. Residue numbers indicated in black correspond to the limits of the α-helices. The arrows indicate the rotation of α-helices between the free and the bound states.
Finally, we compared the three hormones (hPRL, oPL, and hGH) within their complexes with PRLR (supplemental Fig. S5). Although hPRL and oPL are complexed with two rat PRLR-ECDs, whereas hGH is bound to only one human PRLR-ECD, the comparison is relevant as both PRL and GH adopt the same structure in their respective 1:1 and 1:2 complexes (at the exception of the N terminus). Helix α2 from the three complexes was easily superimposed. Most of the differences between the three hormones are localized in the non-interacting regions with the exception of region 52–56PRL (38–47GH) from loop 1, which corresponds to the additional binding site 1 region, called α1′, in the PRL·PRLR2 complex when compared with the GH·GHR2 complex. This region, which moves to accommodate loop L2PRLR, corresponds to the most noticeable difference in GH between the hGH·hGHR2 and hGH·hPRLR2 complexes (46) confirming it is specific to PRLR binding.
The curvature of helix 3 is completely different between the three hormones, but the region involved in the glycine cavity is closely superimposable. This suggests that it is PRLR2 that accommodates the hormone and not the opposite. This would explain why the structure of the hormone is perfectly superimposed in all the 1:1 and 1:2 complexes available for both the PRL and GH families, with the exception of the N terminus for the PRL family, which confirms the importance of this region in the stability of the 1:2 complex. The 1:2 complex structure between GH and PRLR would be necessary to verify this hypothesis.
When comparing the intramolecular hydrogen bonds among the different PRL forms (free or liganded), we found that the conserved helix-interacting residues correspond or are close to α-helix hinges (Fig. 7). One additional conserved interaction involves a residue from loop 1. This link between Thr60PRL and Asp178PRL reinforces the attachment of loop 1 with helix α4 in addition to the disulfide bond between Cys58PRL and Cys174PRL. The hinge positions are conserved in the GH family except for helix α3 where the hinge is localized two residues further. Consequently, movements of α-helix half pieces are coordinated. When PRLR interacts with PRL site 1, it imposes the final structure of segment 178–199PRL from helix α4 and of region 52–56PRL from loop 1. The movement of this second region induces the correlated bending of the “upper part” of the hormone (28–44PRL with 161–178PRL and with 110–125PRL; see Fig. 7). As for the movement of segment 178–199PRL, it drags along the “lower part” of the hormone (regions 15–28PRL and 125–136PRL). Region 15–28PRL reaches its final position only when the second PRLR-ECD interacts at binding site 2.
The intrinsic dynamics of the hormone, favored by the presence of the two long loops, seems to be a prerequisite for its fully functional interaction with the receptor, as hypothesized by Jomain et al. (20) and by the antagonistic properties of the Δ41–52-hPRL (47). As evidenced by the superimposition of the three different hormones in complex with PRLR receptor, the binding of receptor 1 seems to impose the final structure of the hormone, at least at the level of binding site 1. The second receptor then appears to accommodate itself to the 1:1 complex and the N terminus of the hormone locks the 1:2 complex in its final conformation.
CONCLUSION
In this study, we report the first x-ray structure of a 1:2 PRL·PRLR2 complex that involves PRL as the hormone ligand. This structure provides different valuable insights. First, it localizes precisely the two hormone/receptor interfaces (binding sites), allowing to interpret from a structural perspective the site-directed mutagenesis data that has been gathered along the years to determine the hot-spots of each of the two hPRL binding sites. The structure is globally in good agreement with these experimental data, and provides a more exhaustive picture of the residues involved in both receptor/hormone interfaces. In particular, it provides structural support for the key functional role of the glycine 129 pocket in binding site 2, and further clarifies the functional role of the N terminus of hPRL. Second, it allows us to better understand the molecular basis of the properties of the different hormone analogs that have been designed to date, including receptor antagonists, such as Del1–9-G129R-hPRL (2, 20). Third, it suggests that, as described for the GH·GHR system, the binding of the second PRLR molecule to the 1:1 PRL·PRLR complex does not modify the structure of the hormone, suggesting that the receptor adapts to the 1:1 complex rather than the opposite. Altogether, this structure will also be useful for further improving current PRLR antagonists or for designing new ones that may act by novel molecular mechanisms. It will also be a precious tool in attempting to understand the mechanism by which the natural PRLR variant comprising the I146L mutation in its extracellular domain acquires constitutive activity (8).
Supplementary Material
Acknowledgment
We acknowledge the European Synchrotron Radiation Facility for providing access to the ID14 beam line.
This work was supported in part by INSERM, University Paris Descartes, and the Agence Nationale de la Recherche (Grant ANR-PCV07_183953).

The on-line version of this article (available at http://www.jbc.org) contains supplemental text, Figs. S1–S5, Tables 1–3, and Refs. 1–6.
The nucleotide sequence(s) reported in this paper has been submitted to the GenBankTM/EBI Data Bank with accession number(s).
W. L. DeLano (2002) The PyMOL Molecular Graphics System, DeLano Scientific LLC, San Carlos, CA.
- PRL
- prolactin
- GH
- growth hormone
- PL
- placental lactogen
- PRLR
- prolactin receptor
- GHR
- growth hormone receptor
- ECD
- extracellular domain
- h
- human
- r
- rat
- o
- ovine
- WT
- wild type
- SPR
- surface plasmon resonance.
REFERENCES
- 1.Goffin V., Touraine P., Culler M. D., Kelly P. A. (2006) Nat. Clin. Pract. Endocrinol. Metab. 2, 571–581 [DOI] [PubMed] [Google Scholar]
- 2.Bernichtein S., Kayser C., Dillner K., Moulin S., Kopchick J. J., Martial J. A., Norstedt G., Isaksson O., Kelly P. A., Goffin V. (2003) J. Biol. Chem. 278, 35988–35999 [DOI] [PubMed] [Google Scholar]
- 3.Goffin V., Bernichtein S., Touraine P., Kelly P. A. (2005) Endocr. Rev. 26, 400–422 [DOI] [PubMed] [Google Scholar]
- 4.Clevenger C. V., Chang W. P., Ngo W., Pasha T. L., Montone K. T., Tomaszewski J. E. (1995) Am. J. Pathol. 146, 695–705 [PMC free article] [PubMed] [Google Scholar]
- 5.Clevenger C. V., Furth P. A., Hankinson S. E., Schuler L. A. (2003) Endocr. Rev. 24, 1–27 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ginsburg E., Vonderhaar B. K. (1995) Cancer Res. 55, 2591–2595 [PubMed] [Google Scholar]
- 7.Dagvadorj A., Collins S., Jomain J. B., Abdulghani J., Karras J., Zellweger T., Li H., Nurmi M., Alanen K., Mirtti T., Visakorpi T., Bubendorf L., Goffin V., Nevalainen M. T. (2007) Endocrinology 148, 3089–3101 [DOI] [PubMed] [Google Scholar]
- 8.Bogorad R. L., Courtillot C., Mestayer C., Bernichtein S., Harutyunyan L., Jomain J. B., Bachelot A., Kuttenn F., Kelly P. A., Goffin V., Touraine P. (2008) Proc. Natl. Acad. Sci. U.S.A. 105, 14533–14538 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Nicoll C. S., Mayer G. L., Russell S. M. (1986) Endocr. Rev. 7, 169–203 [DOI] [PubMed] [Google Scholar]
- 10.Goffin V., Shiverick K. T., Kelly P. A., Martial J. A. (1996) Endocr. Rev. 17, 385–410 [DOI] [PubMed] [Google Scholar]
- 11.Wells J. A. (1996) Proc. Natl. Acad. Sci. U.S.A. 93, 1–68552582 [Google Scholar]
- 12.Brown R. J., Adams J. J., Pelekanos R. A., Wan Y., McKinstry W. J., Palethorpe K., Seeber R. M., Monks T. A., Eidne K. A., Parker M. W., Waters M. J. (2005) Nat. Struct. Mol. Biol. 12, 814–821 [DOI] [PubMed] [Google Scholar]
- 13.de Vos A. M., Ultsch M., Kossiakoff A. A. (1992) Science 255, 306–312 [DOI] [PubMed] [Google Scholar]
- 14.Fuh G., Cunningham B. C., Fukunaga R., Nagata S., Goeddel D. V., Wells J. A. (1992) Science 256, 1677–1680 [DOI] [PubMed] [Google Scholar]
- 15.Chen W. Y., Wight D. C., Mehta B. V., Wagner T. E., Kopchick J. J. (1991) Mol. Endocrinol. 5, 1845–1852 [DOI] [PubMed] [Google Scholar]
- 16.Kopchick J. J., Parkinson C., Stevens E. C., Trainer P. J. (2002) Endocr. Rev. 23, 623–646 [DOI] [PubMed] [Google Scholar]
- 17.Goffin V., Struman I., Mainfroid V., Kinet S., Martial J. A. (1994) J. Biol. Chem. 269, 32598–32606 [PubMed] [Google Scholar]
- 18.Goffin V., Norman M., Martial J. A. (1992) Mol. Endocrinol. 6, 1381–1392 [DOI] [PubMed] [Google Scholar]
- 19.Goffin V., Kinet S., Ferrag F., Binart N., Martial J. A., Kelly P. A. (1996) J. Biol. Chem. 271, 16573–16579 [DOI] [PubMed] [Google Scholar]
- 20.Jomain J. B., Tallet E., Broutin I., Hoos S., Van Agthoven J., Ducruix A., Kelly P. A., Kragelund B. B., England P., Goffin V. (2007) J. Biol. Chem. 282, 33118–33131 [DOI] [PubMed] [Google Scholar]
- 21.Goffin V., Martial J. A., Summers N. L. (1995) Prot. Eng. 8, 1215–1231 [DOI] [PubMed] [Google Scholar]
- 22.Somers W., Ultsch M., De Vos A. M., Kossiakoff A. A. (1994) Nature 372, 478–481 [DOI] [PubMed] [Google Scholar]
- 23.Elkins P. A., Christinger H. W., Sandowski Y., Sakal E., Gertler A., de Vos A. M., Kossiakoff A. A. (2000) Nat. Struct. Biol. 7, 808–815 [DOI] [PubMed] [Google Scholar]
- 24.Teilum K., Hoch J. C., Goffin V., Kinet S., Martial J. A., Kragelund B. B. (2005) J. Mol. Biol. 351, 810–823 [DOI] [PubMed] [Google Scholar]
- 25.Svensson L. A., Bondensgaard K., Nørskov-Lauritsen L., Christensen L., Becker P., Andersen M. D., Maltesen M. J., Rand K. D., Breinholt J. (2008) J. Biol. Chem. 283, 19085–19094 [DOI] [PubMed] [Google Scholar]
- 26.Gertler A., Grosclaude J., Strasburger C. J., Nir S., Djiane J. (1996) J. Biol. Chem. 271, 24482–24491 [DOI] [PubMed] [Google Scholar]
- 27.Paris N., Rentier-Delrue F., Defontaine A., Goffin V., Lebrun J. J., Mercier L., Martial J. A. (1990) Biotechnol. Appl. Biochem. 12, 436–449 [PubMed] [Google Scholar]
- 28.Schuck P. (2000) Biophys. J. 78, 1606–1619 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.García de la Torre J., Huertas M. L., Carrasco B. (2000) Biophys. J. 78, 719–730 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Potterton E., Briggs P., Turkenburg M., Dodson E. (2003) Acta Crystallogr. D Biol. Crystallogr. 59, 1131–1137 [DOI] [PubMed] [Google Scholar]
- 31.Collaborative Computational Project 4 (1994) Acta Crystallogr. D Biol. Crystallogr. 50, 760–76315299374 [Google Scholar]
- 32.McCoy A. J., Grosse-Kunstleve R. W., Adams P. D., Winn M. D., Storoni L. C., Read R. J. (2007) J. Appl. Crystallogr. 40, 658–674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Brünger A. T., Adams P. D., Clore G. M., DeLano W. L., Gros P., Grosse-Kunstleve R. W., Jiang J. S., Kuszewski J., Nilges M., Pannu N. S., Read R. J., Rice L. M., Simonson T., Warren G. L. (1998) Acta Crystallogr. D Biol. Crystallogr. 54, 905–921 [DOI] [PubMed] [Google Scholar]
- 34.Emsley P., Cowtan K. (2004) Acta Crystallogr. D Biol. Crystallogr. 60, 2126–2132 [DOI] [PubMed] [Google Scholar]
- 35.Adams P. D., Grosse-Kunstleve R. W., Hung L. W., Ioerger T. R., McCoy A. J., Moriarty N. W., Read R. J., Sacchettini J. C., Sauter N. K., Terwilliger T. C. (2002) Acta Crystallogr. D Biol. Crystallogr. 58, 1948–1954 [DOI] [PubMed] [Google Scholar]
- 36.Jones T. A., Zou J. Y., Cowan S. W., Kjeldgaard M. (1991) Acta Crystallogr. Sect. A 47, 110–119 [DOI] [PubMed] [Google Scholar]
- 37.Read R. J., Kleywegt G. J. (2009) Acta Crystallogr. D Biol. Crystallogr. 65, 140–147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Davis I. W., Leaver-Fay A., Chen V. B., Block J. N., Kapral G. J., Wang X., Murray L. W., Arendall W. B., 3rd, Snoeyink J., Richardson J. S., Richardson D. C. (2007) Nucleic Acids Res. 35, W375–W383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kraulis P. J. (1991) J. Appl. Crystallogr. 24, 946–950 [Google Scholar]
- 40.Heinig M., Frishman D. (2004) Nucleic Acids Res. 32, W500–W502 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Walsh S. T., Jevitts L. M., Sylvester J. E., Kossiakoff A. A. (2003) Protein Sci. 12, 1960–1970 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bernichtein S., Jomain J. B., Kelly P. A., Goffin V. (2003) Mol. Cell Endocrinol. 208, 11–21 [DOI] [PubMed] [Google Scholar]
- 43.Kinet S., Bernichtein S., Kelly P. A., Martial J. A., Goffin V. (1999) J. Biol. Chem. 274, 26033–26043 [DOI] [PubMed] [Google Scholar]
- 44.Rozakis-Adcock M., Kelly P. A. (1991) J. Biol. Chem. 266, 16472–16477 [PubMed] [Google Scholar]
- 45.Rozakis-Adcock M., Kelly P. A. (1992) J. Biol. Chem. 267, 7428–7433 [PubMed] [Google Scholar]
- 46.Kossiakoff A. A., Somers W., Ultsch M., Andow K., Muller Y. A., De Vos A. M. (1994) Protein Sci. 3, 1697–1705 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.DePalatis L., Almgren C. M., Patmastan J., Troyer M., Woodrich T., Brooks C. L. (2009) Protein Expr. Purif. 66, 121–130 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.