Abstract
A gene encoding an l-lysine dehydrogenase was identified in the hyperthermophilic archaeon Pyrococcus horikoshii. The gene was overexpressed in Escherichia coli, and its product was purified and characterized. The expressed enzyme is the most thermostable l-lysine dehydrogenase yet described, with a half-life of 180 min at 100 °C. The product of the enzyme's catalytic activity is Δ1-piperideine-6-carboxylate, which makes this enzyme an l-lysine 6-dehydrogenase (EC 1.4.1.18) that catalyzes the reductive deamination of the ϵ- amino group and a type of NAD-dependent amine dehydrogenase. The three-dimensional structure of the enzyme was determined using the mercury-based multiple-wavelength anomalous dispersion method at a resolution of 2.44 Å in the presence of NAD and sulfate ion. The asymmetric unit consisted of two subunits, and a crystallographic 2-fold axis generated the functional dimer. Each monomer consisted of a Rossmann fold domain and a C-terminal catalytic domain, and the fold of the catalytic domain showed similarity to that of saccharopine reductase. Notably, the structures of subunits A and B differed significantly. In subunit A, the active site contained a sulfate ion that was not seen in subunit B. Consequently, subunit A adopted a closed conformation, whereas subunit B adopted an open one. In each subunit, one NAD molecule was bound to the active site in an anti-conformation, indicating that the enzyme makes use of pro-R-specific hydride transfer between the two hydrides at C-4 of NADH (type A specificity). This is the first description of the three-dimensional structure of l-lysine 6-dehydrogenase as an NAD-dependent amine dehydrogenase.
Keywords: Amino Acid, Bacteria, Dehydrogenase, Enzyme Catalysis, Enzyme Structure, Pyrococcus horikoshii, Crystalline Structure, Dehydrogenase, Hyperthermophile, Lysine 6-Dehydrogenase
Introduction
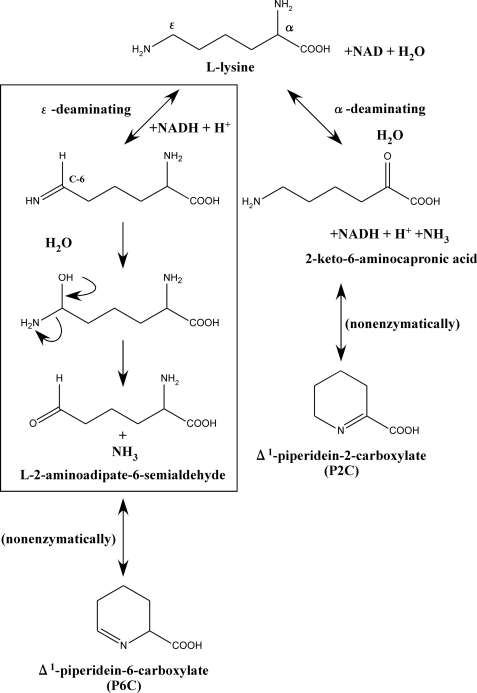
l-Lysine dehydrogenase catalyzes the oxidative deamination of l-lysine in the presence of NAD. To date, two types of l-lysine dehydrogenase have been identified (Fig. 1). The first is l-lysine 6-dehydrogenase (LysDH2; EC 1.4.1.18). This enzyme catalyzes the oxidative deamination of the ϵ-amino group of l-lysine to form l-2-aminoadipate 6-semialdehyde, which in turn nonenzymatically cyclizes to form Δ1-piperideine-6-carboxylate (P6C) (1–3). The second is l-lysine 2-dehydrogenase (EC 1.4.1.15), which catalyzes the oxidative deamination of the α-amino group of l-lysine in the same way other amino acid dehydrogenases do (4). With respect to the latter, there has been one report of an l-lysine 2-dehydrogenase catalyzing the deamination of the α-amino group in humans (5), but because the reaction product of this liver enzyme has not yet been identified, it is still unclear whether or not the enzyme actually catalyzes the oxidative deamination of l-lysine at the α-amino group. By contrast, LysDH has been identified in several microorganisms, and some of those enzymes have been characterized. The first known LysDH was found in Agrobacterium tumefaciens, where it plays a key role in l-lysine metabolism (1). Since then, this enzyme has been extensively characterized (6–9) and utilized for assaying l-lysine (9).
FIGURE 1.
Reaction scheme for the two types of l-lysine dehydrogenase. The proposed mechanism of the P. horikoshii LysDH-catalyzed reaction is boxed.
Recently, the gene encoding LysDH in Geobacillus stearothermophilus was cloned and expressed in Escherichia coli, after which the primary structure of the enzyme was determined (2). Since then, ∼17 different amino acid dehydrogenases have been identified in various organisms (4, 10). Some of these enzymes are useful for enzymatic analysis of amino acids, oxo acids, and special enzymes, as well as for stereospecific synthesis of l-amino acids and their analogs (4, 10, 11). In contrast to the abundant information available for glutamate, alanine, leucine, and phenylalanine dehydrogenases (4), information about the structure and catalytic mechanism of lysine dehydrogenase is still rather limited. LysDH is regarded as a type of NAD(P)-dependent amine dehydrogenase because it catalyzes the dehydrogenation at the ϵ-carbon with deamination, whereas other amino acid dehydrogenases generally catalyze the oxidative deamination of the α-amino group to form the corresponding keto acid. Although quinone- or heme-dependent amine dehydrogenases have been extensively studied (12–15), nothing is currently known about the structural basis for the catalytic mechanism of NAD(P)-dependent amine dehydrogenase.
Within the genomic sequence of an anaerobic hyperthermophilic archaeon, Pyrococcus horikoshii, we found a gene (open reading frame identification number PH1688.1) whose predicted amino acid sequence exhibits 33% identity to that of G. stearothermophilus LysDH. This hyperthermostable LysDH is a potentially useful target for the development of new methods for synthesis of l-pipecolic acid (chiral pharmaceutical intermediate). Up to now, no LysDH has been reported in Archaea, the third domain of life, or in hyperthermophiles. In this study, we expressed the gene, characterized the enzyme produced, and revealed that the enzyme is an extremely thermostable LysDH. We also determined the crystal structure of this enzyme at 2.44-Å resolution in the presence of NAD and sulfate ion. This is the first description of the crystal structure of an NAD(P)-dependent amine dehydrogenase, and it provides new insight into the catalytic mechanism of LysDH.
EXPERIMENTAL PROCEDURES
Cloning and Protein Expression
The gene encoding LysDH (open reading frame identification number PH1688.1; the gene information is available at the DOGAN Genomic Database) was amplified by PCR. The oligonucleotide primers used to amplify the LysDH gene fragment were 5′-GAATCATATGAAAGTCCTTATTTTAGGG-3′, which contains a unique NdeI restriction site overlapping the 5′-initiation codon, and 5′-GTTAAGCTTTTAAGTTCCTTCTATAGATATTCC-3′, which contains a unique HindIII restriction site proximal to the 3′-end of the termination codon. Chromosomal P. horikoshii DNA was isolated as described previously (16) and used as the template. The amplified 1.0-kb fragment was digested with NdeI and HindIII and ligated with the expression vector pColdI (Takara Bio Inc.) linearized with NdeI and HindIII to generate pCLysDH, which was then used to transform E. coli strain Rosetta-gami (DE3) (Novagen). The transformants were cultivated at 37 °C in 2 liters of Luria-Bertani medium until the absorbance at 600 nm reached 0.5. Expression was then induced by addition of 0.1 mm isopropyl β-d-thiogalactopyranoside to the medium, and cultivation was continued for an additional 24 h at 15 °C. P. horikoshii LysDH could be easily obtained also using pET11a or pET15b; however, the pColdI express system was needed to produce an enzyme suitable for crystallization.
Site-directed Mutagenesis
Site-directed mutagenesis was accomplished using a QuikChange XL site-directed mutagenesis kit (Stratagene) according to the manufacturer's instructions. pCLysDH served as the template, and the following oligonucleotide primers were used as the mutagenic primers (with the mutations underlined): 5′-CTAATAGAGCAGTACACAAGGCCTGCT-3′ and 5′-AGCAGGCCTTGTGTACTGCTCTATTAG-3′ for E168Q, 5′-CTAATAGAGGCGTACACAAGGCCTGCT-3′ and 5′-AGCAGGCCTTGTGTACGCCTCTATTAG-3′ for E168A, 5′-GGACGCTAAAGTGGCCTGGACATCTAG-3′ and 5′-CTAGATGTCCAGGCCACTTTAGCGTCC-3′ for R226K, and 5′-CCCCTTTATTTTAAGATAACATGGTCCCCCAGGG-3′ and 5′-CCCTGGGGGACCATGTTATCTTAAAATAAAGGGG-3′ for Y156F.
Purification from the Soluble Fraction
Cells were harvested by centrifugation, suspended in 20 mm Tris-HCl buffer (pH 7.5) containing 5 mm β-mercaptoethanol, and disrupted by sonication. The crude extract was heated at 80 °C for 10 min and then clarified by centrifugation (10,000 × g for 20 min), after which the protein in the supernatant was run on a Ni2+-chelating Sepharose column (Amersham Biosciences) equilibrated with 20 mm Tris-HCl buffer (pH 7.5) containing 0.5 m NaCl, 0.1 m imidazole, and 5 mm β-mercaptoethanol. After washing the column with the same buffer, the enzyme was eluted with 0.5 m imidazole in the same buffer; the active fractions were pooled; and the enzyme solution was dialyzed against 20 mm Tris-HCl (pH 7.5). Purified P. horikoshii LysDH was concentrated using an Amicon Ultra-15 unit (Millipore) and used for crystallization.
Purification from Inclusion Bodies
To purify LysDH from inclusion bodies, E. coli transformants were collected, suspended in 20 mm Tris-HCl buffer (pH 7.5) containing 5 mm β-mercaptoethanol and 4% Triton X-100, and disrupted by sonication. After sonication, the suspension was centrifuged, and the pellet was solubilized in 50 mm Tris-HCl buffer (pH 7.5) containing 5 mm β-mercaptoethanol, 6 m guanidine HCl, 0.2 m NaCl, and 1 mm EDTA. Once solubilized, LysDH was added to refolding buffer containing 0.1 m Tris-HCl (pH 7.5), 5 mm β-mercaptoethanol, 0.4 m arginine, 2 mm EDTA, and 0.1 mm phenylmethylsulfonyl fluoride and incubated for 36 h at 4 °C. Refolded LysDH was heated at 80 °C for 10 min and clarified by centrifugation, after which the protein in the supernatant was run on a Superdex 200 column (Amersham Biosciences) equilibrated with 20 mm Tris-HCl buffer (pH 7.5) containing 5 mm β-mercaptoethanol and 200 mm NaCl, and the resultant protein solution was used for enzymological characterization.
Determination of Enzyme Activity
Enzyme activity was assayed spectrophotometrically using a Shimadzu UV-1600 spectrophotometer equipped with a thermostat. The standard reaction mixture for oxidative deamination consisted of 100 mm glycine-KOH buffer (pH 10.0) containing 10 mm l-lysine, 1.0 mm NAD, and the enzyme in a final volume of 1.0 ml. The pH values of buffer and lysine solutions were adjusted at room temperature and used for the assay without temperature compensation. After warming the reaction mixture by incubation for 3 min at 70 °C without the coenzyme, the reaction was started by addition of the coenzyme. The appearance of NADH was monitored from the absorbance at 340 nm (extinction coefficient (ϵ) of 6.22 mm−1 cm−1). One unit of activity is defined as the amount of enzyme producing 1.0 μmol of NADH/min at 70 °C under the standard assay conditions, and the specific activity is expressed in units/mg of protein. The protein concentration was determined using the Bradford method (17); bovine serum albumin served as the standard.
Identification of the Product
The product of the reaction catalyzed by LysDH was analyzed by high performance liquid chromatography. Two possible products were postulated: Δ1-piperideine-2-carboxylate (P2C) and P6C. As controls, P2C and P6C were therefore prepared by oxidation of l-lysine using l-lysine oxidase from Trichoderma viride (18) and by dehydrogenation of l-lysine using l-lysine dehydrogenase from G. stearothermophilus (2), respectively, as described previously. The reaction mixture (1.0 ml) containing 10 mm l-lysine, 10 mm NAD, 0.2 m Na2CO3 buffer (pH 9.5), and 1.6 mg of enzyme was incubated at 70 °C for 15 min. After addition of 0.2 ml of 10% trichloroacetic acid and centrifugation, the supernatant solution (0.5 ml) was run on a column (4.6 × 120 mm) of TSKgel Aminopak (Tosoh); citrate buffer (0.2 m, pH 3.25) served as the mobile phase at a flow rate of 0.4 ml/min at 55 °C. The eluate was collected in 1.2-ml portions and mixed with 0.2 ml of 50 mm ο-aminobenzaldehyde in 0.2 m potassium phosphate buffer (pH 8.0). The absorbance was then measured at 460 nm after incubation at 37 °C for 2 h.
Stability, pH Optimum, and Kinetic Parameters
To determine the effect of temperature on its stability, the enzyme was incubated at different temperatures in 20 mm Tris-HCl buffer (pH 7.5) containing 5 mm β-mercaptoethanol. The residual activity was determined at appropriate intervals using the standard assay method. To determine the effect of pH on its stability, the enzyme in buffer at various pH values was incubated at 50 °C for 30 min, and the remaining activity was again determined using the standard assay method. The buffers (250 mm) used for these assays were acetate (pH 4.5–6.0), BisTris-HCl (pH 5.5–7.5), Tris-HCl (pH 7.5–9.0), glycine-KOH (pH 8.5–11.0), and Na2HPO4-NaOH (pH 10.5–12.0). The same buffers were also used to determine the optimal pH for enzyme activity at 70 °C. The initial velocity was determined by varying the concentration of one substrate while keeping the concentrations of the other substrates fixed as described previously (19).
Crystallization and Data Collection
The initial screening for crystallization of LysDH was carried out with Wizard 1,2 (Emerald BioStructures) at 20 °C using the sitting-drop vapor diffusion method. The crystal belongs to the orthorhombic space group P212121 with unit cell parameters a = 75.291, b = 113.011, and c = 122.456 Å and α = β = γ = 90°. The crystals were grown in sitting drops composed of 1 μl of enzyme solution (10 mg/ml) containing 1 mm NAD and 1 μl of mother liquor containing 100 mm citrate buffer (pH 5.8) and 2.1 m ammonium sulfate. Data were collected using an ADSC CCD detector system on beamline BL5A at the Photon Factory in Tsukuba, Japan. All diffraction measurements were carried out on crystals cryoprotected with 30% (v/v) glycerol and cooled to 100 K in a stream of nitrogen gas. The native and derivative data were processed using HKL2000 (20).
Phasing, Refinement, and Structure Analysis
Heavy atom derivatives were prepared by soaking the crystals in reservoir solution containing 1 mm thimerosal for 22 h. Phase calculation was carried out using the multiple-wavelength anomalous dispersion (MAD) method with SOLVE (21). The MAD map at 2.57 Å was subjected to maximum-likelihood density modification, followed by autotracing using RESOLVE (22). The resultant model was built using XtalView (23) and refined against the native data set at 2.44-Å resolution using Refmac5 (24) and CNS (25). The NAD molecule and sulfate ion were clearly visible in both the σA-weighted 2Fo − Fc and Fo − Fc density maps and were included in the latter part of the refinement. The R factor and Rfree values for the final model were 22.45 and 23.55%, respectively (Table 1). The current model contains 694 amino acid residues (subunit A, 0–348; and subunit B, 0–157 and 162–348), 135 water molecules, two NAD molecules, and two sulfate ions. The model geometry was analyzed using PROCHECK (26); 94.5% of the non-glycine residues were in the most favored region of the Ramachandran plot, and 5.1% were in the additionally allowed region. In both subunits, Arg209 was found in a disallowed conformation, which may be explained by its location in a sharp turn between α10 and β11. Hydrogen bonds were identified using the program CCP4mg (27, 28). Molecular graphics were created using PyMOL.
TABLE 1.
Statistics of data collection, phase determination, and refinement
FOM, figure of merit; r.m.s.d., root mean square deviation.
| Native | Peak | Edge | Remote | |
|---|---|---|---|---|
| Data collection | ||||
| Wavelength (Å) | 1.0000 | 0.9932 | 1.0090 | 0.99284 |
| Maximum resolution (Å) | 2.44 | 2.57 | 2.57 | 2.57 |
| No. of unique reflections | 39,442 | 63,980 | 63,926 | 63,960 |
| Redundancy | 7.1 | 3.7 | 3.8 | 3.7 |
| Completeness (%)a | 99.6 (96.3) | 99.6 (96.7) | 99.7 (97.4) | 99.4 (95.4) |
| Rsym (%)a,b | 5.9 (26.4) | 6.6 (29.2) | 5.2 (22.7) | 6.3 (30.5) |
| 〈I/σ(I)〉a | 15.5 (5.0) | 14.9 (4.2) | 15.2 (4.2) | 14.6 (3.8) |
| MAD phasing FOM | 0.47 (84.5-2.57) | |||
| Refinement | ||||
| Resolution range (Å) | 47.5–2.44 | |||
| Rcryst (%)c | 22.45 | |||
| Rfree (%)d | 23.55 | |||
| No. of protein atoms | 5563 | |||
| No. of water molecules | 135 | |||
| No. of NAD molecules | 2 | |||
| No. of sulfate ions | 2 | |||
| r.m.s.d. bond length (Å) | 0.010 | |||
| r.m.s.d. bond angle | 1.6° | |||
a Values in parentheses are for the last resolution shell.
b Rsym = ΣhΣi|Ii(h) − 〈I(h)〉|/ΣhΣi|Ii(h)|, where Ii(h) is the intensity measurement for reflection h and 〈I(h)〉 is the mean intensity for this reflection.
c Rcryst = Σh‖Fo| − |Fc‖/Σh|Fo|.
d Rfree was calculated with randomly selected reflections (10%).
RESULTS AND DISCUSSION
Expression of the Gene and Purification of the Recombinant Enzyme
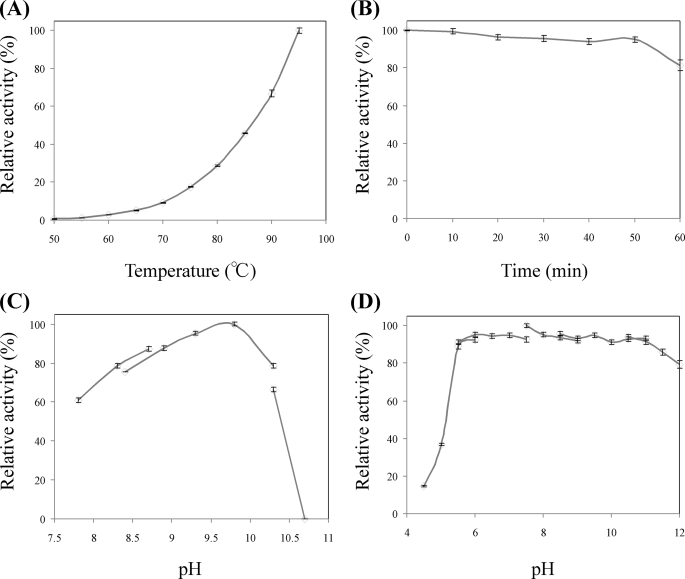
Within the sequence of the P. horikoshii genome (DOGAN Genomic Database), we identified a gene, PH1688.1 (1050 bp at positions 1486245–1485196 of the genome), whose predicted amino acid sequence shows 33% identity to that of G. stearothermophilus LysDH. After transforming E. coli using an expression vector harboring this gene (pCLysDH), the transformant cells exhibited LysDH activity, and the enzyme was readily purified from their crude extract in two simple steps: heat treatment and affinity chromatography. Only 1.7 mg of the purified enzyme was obtained from 7.3 g of cells (wet weight), however; and the pattern on SDS-PAGE showed that P. horikoshii LysDH was present mainly as an inclusion body. For protein purification from the inclusion body, the collected precipitate was dissolved in a 6 m guanidine HCl solution with gentle stirring, after which the dissolved protein was added to a refolding buffer (see “Experimental Procedures”). Once refolded, P. horikoshii LysDH was further purified by heat treatment and gel filtration chromatography. This procedure increased our yield to ∼7.8 mg of the purified enzyme from 7.3 g of cells (wet weight). The specific activities of soluble and refolded LysDHs at 70 °C were estimated to be ∼0.31 and 0.45 units/mg, respectively. At 70 °C, the specific activity of P. horikoshii LysDH was rather less than that of G. stearothermophilus (6.2 units/mg at 50 °C) or A. tumefaciens LysDHs (4.54 units/mg at 37 °C), but as shown in Fig. 2A, the activity of P. horikoshii LysDH dramatically increased with increases in the assay temperature, and the value at 95 °C was 11 times greater than that at 70 °C. Indeed, still greater activity would likely have been observed at the optimum growth temperature (∼104 °C) of this hyperthermophile. The purified enzyme showed a single protein band upon SDS-PAGE, and the N-terminal amino acid sequence of the purified enzyme corresponded to the predicted sequence, including the histidine tag.
FIGURE 2.
A, effect of temperature on LysDH activity. Enzyme activity was measured at various temperatures between 50 and 95 °C. B, effect of temperature on the stability of LysDH. The enzyme was incubated at 100 °C, and the residual activity was measured at the indicated times. C, effect of pH on LysDH activity. Enzyme activity was measured at pH values ranging from 7.8 and 10.7. D, effect of pH on the stability of LysDH. The enzyme was incubated at 50 °C for 30 min in buffers at various pH values, after which residual activity was measured.
Molecular and Catalytic Properties
SDS-PAGE showed the subunit molecular mass of P. horikoshii LysDH to be ∼42.6 kDa, which was consistent with the molecular mass (42,089 Da) calculated from the amino acid sequence. That the native molecular mass determined by gel filtration was ∼97.2 kDa suggests that native LysDH is a homodimer. When we tested the effect of temperature on enzyme activity, we observed maximum activity at ∼95 °C and an extraordinary drop off in activity with reduction in temperature; the activity at 50 °C was only ∼1/170th of that at 95 °C (Fig. 2A). On the other hand, the enzyme was highly thermostable and retained full activity even after incubation for 50 min at 100 °C (pH 7.5). Indeed, the half-life of the enzyme at 100 °C was 180 min (Fig. 2B and supplemental Fig. 1), making P. horikoshii LysDH the most thermostable LysDH described to date.
The enzyme was most active at alkaline pH, with the highest activity at pH ∼10, as is the case with other amino acid dehydrogenases (Fig. 2C). Nonetheless, it was stable over a wide range of pH values, losing no activity when incubated at pH values between 5.0 and 12 for 30 min at 50 °C (Fig. 2D).
Examination of the substrate specificity of P. horikoshii LysDH revealed that l-lysine is the preferred substrate for oxidative deamination in the presence of NAD, although the enzyme showed some activity with S-(β-aminoethyl)-l-cysteine. (With 10 mm substrate, the relative activity was 10.6% of that seen with l-lysine.) d-Lysine, l-ornithine, l-saccharopine, ϵ-amino-n-capronate, α-aminoadipate, Δ-aminovalerate, l-alanine, l-glutamine, l-asparagine, l-arginine, l-glutamate, and l-aspartate were inert. Like A. tumefaciens and G. stearothermophilus LysDHs, P. horikoshii LysDH is highly specific for l-lysine, which would be advantageous for enzymatic detection of l-lysine or for use as an l-lysine biosensor.
Both NAD and NADP were able to serve as the coenzyme for P. horikoshii LysDH, although the reaction rate with NADP (1.0 mm) was only 7.8% of that with NAD (1.0 mm) when l-lysine was the substrate. The A. tumefaciens enzyme is highly specific for NAD (8), whereas the G. stearothermophilus enzyme can utilize both NAD and NADP (2). In this regard, P. horikoshii LysDH is similar to the G. stearothermophilus enzyme. We also examined the effect of various chemicals on the enzyme activity. P. horikoshii LysDH was strongly inhibited by HgCl2 (relative activity with the additive was 17.3% of that without the additive) and ZnCl2 (34.7%) but was unaffected by EDTA, MgCl2, CaCl2, NiCl2, CoCl2, CuCl2, SrCl2, or AlCl3 (each at a concentration of 1 mm). P. horikoshii LysDH showed typical Michaelis-Menten kinetics, and the Km values for NAD and l-lysine were 0.080 and 1.17 mm, respectively.
Identification of the Reaction Product
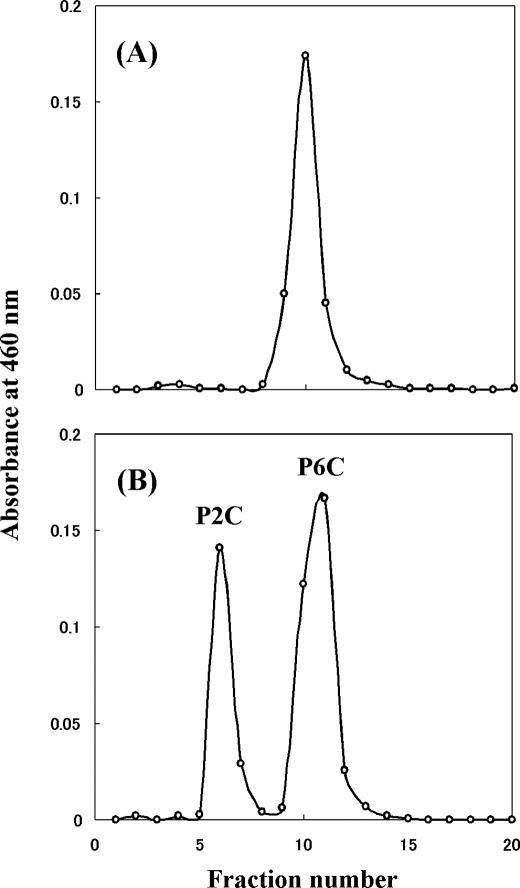
The product of the reaction catalyzed by P. horikoshii LysDH was identified using high performance liquid chromatography. The reaction product from l-lysine was separated as the same peak as authentic P6C (Fig. 3). In addition, orange-colored complexes of the reaction product with ο-aminobenzaldehyde showed the same absorption spectrum as ο-aminobenzaldehyde in complex with authentic P6C, which had an absorption maximum at 460 nm. This was clearly different from the spectrum of ο-aminobenzaldehyde in complex with authentic P2C, which had an absorption maximum at 445 nm (1). Thus, the ϵ-amino group of l-lysine is oxidatively deaminated by the enzyme, yielding l-2-aminoadipate 6-semialdehyde, which spontaneously converts to the dehydrated form, P6C (Fig. 1).
FIGURE 3.
A, ion-exchange chromatography of the reaction product formed from l-lysine; B, authentic P2C and P6C.
These results indicate that the gene PH1688.1 encodes a LysDH, making this the first identification of this enzyme in Archaea. When we investigated the structure of PH1688.1 in the data base, we found that the gene did not cluster with any other genes, and no similar gene was found in other strains of Pyrococcales, suggesting that LysDH may play a specific role during lysine metabolism in P. horikoshii.
Overall Structure
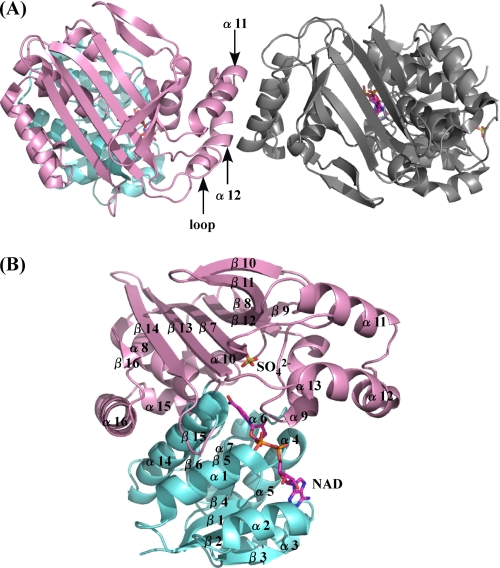
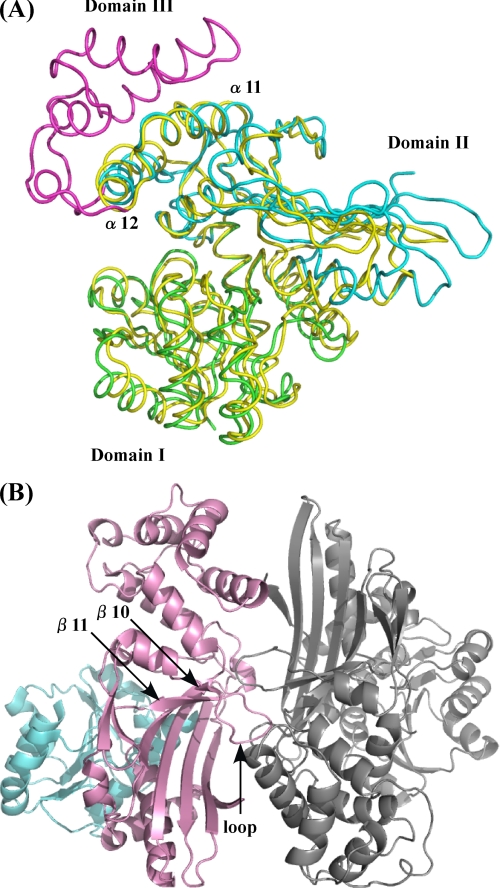
The structure of P. horikoshii LysDH was determined using mercury-based MAD and was refined at a resolution of 2.44 Å (Table 1). The asymmetric unit consisted of one homodimer with a solvent content of 59.5%, which corresponds to a Matthews coefficient (29) of 3.1 Å3/Da. The model of dimeric LysDH consisted of 694 amino acid residues, two NAD molecules, two sulfate ions, and 135 water molecules. The three-dimensional structure of the N-terminal histidine tag region (Met−15 to Arg−1 in both subunits), the C-terminal region (Thr349 in both subunits), and the substrate-binding loop (Ile158–Ser161 in subunit B) could not be modeled in this study because of poor electron density. Well defined electron density for Pro120 and Pro153 indicated a cis-conformation for the peptide bond. The two subunits had approximate dimensions of 49 × 37 × 52 Å and were related by a 2-fold non-crystallographic rotation axis (Fig. 4A). Each monomer consisted of two domains: an N-terminal coenzyme-binding domain (α1–α7, α14, β1–β6, β15), which formed a classical Rossmann fold motif, and a C-terminal domain (α8–α13, α15-α16, β7–β14, β16), within which both the catalytic activity and formation of the homodimer were mediated (Fig. 4B). The dimer associated through hydrophobic interactions between α11, α12, and loop 147–157 of both subunits, which formed a stable four-helix bundle (Fig. 4A). The surface area buried in the dimer interface was 1369 Å2 (calculated using a probe with a radius of 1.4 Å; Protein-Protein Interface Analysis Server), which is 4.5% of the total surface area of the enzyme. Multiple oligomeric structures, including hexameric, dimeric, and tetrameric forms, have been observed for LysDHs from bacteria. The enzyme from G. stearothermophilus adopts a hexameric structure (2), whereas that from A. tumefaciens reportedly forms a dimeric or tetrameric structure and binds 2 mol of NADH/mol of tetrameric enzyme (the dimeric enzyme has one catalytic site) (30). In this regard, P. horikoshii LysDH differs substantially from these two previously described LysDHs.
FIGURE 4.
A, overall structure of P. horikoshii LysDH. The NAD-binding and catalytic domains are shown in cyan and pink, respectively. The adjacent subunit is shown in gray. The region involved in dimer formation is indicated. B, ribbon plot of the P. horikoshii LysDH monomer. NAD (magenta) and the sulfate ion (yellow) are shown as stick models. Oxygen and nitrogen atoms are shown in red and blue, respectively.
Structural Differences between the Two Subunits
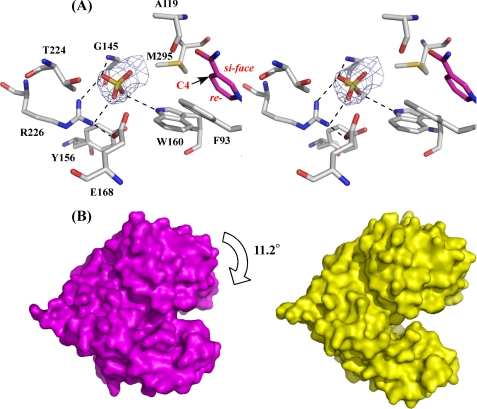
In the electron density map of P. horikoshii LysDH obtained from our preliminary experimental data, we noticed an extra density within the active-site cavity in subunit A and found that a sulfate ion could be modeled into that density after construction and refinement of the peptide chain. The map clearly defined the precise orientation of the sulfate ion (Fig. 5A): its two oxygen atoms formed hydrogen bonds with the side chains of Trp160 and Arg226 and a proton in the main chain amide of Gly145. In subunit B, however, we found no sulfate ion in the active site; instead, it was on the surface of the Rossmann fold domain. Moreover, according to calculations carried out using the DynDom program for protein domain motion analysis (31–33), the Rossmann fold domain in subunit A was rotated 11.2° relative to that in subunit B, which leads to 99% active-site closure in subunit A (Fig. 5B). Structural comparison between LysDH subunits A and B showed that whereas the former assumes a closed conformation, the latter assumes an open one (root mean square deviation of C-α positions between A and B = 1.31 Å). Within the ternary complex, the sulfate in the active site of subunit A occupies a position that corresponds to the Lys carboxylate-binding site in our model (Fig. 5A and see Fig. 9B), so the binding of the sulfate likely results in subunit A being in a closed conformation.
FIGURE 5.
Active site of P. horikoshii LysDH and comparative analysis of subunits A and B. A, schematic representation depicting the interactions between the sulfate ion and the protein in subunit A. The networks of hydrogen bonds are shown as dashed lines. The sulfate ion is shown as a stick model in yellow. The final σA-weighted 2Fo − Fc electron density map for the sulfate ion and the active site in subunit A is shown at the 1σ level. The C-4 atom of the pyridine ring (a hydride acceptor site) and the si- and re-faces are labeled. B, surface representation of subunits A (magenta) and B (yellow).
FIGURE 9.
A, comparison of the active-site pocket of P. horikoshii LysDH with that of M. grisea saccharopine reductase. B, proposed model for l-lysine binding. P. horikoshii LysDH and M. grisea saccharopine reductase are shown in white and green, respectively. l-Lysine (cyan) and saccharopine (yellow) molecules are shown as stick models. Atoms are colored as described for Fig. 3. WAT, water.
Comparison with Known Protein Structures
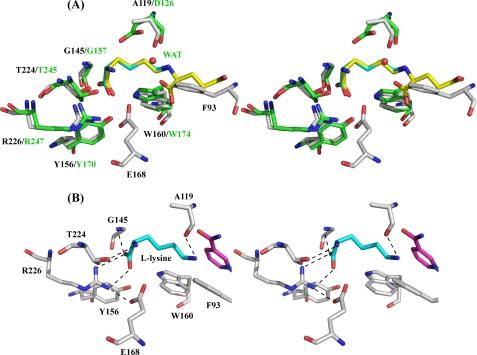
When we sent our model of the LysDH monomer to the Dali server (34) seeking proteins with similar structures, we found that the coenzyme-binding domains (Rossmann fold) of a large number of proteins showed structural similarity to LysDH, although these cases are not discussed here in more detail. With regard to the C-terminal domain, the protein with the highest structural similarity was the saccharopine reductase from Magnaporthe grisea (Protein Data Bank code 1FF9) (35), which had a Z-score of 15.1 and a root mean square deviation of 2.7 Å over 198 C-α positions. The saccharopine reductase folds into three domains: an N-terminal dinucleotide-binding domain with a classical Rossmann fold (Domain I), a C-terminal catalytic domain containing an α/β-fold (Domain II), and an all-helical domain formed by five α-helices (α10–α14) and one short turn helix (γ3) (Domain III). Interestingly, the main chain coordinates of the LysDH monomer were closely related to those of the architecture formed by Domains I and II in the saccharopine reductase (Fig. 6A). Domain III is absent in LysDH, although two α-helices from that domain, α10 and α14, are conserved in LysDH as α11 and α12, respectively. The sequence alignment also showed that 68 residues inserted between α10 and α14 in the M. grisea saccharopine reductase are not present in P. horikoshii LysDH (Fig. 7). A similar deletion of the residues was observed in the sequences of the G. stearothermophilus and A. tumefaciens LysDHs (Fig. 7).
FIGURE 6.
A, comparison of the structures of P. horikoshii LysDH and M. grisea saccharopine reductase. The structure of P. horikoshii LysDH is shown in yellow; that of M. grisea saccharopine reductase is shown in green (Domain I), cyan (Domain II), and magenta (Domain III). B, overall structure of M. grisea saccharopine reductase. The structure is colored as described for Fig. 3. The region involved in dimer formation is indicated.
FIGURE 7.
Amino acid sequence alignment of three LysDHs and saccharopine reductase. PH, P. horikoshii; GS, G. stearothermophilus; AT, A. tumefaciens; MG SPRD, M. grisea saccharopine reductase. Sequences were aligned using ClustalW (38). Asterisks indicate conserved residues. The residues involved in binding the sulfate ion and NAD are shown in blue and red, respectively. The α-helices (α1–α16; yellow) and β-strands (β1–β16; purple) in P. horikoshii LysDH are shown above the alignment.
The quaternary structural arrangement of P. horikoshii LysDH is totally different from that of M. grisea saccharopine reductase, in which dimer interactions are found between residues in the loop connecting β10 and β11 to their 2-fold symmetry-related counterparts (Fig. 6B). By contrast, LysDH dimer interactions were between residues around α11, α12, and loop 147–157 in both subunits, as mentioned above (Fig. 4A). Thus, the presence or absence of Domain III substantially affects the arrangement of the subunits in these two enzymes.
Cofactor Binding
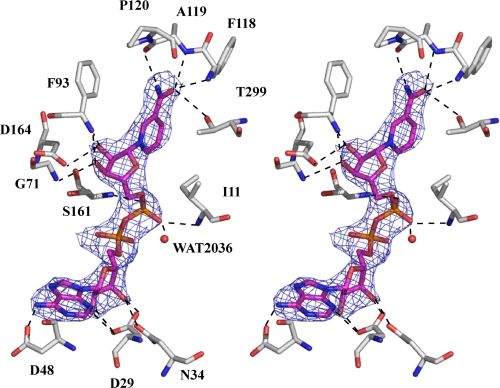
The following description of cofactor binding in the active site is based on the structure of subunit A. The electron density corresponding to the NAD coenzyme bound within the active site was very clear, which enabled us to place the ligand with reasonable accuracy. The map enabled clear positioning of the adenine ring, identification of the C2′-endo-conformation of the two ribose rings, clear definition of the backbone phosphate groups, and assignment of an anti-conformation to the glycosidic bond linking the nicotinamide ring and its associated ribose moiety. N-3 and N-6 of the adenine base form hydrogen bonds with the side chains of Asp29 and Asp48, respectively (Fig. 8), and the hydroxyl groups of the adenine ribose interact with the side chains of Asp29 and Asn34. A glycine-rich motif, GXGXXG, which lies close to the adenine ribose, dictates the nature of the hydrogen bonding between the main chain and the adenine ribose moiety (36). In LysDH, this motif occupies the 7th–12th amino acids from the N terminus (Fig. 7). In addition, the nicotinamide phosphate interacts with Ile11, Ser161, and a water molecule (position 2036). The hydroxyl groups of the nicotinamide ribose interact with the side chain of Asp164 and the main chain amides of Gly71 and Phe93. The pattern of hydrogen bonding between the enzyme and NAD is completed by interactions between the carboxamide moiety of the nicotinamide ring and the main chain amides of Phe118 and Ala119, the main chain oxygen of cis-Pro120, and the side chain of Thr299. These hydrogen bonds cause the glycosidic bond between the nicotinamide ring and its associated ribose moiety to be in the anti-conformation. In general, NAD(P)H-dependent dehydrogenases show ether pro-R or pro-S stereospecificity for hydrogen removal from C-4 of the nicotinamide moiety of NAD(P)H. NAD binds to pro-R-specific enzymes in an anti-conformation and to pro-S-specific enzymes in a syn-conformation (37), which suggests that P. horikoshii LysDH makes use of pro-R-specific hydride transfer (type A), as has been described for A. tumefaciens LysDH (7).
FIGURE 8.
Stereo representation of NAD bound to P. horikoshii LysDH. Residues that interact with NAD are labeled. The networks of hydrogen bonds are shown as dashed lines. The final σA-weighted 2Fo − Fc electron density map for NAD is shown at the 1σ level. Atoms are colored as described for Fig. 3.
Active Site and Insight into the LysDH Reaction
The crystal structure of M. grisea saccharopine reductase in complex with saccharopine and NADPH has been determined. Superposition of this structure (Protein data bank code 1E5Q) onto the structure of LysDH enabled us to compare the amino acid residues involved in substrate binding. On the basis of the orientation of the saccharopine, we modeled the l-lysine molecule into the active site of P. horikoshii LysDH (Fig. 9A) and then minimized the energy of the complex using Swiss-PdbViewer. Within this structure, the carboxyl group of l-lysine forms a salt bridge with the side chain of Arg226 and a hydrogen bond with the main chain amide proton of Gly145, and the C-α amino group is situated within hydrogen-bonding distance of the side chain of Thr224 (Fig. 9B). These three residues are strictly conserved in saccharopine reductase as Arg247, Thr245, and Gly157, respectively, and hold saccharopine in the active site in a similar manner (Fig. 9A). In saccharopine reductase, moreover, the side chain of Trp174 is engaged in a hydrophobic interaction with the carbon skeleton of saccharopine. This residue is also conserved as Trp160 in LysDH. By contrast, Ser99, Tyr100, and Arg224 in saccharopine reductase, which interact with the carboxyl group of the glutamate moiety of saccharopine, are completely absent in LysDH, and superposition of the two enzymes results in a serious steric clash between the glutamate moiety of saccharopine and the side chain of Phe93 in LysDH (Fig. 9A). This is consistent with our observation that LysDH does not utilize saccharopine as a substrate. The most important difference between the two enzymes is that Asp126 in saccharopine reductase is replaced by Ala119 in LysDH. In our model, the main chain oxygen of Ala119 forms a hydrogen bond with the nitrogen atom of the l-lysine ϵ-amino group (Fig. 9B). As a result, the pro-R-hydrogen at prochiral C-6 of l-lysine is placed in front of the re-face of the nicotinamide ring, which enables the pro-R-specific proton transfer to NAD. This observation is in good agreement with the previous finding that A. tumefaciens LysDH abstracts stereospecifically the ϵ-pro-R-hydrogen of l-lysine (6).
In the reverse reaction catalyzed by saccharopine reductase (formation of glutamate), Asp126 is thought to be involved in the hydrolysis of the Schiff base intermediate (22), which requires nucleophilic attack by a water molecule on C-6 of saccharopine, and the nucleophilicity of this water molecule is assumed to be enhanced by interactions with the side chain of Asp126. However, the catalytic residue (Asp126) of M. grisea saccharopine reductase has been identified only from the structural information; it has not yet been confirmed by site-directed mutagenesis. We therefore used site-directed mutagenesis to search for a catalytic residue in the LysDH active site. We found Glu168 to be the only residue close enough to C-6 of l-lysine to serve that purpose (Fig. 9A). Aside from Glu168, there are no other obvious residues in the active site that might facilitate the proton transfer expected to occur during catalysis. For that reason, we constructed E168Q and E168A mutants. Notably, however, the kinetic analysis of the two mutants indicated that their catalytic activities were nearly the same as that of the wild-type enzyme, although the Km values for l-lysine were slightly increased to 2.0–2.3 mm. In addition, Glu168 is conserved as Glu174 in A. tumefaciens LysDH but is replaced by His181 in G. stearothermophilus LysDH (Fig. 7). Apparently, Glu168 is not essential for enzyme catalysis. Among the residues located within the substrate-binding pocket, Tyr156 and Arg226 are strictly conserved in LysDHs and saccharopine reductase. We therefore also constructed Y156F and R226K mutants and observed that these mutations completely abolished the activity of the enzyme. In our model, the side chain hydroxyl group of Tyr156 and the guanidium group of Arg226 (NH2) are estimated to be 7.8 Å from C-6 of l-lysine, whereas the corresponding groups in Tyr170 and Arg247 (NH1) of saccharopine reductase are 7.1–7.5 Å from C-6 of saccharopine. These distances preclude the involvement of these two residues in the hydride transfer. Instead, it appears that Tyr156 and Arg226 play essential roles in the binding of substrates to LysDHs.
Although the mechanism underlying the catalytic reaction of P. horikoshii LysDH remains unclear, this study provides the first structural insight into substrate stereo recognition and hydride transfer by LysDH. The structure of the P. horikoshii LysDH·NAD·substrate analog ternary complex should be a useful focus for further investigation.
Supplementary Material
Acknowledgments
We thank Drs. N. Igarashi, N. Matsugaki, and S. Wakatsuki for help in data collection at Photon Factory beamline BL5A. We also thank Drs. S. Kawamura, T. Araki, and T. Torikata for extremely helpful support.
This work was supported in part by the “National Project on Protein Structural and Functional Analysis” promoted by the Ministry of Education, Science, Sports, Culture, and Technology of Japan and by a grant-in-aid for scientific research (C) from the Japan Society for the Promotion of Science.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Fig. 1.
The atomic coordinates and structure factors (code 3ABI) have been deposited in the Protein Data Bank, Research Collaboratory for Structural Bioinformatics, Rutgers University, New Brunswick, NJ (http://www.rcsb.org/).
- LysDH
- l-lysine 6-dehydrogenase
- P6C
- Δ1-piperideine-6-carboxylate
- P2C
- Δ1-piperideine-2-carboxylate
- BisTris
- 2-[bis(2-hydroxyethyl)amino]-2-(hydroxymethyl)propane-1,3-diol
- MAD
- multiple-wavelength anomalous dispersion.
REFERENCES
- 1.Misono H., Nagasaki S. (1982) J. Bacteriol. 150, 398–401 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Heydari M., Ohshima T., Nunoura-Kominato N., Sakuraba H. (2004) Appl. Environ. Microbiol. 70, 937–942 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ruldeekulthamrong P., Maeda S., Kato S., Shinji N., Sittipraneed S., Packdibamrung K., Misono H. (2008) BMB Rep. 41, 790–795 [DOI] [PubMed] [Google Scholar]
- 4.Ohshima T., Soda K. (2000) in Stereoselective Biocatalysis (Patel R. N. ed) pp. 877–902, Marcel Dekker, Inc., New York [Google Scholar]
- 5.Bürgi W., Richterich R., Colombo J. P. (1966) Nature 211, 854–855 [DOI] [PubMed] [Google Scholar]
- 6.Misono H., Yoshimura T., Nagasaki S., Soda K. (1990) J. Biochem. 107, 169–172 [DOI] [PubMed] [Google Scholar]
- 7.Hashimoto H., Misono H., Nagata S., Nagasaki S. (1989) Agric. Biol. Chem. 53, 1175–1176 [PubMed] [Google Scholar]
- 8.Misono H., Hashimoto H., Uehigashi H., Nagata S., Nagasaki S. (1989) J. Biochem. 105, 1002–1008 [DOI] [PubMed] [Google Scholar]
- 9.Hashimoto H., Misono H., Nagata S., Nagasaki S. (1990) Agric. Biol. Chem. 54, 291–294 [PubMed] [Google Scholar]
- 10.Ohshima T., Soda K. (1989) Trends Biotechnol. 7, 210–214 [Google Scholar]
- 11.Hummel W., Kula M. R. (1989) Eur. J. Biochem. 184, 1–13 [DOI] [PubMed] [Google Scholar]
- 12.Satoh A., Kim J. K., Miyahara I., Devreese B., Vandenberghe I., Hacisalihoglu A., Okajima T., Kuroda S., Adachi O., Duine J. A., Van Beeumen J., Tanizawa K., Hirotsu K. (2002) J. Biol. Chem. 277, 2830–2834 [DOI] [PubMed] [Google Scholar]
- 13.Durham D. R., Perry J. J. (1978) J. Bacteriol. 134, 837–843 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Limburg J., Mure M., Klinman J. P. (2005) Arch. Biochem. Biophys. 436, 8–22 [DOI] [PubMed] [Google Scholar]
- 15.Nozaki M. (1987) Methods Enzymol. 142, 650–655 [DOI] [PubMed] [Google Scholar]
- 16.Murray M. G., Thompson W. F. (1980) Nucleic Acids Res. 8, 4321–4325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bradford M. M. (1976) Anal. Biochem. 72, 248–254 [DOI] [PubMed] [Google Scholar]
- 18.Kusakabe H., Kodama K., Kuninaka A., Yoshino H., Misono H., Soda K. (1980) J. Biol. Chem. 255, 976–981 [PubMed] [Google Scholar]
- 19.Ohshima T., Misono H., Soda K. (1978) J. Biol. Chem. 253, 5719–5725 [PubMed] [Google Scholar]
- 20.Otwinowski Z., Minor W. (1997) Methods Enzymol. 276, 307–326 [DOI] [PubMed] [Google Scholar]
- 21.Terwilliger T. C., Berendzen J. (1999) Acta Crystallogr. Sect. D 55, 849–861 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Terwilliger T. C. (2000) Acta Crystallogr. Sect. D 56, 965–972 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.McRee D. E. (1999) J. Struct. Biol. 125, 156–165 [DOI] [PubMed] [Google Scholar]
- 24.Murshudov G. N., Vagin A. A., Dodson E. J. (1997) Acta Crystallogr. Sect. D 53, 240–255 [DOI] [PubMed] [Google Scholar]
- 25.Brünger A. T., Adams P. D., Clore G. M., DeLano W. L., Gros P., Grosse-Kunstleve R. W., Jiang J. S., Kuszewski J., Nilges M., Pannu N. S., Read R. J., Rice L. M., Simonson T., Warren G. L. (1998) Acta Crystallogr. Sect. D 54, 905–921 [DOI] [PubMed] [Google Scholar]
- 26.Laskowski R. A., MacArthur M. W., Moss D. S., Thornton J. M. (1993) J. Appl. Crystallogr. 26, 283–291 [Google Scholar]
- 27.Potterton E., McNicholas S., Krissinel E., Cowtan K., Noble M. (2002) Acta Crystallogr. Sect. D 58, 1955–1957 [DOI] [PubMed] [Google Scholar]
- 28.Potterton L., McNicholas S., Krissinel E., Gruber J., Cowtan K., Emsley P., Murshudov G. N., Cohen S., Perrakis A., Noble M. (2004) Acta Crystallogr. Sect. D 60, 2288–2294 [DOI] [PubMed] [Google Scholar]
- 29.Matthews B. W. (1968) J. Mol. Biol. 33, 491–497 [DOI] [PubMed] [Google Scholar]
- 30.Hashimoto H., Misono H., Nagata S., Nagasaki S. (1989) J. Biochem. 106, 76–80 [DOI] [PubMed] [Google Scholar]
- 31.Qi G., Lee R., Hayward S. (2005) Bioinformatics 21, 2832–2838 [DOI] [PubMed] [Google Scholar]
- 32.Lee R. A., Razaz M., Hayward S. (2003) Bioinformatics 19, 1290–1291 [DOI] [PubMed] [Google Scholar]
- 33.Hayward S., Berendsen H. J. (1998) Proteins 30, 144–154 [PubMed] [Google Scholar]
- 34.Holm L., Sander C. (1998) Nucleic Acids Res. 26, 316–319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Johansson E., Steffens J. J., Lindqvist Y., Schneider G. (2000) Structure 8, 1037–1047 [DOI] [PubMed] [Google Scholar]
- 36.Baker P. J., Britton K. L., Rice D. W., Rob A., Stillman T. J. (1992) J. Mol. Biol. 228, 662–671 [DOI] [PubMed] [Google Scholar]
- 37.Benner S. A. (1982) Experientia 38, 633–636 [DOI] [PubMed] [Google Scholar]
- 38.Thompson J. D., Higgins D. G., Gibson T. J. (1994) Nucleic Acids Res. 22, 4673–4680 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.