Abstract
The imprinted gene PEG3 confers parenting and sexual behaviors, alters growth and development, and regulates apoptosis. However, a molecular mechanism that can account for the diverse functions of Peg3/Pw1 is not known. To elucidate Peg3-regulated pathways, we performed a functional screen in zebrafish. Enforced overexpression of PEG3 mRNA during zebrafish embryogenesis decreased β-catenin protein expression and inhibited Wnt-dependent tail development. Peg3/Pw1 also inhibited Wnt signaling in human cells by binding to β-catenin and promoting its degradation via a p53/Siah1-dependent, GSK3β-independent proteasomal pathway. The inhibition of the Wnt pathway by Peg3/Pw1 suggested a role in tumor suppression. Hypermethylation of the PEG3 promoter in primary human gliomas led to a loss of imprinting and decreased PEG3 mRNA expression that correlated with tumor grade. The decrease in Peg3/Pw1 protein expression increased β-catenin, promoted proliferation, and inhibited p53-dependent apoptosis in human CD133+ glioma stem cells. Thus, mammalian imprinting utilizes Peg3/Pw1 to co-opt the Wnt pathway, thereby regulating development and glioma growth.
Keywords: Apoptosis, Cancer, Neurobiology/Neuroscience, Oncogene/β-Catenin, Protein/Protein-Protein Interactions, Tumor/Suppressor/p53, Tumor/Oncogene
Introduction
Mammalian imprinting regulates growth and the establishment of parental nurturing behaviors, but the detailed molecular mechanisms by which this occurs are incompletely known. PEG3 (paternally expressed gene 3) is an imprinted gene that is expressed primarily during embryogenesis and in adult ovary, testis, muscle, and brain (1–4). Homozygous deletion of PEG3 in mice leads to growth retardation, impaired maternal nurturing and male sexual behavior, complex olfactory deficits, increased body fat, abnormal metabolism, and a decreased number of oxytocin neurons in the hypothalamus (3, 5–7). Despite numerous studies, however, the mechanism by which Peg3/Pw1 accomplishes this diversity of tasks is incompletely known. Peg3/Pw1 binds TRAF2 to activate NFκB, although it is not essential for this process (8, 9). Peg3/Pw1 also regulates myogenesis via p53-, tumor necrosis factor-α-, and Bax-dependent pathways, but the detailed mechanisms of Peg3/Pw1 actions during muscle development are poorly understood (10–12). In addition, Peg3/Pw1 binds Siah1 to promote Bax translocation during p53-dependent apoptosis (13–15), but again, the molecular mechanism by which Peg3/Pw1 cooperates with Siah1 to promote Bax translocation is not known.
Aberrant methylation of imprinted genes (e.g. IGF2 and CDKN1C) has been implicated in tumorigenesis (16, 17). PEG3 mRNA expression is decreased in established cancer cell lines due to promoter hypermethylation (18, 19). Overexpression of Peg3/Pw1 protein in a cultured glioma cell line decreased tumorigenicity (20), but the mechanism underlying this effect was not determined. Moreover, direct evidence for a role for Peg3/Pw1 in primary human cancers is lacking.
We sought to elucidate the molecular mechanism by which imprinting of PEG3 establishes nurturing behavior, promotes p53-dependent cell death, and regulates tumor growth. Here, we show that Peg3/Pw1 binds β-catenin to inhibit Wnt signaling, thereby regulating development, proliferation, apoptosis, and glioma growth.
EXPERIMENTAL PROCEDURES
Tumor Samples and Cell Lines
Frozen human tissue specimens of World Health Organization Grade II–IV astrocytomas and non-tumor brain samples were obtained from the Brain Tumor Tissue Bank at Brigham and Women's Hospital under the auspices of an institutional review board approved protocol. Histology was confirmed by hematoxylin/eosin. Human U87 glioma cells, human embryonic kidney 293T cells, and HeLa cells were obtained from the American Type Culture Collection. The D566 glioma cells were a gift from D. Bigner. Human glioma-derived stem-like cells were isolated from surgical glioblastoma specimens using a CD133 affinity column (Miltenyi Biotec) and maintained in Dulbecco's modified Eagle's medium/nutrient mixture F-12 containing B-27 supplement, epidermal growth factor (20 ng/ml), and basis fibroblast growth factor (20 ng/ml) as described (21).
Apoptosis Assays
DNA damage was induced by exposure to camptothecin (50 μm) for 24 h. Apoptotic cells were visualized using annexin V (Roche Applied Science) according to the manufacturer's protocol.
mRNA Expression Profiling
Total RNA from glioma or non-tumor brain specimens was reverse-transcribed to generate cDNA, which was then biotinylated and hybridized to Affymetrix human genome U133A expression arrays prior to scanning for quantitation. Statistical comparisons were performed using the t test with corrections for multiple comparisons.
Comparative Genomic Hybridization Microarray Analysis
Genomic DNA isolated from 17 human glioblastoma samples was labeled and hybridized to a spotted oligonucleotide microarray containing ∼385,000 probes with average spacing of 6000 bp (NimbleGen Systems, Inc.). Gains and losses were detected using commercially available software (NimbleGen Systems, Inc.).
Reverse Transcription-PCR, Cloning, and Mutagenesis
Total RNA was isolated from cultured cells or primary human brain tissue samples. Human PEG3 and ZIM2 cDNAs were cloned from non-tumor brain tissues by reverse transcription-PCR such that a hemagglutinin (HA)2 tag was added to the N termini. HA-Peg3SCAN was generated using primers 5′-GCGGCCGCTATGTACCCATACGATGTTCCAGATTACGCTCTTCTGCCTCCAAAGCACTTGTCTG-3′ and 5′-AGATCTGTGGGAGTGGCCATCGTCTTC-3′. HA-Peg3ZF was generated using primers 5′-GCGGCCGCTATGTACCCATACGATGTTCCAGATTACGCTCTTACGCAGGGCCACTCATCAAGATC-3′ and 5′-GAATTCGCCATCCTTCTTAAACTCACC-3′. Mouse PEG3 and dominant-negative PEG3 were the gift of Dr. X. Wu (14). Myc-tagged and HA-tagged SIAH1A cDNAs were the gift of Dr. S. Matsuzawa (22). A human CTNNB1 (β-catenin) cDNA was purchased from OriGene and subcloned into a lentiviral internal ribosome entry site-enhanced green fluorescent protein vector, a pcDNA3 vector, or the pCMV-FLAG-MAT2 vector. β-Catenin lacking phosphorylation sites for GSK3β (mutant β-catenin; S33A, S37A, and T41A) was generated by site-directed mutagenesis and verified by DNA sequencing (23, 24).
Quantitative Real-time PCR
Total RNA (1 μg) isolated from cultured cells or primary frozen tissues was used to generate cDNA. cDNA (20 ng) and ABI TaqMan reagents (Applied Biosystems) for PEG3, CTNNB1, or ACTB were then used to perform quantitative PCR on an ABI 7300 thermocycler PCR machine. At least four replicates were performed for each sample. Statistical significance was calculated using the t test.
Transfection and Lentiviral Transduction Methods
Three PEG3 and one control small hairpin RNA (shRNA) plasmids were purchased from Open Biosystems. Lentiviruses containing these constructs were generated according to the manufacturer's protocol (Invitrogen). Cells were transduced at a multiplicity of infection of 1.
Western Blotting and Immunoprecipitation
The antibodies used were as follows: anti-Myc antibody (clone 9E10, Upstate Biotechnology), anti-β-catenin and anti-phospho-β-catenin (Ser33/Ser37/Thr41) antibodies (Cell Signaling Technology), anti-HA antibody (Abcam), and anti-β-actin and anti-FLAG antibodies (Sigma). Western blotting was performed as described previously (21). For immunoprecipitation, whole cell lysates were immunoprecipitated using 10 μl of anti-HA antibody-coupled beads (Sigma) or anti-β-catenin antibody adsorbed to protein A-Sepharose beads. The density of immunoreactive bands was quantified using a commercially available software program (Adobe Photoshop).
β-Catenin Immunohistochemistry
A tissue array (Accumax) containing 29 paraffin-embedded human glioma samples and four non-tumor brain samples was stained for β-catenin immunoreactivity using the peroxidase method. Control sections were stained using the secondary antibody alone.
3-(4,5-Dimethylthiazol-2-yl)-2,5-diphenyltetrazolium Bromide Cell Growth and Bromodeoxyuridine Cell Proliferation Assays
Cell growth and proliferation were measured using a 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide cell growth assay and a bromodeoxyuridine enzyme-linked immunosorbent assay (Roche Applied Science), respectively, according to the manufacturer's protocol. Eight wells were assayed for each condition, and each experiment was repeated in triplicate. Statistical significance was determined using the t test.
Methylation-specific PCR
Genomic DNA from human glioblastoma specimens, non-tumor brain specimens, or glioma cell lines was treated with sodium bisulfite using the CpGenomeTM DNA modification kit (Chemicon) according to the manufacturer's instructions. Methylation-specific PCR for PEG3 was then performed using the unmethylated DNA PCR primers 5′-CTCATAACACCCAACACCCAACAACA-3′ and 5′-GTTATTTTTTGATGTTAGTTTTTAGTTTTTGT-3′. The methylated DNA primers were 5′-TAACGCCCGACGCCCGACGACG-3′ and 5′-TATTTTTTGACGTTAGTTTTTAGTTTTCGC-3′.
Tcf4/Lef1 Luciferase Reporter Assay
293T cells were cotransfected with a luciferase Tcf4/Lef1-specific reporter vector (pTOPflash, 50 ng) or a vector containing mutated Tcf4/Lef1-binding sites (pFOPflash, 50 ng) (Upstate Biotechnology), an internal β-galactosidase control construct (pCMVβ-gal, 50 ng; Promega), and vectors for the genes of interest. The total amount of DNA in each transfection was kept constant by the addition of an appropriate amount of empty control vector plasmid. Luciferase activity was assayed using a luminometer according to the manufacturer's protocol. β-Galactosidase activity was measured by enzyme-linked immunosorbent assay at 420 nm in the same lysates for normalization. Samples were assayed in triplicate. Statistical significance was determined using the t test.
Functional Screen in Zebrafish Embryos
Human PEG3 cDNA was subcloned into the pCS2+ vector and linearized with NarI. PEG3 mRNA was then transcribed in vitro according to the manufacturer's instructions (Roche Applied Science). Approximately 4–8 ng of PEG3 mRNA or scrambled control mRNA was microinjected into fertilized zebrafish embryos at the single-cell stage, and the embryos were incubated at 28.5 °C in embryo medium for 24 h. The development of intact and dechorionated embryos was evaluated using a dissection microscope.
RESULTS
Peg3/Pw1 Regulates Wnt Signaling in Zebrafish and in Human Cells
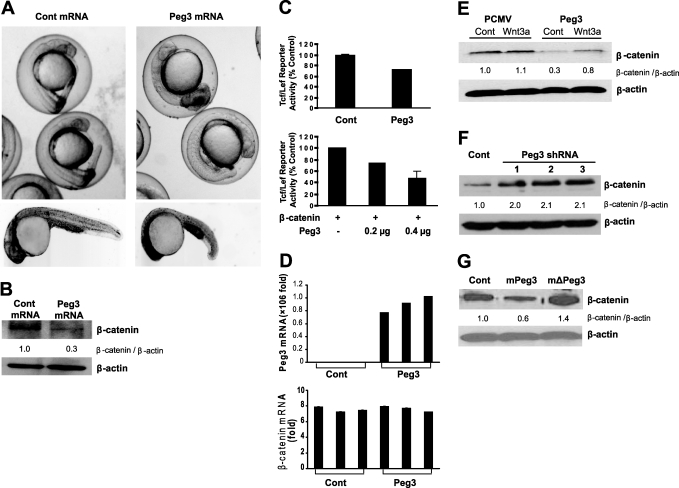
To identify pathways regulated by Peg3/Pw1, we performed a functional screen using zebrafish embryos. Over 90% of zebrafish embryos injected with human PEG3 mRNA at the single-cell stage displayed marked defects in tail development 24 h after injection (Fig. 1A). The axial phenotype of these mutants was similar to that caused by combined Wnt8 and Wnt3a inhibition (25). Western blot analysis of protein extracts obtained from the embryos demonstrated a decrease in β-catenin protein expression after PEG3 mRNA overexpression (Fig. 1B), suggesting that Peg3/Pw1 may interfere with Wnt signaling. To investigate this possibility further, we examined the effect of Peg3/Pw1 on β-catenin-dependent transcription in human cells using an in vitro reporter assay. Peg3/Pw1 protein overexpression in 293T cells decreased β-catenin/Tcf4-dependent transcriptional activity in a dose-dependent manner (Fig. 1C). Quantitative real-time PCR indicated that Peg3/Pw1 overexpression had no effect on β-catenin mRNA expression (Fig. 1D). However, Peg3/Pw1 effectively decreased basal and Wnt3a-stimulated β-catenin protein levels (Fig. 1E). Moreover, knockdown of endogenous Peg3/Pw1 protein using three separate shRNAs increased β-catenin expression in 293T cells (Fig. 1F), and knockdown of endogenous Peg3/Pw1 also increased β-catenin expression in HeLa cells (data not shown). Thus, endogenous Peg3/Pw1 inhibits β-catenin protein expression and transcriptional activity via a post-transcriptional mechanism.
FIGURE 1.
A, zebrafish embryos injected with human PEG3 mRNA at the single-cell stage and allowed to develop for 24 h. Data from intact (upper panels) and dechorionated (lower panels) embryos are shown. Note the defect in tail development after PEG3 mRNA overexpression. B, Western blot analysis illustrating the effect of PEG3 mRNA or noncoding control (Cont) mRNA overexpression on β-catenin protein levels in zebrafish embryos after 24 h of development. β-Actin was used as a loading reference. C, Tcf/Lef1 luciferase reporter assay in 293T cells after Peg3/Pw1 and/or β-catenin overexpression. Data shown are means ± S.E. D, real-time PCR of PEG3 and β-catenin mRNAs after Peg3/Pw1 protein overexpression in 293T cells. Data shown are means ± S.E. E, effect of overexpression of the HA-Peg3/Pw1 vector or a control vector (pCMV) on β-catenin protein expression in 293T cells under control conditions or after Wnt3a (50 ng/ml) exposure. F, Western blot illustrating the effect of Peg3/Pw1 knockdown on β-catenin in 293T cells using three separate Peg3/Pw1 shRNAs. G, Western blot illustrating the effect of mouse Peg3/Pw1 (mPeg3), mouse dominant-negative Peg3/Pw1 (mΔPeg3), or a control vector on endogenous human β-catenin protein expression in HeLa cells.
PEG3 is highly conserved between mice and humans (2). Accordingly, we observed that overexpression of mouse Peg3/Pw1 protein in human cells effectively decreased β-catenin protein levels (Fig. 1G). A C-terminal truncated form of mouse Peg3/Pw1 (ΔPeg3) acts as a dominant-negative and inhibits Peg3/Pw1 actions in several mouse model systems, although the mechanism by which this occurs is not known (5, 8, 9, 11, 13). Overexpressed mouse ΔPeg3 increased human β-catenin protein levels in a manner similar to that observed after endogenous Peg3/Pw1 knockdown (Fig. 1F), suggesting that regulation of β-catenin expression may be the site of action of this mutant form of Peg3/Pw1.
Peg3/Pw1 Interacts with β-Catenin to Regulate Its Expression
To determine whether Peg3/Pw1 interacts with β-catenin, we overexpressed a HA-Peg3/Pw1 fusion protein and immunoprecipitated the protein using a specific anti-HA antibody. β-Catenin co-immunoprecipitated with HA-Peg3/Pw1 (Fig. 2A). Conversely, immunoprecipitation of β-catenin resulted in co-immunoprecipitation of HA-Peg3/Pw1. However, HA-Zim2/Peg3β, which shares the seven 5′-exons with Peg3/Pw1 but has a different set of 3′-exons (26), did not co-immunoprecipitate with β-catenin.
FIGURE 2.
A, immunoprecipitation (IP) of HA-Peg3/Pw1 (upper panels) or endogenous β-catenin (lower panels), followed by Western blotting using anti-HA or anti-β-catenin antibodies. Control (Cont) cells were transfected with empty vector. Both input and immunoprecipitated lysates are shown. Note that HA-Peg3/Pw1 (but not HA-Zim2) co-precipitated with β-catenin. B, schematic diagram of HA-tagged expression constructs for full-length Peg3/Pw1, the N-terminal SCAN domain of Peg3/Pw1 (HA-Peg3SCAN), and the C-terminal zinc finger domains of Peg3/Pw1 (HA-Peg3ZF). C, left panel, anti-HA Western blot (WB) for HA-Peg3/Pw1 (second lane), HA-Peg3SCAN (third lane), and HA-Peg3ZF (fourth lane) illustrating the locations of the respective bands on the gel; right panel, anti-HA Western blot of β-catenin immunoprecipitates after overexpression of HA-Peg3/Pw1 (second lane), HA-Peg3SCAN (third lane), and HA-Peg3ZF (fourth lane). Only HA-Peg3/Pw1 and HA-Peg3SCAN immunoprecipitated with β-catenin. D, Western blot analysis illustrating the effect of HA-Peg3/Pw1, HA-Peg3ZF, HA-Peg3SCAN, Peg3/Pw1 (PlentiPeg3), and control vectors (pCMV and PlentiEGFP) on β-catenin expression in HeLa cells. E, fluorescence micrographs showing immunolocalization of HA-Peg3/Pw1, HA-Peg3ZF, and HA-Peg3SCAN (green) in human 293T cells. Nuclei were counterstained with fluorescein isothiocyanate/4′,6-diamidino-2-phenylindole (FITC/DAPI; blue). Scale bar = ∼4 μm.
Peg3/Pw1 contains a protein-protein interaction SCAN domain (27) near the N terminus, as well as 12 downstream C2H2 zinc finger domains. We generated Peg3/Pw1 deletion constructs containing only the N terminus and SCAN domain (HA-Peg3SCAN) or the C terminus and zinc finger domains (HA-Peg3ZF) of Peg3/Pw1 (Fig. 2B). HA-Peg3SCAN (but not HA-Peg3ZF) co-immunoprecipitated with β-catenin (Fig. 2C). Zim2/Peg3β, in which the Peg3/Pw1 SCAN domain is replaced by a KRAB protein-protein interaction domain, failed to immunoprecipitate with β-catenin in immunoprecipitation assays (Fig. 2A). Importantly, overexpression of HA-Peg3SCAN protein increased β-catenin expression, whereas overexpression of HA-Peg3ZF protein had no effect (Fig. 2D). The stimulatory effect of HA-Peg3SCAN on β-catenin expression resembled that of mouse dominant-negative ΔPeg3 that, like HA-Peg3SCAN, contains a C-terminal truncation but retains the N-terminal SCAN domain.
Localization studies using HA-Peg3/Pw1, HA-Peg3SCAN, and HA-Peg3ZF indicated that the C-terminal zinc finger domain of Peg3/Pw1 was sufficient to specify nuclear localization of the protein (Fig. 2E). In contrast, HA-Peg3SCAN was localized primarily in the cytoplasm.
Peg3/Pw1 Cooperates with Siah1 and p53 to Promote β-Catenin Degradation
β-Catenin can be degraded by GSK3β-dependent and GSK3β-independent proteasomal pathways (28, 29). We therefore examined whether Peg3/Pw1 regulation of β-catenin protein levels requires GSK3β-mediated phosphorylation. 293T cells were transfected to overexpress Peg3/Pw1 or a control vector. The proteasome inhibitor MG132 was used to inhibit β-catenin degradation. Cells were then exposed to the GSK3β inhibitor LiCl for varying amounts of time. As expected, LiCl produced a reversible decline in the level of phosphorylated β-catenin and a concomitant increase in total β-catenin (Fig. 3A). Overexpression of Peg3/Pw1 protein had no effect on the rates of phosphorylation or dephosphorylation of β-catenin by GSK3β.
FIGURE 3.
A, Western blot analysis showing the kinetics of GSK3β phosphorylation of β-catenin under control conditions (PCMV) or after Peg3/Pw1 protein overexpression. LiCl (30 mm) was used to inhibit GSK3β. Phosphorylated β-catenin was detected using a phosphospecific antibody. MG132 (25 μm) was added to inhibit β-catenin degradation. B, Western blot illustrating the effect of Peg3/Pw1 shRNA on expression of FLAG-tagged wild-type (WT) or mutant (MT) β-catenin. C, Western blot illustrating the effect of proteasome inhibition by MG132 (25 μm) on Peg3/Pw1-mediated degradation of β-catenin. D, Western blot illustrating the effect of DNA damage induced by camptothecin (CPT; 50 μm) on β-catenin protein expression in HeLa, 293T, U251, and D566 cells. Cont, control. E, Western blot illustrating the effect of DNA damage induced by camptothecin (50 μm) on β-catenin expression after Peg3/Pw1 knockdown in D566 cells. F, Western blot illustrating the effect of Peg3/Pw1, Peg3/Pw1 shRNA, Siah1, and Siah1 shRNA on β-catenin expression in 293T cells. Note that the combination of Siah1 and Peg3/Pw1 decreased β-catenin to a greater extent compared with Peg3/Pw1 after Siah1 knockdown. G, anti-HA Western blot (WB) of Myc-Siah1 immunoprecipitates (IP) after cotransfection of Myc-Siah1 and HA-Peg3/Pw1 (second lane), HA-Peg3SCAN (third lane), and HA-Peg3ZF (fourth lane) in 293T cells. H, Tcf/Lef luciferase reporter assay in 293T cells after overexpression of the human Peg3/Pw1, Peg3/Pw1 shRNA, and/or Siah1 vector. Data shown are means ± S.E.
We next generated a mutant form of β-catenin lacking phosphorylation sites for GSK3β (S33A, S37A, and T41A) and examined the effect of endogenous Peg3/Pw1 on its expression using RNA interference. shRNA-mediated knockdown of Peg3/Pw1 increased both wild-type and mutant β-catenin protein expression, indicating that the effect of Peg3/Pw1 on β-catenin protein expression was independent of phosphorylation by GSK3β (Fig. 3B). Importantly, MG132 antagonized the Peg3/Pw1-dependent decrease in β-catenin expression (Fig. 3C), suggesting that this process involves proteasome activity.
During p53-mediated apoptosis, Peg3/Pw1 is up-regulated and cooperates with Siah1 to induce Bax translocation and cell death via an unknown mechanism (13–15). Separate studies have indicated that Siah1 and p53 can promote β-catenin degradation via a GSK3β-independent proteasomal pathway (28, 29). To investigate whether Peg3/Pw1-mediated down-regulation of β-catenin expression is linked to p53/Siah1-dependent regulation of β-catenin, we examined three cell lines with intact p53 (293T, HeLa, and D566 glioma cells) and one cell line containing mutant p53 (U251 glioma cells). Activation of p53 by exposure to the DNA-damaging agent camptothecin decreased β-catenin protein expression in HeLa, 293T, and D566 glioma cells, but not in U251 glioma cells (Fig. 3D). Importantly, shRNA-mediated knockdown of endogenous Peg3/Pw1 in 293T cells inhibited the p53-dependent degradation of β-catenin occurring after DNA damage (Fig. 3E), indicating a role for Peg3/Pw1 in this process.
To determine whether Siah1 and Peg3/Pw1 cooperate to induce β-catenin degradation, we overexpressed Peg3/Pw1, Siah1, Peg3/Pw1 shRNA, and/or Siah1 shRNA in human 293T cells. Coexpression of Peg3/Pw1 and Siah1 decreased β-catenin more than expression of either Peg3/Pw1 or Siah1 alone (Fig. 3F). Immunoprecipitation studies confirmed binding of Peg3/Pw1 to Siah1. Moreover, we observed that the interaction of Peg3/Pw1 with Siah1 appeared to be mediated via the SCAN domain (Fig. 3G). Additionally, in vitro luciferase reporter assays indicated that the combination of Peg3/Pw1 and Siah1 inhibited β-catenin-dependent transcriptional activity more than either Peg3/Pw1 or Siah1 alone (Fig. 3H). Taken together, these data indicate that Peg3/Pw1 binds with Siah1 and β-catenin to promote β-catenin degradation via a GSK3β-independent pathway.
Loss of Imprinting of PEG3 in Primary Cancers
β-Catenin acts as an oncogene in many cancers (30), raising the possibility that Peg3/Pw1 may suppress tumor growth by promoting β-catenin degradation. To investigate this matter, we first analyzed PEG3 mRNA expression in 32 Grade IV astrocytomas (glioblastomas; glioblastoma multiforme (GBM)) and five human non-tumor brain samples by microarray analysis. Decreased PEG3 mRNA expression was identified in >80% of surgical GBM specimens analyzed (Fig. 4A), and this result was confirmed by real-time PCR in an independent set of GBM samples (Fig. 4B). Using a panel of World Health Organization Grade II–IV astrocytomas, we observed a correlation between decreased PEG3 mRNA expression and increasing histologic grade (p < 0.04, t test) (Fig. 4C).
FIGURE 4.
A, mRNA microarray analysis of PEG3 mRNA in 32 surgical GBM specimens and five surgical non-tumor brain specimens (NTB) showing decreased PEG3 mRNA expression in GBM (p < 0. 001, t test). B, real-time PCR of PEG3 mRNA expression in five human non-tumor brain specimens and 10 human GBM specimens. Samples were assayed in triplicate. Data shown are means ± S.E. C, mRNA microarray analysis for PEG3 in low-grade oligodendroglioma (LGO; n = 15), low-grade astrocytoma (LGA; n = 15), anaplastic astrocytoma (AA; n = 7), and GBM (n = 32). p < 0.04 for GBM versus low-grade astrocytoma and low-grade oligodendroglioma. D, methylation-specific PCR for methylated (M) and unmethylated (U) PEG3 alleles in primary human non-tumor brain, low-grade astrocytoma, and GBM specimens (upper panels) or in five cultured glioma cell lines (lower panel). E, real-time PCR showing the effect of methyltransferase inhibition using 5-azadeoxycytidine (5-aza-dC) on PEG3 mRNA expression in human U87 glioma cells. F, array comparative genomic hybridization analysis of the PEG3 locus on chromosome 19q13.34 in 17 surgical GBM specimens. DNA copy number is plotted in the upper panel. The genomic location of the PEG3 gene (arrow) is shown in the lower panel.
We next used methylation-specific PCR to determine the methylation status of the PEG3 promoter in primary human glioma specimens and in non-tumor brain tissues. As expected for an imprinted gene, both methylated and unmethylated PEG3 alleles were present in non-tumor brain tissues. In contrast, biallelic PEG3 methylation was detected in six of eight low-grade astrocytomas and five of eight glioblastomas (Fig. 4D). We also identified PEG3 hypermethylation in U87, U343, and T98 human glioma cell lines. The D566 human glioma cell line displayed two unmethylated PEG3 alleles, whereas the U251 human glioma cell line exhibited a normal PEG3 methylation pattern. Exposure of U87 human glioma cells to the cytosine methyltransferase inhibitor 5-azadeoxycytidine increased PEG3 mRNA expression (Fig. 4E), indicating that PEG3 gene expression is regulated by methylation in these cells. High-resolution (∼10 kb) array comparative genomic hybridization using DNA extracted from 17 glioblastoma specimens indicated that only one of the tumors showed evidence of DNA loss in the region of the PEG3 gene (Fig. 4F). Thus, methylation appears to represent the primary genomic mechanism for down-regulating PEG3 mRNA expression in human gliomas.
Peg3/Pw1-dependent Regulation of β-Catenin Alters Tumor Growth
Peg3/Pw1 overexpression has been reported to decrease tumorigenicity in an established glioma cell line (20). To investigate the role of endogenous Peg3/Pw1 in gliomas, we knocked down Peg3/Pw1 protein expression in D566 human glioblastoma cells. Peg3/Pw1 knockdown using either of three separate anti-PEG3 shRNAs increased endogenous β-catenin protein expression (Fig. 5A). Knockdown of Peg3/Pw1 decreased PEG3 mRNA expression, increased DNA synthesis, and increased growth in these cells (Fig. 5B), consistent with the observed effects of Peg3/Pw1 on β-catenin protein levels.
FIGURE 5.
A, effect of Peg3/Pw1 knockdown on β-catenin protein expression in D566 glioma cells using three different Peg3/Pw1 shRNAs. B, left panel, effect of three Peg3/Pw1 shRNAs on PEG3 mRNA expression in D566 glioma cells determined by real-time PCR. Data are means ± S.E. of four replicates (p < 0.01, t test). Middle panel, effect of Peg3/Pw1 knockdown on bromodeoxyuridine incorporation into DNA in D566 glioma cells overexpressing three separate Peg3/Pw1 shRNAs. Data are means ± S.E. of seven replicates (p < 0.03, t test). Right panel, 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide growth assay showing the effect of Peg3/Pw1 knockdown on growth of D566 glioma cells overexpressing three separate Peg3/Pw1 shRNAs. Data are means ± S.E. of seven replicates (p < 0.02, t test). C, primary human CD133+ glioma-derived stem-like cells transduced with a control (Cont) or a β-catenin lentivirus and maintained as tumor spheres for 14 days. Scale bar = 80 μm. D, fluorescence micrographs showing β-catenin inhibition of DNA damage-induced apoptosis in glioma-derived stem-like cells revealed by annexin V staining of apoptotic cells (red). Nuclei were counterstained with 4′,6-diamidino-2-phenylindole (blue). Cells were transduced with either a control lentivirus or a β-catenin lentivirus prior to exposure to camptothecin (CPT; 50 μm). The data are quantitated in right panel. E, β-catenin immunoreactivity in non-tumor brain (NTB) and Grade (GR) I–IV astrocytomas. Lower right panel, quantitation of β-catenin immunoreactivity in 29 astrocytomas of different grades (p < 0.04, t test). F, proposed role of Peg3/Pw1 in a p53/Siah1-dependent β-catenin (β-cat) degradation pathway.
Recent studies suggest that human gliomas contain a subpopulation of CD133+ cancer stem-like cells with different biological properties compared with established cell lines (31). To investigate the effect of increased β-catenin in these cells, we isolated CD133+ glioma-derived stem-like cells from fresh glioblastoma specimens and transduced them using a CTNNB1-internal ribosome entry site-enhanced green fluorescent protein lentivirus. After 14 days, tumor spheres containing glioma stem-like cells overexpressing β-catenin were significantly larger than control tumor spheres (Fig. 5C). In addition, β-catenin inhibited p53-dependent, Peg3/Pw1-dependent apoptosis in CD133+ glioma-derived stem-like cells occurring after DNA damage (Fig. 5D). These findings suggest that much of the inhibitory effect of Peg3/Pw1 on the growth and survival of human glioma cells is mediated through Wnt pathway regulation.
We next investigated whether the observed decrease in PEG3 mRNA expression in higher grade gliomas was associated with increased β-catenin protein expression in these tumors. β-Catenin immunohistochemistry using a tissue array containing 29 human astrocytomas of different grades revealed a clear correlation between increased immunoreactivity for β-catenin and increasing histologic grade (Fig. 5E).
A GSK3β-independent, p53-activated proteasomal pathway for β-catenin degradation has been described in which Siah1 forms a complex with APC, SIP, and Ebi to promote β-catenin degradation (29, 30). On the basis of our findings and other evidence, we propose a model in which Peg3/Pw1 participates in this pathway. In this model, p53 activation induces both Peg3/Pw1 and Siah1. These two proteins interact with each other and bind to β-catenin to promote proteasomal β-catenin degradation that is independent of GSK3β phosphorylation (Fig. 5F). This then leads to decreased β-catenin-dependent transcriptional activity.
DISCUSSION
Regulation of Development and Behavior via Peg3/Pw1-dependent Wnt Inhibition
The previously reported pro-apoptotic activity of Peg3/Pw1 does not easily account for its effects on myogenesis or growth and metabolism and does not explain the observation that PEG3 deletion in mice leads to decreased numbers of hypothalamic neurons. The Peg3/Pw1-dependent regulation of β-catenin expression observed here provides a new framework for interpreting these phenomena. We observed that Peg3/Pw1 decreases β-catenin protein expression and inhibits tail development in zebrafish, a process that is Wnt-dependent (25). We also found that Peg3/Pw1 inhibits the Wnt pathway in human cells. In mammals, the Wnt pathway plays a key role in somitogenesis (32), and Wnt inhibition is necessary for proper development of the hypothalamus (33). Thus, increased β-catenin resulting from the loss of Peg3/Pw1 protein expression in the developing nervous system of PEG3−/− mice may underlie the aberrant hypothalamic development and growth retardation observed in these animals. This latter effect on hypothalamic development could serve as the molecular substrate by which imprinting of PEG3 regulates maternal nurturing and male sexual behavior.
Wnt pathway inhibition promotes apoptosis in many cell systems (34). We and others have shown that injury-induced cell death can occur via a pathway that involves p53, Peg3/Pw1, and Bax, and this process can be inhibited by β-catenin (4, 15, 35–37). Until now, the mechanism by which Peg3/Pw1 cooperates with Siah1 to promote Bax translocation and cell death has been a mystery. Our finding that Peg3/Pw1 inhibits Wnt signaling via a p53-dependent, Siah1-dependent pathway provides an explanation for this effect. Here, we have reported the additional findings that β-catenin protein levels are directly regulated by Peg3/Pw1 and that β-catenin inhibits p53/Peg3-dependent cell death in primary glioma-derived stem-like cells. Taken together, our data suggest that a p53-activated, Peg3/Siah1-dependent decrease in β-catenin promotes p53-dependent cell death.
Peg3/Pw1 Suppression of Tumor Growth
Peg3/Pw1 inhibition of Wnt signaling provides a link between this imprinted gene and a major oncogenic pathway. Previous studies have demonstrated hypermethylation and a loss of imprinting of PEG3 in established cell lines derived from several types of human cancers, but the finding that cell culture can alter the methylation status of genes left the significance of those observations in question. We report here that Peg3/Pw1 inhibits DNA synthesis and promotes apoptosis in human glioblastoma cells. Our finding of the anti-proliferative and pro-apoptotic activity of Peg3/Pw1 in human glioblastoma cells, combined with a loss of imprinting of PEG3 in primary human glioblastoma specimens, reveals a clear growth-suppressive role for this protein in human cancer. This places PEG3 in the company of other imprinted genes such as IGF2 and CDKN1C, where a loss of imprinting promotes tumor growth (16, 17). Peg3/Pw1 thus sits at a crossroads between p53 and β-catenin, two major determinants of tumor development in humans.
Acknowledgment
We greatly appreciate Dr. W. Xia for assistance with the zebrafish studies.
This work was supported, in whole or in part, by National Institutes of Health Grant K08 NS43482 and National Institutes of Health Director's New Innovator Award 1DP2OD002319-01 (to M. D. J.). This work was also supported by an American Brain Tumor Association fellowship (to X. J.), a Sontag Distinguished Scientist award, and a Hagerty Fund research award (to M. D. J.).
- HA
- hemagglutinin
- shRNA
- small hairpin RNA
- GBM
- glioblastoma multiforme.
REFERENCES
- 1.Relaix F., Weng X., Marazzi G., Yang E., Copeland N., Jenkins N., Spence S. E., Sassoon D. (1996) Dev. Biol. 177, 383–396 [DOI] [PubMed] [Google Scholar]
- 2.Kim J., Ashworth L., Branscomb E., Stubbs L. (1997) Genome Res. 7, 532–540 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Li L., Keverne E. B., Aparicio S. A., Ishino F., Barton S. C., Surani M. A. (1999) Science 284, 330–333 [DOI] [PubMed] [Google Scholar]
- 4.Yamaguchi A., Taniguchi M., Hori O., Ogawa S., Tojo N., Matsuoka N., Miyake S., Kasai K., Sugimoto H., Tamatani M., Yamashita T., Tohyama M. (2002) J. Biol. Chem. 277, 623–629 [DOI] [PubMed] [Google Scholar]
- 5.Curley J. P., Pinnock S. B., Dickson S. L., Thresher R., Miyoshi N., Surani M. A., Keverne E. B. (2005) FASEB J. 19, 1302–1304 [DOI] [PubMed] [Google Scholar]
- 6.Swaney W. T., Curley J. P., Champagne F. A., Keverne E. B. (2007) Proc. Natl. Acad. Sci. U.S.A. 104, 6084–6089 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Swaney W. T., Curley J. P., Champagne F. A., Keverne E. B. (2008) Behav. Neurosci. 122, 963–973 [DOI] [PubMed] [Google Scholar]
- 8.Relaix F., Wei X. J., Wu X., Sassoon D. A. (1998) Nat. Genet. 18, 287–291 [DOI] [PubMed] [Google Scholar]
- 9.Ledgerwood E. C., O'Rahilly S., Surani M. A. (2000) Lab. Invest. 80, 1509–1511 [DOI] [PubMed] [Google Scholar]
- 10.Coletti D., Yang E., Marazzi G., Sassoon D. (2002) EMBO J. 21, 631–642 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nicolas N., Marazzi G., Kelley K., Sassoon D. (2005) Dev. Biol. 281, 171–183 [DOI] [PubMed] [Google Scholar]
- 12.Schwarzkopf M., Coletti D., Sassoon D., Marazzi G. (2006) Genes Dev. 20, 3440–3452 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Relaix F., Wei X., Li W., Pan J., Lin Y., Bowtell D. D., Sassoon D. A., Wu X. (2000) Proc. Natl. Acad. Sci. U.S.A. 97, 2105–2110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Deng Y., Wu X. (2000) Proc. Natl. Acad. Sci. U.S.A. 97, 12050–12055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Johnson M. D., Wu X., Aithmitti N., Morrison R. S. (2002) J. Biol. Chem. 277, 23000–23007 [DOI] [PubMed] [Google Scholar]
- 16.Okamoto K., Morison I. M., Taniguchi T., Reeve A. E. (1997) Proc. Natl. Acad. Sci. U.S.A. 94, 5367–5371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pantoja C., de Los Ríos L., Matheu A., Antequera F., Serrano M. (2005) Cancer Res. 65, 26–33 [PubMed] [Google Scholar]
- 18.Maegawa S., Yoshioka H., Itaba N., Kubota N., Nishihara S., Shirayoshi Y., Nanba E., Oshimura M. (2001) Mol. Carcinog. 31, 1–9 [DOI] [PubMed] [Google Scholar]
- 19.Dowdy S. C., Gostout B. S., Shridhar V., Wu X., Smith D. I., Podratz K. C., Jiang S. W. (2005) Gynecol. Oncol. 99, 126–134 [DOI] [PubMed] [Google Scholar]
- 20.Kohda T., Asai A., Kuroiwa Y., Kobayashi S., Aisaka K., Nagashima G., Yoshida M. C., Kondo Y., Kagiyama N., Kirino T., Kaneko-Ishino T., Ishino F. (2001) Genes Cells 6, 237–247 [DOI] [PubMed] [Google Scholar]
- 21.Yu Y., Jiang X., Schoch B. S., Carroll R. S., Black P. M., Johnson M. D. (2007) Cancer Res. 67, 130–138 [DOI] [PubMed] [Google Scholar]
- 22.Santelli E., Leone M., Li C., Fukushima T., Preece N. E., Olson A. J., Ely K. R., Reed J. C., Pellecchia M., Liddington R. C., Matsuzawa S. (2005) J. Biol. Chem. 280, 34278–34287 [DOI] [PubMed] [Google Scholar]
- 23.Sadot E., Conacci-Sorrell M., Zhurinsky J., Shnizer D., Lando Z., Zharhary D., Kam Z., Ben-Ze'ev A., Geiger B. (2002) J. Cell Sci. 115, 2771–2780 [DOI] [PubMed] [Google Scholar]
- 24.Wang Z., Vogelstein B., Kinzler K. W. (2003) Cancer Res. 63, 5234–5235 [PubMed] [Google Scholar]
- 25.Thorpe C. J., Weidinger G., Moon R. T. (2005) Development 132, 1763–1772 [DOI] [PubMed] [Google Scholar]
- 26.Kim J., Bergmann A., Stubbs L. (2000) Genomics 64, 114–118 [DOI] [PubMed] [Google Scholar]
- 27.Edelstein L. C., Collins T. (2005) Gene 359, 1–17 [DOI] [PubMed] [Google Scholar]
- 28.Matsuzawa S. I., Reed J. C. (2001) Mol. Cell 7, 915–926 [DOI] [PubMed] [Google Scholar]
- 29.Liu J., Stevens J., Rote C. A., Yost H. J., Hu Y., Neufeld K. L., White R. L., Matsunami N. (2001) Mol. Cell 7, 927–936 [DOI] [PubMed] [Google Scholar]
- 30.Moon R. T., Kohn A. D., De Ferrari G. V., Kaykas A. (2004) Nat. Rev. Genet. 5, 691–701 [DOI] [PubMed] [Google Scholar]
- 31.Singh S. K., Hawkins C., Clarke I. D., Squire J. A., Bayani J., Hide T., Henkelman R. M., Cusimano M. D., Dirks P. B. (2004) Nature 432, 396–401 [DOI] [PubMed] [Google Scholar]
- 32.Borello U., Berarducci B., Murphy P., Bajard L., Buffa V., Piccolo S., Buckingham M., Cossu G. (2006) Development 133, 3723–3732 [DOI] [PubMed] [Google Scholar]
- 33.Kapsimali M., Caneparo L., Houart C., Wilson S. W. (2004) Development 131, 5923–5933 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Huang M., Wang Y., Sun D., Zhu H., Yin Y., Zhang W., Yang S., Quan L., Bai J., Wang S., Chen Q., Li S., Xu N. (2006) BMC Cancer 6, 221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhang Z., Hartmann H., Do V. M., Abramowski D., Sturchler-Pierrat C., Staufenbiel M., Sommer B., van de Wetering M., Clevers H., Saftig P., De Strooper B., He X., Yankner B. A. (1998) Nature 395, 698–702 [DOI] [PubMed] [Google Scholar]
- 36.Johnson M. D., Yu L. R., Conrads T. P., Kinoshita Y., Uo T., Matthews J. D., Lee S. W., Smith R. D., Veenstra T. D., Morrison R. S. (2004) J. Biol. Chem. 279, 26685–26697 [DOI] [PubMed] [Google Scholar]
- 37.Li H. L., Wang H. H., Liu S. J., Deng Y. Q., Zhang Y. J., Tian Q., Wang X. C., Chen X. Q., Yang Y., Zhang J. Y., Wang Q., Xu H., Liao F. F., Wang J. Z. (2007) Proc. Natl. Acad. Sci. U.S.A. 104, 3591–3596 [DOI] [PMC free article] [PubMed] [Google Scholar]