The crystal structure of a fully active laccase from Trametes sp. AH28-2 is reported.
Keywords: laccases, blue multicopper oxidases, Trametes sp. AH28-2
Abstract
Fungal laccases are oxidoreductases that belong to the multinuclear copper-containing oxidases. They are able to oxidize a wide range of substrates, preferably phenolic compounds, which makes them suitable for employment in the bioremediation of soil and water as well as in other biotechnological applications. Here, the structural analysis of natural laccase B (LacB) from Trametes sp. AH28-2 is presented. This structure provides the opportunity to study the natural post-translational modifications of the enzyme. The overall fold shows a high homology to those of previously analyzed laccases with known three-dimensional structure. However, LacB contains a new structural element, a protruding loop near the substrate-binding site, compared with the previously reported laccase structures. This unique structural feature may be involved in modulation of the substrate recognition of LacB.
1. Introduction
Laccases (benzenediol:oxygen oxidoreductases; EC 1.10.3.2) are copper-containing polyphenol oxidases that can oxidize a wide range of aromatic compounds concomitantly with the reduction of molecular oxygen to water, bypassing the stage of hydrogen peroxide production (Solomon et al., 1996 ▶). They have been found in plants, insects, bacteria and nearly all wood-rotting fungi. In plants, laccases participate in lignin biosynthesis (Bao et al., 1993 ▶). Bacterial laccases are involved in morphogenesis, pigment biosynthesis and copper homeostasis (Suzuki et al., 2003 ▶; Hullo et al., 2001 ▶). In insects, laccases appear to play a role in sclerotization of the cuticle in the epidermis (Dittmer et al., 2004 ▶).
Fungal laccases are the most important group of blue multicopper oxidases (MCOs) with respect to number and extent of characterization. They are involved in the degradation of lignin, pathogenesis and detoxification and also in the development and morphogenesis of fungi (Baldrian, 2006 ▶; Leonowicz et al., 2001 ▶; Eggert et al., 1997 ▶). Most fungal laccases are extracellular monomeric globular proteins with an extent of glycosylation in the range 10–25%. They generally contain four Cu atoms per molecule, with the four Cu atoms classified as one type 1 (T1), one type 2 (T2) and two type 3 (T3). All of the copper ions are apparently involved in the catalytic mechanism (Palmer et al., 2001 ▶; Garavaglia et al., 2004 ▶).
Trametes sp. AH28-2 is a white-rot basidiomycete that selectively degrades lignin when grown on wood and reduces the level of chemical oxygen demand (COD) in the waste water produced in the process of straw pulping. It has been shown to secrete several laccase isozymes (Xiao et al., 2004 ▶, 2006 ▶). Aromatic inducers with different substituted groups can selectively stimulate Trametes sp. AH28-2 to synthesize distinct laccase isozymes. ο-Toluidine induces the expression of laccase A (LacA), while 3,5-dihydroxytoluene mainly stimulates the production of laccase B (LacB). Furthermore, low Cu2+ concentrations (lower than 0.5 mM) can induce laccase C (LacC), which is nearly undetectable in the presence of aromatic compounds. LacA accounts for about 85% of the total activity after induction by kraft lignin. Its molecular mass is 62 kDa and it consists of a monomeric glycoprotein with about 12% carbohydrates and an isoelectric point of 4.2. It shows a good stability from pH 4.2 to pH 8.0 (Xiao et al., 2003 ▶).
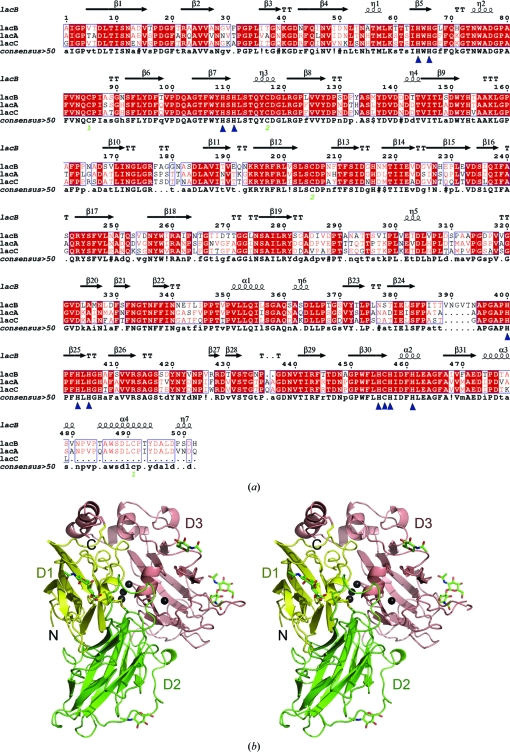
Similar to LacA, LacB is also a typical fungal laccase. It is an extracellular monomeric glycoprotein of approximately 71.5 kDa with a carbohydrate content of about 25% and an isoelectric point of 4.0 (Xiao et al., 2004 ▶). LacB shares 74% and 69% sequence similarity to LacA and LacC, respectively (Fig. 1 ▶ a). Compared with LacA, LacB has a lower optimal reaction temperature and a higher pH optimum. In addition, the thermal stability of LacB is inferior to that of LacA (Xiao et al., 2004 ▶).
Figure 1.
(a) Structure-based sequence alignment of three laccase isozymes from Trametes sp. AH28-2. Conserved residues are enclosed in blue boxes. η1, η2, η3, η4 and η5 are 310-helices. TT indicates tight β-turns. Residues in LacB that interact with copper ions are indicated by blue triangles. The amino-acid sequences were aligned using MultAlin (Corpet, 1988 ▶) and the figure was generated using ESPript (Gouet et al., 1999 ▶). (b) Schematic representation of the Trametes sp. AH28-2 LacB structure (stereoview). Domains 1, 2 and 3 are shown in yellow, green and salmon, respectively. Copper ions are drawn as black spheres. Carbohydrates and disulfide bonds are shown as stick models.
Here, we report the structure of active LacB from Trametes sp. AH28-2 at 3.4 Å resolution. Although the resolution is low, the corresponding electron densities are of good quality, which allowed the fitting of all amino-acid residues, all copper ions and some sugar residues. Thus, this structure provides the opportunity to study the natural post-translational modifications of the enzyme. The overall structure is very similar to the canonical laccase topology. However, one apparent feature of LacB distinguishes it from other laccases of known structure: a protruding loop near the substrate-binding site.
2. Materials and methods
2.1. Purification, crystallization and data collection
The purification of native LacB from Trametes sp. AH28-2 was performed essentially as described previously (Xiao et al., 2004 ▶). The protein used for crystallization was stored at a concentration of 10 mg ml−1 in 10 mM citrate/Na2HPO4 pH 6.0.
Crystallization trials were performed at 283 K by the hanging-drop vapour-diffusion method against 300 µl reservoir solution using the Crystal Screen I and Crystal Screen II reagent kits from Hampton Research. Rod-like crystals, which showed a characteristic blue colour caused by the presence of type 1 copper in the protein, were grown by mixing 2.0 µl protein solution and an equal volume of reservoir solution containing 0.1 M sodium citrate tribasic dihydrate pH 5.2, 25% PEG 4000 and 10% 2-propanol after 20 d. For data collection, the crystal was washed in a cryoprotectant solution consisting of 0.1 M sodium citrate tribasic dihydrate pH 5.2, 25% PEG 4000, 10% 2-propanol and 15% glycerol. A complete data set to 3.4 Å resolution was collected at 100 K on beamline 3W1A of the BSRF (Beijing Synchrotron Radiation Facility) at a wavelength of 1.1000 Å and was processed using HKL-2000 (Otwinowski & Minor, 1997 ▶).
2.2. Structure determination and refinement
The structure of native LacB was solved by molecular replacement using the program MOLREP (Vagin & Teplyakov, 1997 ▶). The TvL structure (T. versicolor laccase; PDB code 1gyc; Piontek et al., 2002 ▶), which has 72% identity to the target structure, was used as the search model. The model was completed by iterative rounds of manual building in Coot (Emsley & Cowtan, 2004 ▶) and refinement in REFMAC (Murshudov et al., 1997 ▶). The final refined model, including all amino-acid residues and eight coppers, had an R factor and R free of 22.3% and 29.2%, respectively. The quality of the model was checked using the program PROCHECK (Laskowski et al., 1993 ▶). The refinement statistics are summarized in Table 1 ▶. The coordinates and structure factors have been deposited in the Protein Data Bank under accession code 3kw7.
Table 1. Data-collection and refinement statistics.
Values in parentheses are for the outer shell.
| Wavelength (Å) | 1.1000 |
| Space group | P41 |
| Molecules in asymmetric unit | 2 |
| Unit-cell parameters (Å, °) | a = b = 98.78, c = 149.96, α = β = γ = 90 |
| Resolution range (Å) | 44.6–3.44 (3.55–3.44) |
| No. of unique reflections | 19312 |
| Solvent content (%) | 62.87 |
| Rmerge† (%) | 16.5 (48) |
| 〈I/σ(I)〉 | 8.0 (2.2) |
| Redundancy | 98.9 (97.4) |
| Refinement summary | |
| R factor‡ (%) | 22.3 |
| Free R factor§ (%) | 29.2 |
| R.m.s.d.¶ bond lengths (Å) | 0.015 |
| R.m.s.d. bond angles (°) | 1.504 |
| No. of atoms in asymmetric unit | 7825 |
| Ramachandran plot | |
| Most favoured (%) | 83.8 |
| Additional allowed (%) | 14.9 |
| Generously allowed (%) | 1.1 |
| Disallowed (%) | 0.2 |
| PDB code | 3kw7 |
R
merge = 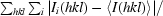
 , where 〈I(hkl)〉 is the mean intensity of the i observations of reflection hkl.
, where 〈I(hkl)〉 is the mean intensity of the i observations of reflection hkl.
R factor = 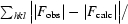
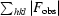 , where |F
obs| and |F
calc| are the observed and calculated structure-factor amplitudes, respectively. The summation includes all reflections used in the refinement.
, where |F
obs| and |F
calc| are the observed and calculated structure-factor amplitudes, respectively. The summation includes all reflections used in the refinement.
Free R factor = 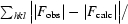
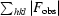 evaluated for a randomly chosen subset of 5% of the diffraction data not included in the refinement.
evaluated for a randomly chosen subset of 5% of the diffraction data not included in the refinement.
Root-mean-square deviation from ideal values.
2.3. Sequence analysis and structural presentation
Amino-acid sequences were aligned using MultAlin (Corpet, 1988 ▶) and the structure-based sequence-alignment figure was generated using ESPript (Gouet et al., 1999 ▶). All illustrations were prepared with PyMOL (DeLano, 2002 ▶).
3. Results and discussion
3.1. Overall structure
We determined the 3.4 Å resolution structure of fully active LacB from Trametes sp. AH28-2 and refined it to R-factor and R free values of 22.4% and 29.2%, respectively (Table 1 ▶). LacB crystallized with two molecules in the asymmetric unit and there was no evidence of higher order oligomeric assemblies in the crystal lattice, which was consistent with the purification of LacB as a monomer by size-exclusion chromatography (Xiao et al., 2004 ▶). The electrostatic surface-potential distribution of LacB reveals a dominance of negative charges, which is in accordance with its acidic pI of about 4.0. The LacB protomer is similar to the canonical globular laccase architecture and consists of three sequentially arranged domains of a β-barrel-type structure (Figs. 1 ▶ a and 1 ▶ b).
Domain 1 (Ala1–Asp128) comprises three short 310-helices (η1, η2 and η3) and two β-sheets. One β-sheet is formed by four antiparallel β-strands (β2, β1, β4 and β6) and the other contains four mixed-type β-strands (β5, β7, β8 and β3). The three 310-helices are in the connecting peptides between the β-strands.
Domain 2 (residues Pro129–Leu308) contains one six-stranded β-sheet of antiparallel type (β10, β9, β12, β17, β14 and β15) and one five-stranded β-sheet of mixed type (β16, β13, β18, β19 and β11) and there are 310-helices in the connecting peptides between domains 1 and 2 and between domains 2 and 3. One 310-helix (η5) forms part of a 40-residue-long extended loop region. Domain 3 (Asp309–His502) consists of four α-helices, two short 310-helices and one β-barrel formed by two five-stranded β-sheets and one β-hairpin (β21 and β22). The first β-sheet is of antiparallel type and contains β-strands β20, β24, β29, β26 and β27, while the other mixed-type β-sheet strand contains β-strands β28, β25, β30, β31 and β23. One 310-helix (η6) and two α-helices (α1 and α2) are located in the connecting regions between the β-strands of the different β-sheets. At the C-terminus of domain 3, α3, α4 and η7 complete the fold. Finally, the structure is further stabilized by two disulfide bridges. One of them (Cys85–Cys491) connects the C-terminal portion to domain 1, whereas the other (Cys117–Cys205) connects domains 1 and 2.
The LacB active site consists of a mononuclear T1 copper site and a trinuclear cluster T2/T3. The T1 copper site is located between domains 2 and 3, whereas the T2/T3 cluster is positioned in the cavity between domains 1 and 3.
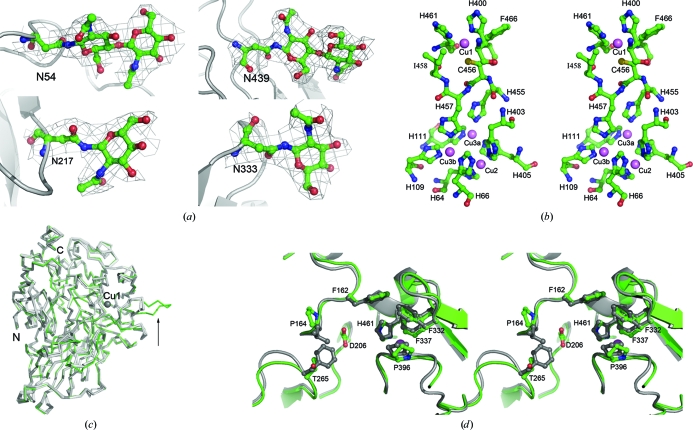
LacB is a monomeric glycoprotein with 25% carbohydrate content (Xiao et al., 2004 ▶). Of the 11 predicted N-glycosylation sites (consensus sequence N-X-T/S), the density maps clearly indicate the presence of glycosylation at Asn54, Asn217, Asn333 and Asn439. The corresponding electron densities were modelled as two N-acetyl-d-glucosamine residues bound to Asn54, one N-acetyl-d-glucosamine residue bound to Asn217, one N-acetyl-d-glucosamine residue bound to Asn333 and a di(N-acetyl-d-glucosamine) moiety bound to Asn439 (Fig. 2 ▶ a). Because of the low-quality corresponding densities, sugar residues could not be modelled in the densities at Asn181, Asn292, Asn341 and Asn377, although the densities suggest that glycosylation probably occurs at these sites.
Figure 2.
(a) OMIT electron-density maps (contoured at 1σ) of sugar residues bound to Asn54, Asn217, Asn333 and Asn439. (b) Stereoview of the four copper sites in LacB from Trametes sp. AH28-2. Copper ions are shown in violet. (c) Backbone-trace representation of LacB (green) compared with other laccases of known structure (grey): ThL (PDB code 3fpx), CmL (Cerrena maxima laccase; PDB code 3div; A. V. Lyashenko, Y. N. Zhukova & A. M. Mikhailov, unpublished work), ToL (Coriolus zonatus laccase; PDB code 2hzh; A. V. Lyashenko, N. E. Zhukhlistova, A. G. Gabdoulkhakov, Y. N. Zhukova, W. Voelter, V. N. Zaitsev, I. Bento, E. V. Stepanova, G. S. Kachalova, O. V. Koroleva, C. Betzel, P. F. Lindley, A. M. Mikhailov, V. I. Tishkov & E. Y. Morgunova, unpublished work), LtL (Lentinus tigrinus laccase; PDB code 2qt6; Ferraroni et al., 2007 ▶), TmL (PDB code 2h5u), TvL (PDB codes 1gyc and 1kya) and TtL (PDB code 2hrg). The unique loop of LacB is marked by a black arrow. (d) Stereo diagram showing the superposition of the substrate-binding pockets of LacB (green) and TvL (grey).
3.2. Copper sites
The T1 copper site is very close to the potential substrate-binding cavity. Similar to other multi-copper oxidases, this copper ion has a planar triangular coordination by two histidines (His400 and His461) and one cysteine (Cys456). Phe466 is in the axial position in the T1 centre and Ile458 is located on the other side. According to the structures of other high-redox-potential laccases such as those from T. hirsute (ThL; PDB code 3fpx; Polyakov et al., 2009 ▶), T. maxima (TmL; PDB code 2h5u; Lyashenko et al., 2006 ▶), T. versicolor (TvL; PDB codes 1gyc and 1kya; Piontek et al., 2002 ▶; Bertrand et al., 2002 ▶) and T. trogii (TtL; PDB code 2hrg; Lyashenko et al., 2006 ▶), the two hydrophobic residues at a distance of about 3.4–3.8 Å from the copper ion are thought to be important for the high redox potential of the site. The type 1 copper site is also termed the ‘blue’ copper site as it confers the typical blue colour to proteins of this family. In the catalytic process the electrons acquired by the T1 copper from the reducing substrates are then transferred to the T2/T3 cluster through the conserved His455-Cys456-His457 triad. This intramolecular electron-transfer pathway starts from the T1 copper ligand Cys456 and subsequently splits between His455 and His457, which bind to the T3(a) and T3(b) copper ions, respectively.
The T2/T3 cluster is located deep inside the molecule, approximately 12.1 Å away from the T1 centre. This cluster is coordinated by His64, His66, His109, His111, His403, His405, His455 and His457. The three-dimensional structure of the copper site is shown in Fig. 2 ▶(b). There is nothing unusual about the Cu-ligand geometry compared with other laccases of known structure.
3.3. Substrate-binding pocket
Based on the analysis of other laccases of known structure, the binding of substrates occurs close to the T1 centre in the substrate-binding region formed by loops 154–165, 203–209, 261–270, 330–339, 384–399 and α2 in LacB. These loops have different lengths and amino-acid compositions in laccases from different organisms. This results in the diversity in the size and shape of the substrate-binding region. Interestingly, a protruding loop comprising amino acids 389–393 forms an extended jut near the substrate-binding site in LacB (Fig. 2 ▶ c). No similar element has been found in previously analyzed laccases with known three-dimensional structures. Therefore, this structural element represents a distinctive feature of LacB. This fragment is also a particular feature of the isoenzymes from Trametes sp. AH28-2.
On the basis of fungal laccase complex structures, namely TvL complexed with xylidine (Piontek et al., 2002 ▶; Bertrand et al., 2002 ▶), TtL complexed with the inhibitor p-toluate (Matera et al., 2008 ▶) and rMaL (Melanocarpus albomyces laccase) complexed with 2,6-DMP (Kallio et al., 2009 ▶), the potential substrate-binding pocket of LacB is a hydrophobic cavity formed by residues Phe162, Pro164, Thr265, Phe332, Phe337, Pro396 and His461. The overall shape of the binding pocket of LacB is very similar to that of TvL. However, it is slightly more spacious than that of TvL because Leu164 and Phe265 in TvL are replaced by Pro164 and Thr265, respectively (Fig. 2 ▶ d). The hydrophilic amino acid Asp206, which is considered to play a key role in the binding of organic substrates to His461, is at the bottom of this cavity.
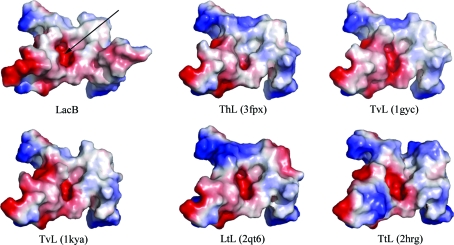
The surface topology and electrostatics of the substrate-binding region are widely divergent between LacB and other fungal laccases of known structure (Fig. 3 ▶). Owing to the protruding loop, LacB possesses more electronegative potential near the substrate-binding pocket than other fungal laccases. This feature is likely to be involved in the modulation of substrate recognition of LacB. This extended jut, as well as the high level of glycosylation, may contribute to the lower molecular stability and lower isoelectric point compared with LacA.
Figure 3.
Comparison of the electrostatic surface plots of substrate-binding sites between LacB and other laccases. The positive surface is drawn in blue and the negative surface in red. The substrate-binding pockets are marked by black arrows.
Supplementary Material
PDB reference: laccase B, 3kw7
Acknowledgments
We thank the staff for their assistance in data collection at the Beijing Synchrotron, People’s Republic of China. This work was supported by Grant No. KJ2008A127 from the Natural Science Foundation of the Department of Education of Anhui Province and by Grant No. 02203105 from the Start-up Foundation for Dr of Anhui University.
References
- Baldrian, P. (2006). FEMS Microbiol. Rev.30, 215–242. [DOI] [PubMed]
- Bao, W., O’Malley, D. M., Whetten, R. & Sederoff, R. R. (1993). Science, 260, 672–674. [DOI] [PubMed]
- Bertrand, T., Jolivalt, C., Briozzo, P., Caminade, E., Joly, N., Madzak, C. & Mougin, C. (2002). Biochemistry, 41, 7325–7333. [DOI] [PubMed]
- Corpet, F. (1988). Nucleic Acids Res.16, 10881–10890. [DOI] [PMC free article] [PubMed]
- DeLano, W. L. (2002). The PyMOL Molecular Viewer. http://www.pymol.org.
- Dittmer, N. T., Suderman, R. J., Jiang, H., Zhu, Y. C., Gorman, M. J., Kramer, K. J. & Kanost, M. R. (2004). Insect Biochem. Mol. Biol.34, 29–41. [DOI] [PubMed]
- Eggert, C., Temp, U. & Eriksson, K. E. (1997). FEBS Lett.407, 89–92. [DOI] [PubMed]
- Emsley, P. & Cowtan, K. (2004). Acta Cryst. D60, 2126–2132. [DOI] [PubMed]
- Ferraroni, M., Myasoedova, N. M., Schmatchenko, V., Leontievsky, A. A., Golovleva, L. A., Scozzafava, A. & Briganti, F. (2007). BMC Struct. Biol.7, 60. [DOI] [PMC free article] [PubMed]
- Garavaglia, S., Cambria, M. T., Miglio, M., Ragusa, S., Iacobazzi, V., Palmieri, F., D’Ambrosio, C., Scaloni, A. & Rizzi, M. (2004). J. Mol. Biol.342, 1519–1531. [DOI] [PubMed]
- Gouet, P., Courcelle, E., Stuart, D. I. & Métoz, F. (1999). Bioinformatics, 15, 305–308. [DOI] [PubMed]
- Hullo, M. F., Moszer, I., Danchin, A. & Martin-Verstraete, I. (2001). J. Bacteriol.183, 5426–5430. [DOI] [PMC free article] [PubMed]
- Kallio, J. P., Auer, S., Jänis, J., Andberg, M., Kruus, K., Rouvinen, J., Koivula, A. & Hakulinen, N. (2009). J. Mol. Biol.392, 895–909. [DOI] [PubMed]
- Laskowski, R. A., MacArthur, M. W., Moss, D. S. & Thornton, J. M. (1993). J. Appl. Cryst.26, 283–291.
- Leonowicz, A., Cho, N. S., Luterek, J., Wilkolazka, A., Wojtas-Wasilewska, M., Matuszewska, A., Hofrichter, M., Wesenberg, D. & Rogalski, J. (2001). J. Basic Microbiol.41, 185–227. [DOI] [PubMed]
- Lyashenko, A. V. et al. (2006). Acta Cryst. F62, 954–957. [DOI] [PMC free article] [PubMed]
- Matera, I., Gullotto, A., Tilli, S., Ferraroni, M., Scozzafava, A. & Briganti, F. (2008). Inorg. Chim. Acta, 361, 4129–4137.
- Murshudov, G. N., Vagin, A. A. & Dodson, E. J. (1997). Acta Cryst. D53, 240–255. [DOI] [PubMed]
- Otwinowski, Z. & Minor, W. (1997). Methods Enzymol.276, 307–326. [DOI] [PubMed]
- Palmer, A. E., Lee, S. K. & Solomon, E. I. (2001). J. Am. Chem. Soc.123, 6591–6599. [DOI] [PubMed]
- Piontek, K., Antorini, M. & Choinowski, T. (2002). J. Biol. Chem.277, 37663–37669. [DOI] [PubMed]
- Polyakov, K. M., Fedorova, T. V., Stepanova, E. V., Cherkashin, E. A., Kurzeev, S. A., Strokopytov, B. V., Lamzin, V. S. & Koroleva, O. V. (2009). Acta Cryst. D65, 611–617. [DOI] [PubMed]
- Solomon, E. I., Sundaram, U. M. & Machonkin, T. E. (1996). Chem. Rev.96, 2563–2606. [DOI] [PubMed]
- Suzuki, T., Endo, K., Ito, M., Tsujibo, H., Miyamoto, K. & Inamori, Y. (2003). Biosci. Biotechnol. Biochem.67, 2167–2175. [DOI] [PubMed]
- Vagin, A. & Teplyakov, A. (1997). J. Appl. Cryst.30, 1022–1025.
- Xiao, Y. Z., Chen, Q., Wu, J., Hang, J., Hong, Y. Z., Shi, Y. Y. & Wang, Y. P. (2004). Mycologia, 96, 26–35. [PubMed]
- Xiao, Y. Z., Hong, Y. Z., Li, J. F., Hang, J., Tong, P. G., Fang, W. & Zhou, C. Z. (2006). Appl. Microbiol. Biotechnol.71, 493–501. [DOI] [PubMed]
- Xiao, Y. Z., Tu, X. M., Wang, J., Zhang, M., Cheng, Q., Zeng, W. Y. & Shi, Y. Y. (2003). Appl. Microbiol. Biotechnol.60, 700–707. [DOI] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
PDB reference: laccase B, 3kw7
PDB reference: laccase B, 3kw7