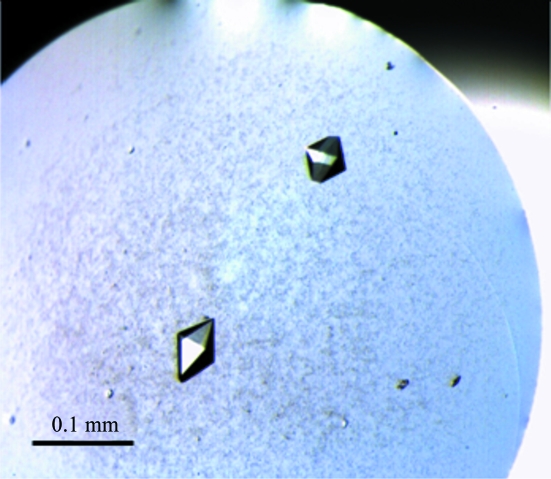
Crystals of B. cepacia IST408 UDP-glucose dehydrogenase (EC 1.1.1.22) were obtained in space group P212121 and diffracted to 2.09 Å resolution.
Keywords: UDP-glucose dehydrogenases, Burkholderia cepacia, BceC
Abstract
Bacteria of the Burkholderia cepacia complex (Bcc) have emerged as important opportunistic pathogens, establishing lung infections in immunocompromised or cystic fibrosis patients. Bcc uses polysaccharide-biofilm production in order to evade the host immune response. The biofilm precursor UDP-glucuronic acid is produced by a twofold NAD+-dependent oxidation of UDP-glucose. In B. cepacia IST408 this enzymatic reaction is performed by the UDP-glucose dehydrogenase BceC, a 470-residue enzyme, the production and crystallization of which are described here. The crystals belonged to the orthorhombic space group P212121 and contained four molecules in the asymmetric unit. Their crystallographic analysis at 2.09 Å resolution and a molecular-replacement study are reported.
1. Introduction
Bacteria belonging to the Burkholderia cepacia complex (Bcc), a group of 17 closely related species, have emerged as highly problematic opportunistic human pathogens in patients with cystic fibrosis (CF). Bcc members often express a mucoid phenotype that is associated with the production of large amounts of exopolysaccharides (EPS), suggesting a possible role of EPS in the persistence of Bcc in the airways of CF patients (Cunha et al., 2004 ▶; Sousa et al., 2007 ▶). Since the sugar compositions of the EPS produced by different members of the Bcc complex are somewhat similar, this type of extracellular polysaccharide was termed cepacian (Sist et al., 2003 ▶). Cepacian is composed of a branched acetylated heptasaccharide-repeat unit with d-glucose, d-rhamnose, d-mannose, d-galactose and d-glucuronic acid in a 1:1:1:3:1 ratio (Cescutti et al., 2000 ▶). The pathway leading to the nucleotide sugar precursors necessary for cepacian biosynthesis has been proposed for the mucoid CF clinical isolate B. cepacia IST408 (Richau et al., 2000 ▶) and the cepacian cluster of genes (named bce) directing its biosynthesis has been identified (Moreira et al., 2003 ▶).
Our interest in B. cepacia IST408 uridine-5′-diphosphoglucose (UDP-glucose) dehydrogenase (encoded by the gene bceC) arises from its pivotal role in providing the UDP-glucuronic acid (UDP-GlcA) precursor necessary for cepacian biosynthesis. UDP-glucose dehydrogenase (UGD; EC 1.1.1.22) catalyzes the twofold NAD+-dependent oxidation of UDP-glucose (UDP-Glc) to UDP-glucuronic acid (UDP-GlcA; Campbell et al., 1997 ▶; Campbell & Tanner, 1999 ▶). UGDs have been the subject of several studies because they are present not only in bacteria but also in multiple prokaryotic and eukaryotic cells, in which UDP-GlcA is the activated donor of d-glucuronic acid, which can be converted into other compounds and serves in many critical roles in different metabolic pathways from mammals to bacteria (Campbell et al., 2000 ▶; Hwang & Horvitz, 2002 ▶; Griffith et al., 2004 ▶; Sommer et al., 2004 ▶; Stewart & Copeland, 2006 ▶). At the time of writing, four X-ray crystal structures of UGD enzymes are publicly available, namely those from the prokaryotes Streptococcus pyogenes (PDB entry 1dli; Campbell et al., 2000 ▶) and Porphyromonas gingivalis (PDB entry 3gg2; J. B. Bonanno, J. Freeman, K. T. Bain, S. Chang, P. Sampathkumar, S. Wasserman, J. M. Sauder, S. K. Burley, S. K. & S. C. Almo, unpublished work) and two eukaryotic representatives from Caenorhabditis elegans (PDB entry 2o3j; Y. Zhang, C. Zhan, Y. Patskovsky, U. Ramagopal, W. Shi, R. Toro, B. C. Wengerter, S. Miltein, M. Vidal, S. K. Burley & S. C. Almo, unpublished work) and from human (PDB entries 2qg4 and 2q3e; K. L. Kavanagh, K. Guo, G. Bunkoczi, P. Savitsky, E. Pilka, C. Bhatia, F. Niesen, C. Smee, G. Berridge, F. Von Delft, J. Wiegelt, C. Arrowsmith, M. Sundstrom, A. Edwards & U. Oppermann, unpublished work). The determination of several three-dimensional structures of UGDs will allow the identification of the conserved stereochemical features within this family of enzymes, which may lead to a deeper understanding of their reaction mechanism.
2. Materials and methods
2.1. Cloning and expression of the bceC gene from B. cepacia IST408
The bceC gene from B. cepacia IST408 was amplified by polymerase chain reaction (PCR) using its genomic DNA as template, Pwo DNA polymerase (Roche Diagnostics, Mannheim, Germany) and the oligonucleotides BPCRbceC (5′-GGGGGATCCATGAATCTGACTAT-3′) and HPCRbceC (5′-GGGAAGCTTGAAACGGGTTAC-3′) designed based on the bceC nucleotide sequence. The PCR product (1413 bp) was digested with BamHI and HindIII (recognition sites are shown in bold) and cloned into the cloning vector pWH844 (Schirmer et al., 1997 ▶), generating pBceC.
This plasmid carries the bceC gene preceded by a sequence coding for six histidines for purification purposes, a glycine residue and a serine residue. The DNA insert cloned in pBceC was sequenced to confirm the fidelity of DNA amplification. The nucleotide sequence of the bceC gene from B. cepacia IST408 has been deposited in the GenBank database with accession No. GQ451909.
Overexpression of the bceC gene was carried out by cultivation of Escherichia coli SURE (Stratagene) transformants harbouring the plasmid pBceC in 1000 ml LB medium supplemented with 100 mg ml−1 ampicillin at 310 K until an OD600nm of 0.6 was reached. The cells were then induced with 0.3 mM isopropyl β-d-1-thiogalactopyranoside (IPTG) for 6 h at 301 K and harvested by centrifugation (10 000g, 30 min, 277 K); the pellets obtained were stored at 253 K.
2.2. BceC purification
The cells were resuspended in 20 mM sodium phosphate pH 7.4, 20 mM imidazole, 1 M NaCl and disrupted in a French press. Crude cell extract was obtained by centrifugation at 27 000g for 40 min at 277 K and the supernatant was applied onto a 5 ml HisTrap column (GE Healthcare) pre-equilibrated with 20 mM sodium phosphate pH 7.4, 20 mM imidazole, 1 M NaCl and connected to an ÄKTA Explorer Instrument (GE Healthcare) according to the manufacturer’s recommendations. The column was washed with buffer A (20 mM sodium phosphate pH 7.4, 20 mM imidazole, 1 M NaCl) to remove any unbound protein and an imidazole concentration gradient (20–500 mM) was applied. The BceC protein eluted at approximately 250 mM imidazole in a symmetrical chromatographic peak. The eluted fractions were immediately pooled and buffer-exchanged with 25 mM Tris–HCl pH 8.3, 50 mM NaCl, 2.5 mM dithiothreitol (DTT), 0.25 mM UDP-GlcA and 0.5 mM oxidized nicotinamide adenine dinucleotide (NAD+) using a PD10 desalting column (GE Healthcare). The composition and purity of the eluted fractions were confirmed by SDS–PAGE analysis and only homogenous fractions were subsequently used. The protein migrated as a single polypeptide with an estimated molecular mass of 52.6 kDa (51.3 kDa from the native protein plus 1.3 kDa corresponding to the His tag). The protein was concentrated to approximately 10 mg ml−1 in a Vivapore 10/20 concentrator (Vivascience Ltd, UK) before storage at 193 K. The protein concentration was determined by the method of Bradford (1976 ▶) using bovine serum albumin fraction V (Sigma, France) as a standard and by direct measurement of the absorbance at 280 nm in a NanoDrop ND-1000 spectrophotometer using an extinction coefficient of 38 120 M −1 cm−1.
2.3. BceC crystallization
Protein crystallization screens were performed by the vapour-diffusion method using a Minibee MicroSys 4000XL Cartesian Dispensing Systems robot (Genomic Solutions, USA). 768 sitting drops consisting of 100 nl protein solution at 10 mg ml−1 plus 100 nl precipitant solution were equilibrated against 100 µl precipitant solution from the Classics, PEGs, MbClass, MbClass II, pHClear, pHClear II, Ammonium Sulfate and MPD crystallization screens from Qiagen Canada Inc. (Montreal, Canada). A crystalline precipitate was found in various drops from different screens, but only the MbClass II and pHClear II screens produced visible single crystals. In particular, solution F7 of MbClass II [2 mM ammonium sulfate, 100 mM sodium acetate pH 4.6 and 12%(w/v) PEG 4K] and solution D8 of pHClear II (100 mM citric acid pH 5.0 and 30% 2-propanol) produced more regularly shaped crystals. Manual reproduction and further optimization of these conditions was attempted in order to decrease nucleation and promote further crystal growth. Finally, only condition condition F7 of MbClass II showed reproducible results on the microlitre scale and the crystals were improved by changing parameters such as the ratio of protein versus precipitant volume and by the use of additives from the 96 solutions of the Additive Screen from Hampton Research (Aliso Viejo, USA). Crystals were only allowed to grow for up to 24 h as longer times led to lower diffraction resolution and to higher instability in the crystal-cryoprotection step. The best crystals (Fig. 1 ▶) were obtained at 293 K from sitting drops consisting of 1 µl protein solution at 5 mg ml−1 (see §2.2) and 1 µl precipitant solution [200 mM ammonium sulfate, 100 mM sodium acetate pH 4.5, 11%(w/v) PEG 4K and 50 mM NaF] equilibrated against 500 µl precipitant solution in the well. Cryoprotection of the crystals was achieved in a two-step procedure by transferring them into a washing (and stabilizing) solution of mother liquor containing 15%(w/v) PEG 4K to remove the precipitated protein surrounding the crystals followed by a quick soak in a cryostabilizing solution consisting of mother liquor containing 25%(v/v) glycerol.
Figure 1.
(a) Native B. cepacia UGD forms octahedron-shaped crystals that reached approximate dimensions of 0.06 × 0.06 × 0.07 mm within 24 h of growing time.
2.4. X-ray diffraction analysis and phase-problem solution
Cryoprotected crystals were flash-cooled in liquid nitrogen and diffraction data were collected at station ID14-1 of the European Synchrotron Radiation Facility (ESRF), Grenoble, France using an ADSC Q210 CCD detector. Reflection intensities were processed with MOSFLM (Leslie, 1992 ▶), scaled and merged together with SCALA and reduced to structure-factor amplitudes using TRUNCATE from the CCP4 suite (Collaborative Computational Project, Number, 1994 ▶). No σ cutoff was applied during data integration and reduction (see Table 1 ▶ for crystal and diffraction data and statistics). An estimation of the number of molecules in the asymmetric unit (Matthews, 1968 ▶), taking into account the resolution-dependent distribution of Matthews coefficients in the PDB (Kantardjieff & Rupp, 2003 ▶), indicated the presence of four molecules in the asymmetric unit with a probability of 72% (three or five molecules corresponded to only 13 or 14% probability, respectively). A molecular-replacement search with Phaser (McCoy et al., 2007 ▶) using a preliminary model of the bacterial UGD from Sphingomonas elodea (Rocha et al., 2010 ▶), which has 42% sequence identity, as the target structure led to translation-function Z scores of 12.0, 9.9, 9.6 and 5.1 for the four molecules and a final log-likelihood gain (LLG) of 293.
Table 1. Crystal and diffraction data statistics.
Values in parentheses are for the outer resolution shell.
| ESRF beamline | ID14-1 |
| Wavelength (Å) | 0.934 |
| Resolution (Å) | 40.23–2.09 (2.20–2.09) |
| Space group | P212121 |
| Unit-cell parameters (Å) | a = 97.49, b = 109.08, c = 187.69 |
| No. of measured reflections | 442772 (61050) |
| No. of unique reflections | 118686 (17111) |
| Redundancy | 3.7 (3.6) |
| 〈I〉/〈σ(I)〉 | 11.8 (4.2) |
| Rp.i.m.† | 0.028 (0.101) |
| Rr.i.m. = Rmeas‡ | 0.054 (0.195) |
| Rmerge§ | 0.046 (0.166) |
| Completeness (%) | 99.8 (99.7) |
| Mosaicity (°) | 0.26 |
| VM (Å3 Da−1) | 2.36 |
| Solvent content (%) | 47.9 |
| Wilson B factor (Å2) | 24.2 |
| No. of molecules in ASU | 4 |
| Monomer molecular weight (kDa) | 52.6 |
R
p.i.m. = 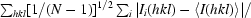
 , where N is the data redundancy, Ii(hkl) is the observed intensity and 〈I(hkl)〉 is the average intensity of multiple observations from symmetry-related reflections. It is an indicator of the precision of the final merged and averaged data set (Weiss, 2001 ▶).
, where N is the data redundancy, Ii(hkl) is the observed intensity and 〈I(hkl)〉 is the average intensity of multiple observations from symmetry-related reflections. It is an indicator of the precision of the final merged and averaged data set (Weiss, 2001 ▶).
R
r.i.m. = R
meas = 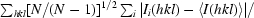
 ), where N is the data redundancy, Ii(hkl) is the observed intensity and 〈I(hkl)〉 is the average intensity of multiple observations of symmetry-related reflections. It is an indicator of the average spread of the individual measurements (Weiss, 2001 ▶).
), where N is the data redundancy, Ii(hkl) is the observed intensity and 〈I(hkl)〉 is the average intensity of multiple observations of symmetry-related reflections. It is an indicator of the average spread of the individual measurements (Weiss, 2001 ▶).
R
merge = 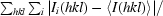
 , where Ii(hkl) is the observed intensity and 〈I(hkl)〉 is the average intensity of multiple observations from symmetry-related reflections.
, where Ii(hkl) is the observed intensity and 〈I(hkl)〉 is the average intensity of multiple observations from symmetry-related reflections.
Acknowledgments
The authors gratefully acknowledge the ESRF, Grenoble for provision of synchrotron radiation. JR is the recipient of a PhD fellowship from Fundação para a Ciência e Tecnologia (FCT), Portugal, SFRH/BD/24216/2005. This work was partially supported by FCT grants POCTI/BME/38859/2001, POCI/BIO/58401/2004 and PTDC/QUI/67925/2006.
References
- Bradford, M. M. (1976). Anal. Biochem.72, 248–254. [DOI] [PubMed]
- Campbell, R. E., Mosimann, S. C., van De Rijn, I., Tanner, M. E. & Strynadka, N. C. (2000). Biochemistry, 39, 7012–7023. [PubMed]
- Campbell, R. E., Sala, R. F., van de Rijn, I. & Tanner, M. E. (1997). J. Biol. Chem.272, 3416–3422. [DOI] [PubMed]
- Campbell, R. E. & Tanner, M. E. (1999). J. Org. Chem.64, 9487–9492.
- Cescutti, P., Bosco, M., Picotti, F., Impallomeni, G., Leitao, J. H., Richau, J. A. & Sá-Correia, I. (2000). Biochem. Biophys. Res. Commun.273, 1088–1094. [DOI] [PubMed]
- Collaborative Computational Project, Number 4 (1994). Acta Cryst. D50, 760–763.
- Cunha, M. V., Sousa, S. A., Leitao, J. H., Moreira, L. M., Videira, P. A. & Sá-Correia, I. (2004). J. Clin. Microbiol.42, 3052–3058. [DOI] [PMC free article] [PubMed]
- Griffith, C. L., Klutts, J. S., Zhang, L., Levery, S. B. & Doering, T. L. (2004). J. Biol. Chem.279, 51669–51676. [DOI] [PubMed]
- Hwang, H. Y. & Horvitz, H. R. (2002). Proc. Natl Acad. Sci. USA, 99, 14224–14229. [DOI] [PMC free article] [PubMed]
- Kantardjieff, K. A. & Rupp, B. (2003). Protein Sci.12, 1865–1871. [DOI] [PMC free article] [PubMed]
- Leslie, A. G. W. (1992). Jnt CCP4/ESF–EACBM Newsl. Protein Crystallogr.26
- Matthews, B. W. (1968). J. Mol. Biol.33, 491–497. [DOI] [PubMed]
- McCoy, A. J., Grosse-Kunstleve, R. W., Adams, P. D., Winn, M. D., Storoni, L. C. & Read, R. J. (2007). J. Appl. Cryst.40, 658–674. [DOI] [PMC free article] [PubMed]
- Moreira, L. M., Videira, P. A., Sousa, S. A., Leitao, J. H., Cunha, M. V. & Sá-Correia, I. (2003). Biochem. Biophys. Res. Commun.312, 323–333. [DOI] [PubMed]
- Richau, J. A., Leitao, J. H. & Sá-Correia, I. (2000). Biochem. Biophys. Res. Commun.276, 71–76. [DOI] [PubMed]
- Rocha, J., Granja, A. T., Sá-Correia, I., Fialho, A. & Frazão, C. (2010). Acta Cryst. F66, 69–72. [DOI] [PMC free article] [PubMed]
- Schirmer, F., Ehrt, S. & Hillen, W. (1997). J. Bacteriol.179, 1329–1336. [DOI] [PMC free article] [PubMed]
- Sist, P., Cescutti, P., Skerlavaj, S., Urbani, R., Leitao, J. H., Sá-Correia, I. & Rizzo, R. (2003). Carbohydr. Res.338, 1861–1867. [DOI] [PubMed]
- Sommer, B. J., Barycki, J. J. & Simpson, M. A. (2004). J. Biol. Chem.279, 23590–23596. [DOI] [PubMed]
- Sousa, S. A., Ulrich, M., Bragonzi, A., Burke, M., Worlitzsch, D., Leitao, J. H., Meisner, C., Eberl, L., Sá-Correia, I. & Doring, G. (2007). Cell. Microbiol.9, 2817–2825. [DOI] [PubMed]
- Stewart, D. & Copeland, L. (2006). Plant Physiol.116, 349–355.
- Weiss, M. S. (2001). J. Appl. Cryst.34, 130–135.