A novel diadenosine 5′,5′′′-P 1,P 4-tetraphosphate phosphorylase from M. tuberculosis H37Rv has been crystallized. The crystal belonged to space group C2 and diffracted X-rays to 1.9 Å resolution.
Keywords: Mycobacterium tuberculosis, phosphorylases, nucleotides
Abstract
A novel diadenosine 5′,5′′′-P 1,P 4-tetraphosphate (Ap4A) phosphorylase (Rv2613c) from Mycobacterium tuberculosis H37Rv has been crystallized by the sitting-drop vapour-diffusion method. The crystal belonged to space group C2, with unit-cell parameters a = 101.5, b = 63.6, c = 79.1 Å, β = 110.9°. The diffraction of the crystals extended to 1.9 Å resolution. The asymmetric unit is expected to contain two molecules of Rv2613c, with a corresponding crystal volume per protein weight (V M) of 2.41 Å3 Da−1 and a solvent content of 49.1%. This is the first report of a crystal of Ap4A phosphorylase.
1. Introduction
Recently, we reported the cloning, purification and molecular characterization of Rv2613c, a protein encoded by the open reading frame Rv2613c of Mycobacterium tuberculosis H37Rv. It was revealed that Rv2613c is a diadenosine 5′,5′′′-P 1,P 4-tetraphosphate (Ap4A) phosphorylase (Mori et al., 2010 ▶). The amino-acid sequence of Rv2613c contained a histidine-triad (HIT) motif consisting of H-ϕ-H-ϕ-H-ϕ-ϕ, where ϕ is a hydrophobic amino acid. This feature is unique among phosphorylases: the HIT motif has previously been reported to be a characteristic structure of nucleotide hydrolases, including yeast Ap4A hydrolase (Bieganowski et al., 2002 ▶; Huang et al., 1995 ▶; Barnes et al., 1996 ▶), rather than phosphorylases, which usually contain an H-X-H-X-Q-ϕ-ϕ motif as reported for yeast Ap4A phosphorylases (Plateau et al., 1989 ▶, 1990 ▶; Guranowski & Blanquet, 1985 ▶; Mulder et al., 1994 ▶). Furthermore, unlike yeast Ap4A phosphorylases, which are composed of approximately 330 amino acids, Rv2613c is composed of 195 amino acids and is comparable to yeast Ap4A hydrolase, which is composed of 182 amino acids. The sequence is also more homologous to Ap4A hydrolase than to Ap4A phosphorylase. Rv2613c shows 28.8% protein-sequence identity and 61.9% similarity to Ap4A hydrolase from Schizosaccharomyces pombe and shows 16.2% protein-sequence identity and 60.7% similarity to Ap4A phosphorylase from Saccharomyces cerevisiae. These observations indicate that Rv2613c is a unique Ap4A phosphorylase with a primary structure that is homologous to that of Ap4A hydrolase rather than typical Ap4A phosphorylases.
Previous analysis of crystal structures revealed that the HIT motif constitutes part of the substrate-binding site (Brenner et al., 1997 ▶) and another study showed by an amino-acid substitution experiment that the His residue at the C-terminal end of the HIT motif plays a critical role in the hydrolysis process of the reaction (Pace et al., 1998 ▶). As described above, Rv2613c contains a HIT motif but exhibits phosphorylase activity rather than hydrolase activity. These results indicate that the structure of the active site of this protein contains a specific difference that confers phosphorylase activity instead of hydrolase activity. In order to reveal the difference in the structure and the mode of action of this protein, elucidation of the crystal structure is required. Here, we report the crystallization and preliminary X-ray crystallographic data of Rv2613c.
2. Materials and methods
2.1. Purification and crystallization
Rv2613c was cloned into pCold I vector (Takara Bio, Shiga, Japan), expressed in Escherichia coli BL21 (DE3) pLysS (Novagen, Wisconsin, USA) and purified to homogeneity by two-step column chromatography as described previously (Mori et al., 2010 ▶). The purity of the Rv2613c preparation was greater than 95% as judged by the results of N-terminal amino-acid sequence analysis and mass spectrometry. The purified Rv2613c was dialyzed against sample buffer (Na HEPES pH 7.6 and 0.5 mM dithiothreitol) and then concentrated to 10 mg ml−1 using an Amicon Ultra-15 (Millipore, Massachusetts, USA) at room temperature.
Sitting-drop crystallization experiments were set up in 96-well Intelli-Plates (Art Robbins, California, USA). Crystallization conditions were screened using the following commercially available crystallization screening kits: Crystal Screen I, Crystal Screen II (Hampton Research, California, USA), JBScreen Classic (Jena Bioscience, Jena, Germany), Wizard I and Wizard II (Emerald BioSystems, Washington, USA). The reservoirs and drop compartments of the 96-well plates were filled with 60 µl and 1 µl reservoir solution, respectively, and 1 µl purified Rv2613c solution was then added to the drop compartment.
2.2. X-ray diffraction data collection and processing
Diffraction data for Rv2613c were collected at the AR-NW12A station of Photon Factory (Tsukuba, Japan) using an ADSC Quantum 210r detector (Area Detector Systems, California, USA). The crystal was harvested using a CryoLoop (Hampton Research), flash-cooled to 100 K under a nitrogen stream and subjected to analysis. 360 consecutive images were collected using an oscillation range of 1° and an exposure time of 2 s at a wavelength of 1.0000 Å. The data from the Rv2613c crystal were processed, merged and scaled using the HKL-2000 program package (DENZO and SCALEPACK; Otwinowski & Minor, 1997 ▶).
3. Results and discussion
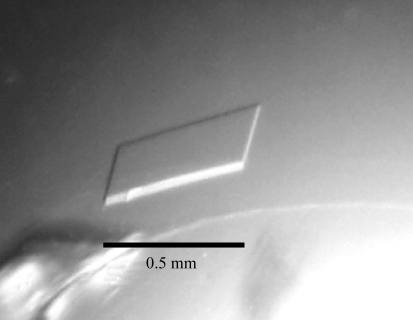
Prismatic colourless Rv2613c crystals formed in about two weeks at 293 K and grew to a maximum dimension of 0.5 mm (Fig. 1 ▶) during the screening process with Wizard II condition No. 12 (0.1 M sodium cacodylate pH 6.5, 0.2 M lithium sulfate and 30% polyethylene glycol 400).
Figure 1.
A native crystal of Rv2613c.
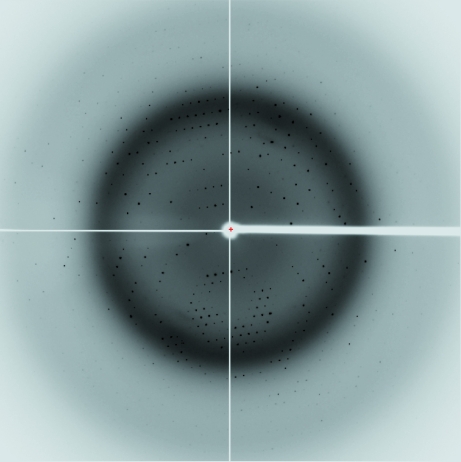
Diffraction data were collected from an Rv2613c crystal using synchrotron radiation. A data set was collected to 1.9 Å resolution from a single crystal (Fig. 2 ▶). Preliminary crystal characterization indicated that it belonged to the monoclinic space group C2, with unit-cell parameters a = 101.5, b = 63.6, c = 79.1 Å, β = 110.9°. 37 314 independent reflections were obtained with an R merge of 6.1%. Data-collection statistics for the crystal are summarized in Table 1 ▶. Rv2613c formed a homotetramer consisting of 25 kDa subunits in solution (Mori et al., 2010 ▶). Assuming the presence of two subunits in the asymmetric unit yielded a V M of 2.41 Å3 Da−1 and a solvent content of 49.1%. The V M and solvent content lie within the range usually found for protein crystals (Matthews, 1968 ▶). The data-collection statistics indicated that the Rv2613c crystal was suitable for solution of the structure.
Figure 2.
X-ray diffraction image from a native Rv2613c crystal. The edge of the detector corresponds to a resolution of 1.8 Å.
Table 1. X-ray diffraction data-collection statistics.
Values in parentheses are for the highest resolution shell.
| Wavelength (Å) | 1.0000 |
| Resolution (Å) | 50.0–1.90 (1.93–1.90) |
| Space group | C2 |
| Unit-cell parameters (Å) | a = 101.5, b = 63.6, c = 79.1, β = 110.9 |
| Unique reflections | 37314 (1826) |
| Average redundancy | 7.4 (6.4) |
| Completeness (%) | 99.8 (98.3) |
| Rmerge† | 0.061 (0.380) |
| Mean I/σ(I) | 42.2 (4.4) |
R
merge = 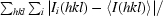
 , where Ii(hkl) is the ith intensity measurement of reflection hkl and 〈I(hkl)〉 is its average.
, where Ii(hkl) is the ith intensity measurement of reflection hkl and 〈I(hkl)〉 is its average.
This is the first report of the crystallization and X-ray preliminary analysis of an Ap4A phosphorylase. Attempts to solve the structure of Rv2613c by either multiple isomorphous replacement or the MAD method using selenomethionine-labelled Rv2613c are in progress.
Acknowledgments
We thank the beamline staff at the AR-NW12A station of Photon Factory for help with data collection. The crystallization of Rv2613c was carried out using a grant (H19-Shinkou-Ippan-006) from the Ministry of Health, Labour and Welfare of Japan. X-ray diffraction data collection and processing was carried out using a Grant-in-Aid for Young Scientists (B) (20780066) from the Ministry of Education, Culture, Sports, Science and Technology of Japan.
References
- Barnes, L. D., Garrison, P. N., Siprashvili, Z., Guranowski, A., Robinson, A. K., Ingram, S. W., Croce, C. M., Ohta, M. & Huebner, K. (1996). Biochemistry, 35, 11529–11535. [DOI] [PubMed]
- Bieganowski, P., Garrison, P. N., Hodawadekar, S. C., Faye, G., Barnes, L. D. & Brenner, C. (2002). J. Biol. Chem.277, 10852–10860. [DOI] [PMC free article] [PubMed]
- Brenner, C., Garrison, P., Gilmour, J., Peisach, D., Ringe, D., Petsko, G. A. & Lowenstein, J. M. (1997). Nature Struct. Biol.4, 231–238. [DOI] [PMC free article] [PubMed]
- Guranowski, A. & Blanquet, S. (1985). J. Biol. Chem.260, 3542–3547. [PubMed]
- Huang, Y., Garrison, P. N. & Barnes, L. D. (1995). Biochem. J.312, 925–932. [DOI] [PMC free article] [PubMed]
- Matthews, B. W. (1968). J. Mol. Biol.33, 491–497. [DOI] [PubMed]
- Mori, S., Shibayama, K., Wachino, J. I. & Arakawa, Y. (2010). Protein Expr. Purif.69, 99–105. [DOI] [PubMed]
- Mulder, W., Scholten, I. H., Roon, H. & Grivell, L. A. (1994). Biochim. Biophys. Acta, 1219, 719–723. [DOI] [PubMed]
- Otwinowski, Z. & Minor, W. (1997). Methods Enzymol.276, 307–326. [DOI] [PubMed]
- Pace, H. C., Garrison, P. N., Robinson, A. K., Barnes, L. D., Draganescu, A., Rösler, A., Blackburn, G. M., Siprashvili, Z., Croce, C. M., Huebner, K. & Brenner, C. (1998). Proc. Natl Acad. Sci. USA, 95, 5484–5489. [DOI] [PMC free article] [PubMed]
- Plateau, P., Fromant, M., Schmitter, J. M. & Blanquet, S. (1990). J. Bacteriol.172, 6892–6899. [DOI] [PMC free article] [PubMed]
- Plateau, P., Fromant, M., Schmitter, J. M., Buhler, J. M. & Blanquet, S. (1989). J. Bacteriol.171, 6437–6445. [DOI] [PMC free article] [PubMed]