Community-acquired respiratory distress syndrome toxin (CARDS TX) from M. pneumoniae was cloned, expressed, purified and crystallized. A diffraction data set from a native CARDS TX crystal was obtained at 2.2 Å resolution.
Keywords: CARDS TX, community-acquired respiratory distress syndrome, Mycoplasma pneumoniae, ADP-ribosyltransferases, ADP-ribosylation
Abstract
Community-acquired respiratory distress syndrome toxin (CARDS TX) is a 591-amino-acid protein with ADP-ribosyltransferase and vacuolating activities that damages the cells lining the respiratory tracts of patients infected with the bacterial pathogen Mycoplasma pneumoniae. Crystals of CARDS TX were grown in space group C2, with unit-cell parameters a = 191.4, b = 107.4, c = 222.1 Å, β = 90.6°. A complete 2.2 Å data set was obtained from a single CARDS TX crystal.
1. Introduction
Mycoplasma pneumoniae infections are responsible for 20–40% of all community-acquired pneumonia cases in humans (Baseman & Tully, 1997 ▶; Waites & Talkington, 2004 ▶) and may also play a role in wheezing syndromes and in chronic asthma (Kraft et al., 1998 ▶; Martin et al., 2001 ▶). MPN372, a 591-amino-acid protein originally annotated as hypothetical (Himmelreich et al., 1996 ▶), was isolated through its ability to bind to human surfactant protein A (SP-A; Kannan et al., 2005 ▶), the major glycoprotein component of pulmonary surfactant (Balis et al., 1985 ▶). Pneumonia patients diagnosed with M. pneumoniae infection demonstrate dramatic seroconversion to MPN372, suggesting it is indeed synthesized in vivo during infection (Kannan & Baseman, 2006 ▶). Analysis of the MPN372 amino-acid sequence revealed that residues 1–239 possess 27% sequence identity to the catalytic subunit of the exotoxin from Bordetella pertussis (Kannan et al., 2005 ▶), a bacterial pathogen that also colonizes the lung and is responsible for whooping cough in humans (Pittman, 1984 ▶). Residues 240–591 of MPN372 demonstrate no detectable sequence similarity to any other reported protein sequence. Because MPN372 appears to be a bona fide M. pneumoniae virulence factor, it is now referred to as community-acquired respiratory distress syndrome toxin (CARDS TX; Kannan & Baseman, 2006 ▶).
Pertussis toxin catalyzes the NAD+-dependent transfer of ADP-ribose to the α subunits of G proteins Gi, Go and Gt, which prevents their association with G-protein-coupled receptors on the cell membrane, thereby interfering with intracellular signaling (Burns, 1988 ▶). Analyses of cell-free extracts derived from CHO and HEp-2 cells incubated with recombinant CARDS TX revealed that it ADP-ribosylates the same substrates as pertussis toxin plus multiple distinct mammalian proteins that are not pertussis-toxin targets (Kannan & Baseman, 2006 ▶). Intact CHO and HeLa cells exposed to exogenous recombinant CARDS TX displayed distinct vacuolization and cell rounding, with disruption of monolayer integrity, eventually leading to cell death (Kannan & Baseman, 2006 ▶). Recombinant CARDS TX was also observed to cause substantial slowing and eventual cessation of cilia movement and disruption of respiratory-cell integrity in baboon tracheal organ cultures in a dose-dependent fashion (Kannan & Baseman, 2006 ▶). Taken together, these data strongly suggest that purified recombinant CARDS TX recapitulates the spectrum of respiratory and extrapulmonary pathologies in patients infected with M. pneumoniae. These characteristics make CARDS TX an attractive candidate for structure–function analysis as well as a candidate for the development of novel therapeutic agents.
2. Protein preparation
Mycoplasmas use both UGA (the ‘universal’ stop codon) and UGG to encode the amino acid tryptophan. The gene encoding CARDS TX possesses eight UGA codons at amino-acid positions 148, 195, 233, 364, 392, 450, 462 and 508 that necessitated polymerase-chain-reaction-mediated site-directed mutagenesis to replace each with UGG prior to expression in Escherichia coli (Kannan & Baseman, 2006 ▶). The modified gene encoding full-length CARDS TX was amplified by polymerase chain reaction and subcloned using the BamHI and NdeI restriction sites of pKA8H, an expression vector derived from pKM265 (Melcher, 2000 ▶) that is designed to fuse an 8×His tag to the N-terminus of CARDS TX such that it can be cleaved using tobacco etch virus (TEV) protease. Transformed E. coli BL21 (DE3) cells were grown in Luria broth medium containing 100 µg ml−1 ampicillin at 310 K. When the A 600nm of the culture reached 0.6, isopropyl β-d-1-thiogalactopyranoside (IPTG) was added to a final concentration of 0.5 mM and incubation was continued at 293 K for 15 h prior to harvesting and storage at 253 K.
Frozen bacterial cell paste from 8 l of culture was thawed, suspended in 150 ml disruption buffer consisting of 50 mM Tris–HCl, 500 mM NaCl pH 8.0 and 0.75 ml protease-inhibitor cocktail (Sigma) and lysed by sonication on ice. The cell lysate was centrifuged at 30 000g for 30 min at 277 K. The pellet was discarded and the clarified supernatant was loaded onto a 5 ml HiTrap Ni2+-affinity column (GE Healthcare) equilibrated with buffer consisting of 50 mM Tris–HCl, 500 mM NaCl, 20 mM imidazole pH 8.0. The column was washed with the same buffer and the protein was eluted using a 20–500 mM imidazole linear gradient. Column fractions were monitored by absorbance at 280 nm and those containing CARDS TX as indicated by SDS–PAGE were pooled. The N-terminal 8×His tag was cleaved in a dialysis bag by incubating TEV protease with CARDS TX at an approximate CARDS TX:TEV protease ratio of ≃ 30:1(w:w) overnight against a buffer consisting of 50 mM Tris pH 8.0, 100 mM NaCl, 1 mM EDTA, 1 mM reduced glutathione (GSH) and 0.1 mM oxidized glutathione (GSSG) at room temperature. The resulting digestion mixture was again loaded onto a 5 ml HiTrap Ni2+-affinity column and the protein was eluted in the flowthrough, while the cleaved His tag and TEV protease, which was engineered to possess its own noncleavable 6×His tag, were retained. Two additional amino acids (Gly-His) remained fused to the CARDS TX N-terminus after removal of the affinity tag. CARDS TX was subsequently dialyzed against buffer consisting of 50 mM sodium acetate pH 5.0, loaded onto a 10 ml Source S ion-exchange column (GE Healthcare) and eluted with a linear 0–500 mM NaCl gradient. The protein eluted when the NaCl concentration reached approximately 180 mM. The yield ranged from 30 to 40 mg of protein per litre of growth medium. The molecular mass of purified CARDS TX was confirmed using electrospray ionization mass spectrometry at the UTHSCSA Mass Spectrometry Laboratory.
3. Analytical ultracentrifugation
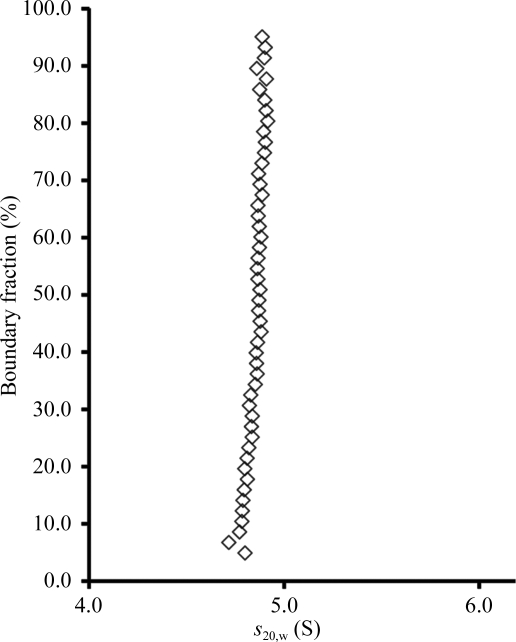
To determine the degree of homogeneity and the oligomeric state of purified CARDS TX in solution, analytical ultracentrifugation sedimentation-velocity experiments were performed in the UTHSCSA Center for Analytical Ultracentrifugation of Macromolecular Assemblies with a Beckman XL-I analytical ultracentrifuge equipped with absorbance optics and double-sector charcoal/Epon-filled centerpieces. CARDS TX at a concentration of 0.42 mg ml−1, corresponding to an absorbance of 0.8 at 280 nm, was centrifuged at 60 000 rev min−1 at 293 K in buffer consisting of 10 mM Tris–HCl pH 7.5, 50 mM NaCl. Velocity data were analyzed using the method of van Holde & Weischet (1978 ▶), as implemented in the UltraScan software package (Demeler, 2005 ▶, 2009 ▶), to determine the sedimentation-coefficient distribution. This analysis method effectively removes the contributions of diffusion to boundary spreading, yielding the integral distribution of s 20,w of all species in the sample. Consequently, a plot of boundary fraction versus s 20,w will be vertical if the sample is homogeneous and will have a positive slope if the sample is heterogeneous (Demeler et al., 1997 ▶). The plot in Fig. 1 ▶ reveals that CARDS TX sediments as a single homogeneous ∼4.8 S species. The molecular weight of CARDS TX as determined by two-dimensional spectrum (Brookes et al., 2009 ▶) and genetic algorithm (Brookes & Demeler, 2006 ▶) analyses in UltraScan was ∼68 kDa, which is in excellent agreement with the molecular mass predicted from the amino-acid sequence for a CARDS TX monomer.
Figure 1.
van Holde and Weischet plot of sedimentation-velocity data probing the oligomeric state and degree of monodispersity in purified CARDS TX. The protein sediments as a homogeneous ∼4.8 S monomeric species.
4. Limited proteolysis and protein crystallization
Initial attempts to crystallize full-length CARDS TX failed. In an effort to identify stable CARDS TX domains that might be amenable to crystallization, limited proteolysis experiments were pursued using chymotrypsin, proteinase K and trypsin. Although treatment with chymotrypsin and proteinase K failed to generate stable CARDS TX domain fragments, incubation with trypsin resulted in a single cleavage event. Purified CARDS TX in storage buffer at 10 mg ml−1 was incubated with trypsin at a ratio of 100:1(w:w) for 30 min at room temperature. Proteolysis was terminated by bringing the solution to 500 µg ml−1 aprotinin (Sigma). The resulting sample was filtered through a 0.2 µm syringe filter and loaded onto a Sephacryl S-100 HR 16/60 column (GE Healthcare) equilibrated with 50 mM Tris–HCl pH 7.8. The protein was eluted with the same buffer at a flow rate of 0.6 ml min−1. The elution profile of the trypsin-treated CARDS TX sample, as well as its migration behavior in a native gel, were subtly different from those of the untreated sample and SDS–PAGE followed by N-terminal sequencing revealed that the trypsin-treated protein had been cleaved after Lys305 and Lys307, leaving noncovalently associated domains defined by amino-acid residues 1–305 and 308–591. This ‘nicked’ CARDS TX protein was dialyzed into a buffer consisting of 20 mM Tris–HCl pH 7.8, 1 mM NAD+, 4 mM TCEP and concentrated to 12 mg ml−1 for crystallization trials using a Phoenix liquid-handling robot (Art Robbins Instruments) and 12 commercially available crystallization suites from Qiagen that provided 1152 crystallization conditions. Screening was performed using the sitting-drop vapor-diffusion method in 96-well IntelliPlates (Art Robbins Instruments) by mixing 200 nl protein solution with 200 nl reservoir solution and equilibrating over 50 µl reservoir solution at 277 and 295 K. Small crystals appeared within three weeks at 295 K in a condition belonging to the Qiagen Cryos Suite and consisting of 0.08 M sodium acetate pH 4.6, 1.6 M ammonium sulfate, 20%(v/v) glycerol. Larger specimens of the size shown in Fig. 2 ▶ were obtained using the hanging-drop vapor-diffusion method in 24-well plates by mixing 1 µl protein solution with 1 µl reservoir solution and equilibrating over 500 µl reservoir solution, except that the reservoir solution contained 1.4 M instead of 1.6 M ammonium sulfate.
Figure 2.
Crystal of nicked CARDS TX used for X-ray data collection. The longest dimension of the crystal is ∼200 µm.
5. Data collection
The CARDS TX crystal shown in Fig. 2 ▶ was grown in the presence of 20%(v/v) glycerol and therefore was simply flash-cooled in liquid nitrogen immediately prior to X-ray data collection. X-ray diffraction data were obtained to 2.2 Å resolution using the UTHSCSA X-ray Crystallography Core Laboratory Rigaku Micromax 007 HF rotating-anode X-ray source equipped with Varimax HiFlux optics and an R-AXIS HTC imaging-plate detector. The diffraction data were processed using the HKL-2000 suite (Otwinowski & Minor, 1997 ▶) and the data-collection statistics are summarized in Table 1 ▶. The asymmetric unit was estimated to contain between five and seven CARDS TX molecules, corresponding to Matthews parameters ranging from 3.4 to 2.4 Å3 Da−1 (Matthews, 1968 ▶).
Table 1. Data-collection statistics for the CARDS TX crystal.
Values in parentheses are for the highest resolution shell.
| Space group | C2 |
| Unit-cell parameters (Å, °) | a = 191.4, b = 107.4, c = 222.1, α = 90, β = 90.6, γ = 90 |
| Wavelength (Å) | 1.54 |
| Resolution (Å) | 30–2.2 |
| Unique reflections | 226771 |
| Rmerge | 0.077 (0.443) |
| Rp.i.m.† | 0.053 (0.298) |
| 〈I/σ(I)〉 | 13.1 (2.3) |
| Completeness (%) | 99.5 (99.7) |
| Redundancy | 2.9 (2.8) |
Weiss & Hilgenfeld (1997 ▶).
Acknowledgments
This work was supported by the Robert A. Welch Foundation Grant AQ-1399 (to PJH), by IH grant U19 AI070412 (to JBB) and by the Kleberg Foundation (to JBB).
References
- Balis, J. U., Paterson, J. F., Paciga, J. E., Haller, E. M. & Shelley, S. A. (1985). Lab. Invest.52, 657–669. [PubMed]
- Baseman, J. B. & Tully, J. G. (1997). Emerg. Infect. Dis.3, 21–32. [DOI] [PMC free article] [PubMed]
- Brookes, E., Cao, W. & Demeler, B. (2009). Eur. Biophys. J., doi:10.1007/s00249-009-0413-5. [DOI] [PubMed]
- Brookes, E. & Demeler, B. (2006). Analytical Ultracentrifugation VIII (Progress in Colloid and Polymer Science), edited by C. Wandrey & H. Cölfen, pp. 78–82. Berlin: Springer.
- Burns, D. L. (1988). Microbiol. Sci.5, 285–287. [PubMed]
- Demeler, B. (2005). Modern Analytical Ultracentrifugation: Techniques and Methods, edited by D. Scott, S. Harding & A. Rowe, pp. 210–229. Cambridge: Royal Society of Chemistry.
- Demeler, B. (2009). UltraScan II. http://www.ultrascan.uthscsa.edu/.
- Demeler, B., Saber, H. & Hansen, J. C. (1997). Biophys. J.72, 397–407. [DOI] [PMC free article] [PubMed]
- Himmelreich, R., Hilbert, H., Plagens, H., Pirkl, E., Li, B.-C. & Herrmann, R. (1996). Nucleic Acids Res.24, 4420–4449. [DOI] [PMC free article] [PubMed]
- Holde, K. E. van & Weischet, W. O. (1978). Biopolymers, 17, 1387–1403.
- Kannan, T. R. & Baseman, J. B. (2006). Proc. Natl Acad. Sci. USA, 103, 6724–6729. [DOI] [PMC free article] [PubMed]
- Kannan, T. R., Provenzano, D., Wright, J. R. & Baseman, J. B. (2005). Infect. Immun.73, 2828–2834. [DOI] [PMC free article] [PubMed]
- Kraft, M., Cassell, G. H., Henson, J. E., Watson, H., Williamson, J., Marmion, B. P., Gaydos, C. A. & Martin, R. J. (1998). Am. J. Respir. Crit. Care Med.158, 998–1001. [DOI] [PubMed]
- Martin, R. J., Kraft, M., Chu, H. W., Berns, E. A. & Cassell, G. H. (2001). J. Allergy Clin. Immunol.107, 595–601. [DOI] [PubMed]
- Matthews, B. W. (1968). J. Mol. Biol.33, 491–497. [DOI] [PubMed]
- Melcher, K. (2000). Anal. Biochem.277, 109–120. [DOI] [PubMed]
- Otwinowski, Z. & Minor, V. (1997). Methods Enzymol.276, 307–326. [DOI] [PubMed]
- Pittman, M. (1984). Pediatr. Infect. Dis.3, 467–486. [DOI] [PubMed]
- Waites, K. B. & Talkington, D. F. (2004). Clin. Microbiol. Rev.17, 697–728. [DOI] [PMC free article] [PubMed]
- Weiss, M. S. & Hilgenfeld, R. (1997). J. Appl. Cryst.30, 203–205.