A multimodular heterotrimeric complex encompassing a trimodular C-terminal fragment of the cellulosomal scaffoldin CipA from C. thermocellum bound to the type II cohesin module of SdbA and the type I dockerin module of CelD has been crystallized by the hanging-drop vapour-diffusion method and initial X-ray diffraction data analysis has been conducted.
Keywords: cellulosome, cohesins, dockerins
Abstract
The multimodular scaffoldin subunit CipA is the central component of the cellulosome, a multienzyme plant cell-wall-degrading complex, from Clostridium thermocellum. It captures secreted cellulases and hemicellulases and anchors the entire complex to the cell surface via high-affinity calcium-dependent interactions between cohesin and dockerin modules termed type I and type II interactions. The crystallization of a heterotrimeric complex comprising the type II cohesin module from the cell-surface protein SdbA, a trimodular C-terminal fragment of the scaffoldin CipA and the type I dockerin module from the CelD cellulase is reported. The crystals belonged to space group P212121, with unit-cell parameters a = 119.37, b = 186.31, c = 191.17 Å. The crystals diffracted to 2.7 Å resolution with four or eight molecules of the ternary protein complex in the asymmetric unit.
1. Introduction
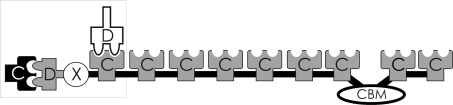
The cellulosome is a large cell-surface-bound multienzyme complex that is responsible for the degradation of cellulose and other plant cell-wall polysaccharides. Originally discovered in the thermophilic anaerobe Clostridium thermocellum (Lamed, Kenig et al., 1983 ▶; Lamed, Setter et al., 1983 ▶), cellulosomes have since been identified in a variety of other anaerobic bacteria, ruminal bacteria and anaerobic fungi (for reviews, see Bayer et al., 1998 ▶, 2004 ▶; Beguin & Lemaire, 1996 ▶; Doi & Kosugi, 2004 ▶; Schwarz, 2001 ▶). The cellulosome of C. thermocellum is composed of three modular protein components: cellulases, hemicellulases and other hydrolytic enzymes, the CipA scaffoldin subunit (Gerngross et al., 1993 ▶) and one of three cell-surface-associated proteins (SdbA, OlpB or Orf2p; Leibovitz & Beguin, 1996 ▶; Leibovitz et al., 1997 ▶; Lemaire et al., 1995 ▶; Fig. 1 ▶). Cellulosome assembly is mediated by two types of high-affinity calcium-dependent interactions between cohesin (Coh) and dockerin (Doc) modules. The type I interaction is responsible for localizing the various enzymes to the scaffoldin, while the type II interaction anchors the scaffoldin to a cell-surface-associated protein. The CipA scaffoldin contains nine type I Coh modules, a type II Doc module, an X-module of unknown function and a cellulose-specific carbohydrate-binding module (CBM), all of which are connected by flexible linkers of varying length (Fig. 1 ▶).
Figure 1.
Schematic of the CipA scaffoldin subunit from C. thermocellum bound to the type II cohesin from SdbA and the type I dockerin from CelD. Cohesins, dockerins, the X-module and the carbohydrate-binding module are labelled C, D, X and CBM, respectively. Type I cohesins and dockerins are shown in grey and white, respectively. Type II cohesins and dockerins are shown in black and grey, respectively. The complex described in the current article is surrounded by a box.
In order to obtain a better structural understanding of the scaffoldin, crystal structures have been obtained for the individual type I and type II Coh and Doc modules (Lytle et al., 2001 ▶; Shimon et al., 1997 ▶; Tavares et al., 1997 ▶) and their complexes (Adams et al., 2006 ▶; Carvalho et al., 2003 ▶, 2007 ▶), the CBM (Tormo et al., 1996 ▶) and the X-module (Adams et al., 2006 ▶). However, few multimodular structures have been determined, which is likely to be a consequence of the inherent flexibility of the linker regions that separate each module. Here, we report the generation, purification, crystallization and preliminary X-ray characterization of a heterotrimeric multimodular complex including the three C-terminal modules of CipA (residues 1533–1853) bound to the type I Doc module (DocI) from the CelD cellulase (residues 549–625) and the type II Coh (CohII) module from the cell-surface protein SdbA (residues 27–200).
2. Expression, purification and complex formation
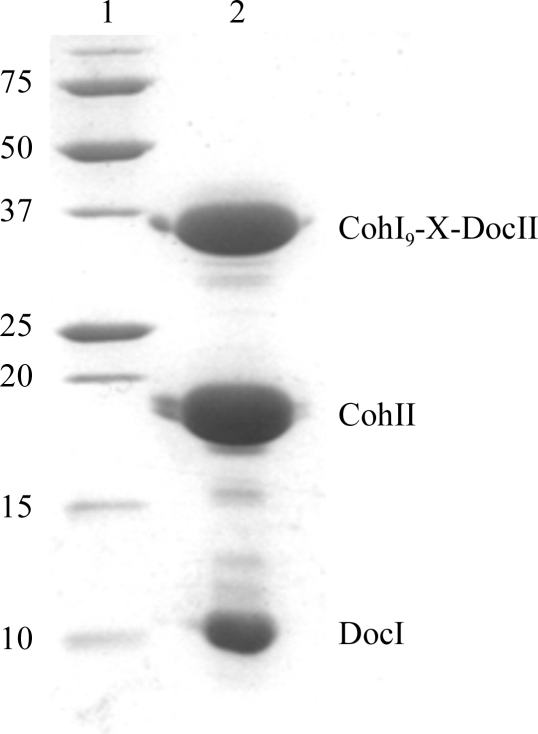
The C-terminal CohI9-X-DocII fragment of CipA with a C-terminal hexahistidine tag, SdbA CohII with an N-terminal hexahistidine tag and CelD DocI with an N-terminal dodecahistidine tag were all recombinantly expressed and purified in a similar manner. Briefly, transformed BL21 (DE3) cells (Novagen) were grown in LB medium supplemented with 100 mg l−1 ampicillin at 310 K while shaking until an OD600 of 0.6 was reached. IPTG was added to a final concentration of 1 mM and growth was continued for an additional 4 h. The cells were harvested by centrifugation (20 min at 3000g), resuspended in 20 ml buffer A (25 mM Tris–HCl pH 7.4, 250 mM NaCl, 8 M urea) and lysed by sonication on ice. The insoluble fraction was removed by centrifugation at 20 000g in a Beckman JA-20 rotor for 20 min. The supernatant was applied onto an Ni2+-charged chelating column pre-equilibrated in buffer A. The column was subsequently washed with buffer A containing 20 mM imidazole and the bound protein was eluted with buffer A containing 400 mM imidazole. Purified CohI9-X-DocII, DocI and CohII were pooled and refolded by dialysis into buffer B (20 mM HEPES pH 7.5, 50 mM NaCl, 1 mM CaCl2 and 1 mM DTT). The refolded complex was separated from excess unbound proteins by size-exclusion chromatography using a HiLoad 16/60 Superdex 200 size-exclusion column (Amersham Pharmacia Biosciences) equilibrated in buffer B. The protein was eluted in 4 ml fractions using buffer B and its purity was confirmed via SDS–PAGE with Coomassie Blue staining (Fig. 2 ▶).
Figure 2.
SDS–PAGE analysis of the purified DocI–CohI9-X-DocII–CohII complex. Lane 1, molecular-weight markers (kDa); lane 2, purified DocI–CohI9-X-DocII–CohII complex.
3. Crystallization
The DocI–CohI9-X-DocII–CohII complex crystals were grown by hanging-drop vapour diffusion with a drop containing 2 µl protein at 28 mg ml−1 and 2–4 µl reservoir solution consisting of 100 mM HEPES pH 7–7.75, 1.3–1.5 M lithium sulfate and 0.5 µl 1 M potassium sodium tartrate. The crystals took 7–10 d to grow at room temperature. The crystals were tetragonal in shape, with dimensions of 0.25 × 0.25 × 0.20 mm (Fig. 3 ▶).
Figure 3.
Example of DocI–CohI9-X-DocII–CohII crystals with typical dimensions of approximately 0.25 × 0.25 × 0.20 mm.
4. Data collection and processing
X-ray data were collected on beamline 9-2 at the Stanford Synchrotron Radiation Lightsource (SSRL) using a MarMosaic 325 CCD detector (MAR USA). Data were collected at 100 K from crystals that had been soaked in reservoir solution containing 20% glycerol as a cryoprotectant and flash-frozen in liquid nitrogen.
The crystals belonged to the primitive orthorhombic space group P212121, with unit-cell parameters a = 119.37, b = 186.31, c = 191.17 Å. Matthews coefficients of 4.08 and 2.04 Å3 Da−1 were obtained with solvent contents of 69.85% and 39.70% for an asymmetric unit containing four and eight heterotrimeric protein complexes, respectively. The data were processed to 2.7 Å resolution with an R merge of 4.6% (Table 1 ▶) using HKL-2000 (Otwinowski & Minor, 1997 ▶). A molecular-replacement strategy based on the X-Doc–CohII structure (Adams et al., 2006 ▶) and the CohI–DocI structures (Carvalho et al., 2003 ▶, 2007 ▶) is being employed to solve the structure of the DocI–CohI9-X-DocII–CohII heterotrimeric complex.
Table 1. Diffraction data statistics for native DocI–CohI9-X-DocII–CohII complex crystals.
Values in parentheses are for the highest resolution shell.
| Space group | P212121 |
| Unit-cell parameters (Å) | a = 119.37, b = 186.31, c = 191.17 |
| Wavelength (Å) | 0.97927 |
| Temperature (K) | 100 |
| Resolution range (Å) | 30.0–2.7 (2.8–2.7) |
| Observed reflections | 113244 |
| Unique reflections | 107440 |
| Data completeness (%) | 95.8 (78.5) |
| Redundancy | 3.6 (2.4) |
| Rmerge† (%) | 4.6 (75.5) |
| 〈I/σ(I)〉 | 30.9 (1.9) |
| Matthews coefficient (Å3 Da−1) | 4.08/2.04‡ |
| Solvent content (%) | 69.85 |
R
merge = 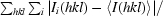
 , where Ii(hkl) and 〈I(hkl)〉 represent the diffraction-intensity values of the individual measurements and the corresponding mean values.
, where Ii(hkl) and 〈I(hkl)〉 represent the diffraction-intensity values of the individual measurements and the corresponding mean values.
Four/eight heterotrimeric protein complexes per asymmetric unit.
Acknowledgments
We would like to acknowledge SSRL for synchrotron data collection. ZJ is a Canada Research Chair in Structural Biology. SPS is a Canadian Institutes of Health Research New Investigator. This research was supported by a Canadian Institutes of Health Research Operating Grant (ZJ) and a National Science and Engineering Research Council of Canada Discovery Grant (SPS).
References
- Adams, J. J., Pal, G., Jia, Z. & Smith, S. P. (2006). Proc. Natl Acad. Sci. USA, 103, 305–310. [DOI] [PMC free article] [PubMed]
- Bayer, E. A., Belaich, J. P., Shoham, Y. & Lamed, R. (2004). Annu. Rev. Microbiol.58, 521–554. [DOI] [PubMed]
- Bayer, E. A., Chanzy, H., Lamed, R. & Shoham, Y. (1998). Curr. Opin. Struct. Biol.8, 548–557. [DOI] [PubMed]
- Beguin, P. & Lemaire, M. (1996). Crit. Rev. Biochem. Mol. Biol.31, 201–236. [DOI] [PubMed]
- Carvalho, A. L., Dias, F. M., Nagy, T., Prates, J. A., Proctor, M. R., Smith, N., Bayer, E. A., Davies, G. J., Ferreira, L. M., Romão, M. J., Fontes, C. M. & Gilbert, H. J. (2007). Proc. Natl Acad. Sci. USA, 104, 3089–3094. [DOI] [PMC free article] [PubMed]
- Carvalho, A. L., Dias, F. M., Prates, J. A., Nagy, T., Gilbert, H. J., Davies, G. J., Ferreira, L. M., Romão, M. J. & Fontes, C. M. (2003). Proc. Natl Acad. Sci. USA, 100, 13809–13814. [DOI] [PMC free article] [PubMed]
- Doi, R. H. & Kosugi, A. (2004). Nature Rev. Microbiol.2, 541–551. [DOI] [PubMed]
- Gerngross, U. T., Romaniec, M. P., Kobayashi, T., Huskisson, N. S. & Demain, A. L. (1993). Mol. Microbiol.8, 325–334. [DOI] [PubMed]
- Lamed, R., Kenig, R., Setter, E. & Bayer, E. A. (1983). Biotechnol. Bioeng. Symp.13, 163–181.
- Lamed, R., Setter, E. & Bayer, E. A. (1983). J. Bacteriol.156, 828–836. [DOI] [PMC free article] [PubMed]
- Leibovitz, E. & Beguin, P. (1996). J. Bacteriol.178, 3077–3084. [DOI] [PMC free article] [PubMed]
- Leibovitz, E., Ohayon, H., Gounon, P. & Beguin, P. (1997). J. Bacteriol.179, 2519–2523. [DOI] [PMC free article] [PubMed]
- Lemaire, M., Ohayon, H., Gounon, P., Fujino, T. & Beguin, P. (1995). J. Bacteriol.177, 2451–2459. [DOI] [PMC free article] [PubMed]
- Lytle, B. L., Volkman, B. F., Westler, W. M., Heckman, M. P. & Wu, J. H. (2001). J. Mol. Biol.307, 745–753. [DOI] [PubMed]
- Otwinowski, Z. & Minor, W. (1997). Methods Enzymol.276, 307–326. [DOI] [PubMed]
- Schwarz, W. H. (2001). Appl. Microbiol. Biotechnol.56, 634–649. [DOI] [PubMed]
- Shimon, L. J., Bayer, E. A., Morag, E., Lamed, R., Shoham, Y. & Frolow, F. (1997). Structure, 3, 381–390. [DOI] [PubMed]
- Tavares, G. A., Beguin, P. & Alzari, P. M. (1997). J. Mol. Biol.273, 701–713. [DOI] [PubMed]
- Tormo, J., Lamed, R., Chirino, A. J., Morag, E., Bayer, E. A., Shoham, Y. & Steitz, T. A. (1996). EMBO J.15, 5739–5751. [PMC free article] [PubMed]