1-Deoxy-d-xylulose 5-phosphate reductoisomerase from P. falciparum has been crystallized in the presence of NADPH. Diffraction data to 1.85 Å resolution have been collected using synchrotron radiation.
Keywords: 1-deoxy-d-xylulose 5-phosphate reductoisomerase, malaria, nonmevalonate pathway
Abstract
The nonmevalonate pathway of isoprenoid biosynthesis present in Plasmodium falciparum is known to be an effective target for antimalarial drugs. The second enzyme of the nonmevalonate pathway, 1-deoxy-d-xylulose 5-phosphate reductoisomerase (DXR), catalyzes the transformation of 1-deoxy-d-xylulose 5-phosphate (DXP) to 2-C-methyl-d-erythritol 4-phosphate (MEP). For crystallographic studies, DXR from the human malaria parasite P. falciparum (PfDXR) was overproduced in Escherichia coli, purified and crystallized using the hanging-drop vapour-diffusion method in the presence of NADPH. X-ray diffraction data to 1.85 Å resolution were collected from a monoclinic crystal form belonging to space group C2 with unit-cell parameters a = 168.89, b = 59.65, c = 86.58 Å, β = 117.8°. Structural analysis by molecular replacement is in progress.
1. Introduction
The universal terpenoid precursors isopentenyl diphosphate (IPP) and dimethylallyl diphosphate (DMAPP) are synthesized either via the classical mevalonate (MVA) pathway or the novel nonmevalonate pathway. In the MVA pathway, IPP is synthesized from three molecules of acetyl-CoA via the intermediate MVA. It has long been assumed that the MVA pathway is commonly used for isoprenoid biosynthesis in all organisms. Recently, however, the existence of a second mevalonate-independent pathway for the biosynthesis of IPP and DMAPP has been detected in eubacteria, higher plants, algae, cyanobacteria and diatoms (Rohmer, 1999 ▶; Lichtenthaler, 2000 ▶). This novel nonmevalonate pathway also occurs in the apicoplast of the human malaria parasite Plasmodium falciparum (Jomaa et al., 1999 ▶). Since the nonmevalonate pathway is not found in animals, it is an ideal target for the development of herbicides and antibacterial drugs.
The second enzyme of the nonmevalonate pathway, 1-deoxy-d-xylulose 5-phosphate reductoisomerase (DXR; EC 1.1.1.267), catalyzes the NADPH and divalent cation (Mg2+ or Mn2+) dependent transformation of 1-deoxy-d-xylulose 5-phosphate (DXP) into 2-C-methyl-d-erythritol 4-phosphate (MEP). In 1999, Jomaa and coworkers reported that DXR from P. falciparum (PfDXR) was inhibited by fosmidomycin and its derivative FR-900098 and that mice infected with P. vinckei, a relative of the human malaria parasite, could be cured using these drugs (Jomaa et al., 1999 ▶). These data established PfDXR as a promising antimalarial target; indeed, clinical phase II studies using fosmidomycin in combination with clindamycin in Gabon and Thailand proved that PfDXR is an effective target for the chemotherapy of malaria (Borrmann et al., 2004 ▶).
To date, several crystal structures of DXR from Escherichia coli (Reuter et al., 2002 ▶; Yajima et al., 2002 ▶, 2004 ▶, 2007 ▶; Steinbacher et al., 2003 ▶; MacSweeney et al., 2005 ▶), from Zymomonas mobilis (Ricagno et al., 2004 ▶) and from Mycobacterium tuberculosis (Henriksson et al., 2006 ▶, 2007 ▶) have been reported. However, the crystal structure of PfDXR has not yet been reported. Here, we report the crystallization of PfDXR in the presence of NADPH. The structural study of PfDXR should be useful for the development of novel PfDXR inhibitors.
2. Materials and methods
2.1. Overproduction and purification
Since the first 30 amino acids of PfDXR resemble an endoplasmic reticulum signal sequence and the following 44 amino acids exhibit the characteristics of a plastidial targeting sequence (Jomaa et al., 1999 ▶), the DNA encoding residues Lys75–Ser488 (C-terminus) of P. falciparum DXR was obtained by reverse-transcription PCR. Reverse transcription was carried out using SuperScriptII reverse transcriptase as described in the user’s manual (Invitrogen) with the total RNA of P. falciparum (FCR-3) as the template. The target DNA was PCR-amplified from the reverse-transcription products using AccuPrime Pfx DNA polymerase (Invitrogen) with 5′-CGCGGAT CCAAGAAACCAATTAATGTAGC-3′ and 5′-CGCAAGCTTTCATGAAGAATTATGTTTGTT-3′ as the forward and reverse primers, respectively. The PCR product was cloned into pQE30 expression plasmid (Qiagen) with the BamHI and HindIII cloning sites (bold). The construct was verified by sequencing.
E. coli BL21 (DE3) cells (Novagen) harbouring the expression plasmid were grown in LB medium (3 l shake flask containing 1 l medium) at 310 K to an OD600 of 0.6. Overproduction of PfDXR was induced by 0.5 mM IPTG for 20 h at 293 K. After this period, cells were harvested by centrifugation at 8000g for 15 min, suspended in buffer A (10 mM Tris–HCl pH 8.0, 100 mM NaCl, 2 mM DTT, 20 mM imidazole) and disrupted using ultrasonication on ice for 5 × 30 s. The cell extract was obtained by centrifugation at 15 000g for 15 min and was applied onto a 1 ml HisTrap HP column (GE Healthcare) equilibrated with buffer B (10 mM Tris–HCl pH 8.0, 100 mM NaCl, 2 mM DTT). The column was washed with 50 column volumes of wash buffer (100 mM imidazole in buffer B). After washing, PfDXR was eluted with ten column volumes of elution buffer (500 mM imidazole in buffer B). The PfDXR was further purified by gel chromatography using a Superdex 200pg column (GE Healthcare) equilibrated with buffer C (50 mM Tris–HCl pH 7.8, 2 mM DTT). The fractions containing PfDXR were pooled and concentrated to 10 mg ml−1 using a Centricon-30 (Millipore).
2.2. Crystallization
The protein solution was mixed with 6 mM NADPH dissolved in buffer C at a volume ratio of 1:1. Initial sparse-matrix crystal screening (Jancarik & Kim, 1991 ▶) was conducted using Crystal Screen I (Hampton Research, USA), Wizard I, II and III and Cryo I and II (Emerald BioSystems, USA). Crystallization was carried out by the hanging-drop method, in which 1 µl protein solution (5 mg ml−1 protein and 3 mM NADPH) was mixed with the same volume of reservoir solution and incubated at 293 K. The drops were suspended over 200 µl reservoir solution in 48-well plates.
2.3. X-ray data collection
For data collection under cryogenic conditions, the crystals in a droplet were directly transferred to harvesting solution [3 mM NADPH, 0.3 M KCl, 20%(m/v) PEG 3350 and 20%(v/v) glycerol in 0.1 M Tris–HCl pH 8.0] for 1 min. Crystals were mounted in nylon loops and flash-cooled in a cold nitrogen-gas stream at 100 K just prior to data collection. Data collection was performed by the rotation method at 100 K using an ADSC Q270 CCD detector with synchrotron radiation [λ = 1.000 Å on beamline 17A of the Photon Factory (PF), Tsukuba, Japan]. The Laue group and unit-cell parameters were determined using the HKL-2000 package (Otwinowski & Minor, 1997 ▶).
3. Results and discussion
3.1. Overproduction, purification and crystallization
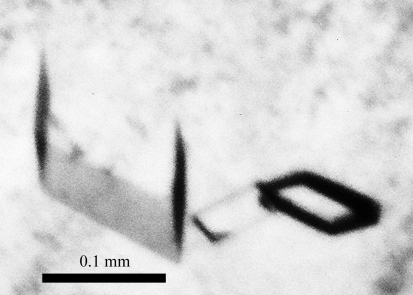
PfDXR was successfully cloned, overproduced and purified to homogeneity. SDS–PAGE of the purified enzyme revealed a single 47 kDa protein band by Coomassie Brilliant Blue staining. Initial crystal screening produced several microcrystals within one week. Rhomboidal crystals grew from condition Nos. 6 and 8 of Wizard III [No. 6, 20%(m/v) PEG 3350 and 0.2 M potassium formate; No. 8, 20%(m/v) PEG 3350 and 0.2 M potassium nitrate]. Trials to improve the crystallization conditions were performed by varying the pH, the buffer system and the concentration of the crystallizing agent. To obtain crystals suitable for X-ray analysis, a droplet was prepared by mixing equal volumes (2 µl + 2 µl) of the working solution described above and reservoir solution [0.3 M KCl and 20%(m/v) PEG 3350 in 0.1 M Tris–HCl pH 8.0] and suspended over 500 µl reservoir solution in 24-well plates. Rhomboidal crystals with typical dimensions of approximately 0.1 × 0.1 × 0.1 mm grew within one week (Fig. 1 ▶).
Figure 1.
Monoclinic crystals of PfDXR.
3.2. Data collection
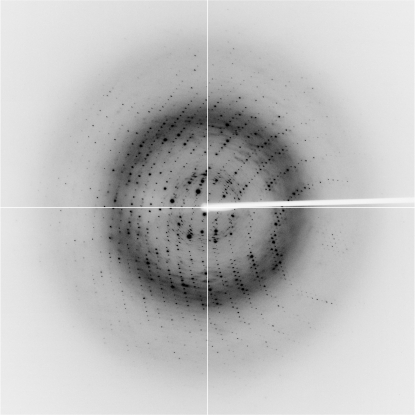
The Laue group of the PfDXR crystals was found to be 2/m and the unit-cell parameters were a = 168.89, b = 59.65, c = 86.58 Å, β = 117.8°. Only reflections with h + k = 2n were observed for hkl reflections, indicating the monoclinic space group C2. Assuming the presence of two subunits (one dimer) per asymmetric unit led to an empirically acceptable V M value of 2.03 Å3 Da−1, corresponding to a solvent content of 39.4% (Matthews, 1968 ▶). The current best diffraction data from a PfDXR crystal were collected to 1.85 Å resolution (Fig. 2 ▶). Data-collection statistics are summarized in Table 1 ▶.
Figure 2.
X-ray diffraction image from a PfDXR crystal. The edge of the detector corresponds to a resolution of 1.7 Å.
Table 1. Data-collection statistics for PfDXR.
Values in parentheses are for the outer shell.
| Space group | C2 |
| Unit-cell parameters (Å, °) | a = 168.89, b = 59.05, c = 86.58, β = 117.8 |
| No. of subunits per asymmetric unit | 2 [one dimer] |
| Solvent content (%) | 39.4 |
| X-ray source | PF BL17A |
| Detector | ADSC Q270 |
| Wavelength (Å) | 1.000 |
| Resolution range (Å) | 50–1.85 (1.92–1.85) |
| No. of observed reflections | 227987 |
| No. of unique reflections | 63420 |
| Multiplicity | 3.6 (2.6) |
| Mean I/σ(I) | 29.8 (2.5) |
| B factor (Wilson plot) (Å2) | 24.4 |
| Rmerge† (%) | 5.8 (32.4) |
| Completeness (%) | 98.9 (91.3) |
R
merge = 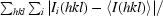
 , where I
i(hkl) is the ith measurement and 〈I(hkl)〉 is the weighted mean of all measurements of I(hkl).
, where I
i(hkl) is the ith measurement and 〈I(hkl)〉 is the weighted mean of all measurements of I(hkl).
3.3. Initial phase determination
Initial phase determination was performed by molecular replacement (MR) using the coordinate set of the E. coli DXR dimer (PDB code 1onn; Steinbacher et al., 2003 ▶), which shares approximately 30% amino-acid sequence identity with PfDXR, as a search model. The bound water molecules were removed from the search model. Cross-rotation and translation functions were calculated using the program AMoRe (Navaza, 1994 ▶) from the CCP4 suite (Collaborative Computational Project, Number 4, 1994 ▶). The results showed a clear solution [correlation coefficient of 0.369 (the first noise solution was 0.278) and R factor of 0.515 (the first noise solution was 0.546) in the resolution range 15.0–3.0 Å] and a reasonable molecular arrangement of PfDXR in the asymmetric unit. The MR solution was supported by the observation that the directions of the noncrystallographic twofold axes determined by the self-rotation function (data not shown) were consistent with the MR solution obtained. Automatic model building and refinement using the programs ARP/wARP (Lamzin & Wilson, 1993 ▶) and REFMAC5 (Murshudov et al., 1997 ▶) and further iterative manual model building and refinement with the programs XtalView (McRee, 1999 ▶) and REFMAC5 are currently in progress. In parallel with refinement, we are preparing crystals of PfDXR in complex with metal ions (Mg2+ or Mn2+), NADPH and fosmidomycin in order to study their mode of interaction with the enzyme.
Acknowledgments
We thank Drs Y. Yamada, N. Matsugaki and N. Igarashi of the Photon Factory for their help with data collection at the synchrotron facility. This work was supported in part by a grant from the Kato Memorial Bioscience Foundation (to NT) and a Grant-in-Aid for Young Scientists B No. 17770109 from the MEXT of Japan (to MN).
References
- Borrmann, S., Issifou, S., Esser, G., Adegnika, A. A., Ramharter, M., Matsiegui, P.-B., Oyakhirome, S., Mawili-Mboumba, D. P., Missinou, M. A., Kun, J. F. J., Jomaa, H. & Kremsner, P. G. (2004). J. Infect. Dis.190, 1534–1540. [DOI] [PubMed]
- Collaborative Computational Project, Number 4 (1994). Acta Cryst. D50, 760–763.
- Henriksson, L. M., Björkelid, C., Mowbray, S. L. & Unge, T. (2006). Acta Cryst. D62, 807–813. [DOI] [PubMed]
- Henriksson, L. M., Unge, T., Carlsson, J., Aqvist, J., Mowbray, S. L. & Jones, T. A. (2007). J. Biol. Chem.282, 19905–19916. [DOI] [PubMed]
- Jancarik, J. & Kim, S.-H. (1991). J. Appl. Cryst.24, 409–411.
- Jomaa, H., Wiesner, J., Sanderbrand, S., Altincicek, B., Weidemeyer, C., Hintz, M., Tubachova, I., Eberl, M., Zeidler, J., Lichtenthaler, H. K., Soldati, D. & Beck, E. (1999). Science, 285, 1573–1576. [DOI] [PubMed]
- Lamzin, V. S. & Wilson, K. S. (1993). Acta Cryst. D49, 129–147. [DOI] [PubMed]
- Lichtenthaler, H. K. (2000). Biochem. Soc. Trans.28, 785–789. [PubMed]
- MacSweeney, A., Lange, R., Fernandes, R. P., Shultz, H., Dale, G. E., Douangamath, A., Proteau, P. J. & Oefner, C. (2005). J. Mol. Biol.345, 115–127. [DOI] [PubMed]
- Matthews, B. W. (1968). J. Mol. Biol.33, 491–497. [DOI] [PubMed]
- McRee, D. E. (1999). J. Struct. Biol.125, 156–165. [DOI] [PubMed]
- Murshudov, G. N., Vagin, A. A. & Dodson, E. J. (1997). Acta Cryst. D53, 240–255. [DOI] [PubMed]
- Navaza, J. (1994). Acta Cryst. A50, 157–163.
- Otwinowski, Z. & Minor, W. (1997). Methods Enzymol.276, 307–326. [DOI] [PubMed]
- Reuter, K., Sanderbrand, S., Jomaa, H., Wiesner, J., Steinbrecher, I., Beck, E., Hintz, M., Klebe, G. & Stubbs, M. T. (2002). J. Biol. Chem.277, 5378–5384. [DOI] [PubMed]
- Ricagno, S., Grolle, S., Bringer-Meyer, S., Sahm, H., Lindqvist, Y. & Schneider, G. (2004). Biochim. Biophys. Acta, 1698, 37–44. [DOI] [PubMed]
- Rohmer, M. (1999). Nat. Prod. Rep.16, 565–574. [DOI] [PubMed]
- Steinbacher, S., Kaiser, J., Eisenreich, W., Huber, R., Bacher, A. & Rohdich, F. (2003). J. Biol. Chem.278, 18401–18407. [DOI] [PubMed]
- Yajima, S., Hara, K., Iino, D., Sasaki, Y., Kuzuyama, T., Ohsawa, K. & Seto, H. (2007). Acta Cryst. F63, 466–470. [DOI] [PMC free article] [PubMed]
- Yajima, S., Hara, K., Sanders, J. M., Yin, F., Ohsawa, K., Wiesner, J., Jomaa, H. & Oldfield, E. (2004). J. Am. Chem. Soc.126, 10824–10825. [DOI] [PubMed]
- Yajima, S., Nonaka, T., Kuzuyama, T., Seto, H. & Ohsawa, K. (2002). J. Biochem.131, 313–317. [DOI] [PubMed]