Abstract
We describe a mutant Escherichia coli RNA polymerase (RNAP) that forms stable open promoter complexes even at −20°C but with a shortened melted region that extends downstream to only position −7. In the presence of initiating transcription substrates, the mutant RNAP undergoes a temperature-dependent isomerization, resulting in a promoter complex that is indistinguishable from the wild-type RNAP–promoter complex, with the melted region extended downstream to position +4. We propose that the open complex formed by the mutant RNAP represents an intermediate on the normal promoter-opening pathway and that our results support earlier findings that initial promoter opening occurs in the upstream region of the −10 promoter consensus element and subsequently extends downstream to encompass the transcription start site.
Binding of RNA polymerase (RNAP) to promoter DNA occurs through the initial formation of a specific closed complex, followed by isomerization to a transcription-competent open complex in which the DNA from approximately −12 to +2 (with respect to the transcription start site at +1) becomes accessible to single-strand specific chemical probes such as potassium permanganate, KMnO4 (1), and is presumed to be melted. Formation of the open complex occurs highly cooperatively at temperatures above ≈15°C (2).
Kinetic and thermodynamic studies of the transition from the initial, closed complex to the final, transcription-competent open complex have indicated the existence of intermediate complexes before the formation of the final open complex (3). A major conformational change in RNAP occurs before the formation of the final open complex, which is then followed by formation of the transcription bubble (4, 5). The detailed pathway of strand separation to form the transcription bubble is promoter-dependent but must fall between two extremes, an all-or-none phenomenon in which the transcription bubble forms in a concerted manner or a stepwise phenomenon in which the transcription bubble nucleates in one region, then propogates in one or two directions to form the complete transcription bubble (6). The molecular mechanism by which the open complex is formed has not been established. It is clear that DNA melting requires extensive protein–DNA interactions, but RNAP sequences participating in these interactions are not known. The σ subunit plays a critical role in the melting process based on the following observations: (i) σ is necessary for promoter recognition; (ii) genetic and biochemical data indicate that evolutionarily conserved segments of σ contact the −10 consensus element of the promoter, which becomes melted in the open complex (7, 8); and (iii) mutations in σ have been isolated that lead to aberrant promoter melting (9). However, it is clear that the σ subunit is not the only determinant of DNA melting by RNAP. First, σ alone is unable to form open complexes. Second, strand separation remains in the elongation complex after σ release. Third, mutations in the β and β′ subunits of RNAP can lead to altered promoter melting temperatures (10). None of these mutations in the large subunits has been characterized in detail.
A set of extensive deletions in the β subunit, in a region of evolutionarily variable sequence centered at amino acid position 235, has been shown to assemble into functional RNAP in vitro and in vivo (11). While studying transcription initiation by mutant RNAPs, we observed that promoter complexes formed by enzymes harboring β deletions are highly unusual. Promoter complex formation by mutant RNAP harboring the largest deletion, βΔ(186–433), is the focus of this work.
MATERIALS AND METHODS
RNAP Purification.
The portion of rpoB harboring deletions was recloned from the original pMKSe2 background (11) into pET15b-β. In this plasmid, a complete rpoB is cloned between the NdeI and BamHI sites of pET15b (Novagen). The β subunit expressed by pET15b-β or its derivatives contain an N-terminal hexahistidine tag specified by pET15b. pET15b-β and pET15b-βΔ(186–433) were transformed into BL21(DE3) Escherichia coli cells, and transformants were grown in 4 liters of Luria–Bertani broth without addition of isopropyl β-d-thiogalactoside until late log phase. The cells were collected, and RNAP was purified by a combination of the standard purification procedure and metal ion affinity chromatography (12), concentrated by filtration through a C-100 concentrator (Amicon) to ≈1 mg/ml, and stored in 50% glycerol storage buffer at −20°C.
Footprinting and Transcription Reactions.
The 106-bp EcoRI DNA fragment containing the T7 A2 promoter (−84 to +32) was prepared as described (13). The fragment was 32P-end-labeled by filling in EcoRI sticky ends with Klenow enzyme in the presence of α-[ 32P] dATP. The fragment was then treated with AccI (cuts at position −70) to obtain a top-strand labeled fragment or with HincII (cuts at position +22) to obtain a bottom-strand labeled fragment. Promoter complexes were formed in 20-μl reactions containing 200 nM wild-type (wt) or mutant RNAP, 100 nM 32P-end-labeled DNA fragment, 40 mM Tris⋅HCl (pH 7.9), 40 mM KCl, and 10 mM MgCl2. Reactions were preincubated for 15 min at 37°C. DNase footprinting reaction was initiated by addition of DNase I (2 μg/ml DNase I, Worthington). The reaction proceeded for 30 s at 37°C and was terminated by addition of EDTA to 15 mM followed by phenol extraction and ethanol precipitation. For KMnO4 probing, promoter complexes were treated with KMnO4 (1 mM) for 15 s at 37°C. Reactions were terminated by addition of β-mercaptoethanol to 300 mM followed by phenol extraction, ethanol precipitation, and 10% piperidine treatment.
To footprint promoter complexes formed at different temperatures, RNAP and 32P-end-labeled T7 A2 promoter-containing fragments were combined on ice, incubated for 15 min on ice, transferred to the assay temperature, and incubated for an additional 15 min, and the footprinting reaction was performed. Alternatively, promoter complexes were allowed to form for 15 min at 37°C, followed by an additional 15-min incubation at the assay temperature and subsequent footprinting. The same conditions were used for KMnO4 probing at different temperatures. Control experiments demonstrated that KMnO4 modification was complete after 15 s at all temperatures used. For DNase footprinting, the following concentrations of DNase were used in a standard 30-s footprinting reaction: 25, 12.5, and 5 μg/ml DNase I at 0, 5, and 10°C, respectively. To probe promoter complexes with KMnO4 at −20°C, standard 20-μl reactions containing 50% glycerol were formed on ice, transferred to a Stratacooler (Stratagene), kept in a −20°C freezer, incubated for 1 h, and treated with KMnO4 without being removed from the Stratacooler. Control experiments demonstrated that glycerol at this concentration did not influence promoter melting by the wt and mutant RNAPs at ambient temperature.
To footprint promoter complexes in the presence of transcription substrates, a standard reaction was supplemented with CpG (0.5 mM) and individual NTPs (50 μM) as indicated in the figure legends. After a 15-min incubation at 37°C, the footprinting reaction was performed. In the experiment shown in Fig. 4, reactions also were supplemented with 10 μCi of [α-32P] CTP (3000 Ci/mmol, Amersham).
Figure 4.
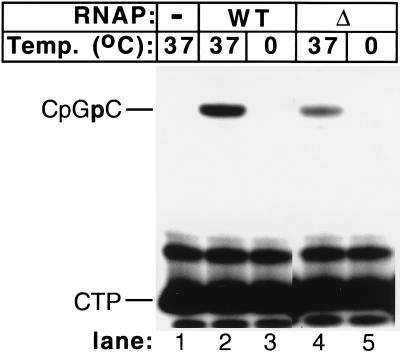
The mutant RNAP is unable to synthesize RNA at low temperatures. Transcription complexes were formed at the indicated temperature in the presence of 0.5 mM CpG and 50 μM α-[32P] CTP (30 Ci/mmol). Transcription reactions proceeded for 15 min. The reaction products were separated by denaturing electrophoresis in a 20% polyacrylamide gel and subsequently were autoradiographed.
Products of footprinting reactions were analyzed by urea/PAGE electrophoresis (7 M urea, 6% polyacrylamide), followed by autoradiography. Products of abortive initiation reactions were analyzed in the same way but with 20% polyacrylamide gels.
To quantify the extent of promoter opening, KMnO4 footprinting reactions were separated by urea/PAGE, and the gels then were quantitated on a PhosphorImager (Molecular Dynamics). The values for normalized promoter opening (%) were calculated by using imagequant software (Molecular Dynamics) by determining the cumulative intensity of KMnO4-reactive thymines in the transcription bubble area (−12 to −3 for the wt; −12 to −7 for the mutant), dividing this value by the total intensity in the lane (a sum of the signal from the transcription bubble and from full-sized DNA at the top of the lane) and multiplying by 100.
RESULTS
Open Promoter Complexes Formed by βΔ(186–433) RNAP Are Shortened in the Downstream Direction.
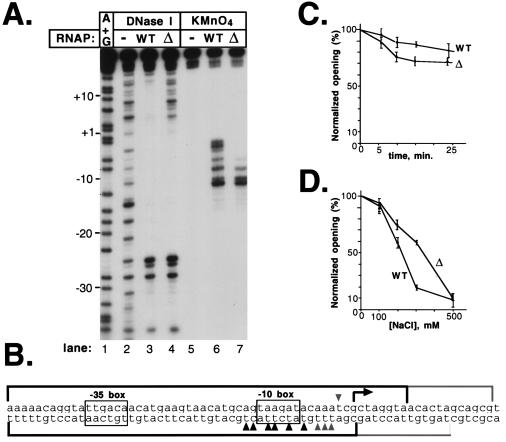
Promoter complexes formed by the mutant and the wt RNAP were studied by DNA footprinting (Fig. 1A). The DNase I footprint of the wt RNAP on the A2 promoter of bacteriophage T7 was typical for open complexes on σ70-promoters. Upstream of the transcription start site, protection started at approximately position −40, followed by a region of hypersensitivity at approximately position −25. The −10 promoter region was protected completely from DNase I attack, and this protection extended through the transcription start site to approximately position +20 (Fig. 1A, lane 3). With the mutant RNAP, an identical footprint was observed upstream of the transcription start site, position +1 was only partially protected, and there was no protection beyond position +5 (Fig. 1A, lane 4).
Figure 1.
The mutant RNAP forms stable promoter complexes that lack protein–DNA contacts downstream of the transcription initiation start point. (A) 100 nM of a 106-bp DNA fragment containing the T7 A2 promoter (−84 to +32) 32P-end-labeled on the bottom strand was combined with 200 nM wt or mutant RNAP in 20-μl reactions containing 40 mM Tris⋅HCl (pH 7.9), 40 mM KCl, and 10 mM MgCl2. Reactions were preincubated for 15 min at 37°C and footprinted with DNase I or probed with KMnO4. Reaction products were resolved on a 6% sequencing gel and visualized by autoradiography. (B) Summary of the RNAP–DNA footprinting experiments on the top and bottom strands of the T7 A2 promoter. Reactive thymines are indicated by arrows. DNase I-protected and KMnO4-sensitive sites found in the mutant and the wt promoter complexes are shown in black. DNase I-protected and KMnO4-sensitive sites specific for the wt complex are shown in gray. (C) Heparin resistance of RNAP-T7 A2 promoter complexes. Promoter complexes were formed as described above, and heparin was added to the final concentration 200 μg/ml (time 0), and incubation at 37°C was continued. At various time points, reaction aliquots were withdrawn, probed with KMnO4, resolved by denaturing PAGE, and quantified by using a PhosphorImager (Molecular Dynamics). The mean values of three independent measurements are given. The error bars represent the SD of the measurements. The normalized opening (%) value was calculated as described in Materials and Methods. (D) Reactions were set up as described above in a buffer containing the indicated concentrations of NaCl. Reactions were incubated for 15 min at 37°C and analyzed as in C.
Open complex formation by the wt and mutant RNAPs was investigated by using KMnO4 probing. In the wt open complex, thymines at −15, −12, −11, −9, −7, −5, −4, and −3, as well as cytosine at −14, were modified by KMnO4 (Fig. 1A, lane 6). In the mutant RNAP complex, the KMnO4-reactive region did not extend as far downstream; only thymines at positions −15, −12, −11, and −9 and cytosine at −14 were modified by KMnO4 (Fig. 1A, lane 7). The thymine at position −7 also was modified in the mutant complex but five times less efficiently than in the wt complex. In addition, modification of thymine at position −11 in the mutant complex was twice as efficient as in the wt complex (T−12 modification efficiency was used as a standard in these measurements). A summary of the footprinting results for both DNA strands of the T7 A2 promoter is schematically presented in Fig. 1B. Similar results were obtained when complexes on the T7 A1, deoP1, and galP1 promoters were studied (data not shown).
The nature of the mutant promoter complex was investigated by challenging it with heparin and elevated concentrations of NaCl. Despite the lack of downstream DNA contacts and the shortened region of KMnO4 sensitivity, the mutant complexes were nearly as stable as the wt open complexes toward the heparin challenge (Fig. 1C) and were slightly more stable than the wt open complexes to elevated salt concentrations (Fig. 1D).
βΔ(186–433) RNAP Promoter Complexes Form at Temperatures as Low as −20°C.
A possible interpretation of the shortened footprint observed for the promoter complex of the mutant RNAP is that the complex corresponds to an intermediate of the opening process that has been trapped by the mutation. It has been proposed that complexes obtained on the λ PR promoter in the absence of Mg2+ ions may correspond to opening intermediates defined by kinetic analyses (14). Footprinting experiments conducted with the wt and mutant RNAPs on the T7 A2 promoter in the presence or absence of Mg2+ demonstrated that the observed difference between the mutant and wt footprints was not affected by Mg2+ ions (data not shown).
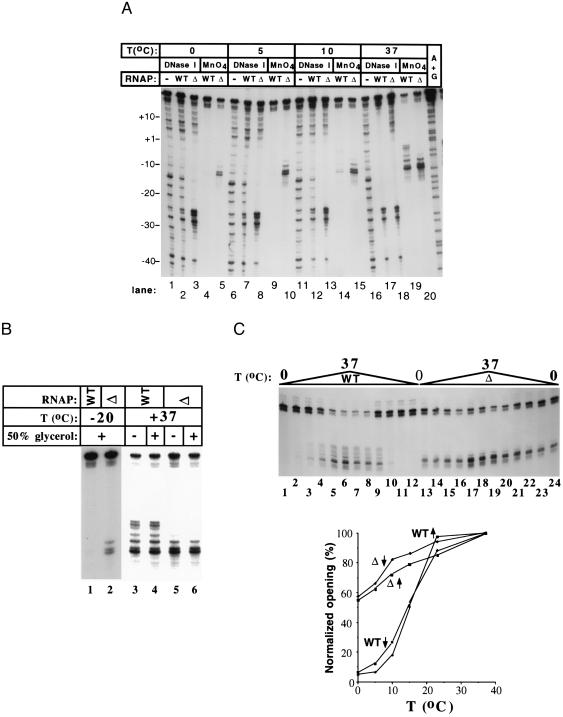
Lowering the reaction temperature also can cause accumulation of promoter complexes “frozen” at different stages of the promoter opening pathway (15). Accordingly, we reasoned that complexes formed by the wt enzyme at lower temperatures may resemble the mutant complexes at 37°C. DNase I and KMnO4 probing of promoter complexes formed over a wide range of temperatures indicates that this was not the case. When the reaction temperature was lowered, the wt complex “closed,” and no intermediate complexes were observed (Fig. 2C). The inability to obtain DNase I footprints with the wt enzyme at low temperatures was presumably caused by the low stability of the complex under these conditions (16). Surprising to note, the footprints of the mutant complex remained essentially unchanged between 37 and 0°C (Fig. 2A, lanes 20, 15, 10, and 5 and lanes 18, 13, 8, and 5, respectively). The only observable difference in the DNase I footprint was a 2-fold increase in the relative intensity of the hypersensitive bands in the −25 region.
Figure 2.
The mutant RNAP promoter complexes are stable at low temperatures. (A) DNase I footprinting and KMnO4 probing of promoter complexes at different temperatures (bottom strand). Promoter complexes were formed at the indicated temperatures and followed by DNase I and KMnO4 treatment. (B) KMnO4 probing of promoter complexes formed at −20°C in 50% glycerol to prevent freezing. The 37°C controls were conducted in the presence (lanes 4 and 6) or the absence (lanes 3 and 5) of 50% glycerol in the reaction buffer. The results indicate that open complex formation and KMnO4 sensitivity were not affected by the glycerol. See Materials and Methods for experimental details. (C) Quantitative KMnO4 probing of promoter complexes formed at different temperatures. Promoter complex formation reactions were set up on ice or 37°C, brought to the assay temperature, incubated for 15 min, and probed with KMnO4. The reaction products were separated by denaturing PAGE (Top) and quantified as described in Materials and Methods (Bottom). KMnO4 probing was performed at 0 (lanes 1, 12, 13, and 24); 5 (lanes 2, 11, 14, and 23); 10 (lanes 3, 10, 15, and 22); 15 (lanes 4, 9, 16, and 21); 23 (lanes 5, 8, 17, and 20); and 37°C (lanes 6, 7, 18, and 19). The upward vertical arrows indicate an up-temperature shift from 0°C to the assay temperature; downward arrows indicate a down-temperature shift from 37°C to the assay temperature.
KMnO4 probing revealed that the mutant RNAP formed significant amounts of “open” complex even at −20°C (Fig. 2B, lane 2). These experiments were conducted in 50% glycerol to prevent the reactions from freezing. At these conditions, a low signal at T−12 and T−11 also was detected in the wt complex (Fig. 2B, lane 1). These low levels of thymine sensitivity also were detected reproducibly when the wt complex was probed in the absence of glycerol at 0 and 5°C (Fig. 2A, lanes 4 and 9). This signal was protein-dependent; no T−12–T−11 sensitivity was observed when RNAP was omitted from the reaction (see, for example, Fig. 1A, lane 5). Control experiments established that glycerol at this concentration did not affect the extent of promoter melting at 37°C (Fig. 2B).
Additional experiments demonstrated that the mutant complex footprints did not extend further downstream when the reaction temperature was raised up to 60°C (irreversible denaturation of the wt and mutant RNAPs occurred at higher temperatures) or when supercoiled templates were used to form promoter complexes (data not shown).
Quantitative KMnO4 probing revealed that, as expected, the wt complex underwent a cooperative closed-to-open transition at ≈15°C (Fig. 2C). In contrast, the amount of open complex formed by the mutant RNAP decreased linearly in the temperature range from 37 to 0°C. The amount of open complexes formed at a given temperature was the same whether the reactions were set up on ice and then brought to the assay temperature or complexes were preformed at 37°C and then transferred to the assay temperature, demonstrating that the reactions were in equilibrium at the time of footprinting.
βΔ(186–433) RNAP Promoter Complex Undergoes a Temperature-Dependent Conformational Change in the Presence of Transcription Substrates.
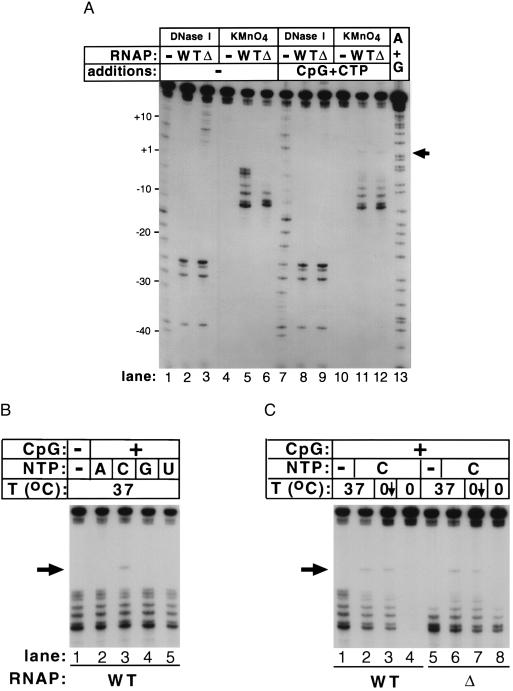
The mutant RNAP is transcriptionally active (11), yet it forms promoter complexes that barely protect the transcription start site from DNase I attack and does not form a transcription bubble encompassing the start site. To investigate this apparent paradox, we studied promoter complexes formed by the wt and mutant enzymes undergoing abortive initiation. Transcription on the T7 A2 promoter can be initiated by the dinucleotide CpG. In the presence of CTP, catalytic amounts of the abortive product CpGpC are formed. The presence of transcription substrates CpG (0.5 mM) or CTP (50 μM) alone did not result in any changes in the DNase I or KMnO4 footprints of either wt or mutant RNAP (data not shown). However, in the presence of 0.5 mM CpG and 50 μM CTP together, the DNase I footprint of the mutant RNAP underwent a dramatic downstream extension and became indistinguishable from the wt footprint (Fig. 3A, compare lanes 3 and 9). The changes in the DNase I footprint of the mutant RNAP (an extension of ≈12 bases in the downstream direction) were accompanied by the appearance of greatly increased KMnO4 reactivity at T−7 and new reactivity at T+4 (indicated by an arrow on Fig. 3, compare lanes 6 and 12), an extension of the KMnO4 footprint by 11 bases in the downstream direction. We note that a change also was evident in the wt complex, as judged by decreased KMnO4 reactivity at positions of sensitive positions −5, −4, and −3 and the appearance of reactivity at position +4. A less dramatic downstream extension of a KMnO4 footprint in the presence of initiating substrates of ≈4 bases has been observed with Bacillus subtilis RNAP with the σD alternative σ factor at the Phag promoter (18).
Figure 3.
Promoter complexes engaged in abortive RNA synthesis undergo a temperature-dependent conformational change. (A) Promoter complexes were formed at 37°C in the presence or absence of 0.5 mM CpG and 50 μM CTP, incubated for 15 min, and footprinted as described in the legend of Fig. 1. (B) KMnO4 probing of promoter complexes formed at 37°C in the presence of CpG (0.5 mM) and different individual NTPs (50 μM each). (C) KMnO4 probing of promoter complexes formed at low temperature in the presence of CpG and CTP. Promoter complexes were formed on ice or at 37°C and incubated for 15 min. Reactions shown in lanes 2–4 and 6–8 were supplemented with 0.5 mM CpG and 50 μM CTP, incubated for 15 min, and probed with KMnO4 (lanes 2, 4, 6, and 8). Complexes shown in lanes 3 and 7 were formed at 37°C, incubated in the presence of transcription substrates, transferred on ice, and incubated for an additional 15 min before KMnO4 probing. Lanes 1 and 5 show complexes formed in the absence of transcription substrates at 37°C.
The substrate-induced isomerization of the RNAP–promoter complex occurred only when the correct combination of initiating transcription substrates (CpG and CTP) was provided (Fig. 3B). Apparently, either simultaneous occupancy of the two RNAP substrate-binding sites or phosphodiester bond formation was required to induce the isomerization. The wt RNAP–promoter complex that underwent substrate-induced rearrangement at 37°C was markedly stabilized to lower temperatures (Fig. 3C, lane 3). Most importantly, the mutant complex could only rearrange at 37°C, not at 0°C (Fig. 3C, compare lanes 6 and 8).
βΔ(186–433) RNAP–Promoter Complex Formed at 0°C Is Transcriptionally Inactive.
The results of the previous experiment led us to suspect that promoter complexes formed by the mutant RNAP at 0 and 37°C may differ in their ability to transcribe DNA despite the fact that they appear indistinguishable in the footprinting experiments. The experiment presented in Fig. 4 demonstrates that this was indeed the case. Both wt and mutant RNAP synthesized abortive CpGpC product at 37°C but not at 0°C. In fact, when transcription by the two enzymes was studied over the whole temperature range, their behavior was identical, and the transcription activity of the two enzymes followed exactly the curve of promoter opening for the wt enzyme shown in Fig. 2C (data not shown).
DISCUSSION
The βΔ(186–433) RNAP exhibits the most dramatic promoter melting defect yet reported for a mutant in a core RNAP subunit. The mutant enzyme forms promoter complexes that lack the extensive protein–DNA contacts downstream of the transcription initiation start point and form a transcription bubble that also is shortened from the downstream end relative to the wt complex, as defined by KMnO4 sensitivity. It should be noted that, although KMnO4 sensitivity is presumed to indicate strand separation, structural distortions in the absence of strand separation also can lead to KMnO4 sensitivity. For the purposes of this study, KMnO4 sensitivity was used to assess open complex formation. Whether this indicates strand separation or not is somewhat irrelevant to our conclusions. Most surprising to note, unlike the wt complex, the mutant complex was stable at temperatures as low as −20°C. Finally, the mutant complex was able to isomerize in a temperature-dependent manner into a fully open complex only in the presence of substrates sufficient for the synthesis of the first phosphodiester bond.
It is tempting to speculate that the observed defects of βΔ(186–433) RNAP are the direct consequence of the absence of interactions between the deleted, functionally dispensable region of the β subunit and promoter DNA. This interpretation would be consistent with the data of Nudler et al. (17), who demonstrated that a site of the β subunit between amino acids 126 and 239 is in intimate contact with the downstream DNA in the E. coli RNAP elongating complex. On the other hand, our data show that mutant RNAPs with substantially smaller deletions in the same region of β–βΔ(193–248), βΔ(195–292), and βΔ[(166–328) produced footprints that were indistinguishable from the footprint of βΔ(186–433) (data not shown). Moreover, the downstream DNA contacts were observed in footprints of the mutant RNAP–promoter complex in the presence of transcription substrates (Fig. 3). Thus, the downstream extension of the footprints observed for the wt RNAP–promoter complex was not due to direct interaction of β(186–433) with downstream DNA but rather was due to an allosteric mechanism affecting the conformation of the RNAP. Nevertheless, the results clearly demonstrate that, in addition to the σ subunit, profound defects in promoter opening can be caused by mutations in the β subunit.
Analysis of promoter complexes formed by the mutant enzyme leads to several important insights into the early steps of open promoter complex formation by E. coli RNAP: 1. The stability of the mutant complex, as demonstrated by increased resistance to lowered temperature (Fig. 2A) and resistance to heparin and NaCl challenges (Fig. 1 C and D), demonstrates that, in contrast to the recent finding with elongating RNAP (17), protein–DNA contacts downstream of the transcription start site may not contribute significantly to open complex stability. 2. Because of the apparently normal transcription activity of the mutant RNAP and the wt pattern of protection of the upstream promoter DNA, promoter complexes formed by the mutant RNAP in the absence of transcription substrates likely represent an intermediate in the pathway of open complex formation that is normally not observed because of its short lifetime. Thus, it appears, at least for the promoters investigated here, that strand separation occurs initially in the downstream region from approximately −12 to −7 and subsequently extends further downstream to include the transcription start site. This conclusion is in agreement with several previous studies that used altered conditions such as temperature or Mg2+ concentration to probe potential intermediates in the DNA-melting pathway (14, 18–21). Unlike these previous studies, however, the downstream extension of our mutant RNAP KMnO4 footprint was accompanied by a large downstream extension of the DNase I footprint as well. In the previous studies cited, the downstream extension of the transcription bubble with increasing temperature or Mg2+ concentration occurred within the context of a constant DNase I footprint. 3. Appearance of the downstream protected region, and of the downstream open region, in the mutant RNAP complexes in the presence of transcription substrates suggests that a conformational change occurs in the RNAP at this step. We do not know if the conformational change in this case requires only the binding energy of the two transcription substrates or actually requires phosphodiester bond formation. 4. The apparent loss of cooperativity of promoter opening in the mutant complex leads us to propose that the observed cooperativity of promoter melting in the wt complex is not driven by the intrinsic thermodynamics of DNA melting in the A-T-rich −10 promoter consensus element (22, 23). Formation of the intermediate open complex even at −20°C by the mutant RNAP indicates that β residues 186–433 represent a barrier to initial open complex formation, perhaps by inhibiting conformational changes in the RNAP. This barrier, however, is overcome by wt RNAP at sufficiently high temperatures.
Acknowledgments
This work was initiated by K.S. during the 1995 European Molecular Biology Organization Practical Course “DNA Probing by Metal Complexes,” led by P. E. Nielsen. K.S. was supported by a Jane Coffin Childs Memorial Fund for Medical research postdoctoral fellowship and by a Career Award in Biomedical Sciences from the Burroughs Wellcome Fund for Biomedical Research. S.A.D. is a Pew Scholar in The Biomedical Sciences. This work was supported in part by grants from the Irma T. Hirschl Trust and the Pew Foundation and by Grant GM53759 from the National Institutes of Health (to S.A.D.).
Footnotes
This paper was submitted directly (Track II) to the Proceedings Office.
Abbreviations: RNAP, RNA polymerase; wt, wild-type.
References
- 1.Borowiec J A, Zhang L, Sasse-Dwight S, Gralla J D. J Mol Biol. 1987;196:101–111. doi: 10.1016/0022-2836(87)90513-4. [DOI] [PubMed] [Google Scholar]
- 2.Kirkegaard K, Buc H, Spassky A, Wang J. Proc Natl Acad Sci USA. 1983;80:2544–2548. doi: 10.1073/pnas.80.9.2544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Buc H, McClure W R. Biochemistry. 1985;24:2712–2723. doi: 10.1021/bi00332a018. [DOI] [PubMed] [Google Scholar]
- 4.Roe J-H, Record M T., Jr Biochemistry. 1985;24:4721–4726. doi: 10.1021/bi00339a002. [DOI] [PubMed] [Google Scholar]
- 5.Roe J-H, Burgess R R, Record M T., Jr J Mol Biol. 1985;184:441–453. doi: 10.1016/0022-2836(85)90293-1. [DOI] [PubMed] [Google Scholar]
- 6.deHaseth P L, Helmann J D. Mol Microbiol. 1995;16:817–824. doi: 10.1111/j.1365-2958.1995.tb02309.x. [DOI] [PubMed] [Google Scholar]
- 7.Waldburger C, Gardella T, Wong R, Susskind M M. J Mol Biol. 1990;215:267–276. doi: 10.1016/s0022-2836(05)80345-6. [DOI] [PubMed] [Google Scholar]
- 8.Severinova E, Severinov K, Fenyo D, Marr M, Brody E N, Roberts J W, Chait B T, Darst S A. J Mol Biol. 1996;263:637–647. doi: 10.1006/jmbi.1996.0604. [DOI] [PubMed] [Google Scholar]
- 9.Juang Y-L, Helmann J D. J Mol Biol. 1994;235:1470–1488. doi: 10.1006/jmbi.1994.1102. [DOI] [PubMed] [Google Scholar]
- 10.Gragerov A I, Kocherginskaya S A, Larionov O A, Kalyaeva E S, Nikiforov V G. Mol Gen Genet. 1980;180:399–403. doi: 10.1007/BF00425854. [DOI] [PubMed] [Google Scholar]
- 11.Severinov K, Kashlev M, Severinova E, Bass I, McWilliams K, Kutter E, Nikiforov V, Snyder L, Goldfarb A. J Biol Chem. 1994;269:14254–14259. [PubMed] [Google Scholar]
- 12.Kashlev M, Nudler E, Severinov K, Borukhov S, Komissarova N, Goldfarb A. Methods Enzymol. 1996;274:326–334. doi: 10.1016/s0076-6879(96)74028-4. [DOI] [PubMed] [Google Scholar]
- 13.Godovikova T S, Grachev M A, Kutyavin I V, Tsarev I G, Zarytova V F, Zaychikov E F. Eur J Biochem. 1987;166:611–616. doi: 10.1111/j.1432-1033.1987.tb13557.x. [DOI] [PubMed] [Google Scholar]
- 14.Suh W-C, Ross W, Record M T., Jr Science. 1993;259:358–361. doi: 10.1126/science.8420002. [DOI] [PubMed] [Google Scholar]
- 15.Cowing D W, Mecsas J, Record M T, Jr, Gross C A. J Mol Biol. 1989;210:521–530. doi: 10.1016/0022-2836(89)90128-9. [DOI] [PubMed] [Google Scholar]
- 16.Spassky A, Kirkegaard K, Buc H. Biochemistry. 1985;24:2723–2731. doi: 10.1021/bi00332a019. [DOI] [PubMed] [Google Scholar]
- 17.Nudler E, Avetissova E, Markovtsev V, Goldfarb A. Science. 1996;273:211–217. doi: 10.1126/science.273.5272.211. [DOI] [PubMed] [Google Scholar]
- 18.Chen Y-F, Helmann J D. J Mol Biol. 1997;267:47–59. doi: 10.1006/jmbi.1996.0853. [DOI] [PubMed] [Google Scholar]
- 19.Burns H, Minchin S. Nucleic Acids Res. 1994;22:3840–3845. doi: 10.1093/nar/22.19.3840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Meier T, Schickor P, Wedel A, Cellai L, Heumann H. Nucleic Acids Res. 1995;23:988–994. doi: 10.1093/nar/23.6.988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zaychikov E, Denissova L, Meier T, Götte M, Heumann H. J Biol Chem. 1997;272:2259–2267. doi: 10.1074/jbc.272.4.2259. [DOI] [PubMed] [Google Scholar]
- 22.Tachibana H, Ishihama A. Nucleic Acids Res. 1985;13:9031–9042. doi: 10.1093/nar/13.24.9031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Margalit H, Shapiro B A, Nussinov R, Owens J, Jernigan R L. Biochemistry. 1988;27:5179–5188. doi: 10.1021/bi00414a035. [DOI] [PubMed] [Google Scholar]