Abstract
Curcumin has shown some promise in prevention of oral carcinogenesis by mechanism(s) that are still not completely resolved. Messenger-RNA translation is mediated in eukaryotes by the eIF4F complex comprised of eukaryotic translation initiation factors eIF4E, eIF4G, and eIF4A. Over expression of some of these components or inactivation of initiation repressor proteins (4E-BP1) has been implicated in cancer development including oral carcinogenesis by affecting cell survival, angiogenesis, and tumor growth and invasion. In this study, we examined the possibility that curcumin affects the translational machinery differently in normal, immortalized normal, leukoplakia and malignant cells. Curcumin treatment in vitro inhibited the growth of immortalized oral mucosa epithelial cells (NOM9-CT) and the leukoplakia cells (MSK-Leuk1s) as well as in the UMSCC22B and SCC4 cells derived from head and neck squamous cell carcinoma. Curcumin only exerted minor effects on the growth of normal oral epithelial cells (NOM9). In the immortalized, leukoplakia and cancer cells, curcumin inhibited cap-dependent translation by suppressing the phosphorylation of 4E-BP1, eIF4G, eIF4B and Mnk1, and also reduced the total levels of eIF4E and Mnk1. Our findings demonstrate that immortalized normal, leukoplakia and malignant oral cells are more sensitive to curcumin and show greater modulation of protein translation machinery than the normal oral cells indicating that targeting this process may be an important approach to chemoprevention in general and for curcumin in particular.
Keywords: Immortalized cells, oral carcinogenesis model, translation control
Introduction
Gene expression is regulated at multiple stages including the process of transcription to modulate mRNA levels and the step of translation to modulate protein levels. Other mechanisms include mRNA stability and protein degradation. Deregulation of any of these steps causes aberrant gene expression leading to abnormal cell growth and development of premalignant and malignant lesions. In eukaryotes, mRNA translation is mainly regulated at the initiation, the rate-limiting step of translation, and involves a large multiprotein complex centered on the trimer eIF4F, which in turn is composed of the cap-binding protein eIF4E, the RNA helicase eIF4A and the multidomain adaptor protein eIF4G (1). The regulation of gene expression at the level of translation initiation is critical for proper control of cellular growth, proliferation, differentiation and apoptosis.
There is a vast difference in the amount of eIF4F required by cellular mRNAs to be translated efficiently and translation depends upon the composition of their 5′-UTRs, with higher eIF4F levels resulting in increased translation of growth factor mRNAs (2). The accessibility of the cap for eIF4E as well as the accessibility of eIF4E for eIF4G is tightly regulated by eIF4E-binding protein 1 (4E-BP1) belonging to a family of repressor proteins. The activity of 4E-BP1 is regulated by hierarchical phosphorylation of a set of conserved serine and threonine residues; hyperphosphorylated forms bind much weaker to eIF4E than hypophosphorylated forms (3). Dysregulated cap-dependent translation due to inappropriate eIF4F activation has recently been shown to play an important role in various human diseases and is emerging as one of the most promising approaches for cancer intervention.
Head and neck squamous cell carcinomas (HNSCC) are often characterized by overexpression of epidermal growth factor receptor (EGFR), c-Met as well as cytokine and G protein-coupled receptors, which converge on PI3K/Akt pathway in turn activating mTOR and positively regulating cap-dependent translation (4). In most HNSCCs, eIF4E is overexpressed and related to the disease progression (5). A previous study from our group has shown that siRNA against eIF4E can suppress the growth of a HNSCC cell line (6). Furthermore, phosphorylation of 4E-BP1 and eIF4G has been correlated with high rate of tumor proliferation in other cancers (7, 8). Such studies have indicated that eIF4F complex is an attractive target for cancer therapy in head and neck tumorigenesis. Recently, various approaches for targeting eIF4F complex have been described including the use of small peptide, antiviral drug, and a natural compound silibinin (9–11).
Curcumin, a derivative of the East Indian plant Curcuma longa, has been shown to exert anticancer effects against a broad range of cancers including HNSCC. It affects every major hallmark of cancer including cellular proliferation, growth, survival, angiogenesis and metastasis. Depending upon the cell type, curcumin can employ different molecular mechanisms such as inhibition of NFκB, downregulation of c-myc, cyclin D1, and protein tyrosine kinases (12). Interestingly, expression of cyclin D1 is also downregulated by the inhibition of eIF4E in head and neck cancer cells (6). Earlier studies have shown that curcumin treatment resulted in suppression of HNSCC growth both in vitro (13) and in vivo (14). However, it is not known whether curcumin affects cap-dependent translation differentially in normal, immortalized normal, leukoplakia and malignant cells, which is important in the context of cancer chemoprevention.
To address this question, we used an in vitro oral carcinogenesis model system that consists of normal, immortalized normal, leukoplakia and malignant human oral epithelial cells to evaluate the effects and molecular mechanism of action of curcumin on cap-dependent translation in oral epithelial cells representing different stages of oral carcinogenesis. Here, we report that curcumin inhibits the growth of these cells differentially and that the extent of growth inhibition depends, at least in part, on the disruption of eIF4F complex leading to diminished levels of critical cell cycle regulatory proteins including cyclin D1.
Material and Methods
Reagent and antibodies
Curcumin, with purity greater than 98%, was purchased from LKT laboratories (St. Paul, MN), dissolved in dimethyl sulfoxide (DMSO) and then further diluted in cell culture medium. All manipulations with drugs were carried out under subdued lighting. The antibody against ODC and β-actin (Sigma, St. Louis, MO) and the rest of the antibodies used were procured from Cell Signaling Technology (Danvers, MA).
Cell culture
Normal oral mucosa (NOM9) epithelial cells were derived from histologically normal buccal mucosa specimen using the method described by Xu et al. (15). The specimen was obtained from oral cancer patient according to a protocol (LAB03-0743) approved by the Institutional Review Board. All experiments were performed on second passage cells with the NOM9 cells. Immortalized but non-tumorigenic normal oral epithelial cells designated NOM9-CT were established from NOM9 cells by overexpressing Cdk4 (C) and hTERT (T) using methods described by Ramirez et al., (16). All of these cells were routinely maintained in serum-free keratinocyte growth medium (KGM®, Lonza Inc., Walkersville, MD). MSK-leuk1s a progressive variant of the cell line established from a dysplastic leukoplakia lesion (17) was kindly provided by Dr. P.G. Sacks (Memorial Sloan-Kettering Cancer Center, New York). The UMSCC22B cell line derived from a lymph node metastasis of a squamous cell carcinoma of the hypopharynx (18) was obtained from Dr. Thomas Carey (University of Michigan, Ann Arbor, MI) and SCC4 derived from a squamous cell carcinoma of the tongue (19), were grown in monolayer culture in a 1:1 (v/v) mixture of DMEM and Ham’s F12 medium supplemented with 5% fetal bovine serum in an incubator with a humidified atmosphere of 95% air and 5% CO2 maintained at 37°C.
Western blot analysis
Cells were lysed in cell lysis buffer consisting of 25 mM Tris-HCl pH 7.6, 150 mM NaCl, 1% NP-40, 1% Sodium deoxycholate and 0.1 % SDS (Thermo Scientific, Rockford, IL). After centrifugation, protein concentrations of the supernatants were determined (Protein Assay Kit, Bio-Rad Laboratories, Hercules, CA) and diluted with 2x Laemmli sample buffer (62.5 mM Tris-HCl [pH 6.8], 2% SDS, 25% glycerol, 710 mM β-mercaptoethanol, 0.01% bromophenol blue) to obtain equal protein amounts. Samples were boiled for 5 min at 95°C and separated on an 8% or 12% polyacrylamide gels containing SDS. For immunoblotting, proteins were transferred to a PVDF membrane (Millipore, Bedford, MA). Staining the membranes with Ponceau was used to assess the loading and transfer of proteins in the different lanes of the gel. The membrane was then incubated in blocking buffer (Tris-buffered saline [pH 7.6], 0.1% Tween-20, and 5% nonfat dry milk) for 1 hr, followed by overnight incubation with primary antibodies in blocking buffer at 4°C. The membranes were washed in the above buffer without the milk. The bound primary antibodies were then detected using IgG-horseradish peroxidase-conjugate (GE Healthcare, Piscataway, NJ) and visualized with an enhanced chemiluminescence detection system (GE Healthcare, Piscataway, NJ).
Transient transfection and in vitro translation assay
Cells were electroporated with bicistronic reporter plasmid (20) or control plasmid as described previously (21). Briefly, 1.2 × 106 cells were reconstituted in 100 μl of electroporation transfection solution or Nucleofector solution V (Amaxa Biosystems, Cologne, Germany). A bicistronic reporter or control plasmid was added to the cells and the mixtures were transferred to electroporation cuvettes and subjected to electroporation according to the manufacturer’s programs and instructions. The electroporated and transfected cell suspensions were immediately mixed with 500 μl of pre-warmed RPMI medium supplemented with 5% FBS. The cells were then transferred to 12-well plates for luciferase assay containing pre-warmed KGM medium supplemented with 5% FBS. Cells were incubated for 24 h after which they were incubated in serum-free KGM medium for another 24 h prior to treatment with curcumin. An in vitro translation assay of the bicistronic reporter assay system was done using the Dual-LuciferaseR reporter assay system (Promega, Madison, WI) as recommended by the manufacturer. After treatment with curcumin for 8 hrs cells were lysed with 250 μl/well of Passive Lysis Buffer (Promega, Madison, WI) by scraping vigorously with a rubber policeman and cell lysates were subjected to 1 or 2 freeze-thaw cycles to accomplish complete lysis of cells. Afterwards, 20 μl of each cell lysate were assayed for Renilla and firefly luciferase activity with a luminometer (EG & G Berthold).
Analysis of cell growth inhibition
For cell growth analysis, cells were seeded in 96-well plates at a density of 15,000 cells per well before treatment for 24 hours. Cells were treated with various doses of curcumin (0–50 μM) and controls were treated with the carrier (DMSO) alone. As described earlier (22) crystal violet assay was used to determine the percentage of growth inhibition.
Results
Curcumin differentially inhibits proliferation in oral normal, immortalized normal, leukoplakia and cancer cells
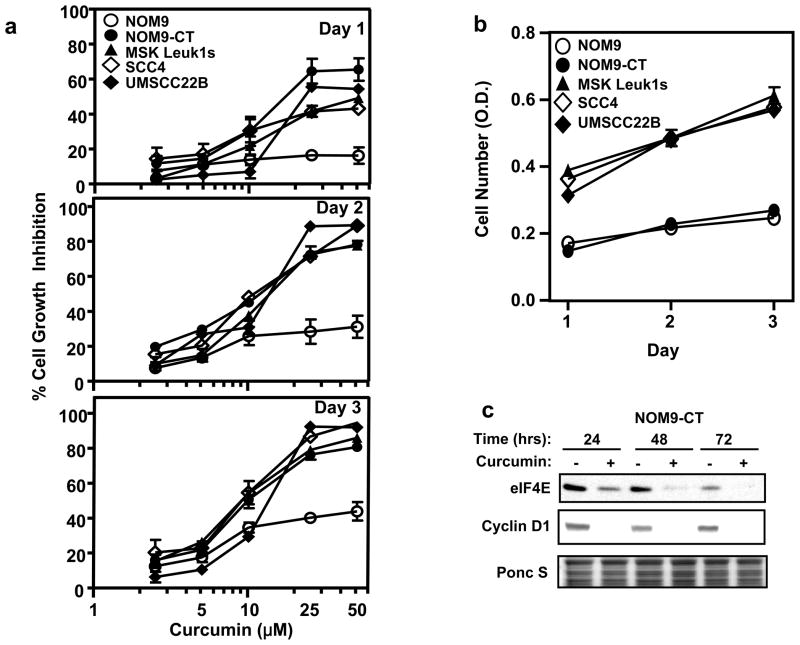
Previously, others and we have shown that curcumin inhibit the growth of HNSCC cell lines (13, 14), however, there is no report on the effect of curcumin on normal and immortalized or leukoplakia-derivded oral keratinocytes. We found that curcumin treatment of normal, immortalized normal, leukoplakia and malignant oral keratinocytes inhibited differentially cellular proliferation in dose- and time-dependent manner (Fig. 1a). The immortalized normal, leukoplakia and malignant cells appeared to be more sensitive to curcumin than the normal epithelial cells. This differential effect is not merely a reflection of a lower growth rate of the NOM9 cells as NOM9-CT cells had one and a half fold higher growth rate than NOM9. The MSK-leuk1s, UMSCC22B and SCC4 cells had a higher plating efficiency and three fold faster growth rate than the NOM9 cells (Fig. 1b). The similar inhibition of NOM9-CT, MSK-leuk1s, SCC4 and UMSCC22B despite their different growth rates suggests also that the effect of curcumin is not dependent tightly, if at all, on growth rate.
Figure 1.
Differential effects of curcumin on cell growth in normal, immortalized oral keratinocytes, leukoplakia and head and neck squamous cell carcinoma. (a) Each well of a 96-well plate was seeded with 15,000 cells and the plates were incubated overnight before treatment with DMSO control or the indicated curcumin concentrations for various time periods. Cell growth was determined by using the crystal violet assay. Data are shown as mean ± SD (n = 3); (b) In 96-well plate 15,000 cells per well were seeded and the plates were incubated for various time periods. Cell growth was determined by using the crystal violet assay. Data are shown as mean ± SD (n = 3); (c) NOM9-CT cells were treated with DMSO or 50 μM curcumin for 24, 48 and 72 hrs. Total cell lysates were prepared and subjected to 10% PAGE in the presence of SDS followed by sequential immunoblotting with antibodies against eIF4E and cyclin-D1.
To gain some insight into the mechanism/s by which curcumin induces cell growth inhibition we studied the NOM9-CT cells and found that there was a marked decrease in the cyclin D1 protein after curcumin treatment. Interestingly, several cellular protein targeted by curcumin viz. c-myc, cyclin D1, are also regulated by elevated levels of eIF4E. Previously, we have shown that silencing eIF4E in head and neck carcinoma cells using siRNA leads to decreased cyclin D1 levels and cell growth inhibition (6). Interestingly, levels of total eIF4E protein were also downregulated in curcumin treated cells correlating with the changes in total cyclin D1 protein levels (Fig. 1c). We therefore analyzed curcumin’s role in the modulation of protein translation in human normal, immortalized, immortalized normal, leukoplakia and cancer oral cells.
Differential effects of curcumin on protein translation initiation complex in normal, immortalized normal, leukoplakia and malignant oral cells
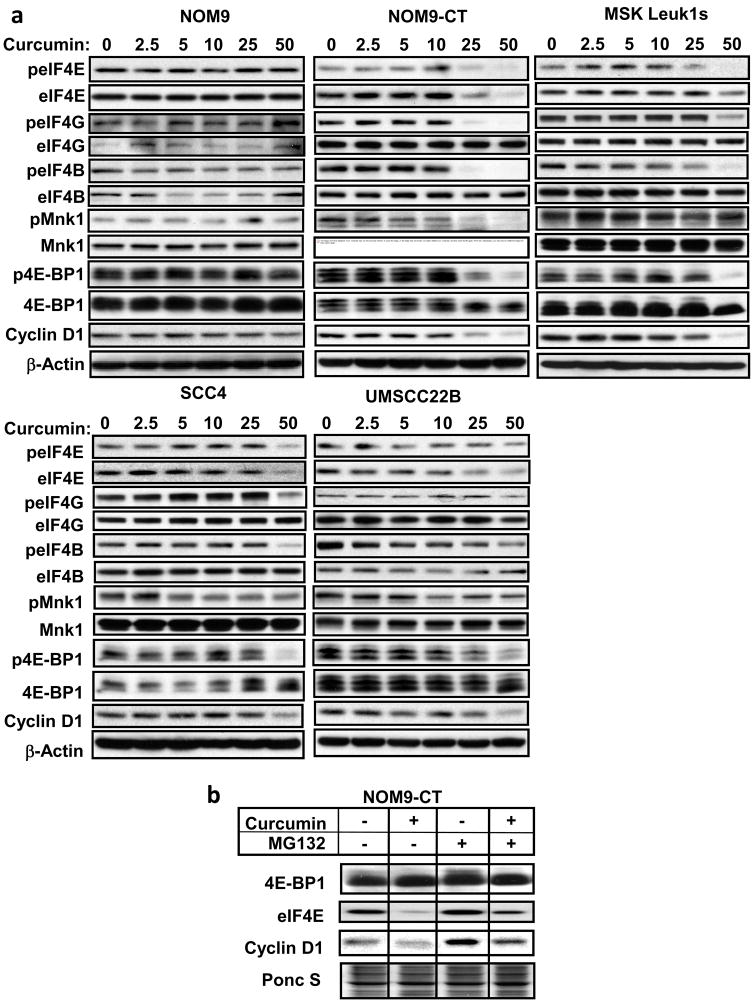
Different components of the eIF4F complex as well as proteins that either activate (Mnk1) or inhibit (4E-BP1) the translational machinery were affected differentially by curcumin (Fig. 2a). Also, the immortalized normal (NOM9-CT), leukoplakia (MSK-leuk1s) and cancer (SCC4, UMSCC22B) cells had high levels of constitutively active Mnk1 in comparison to the normal keratinocytes (NOM9). While NOM9 cells showed minor changes, NOM9-CT cells showed marked and dose-dependent decreases in peIF4E, pMnk1 along with total eIF4E and Mnk1 levels, whereas decreased levels of peIF4G, peIF4B and p4E-BP1 were observed without affect on their respective total protein levels. Similarly, MSK-leuk1s and malignant cells showed decreased levels of peIF4E along with total eIF4E, peIF4G, peIF4B, pMnk1, and p4E-BP1 also showed dose-dependent decrease. In response to the higher doses of curcumin, the levels of cyclin D1 in various cell lines decreased markedly corresponding with the decrease in translation machinery components. However, only a modest decrease in cyclin D1 protein was observed in curcumin-treated NOM9 cells.
Figure 2.
Effects of curcumin on components of the translational machinery. (a) NOM9, NOM9-CT, MSK-Leuk1s, SCC4 and UMSCC22B cells were treated with DMSO or various doses of curcumin for 30 min. Whole cell lysates were then prepared and Western blotting was performed using antibodies against the proteins (total protein and phosphorylated forms identified with the letter “p” listed to the left of each row. (b) NOM-9CT cells were pretreated with 5 μM MG132 for 24 hrs before treating with 50 μM curcumin for 30 min. The cell lysates were subjected to western blot analysis on 12% PAGE in the presence of SDS using antibodies against 4E-BP1, eIF4E and cyclin D1. The data are from one experiment, which was repeated with similar results.
Curcumin decreased the stability of eIF4E protein via proteasomal pathway
Real-time quantitative-PCR analysis was done to determine whether the decrease in the levels of eIF4E and other eIFs was due to their decreased transcription because curcumin modulates most of its target genes via inhibiting this process. Treatment of NOM9-CT cells with curcumin for 30 min did not alter the mRNA levels of 4E-BP1, eIF4E and cyclin D1 (data not shown), suggesting the involvement of posttranscriptional mechanisms in the process. Treatment with the proteasome inhibitor MG132 increased the levels of eIF4E (Fig. 2b), however, there was no effect on total 4E-BP1 proteins in MG132 pretreated cells. These results demonstrate that eIF4E is downregulated by curcumin in a proteasome-dependent manner.
Curcumin inhibits cap-dependent translation and expression of cell growth promoting proteins
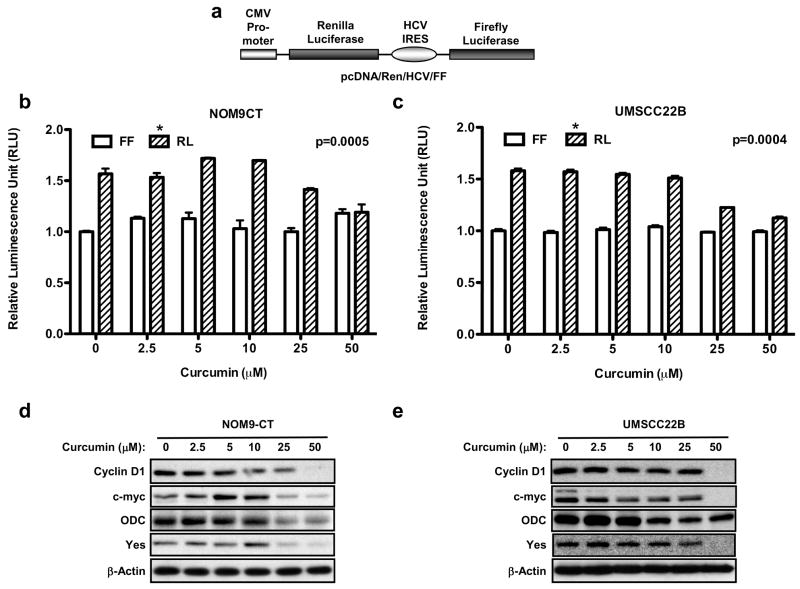
To test whether cap-dependent translation is a relevant biological target of curcumin in vivo, a transient expression assay was used in which cells were transfected with the bicistronic reporter and measured luciferase production in response to increasing doses of curcumin. The reporter construct consisted of two cistrons separated by the hepatitis C virus (HCV) internal ribosome entry site (IRES) (Fig. 3a), translation of the renilla luciferase (RL) cistron is cap-dependent, whereas that of the firefly luciferase (FL) cistron is directed by the hepatitis C virus IRES and is therefore cap-independent. Expression of renilla luciferase (RL) was significantly inhibited by curcumin in dose-dependent manner in both NOM9-CT (p=0.0005) and UMSCC22B cells (p=0.0004), whereas there was no significant effect on the levels of firefly luciferase (FL) (Fig. 3b, c). This shows that curcumin significantly inhibits in vivo cap-dependent translation.
Figure 3.
Effect of curcumin on cap-dependent translation. (a) Schematic diagram of Ren-HCV-FF mRNA; Graphical representation of the effects of various doses of curcumin on cap-dependent translation (Renilla luciferase) and HCV IRES-mediated translation (Firefly luciferase) in intact (b) NOM9-CT, and (c) UMSCC22B cells. Luciferase activity in treated cells is expressed relative to cells treated with vehicle (DMSO) alone. The results are the average of three experiments; error bars, s.e.m., significantly different (NOM9-CT, p=0.0005 and UMSCC22B, p=0.0004) compared with DMSO-treated control by one-way ANOVA; (d) NOM9-CT and (e) UMSCC22B cells were treated for 8 hrs with DMSO or various doses of curcumin. The whole cell lysates were resolved on 10% PAGE in the presence of SDS and subjected to Western blot analysis using antibodies against cyclin-D1, c-Myc, ODC, Yes and β-actin.
We proceeded to examine the effect of curcumin on expression of cell growth promoting proteins in NOM9-CT and UMSCC22B cells, to test the hypothesis that curcumin would downregulate translation of weak mRNAs while having a little effect on strong mRNAs. Indeed, Western blotting data shown in Figs. 3d and 3e demonstrate that the levels of β-actin (which has a short, unstructured 5′-UTRs) was unaffected, while expression of cyclin D1, c-myc, ornithine decarboxylase (ODC) and Yes (all of which have long, highly structured 5′-UTRs) was markedly decreased, suggesting that curcumin inhibited cap-dependent translation of weak mRNAs encoding proteins.
Discussion
In this study, we have used an in vitro oral carcinogenesis model for examining the hypothesis that curcumin’s anti-proliferative effects may be mediated at least in part by inhibition of cap-dependent translation. Our results demonstrate that, in immortalized, immortalized normal, leukoplakia and malignant cells, curcumin disrupts the translational machinery resulting in inhibition of protein translation. We propose that these effects contribute to the preferential inhibition by curcumin of the proliferation of immortalized, leukoplakia and cancerous cells in comparison to normal cells.
eIF4E is a rate-limiting factor required for the initiation of translation. eIF4E inactivation is mediated by inhibition of 4E-BP1 phosphorylation, which results in a shift of equilibrium from eIF4E/eIF4G complexes to eIF4E/4E-BP1 complexes. Our data show that curcumin inhibits the phosphorylation of 4E-BP1 and thus enhancing it’s activity. Our data is consistent with an ancillary observation of a decrease in the phosphorylation of 4E-BP1 made during a study on effects of curcumin on the mTOR pathway (23, 24). The inhibition of p4E-BP1 and eIF4E in curcumin-treated cells was associated with a reduction in cyclin D1 protein level, which could explain the inhibitory effect of curcumin on cell proliferation. In this respect, the effect of curcumin was similar to the effect of siRNA targeting eIF4E (6, 25) or the overexpression of 4E-BP1(26).
One of the key findings of this study was constitutive high level of activated Mnk1 in immortalized, leukoplakia and malignant cells in comparison with the normal oral keratinocytes. Mnk1, an eIF4E Ser 209 kinase, also interacts with the carboxy-terminal part of eIF4GI to form eIF4E eIF4GI Mnk-1 trimeric complex (27). Earlier it has been shown that constitutively activated Mnk1 promotes tumorigenesis (28), whereas, it is dispensable for normal growth and development in mammals (29). We have shown here for the first time that curcumin inhibits the phosphorylation of Mnk1 in immortalized normal oral keratinocytes, leukoplakia and malignant cells. This provides a novel chemopreventive approach for targeting Mnk1 kinase by curcumin.
eIF4G acts as a scaffolding molecule that interacts with several other components of the ribosomal initiation complex and its association with eIF4E strongly enhances the binding of the latter to 5′mRNA cap structures (30). Overexpression of eIF4G causes malignant transformation of NIH3T3 cells and is also a general event in lung carcinogenesis (31). In this study, we found that the basal levels of total eIF4G protein were low in normal oral keratinocytes (NOM9) in comparison to the other cell lines analyzed. Furthermore, we found that except in normal cells curcumin inhibits activation of eIF4G protein. In a recent study by Yu et al., (32) curcumin was shown to inhibit the phosphorylation of eIF4G in prostate cancer cells via activation of an unspecified protein phosphatase, thus disrupting the activity of eIF4F complex may be one of the hitherto underappreciated mechanisms of curcumin induced cell growth inhibition.
eIF4B stimulates eIF4F activity by potentiating the eIF4A RNA helicase activity (33). It also interacts directly with the eukaryotic translation initiation factor 3 (eIF3) (34) with which eIF4G interacts thus bridging the mRNA with the ribosome (35). Phosphorylation of eIF4B on Ser 422 is physiologically significant as it increases the interaction of eIF4B with eIF3 and is modulated by S6 kinase, implying that eIF4B may mediate some of the effects of S6Ks on translation (36). Earlier, curcumin has been shown to inhibit activated S6K (37) in rhabdomyosarcoma cell lines. Interestingly, in the current study curcumin also inhibited Ser 422 phosphorylation of eIF4B (Fig. 2a) that is critical for recruiting 40S ribosomal subunit to initiate scanning to the initiation codon and it’s joining with the 60S subunit.
Taken together, our results provide novel infomation that curcumin targets eIF4F complex thus inhibiting the protein translation and downregulating cyclin D1 and c-Myc proteins involved in cell cycle and proliferation. Whether the effective doses of curcumin used in the current study are achievable in vivo is not clear. Phase I clinical trials have shown that up to 8g/day of curcumin can be delivered to patients with virtually no side effects (38). Metabolism of the drug may differ from in vitro to that in vivo making it difficult to correlate its bioavailability. Recently it has been suggested that heat-solubilized curcumin could be used for increasing its bioavailability (39). Earlier, use of serum albumin as a carrier for curcumin has been reported (40) and is shown recently to stabilize curcumin in serum (41). Also, higher concentrations of curcumin could be delivered by means of liposomal delivery (42) and this formulation suppresses HNSCC growth both in vitro and in vivo (43). The results suggest that liposomal curcumin is a potentially useful nontoxic therapeutic agent for HNSCC. Another group (14) has shown that curcumin paste could be used topically for oral lesions and indeed, has been found to be very effective in the growth suppression of HNSCC xenograft tumors. Lastly, recent reports described the potentiation of the effects of low doses of curcumin on growth and apoptosis of immortalized human skin keratinocytes (HaCaT cells) (44) and epidermoid carcinoma cells (A431) (45) by combination with UV or visible light. All of these observations along with curcumin’s well-established pharmacological safety provide a rationale for clinical testing of curcumin as a chemopreventive agent in oral carcinogenesis.
Acknowledgments
Financial Support: This study was supported in part by a PO1 grant CA106451 (PI, Scott M. Lippman) from the National Cancer Institute and by the Irving and Nadine Mansfield and Robert David Levitt Cancer Research Chair (RL).
References
- 1.von der Haar T, Gross JD, Wagner G, McCarthy JE. The mRNA cap-binding protein eIF4E in post-transcriptional gene expression. Nat Struct Mol Biol. 2004;11:503–11. doi: 10.1038/nsmb779. [DOI] [PubMed] [Google Scholar]
- 2.Pickering BM, Willis AE. The implications of structured 5′ untranslated regions on translation and disease. Semin Cell Dev Biol. 2005;16:39–47. doi: 10.1016/j.semcdb.2004.11.006. [DOI] [PubMed] [Google Scholar]
- 3.Gingras AC, Raught B, Gygi SP, et al. Hierarchical phosphorylation of the translation inhibitor 4E-BP1. Genes Dev. 2001;15:2852–64. doi: 10.1101/gad.912401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Shaw RJ, Cantley LC. Ras, PI(3)K and mTOR signalling controls tumour cell growth. Nature. 2006;441:424–30. doi: 10.1038/nature04869. [DOI] [PubMed] [Google Scholar]
- 5.Nathan CA, Franklin S, Abreo FW, Nassar R, De Benedetti A, Glass J. Analysis of surgical margins with the molecular marker eIF4E: a prognostic factor in patients with head and neck cancer. J Clin Oncol. 1999;17:2909–14. doi: 10.1200/JCO.1999.17.9.2909. [DOI] [PubMed] [Google Scholar]
- 6.Oridate N, Kim HJ, Xu X, Lotan R. Growth inhibition of head and neck squamous carcinoma cells by small interfering RNAs targeting eIF4E or cyclin D1 alone or combined with cisplatin. Cancer Biol Ther. 2005;4:318–23. doi: 10.4161/cbt.4.3.1504. [DOI] [PubMed] [Google Scholar]
- 7.Rojo F, Najera L, Lirola J, et al. 4E-binding protein 1, a cell signaling hallmark in breast cancer that correlates with pathologic grade and prognosis. Clin Cancer Res. 2007;13:81–9. doi: 10.1158/1078-0432.CCR-06-1560. [DOI] [PubMed] [Google Scholar]
- 8.Petricoin EF, 3rd, Espina V, Araujo RP, et al. Phosphoprotein pathway mapping: Akt/mammalian target of rapamycin activation is negatively associated with childhood rhabdomyosarcoma survival. Cancer Res. 2007;67:3431–40. doi: 10.1158/0008-5472.CAN-06-1344. [DOI] [PubMed] [Google Scholar]
- 9.Assouline S, Culjkovic B, Cocolakis E, et al. Molecular targeting of the oncogene eIF4E in acute myeloid leukemia (AML): a proof-of-principle clinical trial with ribavirin. Blood. 2009;114:257–60. doi: 10.1182/blood-2009-02-205153. [DOI] [PubMed] [Google Scholar]
- 10.Ko SY, Guo H, Barengo N, Naora H. Inhibition of ovarian cancer growth by a tumor-targeting peptide that binds eukaryotic translation initiation factor 4E. Clin Cancer Res. 2009;15:4336–47. doi: 10.1158/1078-0432.CCR-08-2924. [DOI] [PubMed] [Google Scholar]
- 11.Lin CJ, Sukarieh R, Pelletier J. Silibinin inhibits translation initiation: implications for anticancer therapy. Mol Cancer Ther. 2009;8:1606–12. doi: 10.1158/1535-7163.MCT-08-1152. [DOI] [PubMed] [Google Scholar]
- 12.Aggarwal BB, Kumar A, Bharti AC. Anticancer potential of curcumin: preclinical and clinical studies. Anticancer Res. 2003;23:363–98. [PubMed] [Google Scholar]
- 13.Chakravarti N, Myers JN, Aggarwal BB. Targeting constitutive and interleukin-6-inducible signal transducers and activators of transcription 3 pathway in head and neck squamous cell carcinoma cells by curcumin (diferuloylmethane) Int J Cancer. 2006;119:1268–75. doi: 10.1002/ijc.21967. [DOI] [PubMed] [Google Scholar]
- 14.LoTempio MM, Veena MS, Steele HL, et al. Curcumin suppresses growth of head and neck squamous cell carcinoma. Clin Cancer Res. 2005;11:6994–7002. doi: 10.1158/1078-0432.CCR-05-0301. [DOI] [PubMed] [Google Scholar]
- 15.Xu L, Schantz SP, Edelstein D, Sacks PG. A simplified method for the routine culture of normal oral epithelial (NOE) cells from upper aerodigestive tract mucosa. Methods in Cell Science. 1996;18:31–9. [Google Scholar]
- 16.Ramirez RD, Sheridan S, Girard L, et al. Immortalization of human bronchial epithelial cells in the absence of viral oncoproteins. Cancer Res. 2004;64:9027–34. doi: 10.1158/0008-5472.CAN-04-3703. [DOI] [PubMed] [Google Scholar]
- 17.Sacks PG. Cell, tissue and organ culture as in vitro models to study the biology of squamous cell carcinomas of the head and neck. Cancer Metastasis Rev. 1996;15:27–51. doi: 10.1007/BF00049486. [DOI] [PubMed] [Google Scholar]
- 18.Krause CJ, Carey TE, Ott RW, Hurbis C, McClatchey KD, Regezi JA. Human squamous cell carcinoma. Establishment and characterization of new permanent cell lines. Arch Otolaryngol. 1981;107:703–10. doi: 10.1001/archotol.1981.00790470051012. [DOI] [PubMed] [Google Scholar]
- 19.Rheinwald JG, Beckett MA. Tumorigenic keratinocyte lines requiring anchorage and fibroblast support cultures from human squamous cell carcinomas. Cancer Res. 1981;41:1657–63. [PubMed] [Google Scholar]
- 20.Poulin F, Gingras AC, Olsen H, Chevalier S, Sonenberg N. 4E-BP3, a new member of the eukaryotic initiation factor 4E-binding protein family. J Biol Chem. 1998;273:14002–7. doi: 10.1074/jbc.273.22.14002. [DOI] [PubMed] [Google Scholar]
- 21.Kadara H, Lacroix L, Lotan D, Lotan R. Induction of endoplasmic reticulum stress by the pro-apoptotic retinoid N-(4-hydroxyphenyl)retinamide via a reactive oxygen species-dependent mechanism in human head and neck cancer cells. Cancer Biol Ther. 2007;6:705–11. doi: 10.4161/cbt.6.5.3963. [DOI] [PubMed] [Google Scholar]
- 22.Gillenwater A, Zou CP, Zhong M, Lotan R. Effects of sodium butyrate on growth, differentiation, and apoptosis in head and neck squamous carcinoma cell lines. Head Neck. 2000;22:247–56. doi: 10.1002/(sici)1097-0347(200005)22:3<247::aid-hed7>3.0.co;2-o. [DOI] [PubMed] [Google Scholar]
- 23.Bauer C, Diesinger I, Brass N, Steinhart H, Iro H, Meese EU. Translation initiation factor eIF-4G is immunogenic, overexpressed, and amplified in patients with squamous cell lung carcinoma. Cancer. 2001;92:822–9. doi: 10.1002/1097-0142(20010815)92:4<822::aid-cncr1388>3.0.co;2-a. [DOI] [PubMed] [Google Scholar]
- 24.Beevers CS, Chen L, Liu L, Luo Y, Webster NJ, Huang S. Curcumin disrupts the Mammalian target of rapamycin-raptor complex. Cancer Res. 2009;69:1000–8. doi: 10.1158/0008-5472.CAN-08-2367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Graff JR, Konicek BW, Vincent TM, et al. Therapeutic suppression of translation initiation factor eIF4E expression reduces tumor growth without toxicity. J Clin Invest. 2007;117:2638–48. doi: 10.1172/JCI32044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Averous J, Fonseca BD, Proud CG. Regulation of cyclin D1 expression by mTORC1 signaling requires eukaryotic initiation factor 4E-binding protein 1. Oncogene. 2007 doi: 10.1038/sj.onc.1210715. [DOI] [PubMed] [Google Scholar]
- 27.Pyronnet S, Imataka H, Gingras AC, Fukunaga R, Hunter T, Sonenberg N. Human eukaryotic translation initiation factor 4G (eIF4G) recruits mnk1 to phosphorylate eIF4E. Embo J. 1999;18:270–9. doi: 10.1093/emboj/18.1.270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wendel HG, Silva RL, Malina A, et al. Dissecting eIF4E action in tumorigenesis. Genes Dev. 2007;21:3232–7. doi: 10.1101/gad.1604407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ueda T, Watanabe-Fukunaga R, Fukuyama H, Nagata S, Fukunaga R. Mnk2 and Mnk1 are essential for constitutive and inducible phosphorylation of eukaryotic initiation factor 4E but not for cell growth or development. Mol Cell Biol. 2004;24:6539–49. doi: 10.1128/MCB.24.15.6539-6549.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.von Der Haar T, Ball PD, McCarthy JE. Stabilization of eukaryotic initiation factor 4E binding to the mRNA 5′-Cap by domains of eIF4G. J Biol Chem. 2000;275:30551–5. doi: 10.1074/jbc.M004565200. [DOI] [PubMed] [Google Scholar]
- 31.Bauer C, Brass N, Diesinger I, Kayser K, Grasser FA, Meese E. Overexpression of the eukaryotic translation initiation factor 4G (eIF4G-1) in squamous cell lung carcinoma. Int J Cancer. 2002;98:181–5. doi: 10.1002/ijc.10180. [DOI] [PubMed] [Google Scholar]
- 32.Yu S, Shen G, Khor TO, Kim JH, Kong AN. Curcumin inhibits Akt/mammalian target of rapamycin signaling through protein phosphatase-dependent mechanism. Mol Cancer Ther. 2008;7:2609–20. doi: 10.1158/1535-7163.MCT-07-2400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pestova TV, Kolupaeva VG, Lomakin IB, et al. Molecular mechanisms of translation initiation in eukaryotes. Proc Natl Acad Sci U S A. 2001;98:7029–36. doi: 10.1073/pnas.111145798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Methot N, Song MS, Sonenberg N. A region rich in aspartic acid, arginine, tyrosine, and glycine (DRYG) mediates eukaryotic initiation factor 4B (eIF4B) self-association and interaction with eIF3. Mol Cell Biol. 1996;16:5328–34. doi: 10.1128/mcb.16.10.5328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Etchison D, Milburn SC, Edery I, Sonenberg N, Hershey JW. Inhibition of HeLa cell protein synthesis following poliovirus infection correlates with the proteolysis of a 220,000-dalton polypeptide associated with eucaryotic initiation factor 3 and a cap binding protein complex. J Biol Chem. 1982;257:14806–10. [PubMed] [Google Scholar]
- 36.Raught B, Peiretti F, Gingras AC, et al. Phosphorylation of eucaryotic translation initiation factor 4B Ser422 is modulated by S6 kinases. EMBO J. 2004;23:1761–9. doi: 10.1038/sj.emboj.7600193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Beevers CS, Li F, Liu L, Huang S. Curcumin inhibits the mammalian target of rapamycin-mediated signaling pathways in cancer cells. Int J Cancer. 2006;119:757–64. doi: 10.1002/ijc.21932. [DOI] [PubMed] [Google Scholar]
- 38.Cheng AL, Hsu CH, Lin JK, et al. Phase I clinical trial of curcumin, a chemopreventive agent, in patients with high-risk or pre-malignant lesions. Anticancer Res. 2001;21:2895–900. [PubMed] [Google Scholar]
- 39.Kurien BT, Scofield RH. Heat-solubilized curcumin should be considered in clinical trials for increasing bioavailability. Clin Cancer Res. 2009;15:747. doi: 10.1158/1078-0432.CCR-08-1957. author reply. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kunwar A, Barik A, Pandey R, Priyadarsini KI. Transport of liposomal and albumin loaded curcumin to living cells: an absorption and fluorescence spectroscopic study. Biochim Biophys Acta. 2006;1760:1513–20. doi: 10.1016/j.bbagen.2006.06.012. [DOI] [PubMed] [Google Scholar]
- 41.Leung MH, Kee TW. Effective stabilization of curcumin by association to plasma proteins: human serum albumin and fibrinogen. Langmuir. 2009;25:5773–7. doi: 10.1021/la804215v. [DOI] [PubMed] [Google Scholar]
- 42.Li L, Braiteh FS, Kurzrock R. Liposome-encapsulated curcumin: in vitro and in vivo effects on proliferation, apoptosis, signaling, and angiogenesis. Cancer. 2005;104:1322–31. doi: 10.1002/cncr.21300. [DOI] [PubMed] [Google Scholar]
- 43.Wang D, Veena MS, Stevenson K, et al. Liposome-encapsulated curcumin suppresses growth of head and neck squamous cell carcinoma in vitro and in xenografts through the inhibition of nuclear factor kappaB by an AKT-independent pathway. Clin Cancer Res. 2008;14:6228–36. doi: 10.1158/1078-0432.CCR-07-5177. [DOI] [PubMed] [Google Scholar]
- 44.Dujic J, Kippenberger S, Hoffmann S, et al. Low concentrations of curcumin induce growth arrest and apoptosis in skin keratinocytes only in combination with UVA or visible light. J Invest Dermatol. 2007;127:1992–2000. doi: 10.1038/sj.jid.5700801. [DOI] [PubMed] [Google Scholar]
- 45.Dujic J, Kippenberger S, Ramirez-Bosca A, et al. Curcumin in combination with visible light inhibits tumor growth in a xenograft tumor model. Int J Cancer. 2009;124:1422–8. doi: 10.1002/ijc.23997. [DOI] [PubMed] [Google Scholar]