Abstract
The ferredoxin-dependent nitrite reductase from the green alga Chlamydomonas reinhardtii has been cloned, expressed in Escherichia coli as a His-tagged recombinant protein, and purified to homogeneity. The spectra, kinetic properties and substrate-binding parameters of the C. reinhardtii enzyme are quite similar to those of the ferredoxin-dependent spinach chloroplast nitrite reductase. Computer modeling, based on the published structure of spinach nitrite reductase, predicts that the structure of C. reinhardtii nitrite reductase will be similar to that of the spinach enzyme. Chemical modification studies and the ionic-strength dependence of the enzyme’s ability to interact with ferredoxin are consistent with the involvement of arginine and lysine residues on C. reinhardtii nitrite reductase in electrostatically-stabilized binding to ferredoxin. The C. reinhardtii enzyme has been used to demonstrate that hydroxylamine can serve as an electron-accepting substrate for the enzyme and that the product of hydroxylamine reduction is ammonia, providing the first experimental evidence for the hypothesis that hydroxylamine, bound to the enzyme, can serve as a late intermediate during the reduction of nitrite to ammonia catalyzed by the enzyme.
Keywords: Nitrite reductase, NII1, PETF, ferredoxin, hydroxylamine reduction, tertiary structure
Introduction
The initial portion of the pathway for nitrite assimilation in photosynthetic organisms, independent of the nitrogenase-catalyzed reduction of nitrogen to ammonia that occurs in organisms capable of nitrogen fixation, involves three reductive steps – the 2-electron reduction of nitrate to nitrite, the 6-electron reduction of nitrite to ammonia and the 2-electron reductive transfer of an amido-group from one molecule of glutamine to one molecule of 2-oxoglutarate to form 2 molecules of glutamate (Hase et al. 2006). The second of these reactions, i.e., the reduction of nitrite to ammonia, is catalyzed by the enzyme ferredoxin:nitrite oxidoreductase (EC 1.7.7.1, hereafter referred to as nitrite reductase), which uses reduced ferredoxin as its only physiological electron donor (Hase et al. 2006). These ferredoxin-dependent nitrite reductases, which are located in the chloroplast stroma in photosynthetic eukaryotes, are soluble enzymes with molecular masses of approximately 65 kDa and contain a unique prosthetic group arrangement in which a [4Fe-4S] cluster and a siroheme are coupled by a bridging sulfur from a cysteine, which serves simultaneously as a ligand to one of the cluster irons and as an axial ligand to the heme iron (Swamy et al. 2005; Hase et al. 2006). The spinach chloroplast enzyme has been extensively characterized and much is known about the oxidation-reduction and spectroscopic properties of its prosthetic group (Kuznetsova et al, 2004a; Kuznetsova et al. 2004b; Hase et al. 2006). A 2.8 Å resolution x-ray crystal structure is available for a His-tagged recombinant form of the spinach enzyme (Swamy et al. 2005) and the single ferredoxin-binding site on the enzyme has been modeled using this structure (Swamy et al. 2005; Hirasawa et al. 2009). The fact that “plant-type” ferredoxins are one-electron donors, coupled with the presence of only a single ferredoxin-binding site on nitrite reductase (Mikami and Ida 1980; Swamy et al. 2005; Hirasawa et al. 2009), makes elucidating the mechanism of the 6-electron reduction of nitrite to ammonia catalyzed by the enzyme particularly challenging, as no partially-reduced intermediates are released to any significant extent during the catalytic cycle and the enzyme can store only two electrons (Kuznetsova et al, 2004a; Kuznetsova et al. 2004b; Hase et al. 2006; Sétif et al. 2009).
Chlamydomonas reinhardtii is a chlorophyte alga in the green plant lineage, separated from the streptophytes by about a billion years (Merchant et al, 2007). Chloroplast metabolism, especially the photosynthetic apparatus, but also nitrogen assimilation, is highly conserved between land plant chloroplasts and C. reinhardtii (Fernández and Galvan 2008). As a microorganism, C. reinhardtii affords special advantages for the study of regulation of nutrient assimilation because of the facility with which the growth medium can be manipulated and the possibility of genetic dissection of metabolic pathways (Grossman et al. 2007; Gonzalez-Ballester and Grossman 2009; Hanikenne et al. 2009; Harris 2009; Moseley and Grossman 2009). The availability of the full genome sequence, with more than 15,000 gene/transcript models for this green alga (Merchant et al. 2007) has also made it a highly attractive target for such studies. Accordingly, there is a substantial literature on the biochemistry and expression of enzymes required for nitrogen, phosphorus and sulfur assimilation in C. reinhardtii (Fernández and Galvan 2008; Gonzalez-Ballester and Grossman 2009; Moseley and Grossman 2009). The NII1 gene encoding nitrite reductase of C. reinhardtii was identified because of its linkage to loci required for growth of C. reinhardtii on nitrate as the sole nitrogen source (Quesada et al. 1998) and its availability has enabled us to carry out an extensive biochemical characterization of this important enzyme in the nitrogen cycle.
While the chloroplastic, ferredoxin-dependent nitrite reductases from spinach and other vascular plants have been extensively characterized (Kuznetsova et al, 2004a; Kuznetsova et al. 2004b; Hase et al. 2006 and references cited therein), much less information is available on ferredoxin-dependent nitrite reductases from cyanobacteria (Flores et al. 2005) and even less is known about the algal enzymes. Although ferredoxin-dependent nitrite reductases have been purified from C. reinhardtii cells (Romero et al. 1987; Romero et al. 1989) and from other green algae (Zumft 1972; Vigara et al. 2002) and partially characterized, no system for expressing the C. reinhardtii enzyme in Escherichia coli so that site-directed mutagenesis studies can be carried out and the large amounts of enzyme needed for biophysical and structural studies can be produced has been reported to date. Below we describe a system for expressing and purifying a His-tagged, recombinant form of C. reinhardtii nitrite reductase from E. coli and report on the properties of the recombinant enzyme, including evidence from electron paramagnetic resonance (EPR) measurements for the presence of a [4Fe-4S] cluster in the enzyme. We have also used this form of C. reinhardtii nitrite reductase to provide the first evidence that hydroxylamine, which had long been postulated to be a possible intermediate in the 6-electron reduction of nitrite to ammonia that is catalyzed by the enzyme (Kuznetsova et al. 2004a; Kuznetsova et al. 2004b; Hase et al. 2006), can serve as a substrate for the enzyme and is reduced to ammonia.
Methods
A full-length cDNA, corresponding to clone ID 1112104D10, was identified among the ESTs (accessions BU653098 and BU653097) sequenced for Chlamydomonas (Jain et al. 2007). The insert in pBluescript II SK was sequenced to verify its identity and length. The open reading frame encoding the mature form of C. reinhardtii nitrite reductase was sub-cloned into the NdeI and BamHI sites of pET-28b vector (Novagen Inc.). The nucleotide sequence of the resulting construct was confirmed by sequencing in the Biotechnology and Genomics Core Facility at Texas Tech University. The mature form of protein excludes the 28 amino acids at the N-terminus that likely serves as the chloroplast targeting peptide of the precursor form of the enzyme and starts with an alanine. The open reading frame begins with, ATG GGC AGC AGC CAT CAT CAT CAT CAT CAC AGC AGC GGC CTG GTG CCG CGC GGC AGC CAT ATG, so that protein expressed using this construct contains 21 additional amino acids, including six consecutive histidines at the N-terminus. E. coli cells were transformed with the resulting recombinant clone, pET28b-NiR, according to the procedure described previously for recombinant spinach nitrite reductase (Tripathy et al. 2007). As was described previously for the case of recombinant spinach nitrite reductase (Tripathy et al. 2007), in order to insure that the biosynthesis of siroheme in E. coli did not limit the production of nitrite reductase holoenzyme, the cells were always co-transformed with a plasmid containing the E. coli cysG gene encoding the enzyme that catalyzes the rate-limiting step in siroheme biosynthesis, an S-adenosyl-l-methionine-dependent uroporphyrinogen III methyltransferase (Leustek et al. 1997). While the inclusion of the heme biosynthesis precursor ∂-aminolevulinic acid did not affect our expression yields in E. coli of recombinant spinach nitrite reductase (Tripathy et al. 2007), addition of ∂-aminolevulinic acid increased the yield of expression of C. reinhardtii nitrite reductase as had also been reported for other nitrite reductase expression systems (Bellissimo and Privalle 1995). Thus, to maximize the yield of active enzyme, ∂-aminolevulinic acid was included, at a concentration of 0.2 mM, in the E. coli growth medium routinely used for expression of the C. reinhardtii enzyme. The over-expression and purification of the His-tagged C. reinhardtii nitrite reductase was carried out using the procedure described previously for recombinant spinach nitrite reductase (Tripathy et al. 2007). The yield was approximately 10 mg of purified nitrite reductase from 12 liters of E. coli cells. Recombinant spinach nitrite reductase was prepared and purified as described previously (Tripathy et al. 2007). Nitrite reductase concentration was measured as described previously (Hirasawa and Knaff 1985).
Polyacrylamide gel electrophoresis in the presence of sodium dodecylsulfate as a denaturing detergent (SDS-PAGE) was carried out under conditions designed to reduce disulfide bonds. Approximately 20 µg of protein was incubated at 90 °C for 3 minutes in a solution containing 6-fold concentrated loading dye (BIO-RAD) and 0.1 M dithiothreitol. The mixture was then subjected to electrophoresis in a 10% Tris-HCl gel (BIO-RAD) for 1 hour 30 minutes at constant electric field strength of 100 volts. The gel was stained with Coomassie Brilliant Blue overnight and de-stained with 40% methanol and 10% acetic acid. Immuno-blot analysis was carried out using a monoclonal antibody against the (His)6 tag epitope (Novagen) and analytical reagents obtained from Novagen, following procedures supplied by the vendor. Briefly, the protein was electrophoretically transferred to a polyvinylidene fluoride (PVDF) membrane (Millipore) using standard transfer buffer (192 mM glycine, 25 mM Tris base, 20% methanol, pH 8.0). After incubation with blocking solution the blot was incubated for one hour with His-tag monoclonal antibody (1:1000 dilution with blocking solution) and then for 1 hr (hour) with Goat Anti-Mouse IgG AP conjugate (1:1500 dilution in blocking solution). Bands containing the (His)6 antigen were visualized by incubating the membrane in a color developing solution contained in the Novagen kit.
Mass spectrometry (MALDI-ToF-ToF) was carried out as follows: For analysis of the intact protein, one µl of protein solution (10 µg/ul in 20 mM ammonium bicarbonate buffer) was mixed with 5 µl of 30 mM sinapinic acid and from this mixture 0.5 µl of sample was spotted onto the target plate according to the manufacturer’s instructions and was analyzed using a MALDI-ToF/ToF 4800 Plus mass spectrometer (Applied Biosystems, Framingham, MA) equipped with a 355-nm Nd-YAG laser. Analysis was done in a positive ion linear mode and the external calibration was carried out using bovine serum albumin and lysozyme as standards. For mass spectrometry of tryptic peptides, a small Coomassie-stained band was carefully excised from the gel. Eppendorf tubes containing the protein band were placed in a Thermomixer Comfort (Eppendorf) and washed with 100 µl of H2O for five minutes at 37 °C and at 600 rpm. The sample was then washed with 100 µl of a H2O /acetonitrile (50/50) solution for 5 minutes at 37 °C and at 600 rpm. This step was repeated until the dye was completely removed, and the band was then incubated for 1 hour in 50 µl of a 10 mM DTT solution at 56 °C and 600 rpm. Alkylation was performed using a 55 mM iodoacetamide (IA) solution (in 40 mM ammonium bicarbonate) for 45 minutes in the dark, after which the sample was washed with H2O /acetonitrile (50/50). A spot of this sample was in 100 µl of 100% acetonitrile for one minute and, after removal of the acetonitrile, the protein band was dried for 15 minutes. Digestion was performed by the addition of 10 µL of an aqueous ammonium hydrogen carbonate (30 mM) containing 200 ng of trypsin (Promega Sequencing Grade Modified Trypsin, Promega Corporation, Madison, USA) at 37 °C overnight, and the extracted peptide solution was vacuum-dried and kept at −20 °C for further analysis by the MALDI-ToF-ToF mass spectrometry. Prior to analysis, peptides were resuspended in H2O/acetonitrile (50/50) containing 0.1% trifluoroacetic acid and 0.5 µl samples of the peptide-containing solution was deposited on the MALDI-plate, followed by addition of 0.5 µl of matrix solution. The matrix solution consisted of 5 mg/ml of α-cyano-4-hydroxycinnamic in H2O/acetonitrile (50/50) containing 0.1% trifluoroacetic acid. Database searching against the NCBInr database (taxonomy: all entries) was performed using the Mascot search engine (Matrix Science, London, UK). A peptide mass tolerance of ± 50 ppm was used and a ± 250 ppm tolerance was used for MS/MS fragments ions. One missed cleavage site was allowed and cysteine-carbamidomethylation and methionine oxidation were allowed as variable modifications.
C. reinhardtii ferredoxin 1 (the product of the petF gene) was purified from C. reinhardtii cells as described previously (Terauchi et al. 2009), with the following modifications - Chromatography was carried out using, in order, ion exchange chromatography on DE-52 DEAE-cellulose (Whatman), followed by Q-Sepharose (General Electric Healthcare) and then gel filtration chromatography on Sephacryl S100 (General Electric Healthcare). Linear NaCl gradients from 50 mM to 600 mM were used during the ion exchange steps. All proteins used showed a single Coomassie Blue-staining band after polyacrylamide gel electrophoresis in the presence of sodium dodecyl sulfate. Ferredoxin concentration was determined as described previously (Hirasawa and Knaff 1985). Although it has recently been shown that C. reinhardtii ferredoxin 2 may be a more efficient electron donor to C. reinhardtii nitrite reductase than ferredoxin 1 (Terauchi et al. 2009), the difference in catalytic efficiencies between these two C. reinhardtii chloroplast ferredoxins is small and the greater availability of ferredoxin 1 resulted in its being used for all of the experiments described below.
Nitrite reductase activity with either reduced methyl viologen serving as the electron donor or reduced ferredoxin serving as the electron donor was measured as described previously (Hirasawa et al. 1993). The C. reinhardtii enzyme displays a fairly broad activity vs. pH profile with an optimum at pH 7.5, similar to pH profiles exhibited by other plant nitrite reductases (Hicklesby et al. 1976; Vega and Kamin 1977; Hirasawa and Tamura 1980; Ida and Mikami 1986; Ida et al. 1989; Romero et al. 1989; Ip et al. 1990) and so activity was assayed at this pH. Chemical modification of nitrite reductase arginine and lysine side chains, with either phenylglyoxal or N-acetylsuccinimide, was carried out as described previously (Hirasawa et al. 1993). NADPH oxidation, coupled to the reduction of either nitrite or hydroxylamine in a reaction mixture containing ferredoxin, ferredoxin:NADP+ oxidoreductase (hereafter abbreviated as FNR) and nitrite reductase, was measured as described by Yonekura-Sakakihara et al. (Yonekura-Sakakihara et al. 2000) for the coupling of NADPH oxidation to the sulfite-reductase catalyzed reaction, except that nitrite reductase replaced sulfite reductase and spinach leaf FNR (Shin and Oshino 1978) replaced maize FNR. Ammonia production was measured using a Conway microdiffusion unit (Conway 1957).
Absorbance spectra in the visible and ultraviolet regions were measured at 0.5 nm spectral resolution using a Shimadzu Model 2401PC spectrophotometer. Complex formation between nitrite reductase and either ferredoxin or nitrite was measured using the previously described (Hirasawa and Knaff 1985) spectral perturbation method. Electron paramagnetic resonance (EPR) spectra were recorded on frozen samples with a Bruker ESP300E spectrometer equipped with an ER-4116 dual-mode cavity and an Oxford instruments ESR-9 flow cryostat as described previously (Hirasawa et al. 2009). Spectra were typically recorded at 15 K with a microwave power of 2 mW, a modulation frequency of 100 KHz, and a modulation amplitude of 6.4 Gauss. A 10-fold molar excess of KCN was added to oxidized samples prior to dithionite reduction in order observed the EPR spectrum of the magnetically isolated [4Fe-4S]+ cluster in reduced nitrite reductase samples. Total iron (Miller and Massey 1965), acid-labile sulfide (Siegel et al. 1973) and siroheme (Siegel et al. 1973) contents were measured using standard methods, with spinach ferredoxin used as the standard for the sulfide determination. Spectroelectrochemical oxidation-reduction titrations were carried out as described previously (Smith et al. 1981).
A model for the three-dimensional structure of the C. reinhardtii nitrite reductase was created with the three-dimensional structure of spinach chloroplast nitrite reductase (Swamy et al. 2005) used as the starting model, using the program Chimera (Pettersen et al. 2004). Based upon a comparison of the two sequences, non-conserved amino were replaced with the most likely rotomers that avoid steric interactions. The modified model was then subjected to energy minimization.
Results
The purified C. reinhardtii nitrite reductase showed only a single Coomassie-staining band after SDS-PAGE, even at high total protein concentrations, with an approximate molecular mass of 63 kDa. An immuno-blot, using an antibody against the His-tag epitope, also showed only a single band at this molecular mass, confirming the presence of the (His)6 tag at the N-terminus of the expressed protein. The MALDI-ToF-ToF mass spectrum also showed only a single peak, although the peak did show some asymmetric broadening on the larger molecular mass side. These analyses indicate that the protein is at least 95% pure. Amino acid sequences, obtained by mass spectrometry, of several tryptic peptides derived from the C. reinhardtii enzyme gave an exact match to the sequences for these segments of the enzyme derived from translation of the base sequence of the gene (data not shown).
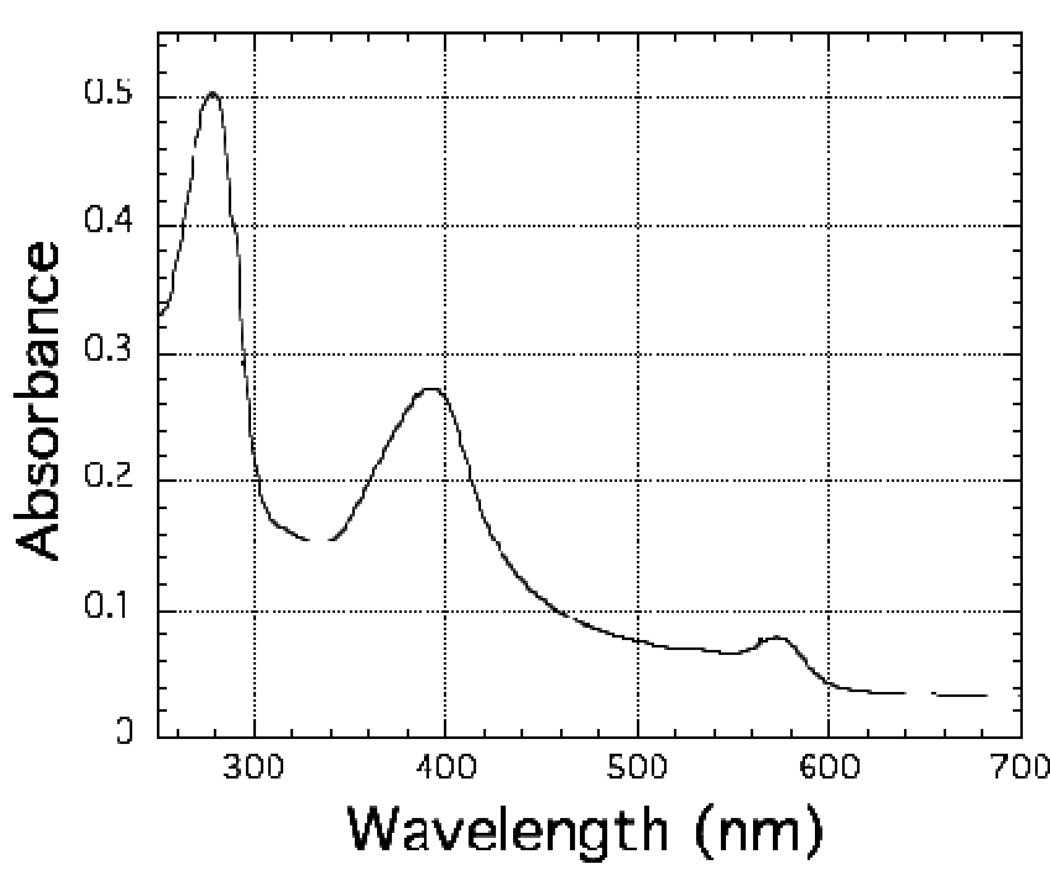
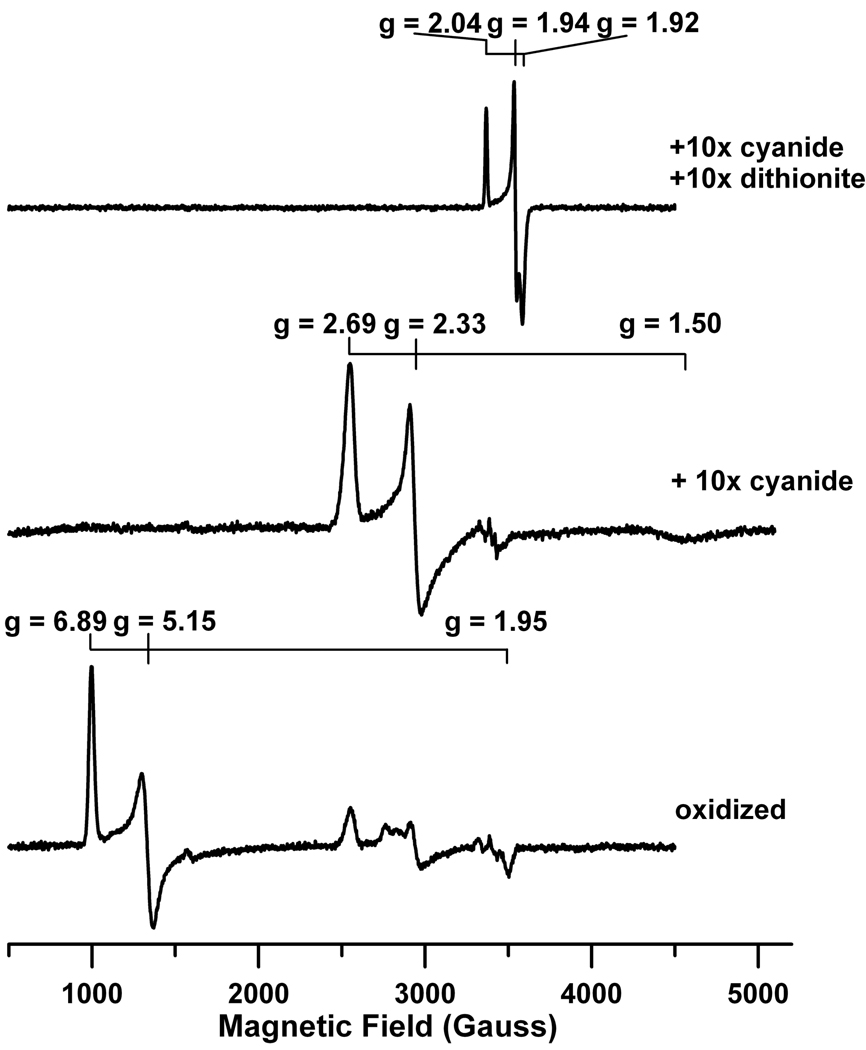
The purified C. reinhardtii enzyme exhibited absorbance maxima at 279 nm, 393 nm, 573 nm and 690 nm (Figure 1). Its spectrum is quite similar to that reported previously for the enzyme isolated directly from C. reinhardtii cells (Romero et al. 1987) and those of other ferredoxin-dependent, nitrite reductases (Zumft 1972; Hucklesby et al. 1976; Vega and Kamin 1977; Hirasawa and Tamura 1980; Ida and Mikami 1986; Ida et al. 1989; Ip et al. 1990; Bellissimo and Privalle 1995; Curdt et al. 2000; Vigara et al. 2002; Tripathy et al. 2007) and suggested that, like other nitrite reductases (Hase et al. 2006), it contains a siroheme and a [4Fe-4S] cluster as prosthetic groups. EPR spectra (Figure 2) of the as-isolated, oxidized enzyme, which contains a high-spin (S = 5/2) Fe3+ siroheme and an EPR-silent (S = 0) [4Fe-4S]2+ cluster (Aparicio et al. 1975; Stoller et al. 1977; Cammack et al. 1978; Lancaster et al. 1979; Ip et al. 1990; Kuznetsova et al. 2004a; Kuznetsova et al. 2004b; Tripathy et al. 2007) and of its cyanide adduct, in which the [4Fe-4S] cluster remains in the oxidized 2+ redox state and the high spin (S = 5/2) Fe3+ siroheme has been converted to a low-spin (S =1/2) Fe3+ siroheme cyanide complex (Aparicio et al. 1975; Cammack et al. 1978; Lancaster et al. 1979; Wilkerson et al. 1983; Hirasawa et al. 1987; Kuznetsova et al. 2004b), confirm the presence of the siroheme, as does pyridine hemochromagen analysis of an acidic-acetone extract of the protein. As is also- shown in Figure 2, addition of dithionite to the cyanide-adduct, reduces both prosthetic groups, resulting in an EPR-silent low-spin (S = 0) Fe2+ siroheme and the characteristic spectrum of a magnetically isolated (S = 1/2) reduced [4Fe-4S]1+ cluster (Ip et al. 1990; Aparicio et al. 1975; Stoller et al. 1977; Cammack et al. 1978; Lancaster et al. 1979; Wilkerson et al. 1983; Kuznetsova et al. 2004a). Analyses for non-heme iron and acid-labile sulfide are also consistent with the presence of an iron-sulfur cluster.
Figure 1.
The spectrum of His-tagged, recombinant C. reinhardtii nitrite reductase. Nitrite reductase was present at a concentration of 3.13 µM in 10 mM potassium phosphate buffer, pH 7.7. The spectrum was recorded at ambient temperature in a 1.0 cm optical pathlength cuvette.
Figure 2.
X-band EPR spectra of oxidized, cyanide-bound and dithionite-reduced cyanide-bound wild type C. reinhardtii nitrite reductase. The cyanide-bound sample was prepared by treating the oxidized sample with a 10-fold molar excess of KCN in 250 mM potassium phosphate buffer at pH 7.8. The cyanide-bound nitrite reductase sample was anaerobically reduced using a 10-fold molar excess of sodium dithionite. EPR spectra were recorded at 15 K with a microwave frequency of 9.604 GHz and a modulation amplitude of 6.4 Gauss. A microwave power of 0.02 mW, 2 mW and 20 mW was applied to the dithionite-reduced cyanide-bound, oxidized and cyanide-bound C. reinhardtii nitrite reductase samples, respectively.
The amino acid sequence of the C. reinhardtii nitrite reductase, deduced from the nucleotide sequence of the gene encoding this enzyme, in the region containing the four cysteines (shown in bold-face) associated with prosthetic group binding (SCTGNQFCGFGLAETKAKAV-TGCPNSCGQA) is very similar to the consensus sequence of the prosthetic group binding domains of eleven other ferredoxin-dependent, cyanobacterial and plant nitrite reductases (ACTGSQFCGQAIIETKARAL-TGCPNSCGQV). The high degree of homology is consistent with the hypothesis that the prosthetic group content and arrangement is identical in all of these nitrite reductases.
Data obtained during a spectroelectrochemical oxidation-reduction titration of the enzyme (not shown) revealed the presence of two separate one-electron redox couples with Em values of −255 ±10 mV and −390 ±10 mV, respectively. The difference spectrum associated with the Em = −255 mV component is similar to that reported previously for siroheme reduction in spinach nitrite reductase (Hirasawa et al. 1994) and the difference spectrum associated with the Em = −390 mV components is similar to that reported previously for reduction of the [4Fe-4S]2+,1+ cluster of spinach nitrite reductase (Hirasawa et al. 1994) and on that basis have been assigned to the siroheme and [4Fe-4S] cluster, respectively, of the C. reinhardtii enzyme. The Em value measured for the siroheme of C. reinhardtii nitrite reductase is slightly more positive than the −290 mV value reported for the siroheme in the ferredoxin-dependent nitrite reductase isolated from spinach leaf, while the −390 mV Em value obtained for the [4Fe-4S]2+,1+ cluster of C. reinhardtii nitrite reductase is slightly more negative than the −365 mV value determined for the spinach leaf enzyme (Hirasawa et al. 1994). It should be pointed out that the Em value of the siroheme in a recombinant, His-tagged form of spinach nitrite reductase appears to be shifted by approximately 50 mV to the negative compared to the value obtained for enzyme isolated directly from spinach leaf (Sétif et al. 2009).
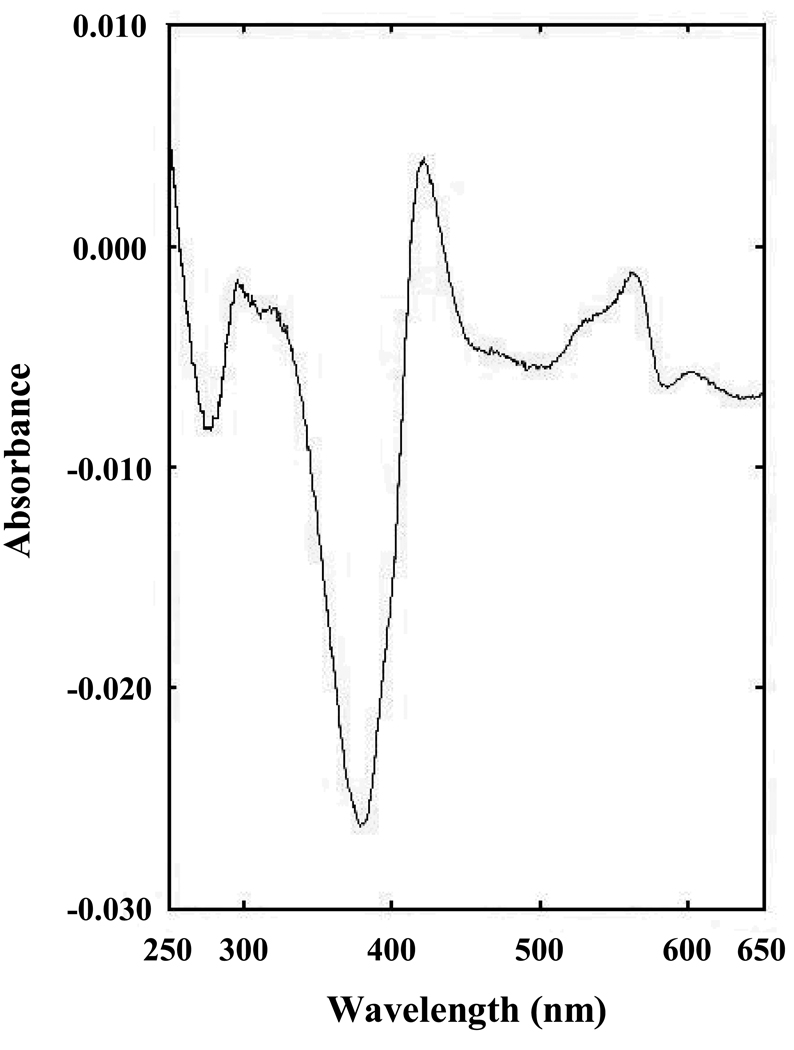
Figure 3 shows the difference spectrum that arises from complex formation between the oxidized, as isolated, C. reinhardtii nitrite reductase and nitrite. The absorbance maxima (421 nm and 564 nm) and minima (379 nm, 586 nm and 693 nm) are similar to those reported previously for the nitrite reductase isolated from C. reinhardtii cells (Romero et al. 1987) and those reported for other ferredoxin-dependent nitrite reductases (Vega and Kamin 1977; Hirasawa and Tamura 1980; Hirasawa et al. 1987; Mikami and Ids 1989; Bellissimo and Privalle 1995; Kuznetsova et al. 2004b). A plot of the absorbance difference at 421 nm minus 379 nm arising from formation of this complex vs. nitrite concentration (not shown) were hyperbolic, consistent with the presence of a single binding site with a Kd value of 50 µM. This Kd value is significantly higher than the 15 µM value measured in our laboratory under identical conditions for a His-tagged, recombinant form of spinach nitrite reductase (Tripathy et al. 2007) and the 10 µM value reported for the enzyme isolated from spinach leaf (Mikami and Ida 1989). Indirect measurements, based on the ability of sodium nitrite to protect the spinach leaf enzyme against denaturation, have been used to estimate Kd values for nitrite binding ranging from 60 µM to 150 µM, depending on the ionic strength (Privalle et al. 1985).
Figure 3.
Spectral perturbations arising from complex formation between C. reinhardtii nitrite reductase and nitrite. C. reinhardtii nitrite reductase was present at a concentration of 3.0 µM in 250 mM potassium phosphate buffer (pH 7.7). Sodium nitrite was present at a concentration of 32 µM. The difference spectrum shown is that of the complex minus that of nitrite reductase alone. All spectra were measured at ambient temperature in a 1.0 cm optical pathlength cuvette.
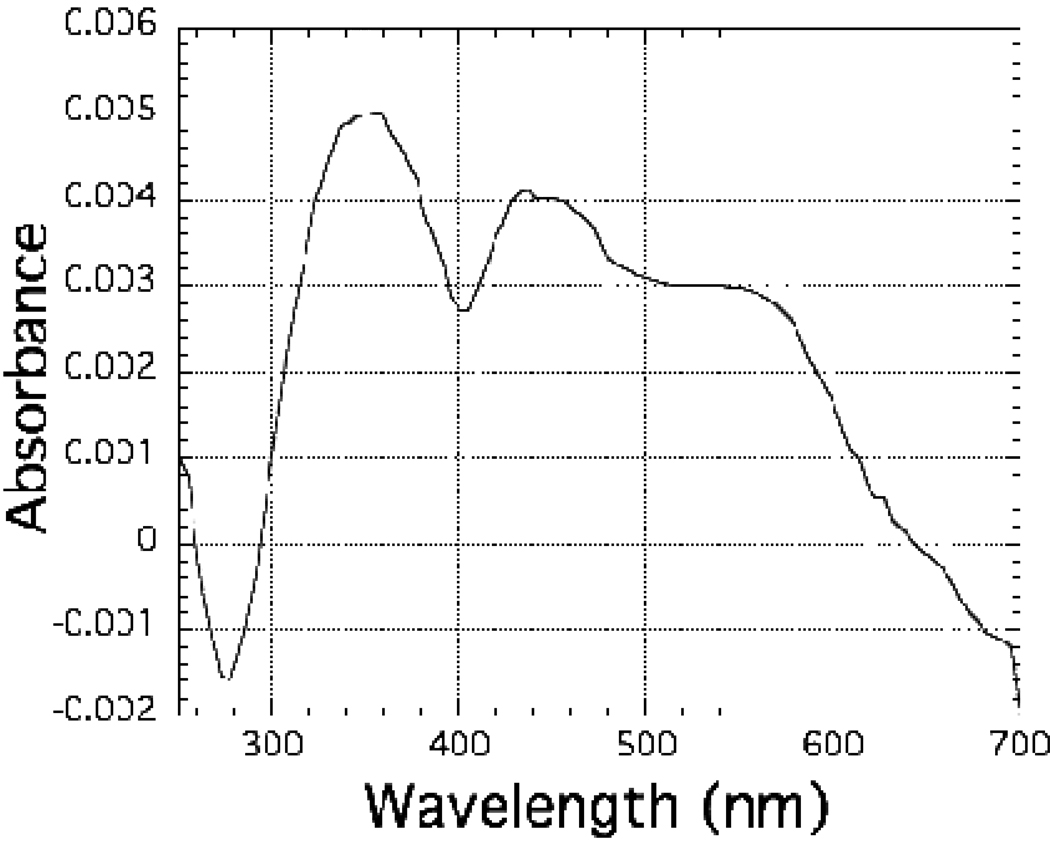
The observation that C. reinhardtii nitrite reductase binds to a ferredoxin affinity column, in which ferredoxin from the cyanobacterium Synechocystis sp. PCC 6803 has been covalently attached to a cyanogen bromide-activated Sepharose 4B matrix, provided the initial indication that the two proteins can form a complex of at least moderately high affinity. If electrostatic forces play a significant role in the interaction between C. reinhardtii nitrite reductase and ferredoxin, one might expect that the binding of the enzyme to the ferredoxin-affinity column would diminish at high ionic strength, where electrostatic forces weaken, and in fact the enzyme does elute from the ferredoxin affinity column during the application of 250 mM potassium phosphate buffer. As shown in Figure 4, complex formation between C. reinhardtii nitrite reductase and ferredoxin can also be monitored by the spectral perturbations that arise as a result of the interactions between the two proteins. In a control experiment (not shown), no spectral perturbations were observed when bovine serum albumin was added to C. reinhardtii nitrite reductase instead of ferredoxin. The difference spectrum for the complex between C. reinhardtii nitrite reductase and C. reinhardtii ferredoxin 1, although it differs somewhat from those obtained with nitrite reductase isolated directly from spinach leaf (Hirasawa and Knaff 1985; Mikami and Ida 1989), is very similar to that observed with a His-tagged recombinant form of spinach nitrite reductase (Tripathy et al. 2007). The difference spectrum shown in Figure 4, which was recorded on a mixture of proteins in 10 mM potassium phosphate buffer, is not seen in 250 mM potassium phosphate buffer and is eliminated by the addition of either NaCl, KCl or Na2SO4 to samples present in 10 mM sodium phosphate buffer, all of which are also consistent with a significant contribution of electrostatic forces to the interaction between the two proteins. Double-reciprocal plots of the magnitude of the absorbance changes at 432 nm minus 401 nm arising from the interaction of C. reinhardtii nitrite reductase with C. reinhardtii ferredoxin 1 as a function of ferredoxin concentration were linear and could be used to estimate a value of 10 µM for the Kd for the complex in 10 mM potassium phosphate buffer (pH 7.7). Kd values for the complex formed between nitrite reductase and ferredoxin isolated from spinach leaf ranging from 0.7 µM to 10 µM have been reported for measurements made using a variety of low ionic strength buffers (Hirasawa and Knaff 1985; Privalle et al. 1985; Mikami and Ida 1989) and a value of 12 µM has been reported for the complex between spinach leaf ferredoxin and a His-tagged recombinant version of spinach nitrite reductase under the conditions used to carry out the experiment that yielded the data for Figure 4 (Tripathy et al. 2007).
Figure 4.
Spectral perturbations arising from complex formation between C. reinhardtii nitrite reductase and ferredoxin. C. reinhardtii nitrite reductase was present at a concentration of 3.13 µM and C. reinhardtii ferredoxin 1 was present at a concentration of 10.0 µM in 10 mM potassium phosphate buffer (pH 7.7). The difference spectrum shown is that of the mixture of the two proteins, from which has been subtracted the sum of the spectra of the two separate proteins. All spectra were measured at ambient temperature in a 1.0 cm optical pathlength cuvette.
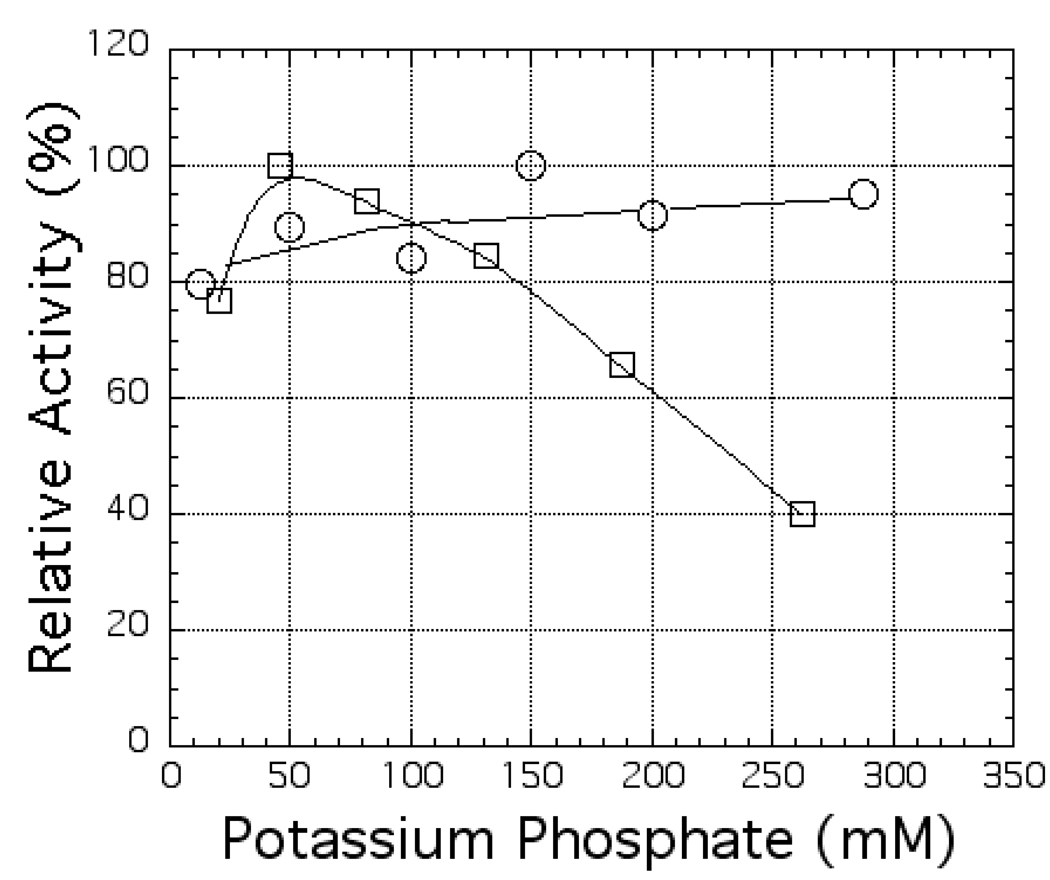
Figure 5 shows that an ionic strength effect was also observed for the activity of C. reinhardtii nitrite reductase when reduced ferredoxin serves as the electron donor while, in a control experiment, increasing the ionic strength over the same range had virtually no effect on the activity with the non-physiological electron donor, reduced methyl viologen. In 250 mM potassium phosphate buffer, the activity observed with C. reinhardtii ferredoxin 1 was only ca. 40% of that observed at low ionic strength, while the activity observed with methyl viologen at this ionic strength was identical, within the experimental uncertainties of the measurements, to that seen at low ionic strength. If the decrease in activity at high ionic strength were due to an ionic strength effect on catalytic activity per se, one might expect a significant inhibition of the activity observed with methyl viologen as the electron donor. However, the results shown in Figure 5 suggest that it is some specific aspect of the ability of the C. reinhardtii nitrite reductase to interact with C. reinhardtii ferredoxin 1 that is inhibited as the ionic strength is increased.
Figure 5.
The effect of ionic strength on the activity of C. reinhardtii nitrite reductase. The ferredoxin-dependent (open squares) and methyl viologen-dependent activities (open circles) were assayed as described in the Methods section except that the concentration of potassium phosphate buffer present in the assay mixture was varied as indicated on the x-axis.
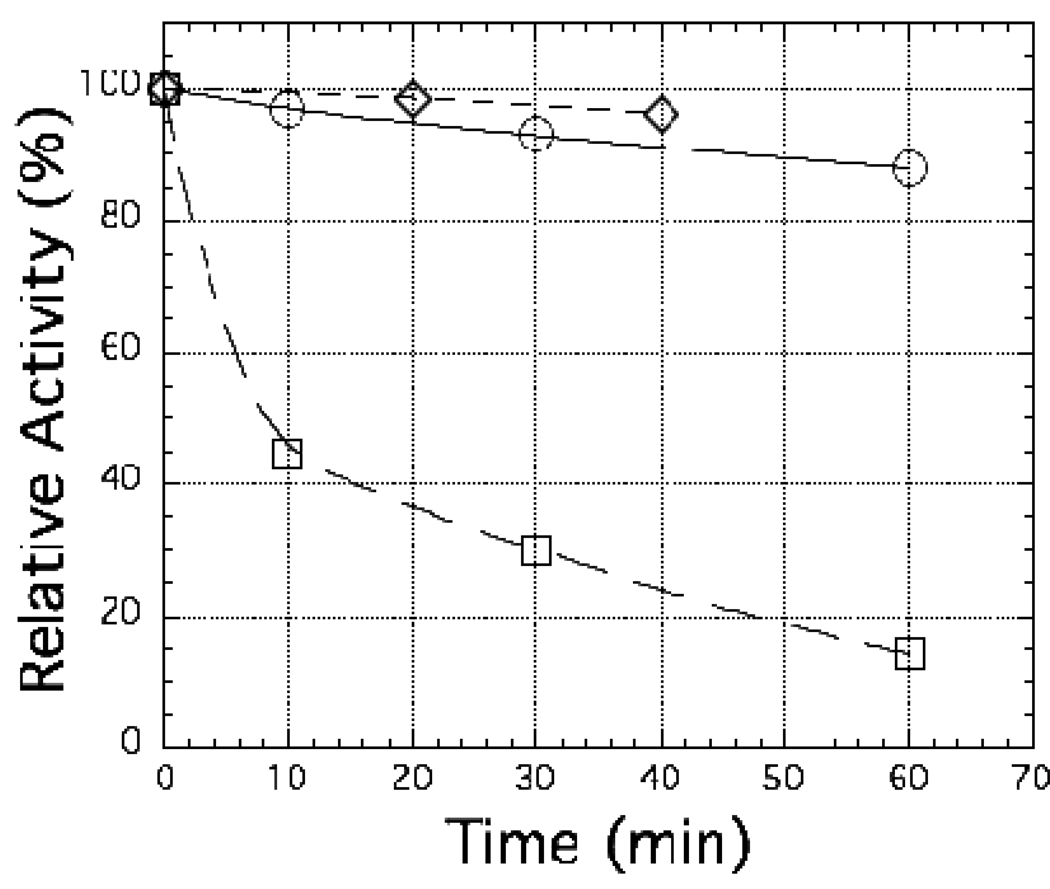
Previous experiments with spinach nitrite reductase have shown that the ability of the enzyme to interact productively with ferredoxin was significantly decreased if the enzyme were treated with either the arginine side-chain modifier phenylglyoxal or the lysine side-chain modifier N-acetylsuccinimide (Hirasawa et al. 1993). Figure 6 shows that treatment of C. reinhardtii nitrite reductase with N-acetylsuccinimide results in a loss of 86% of its activity when C. reinhardtii ferredoxin 1 serves as the electron donor (see the open squares in Figure 6) under conditions where the activity with methyl viologen as the electron donor is only inhibited by 13% (see the open circles in Figure 6). This observation, plus the fact that pre-formation of the complex between C. reinhardtii nitrite reductase and ferredoxin 1 gives almost complete protection against subsequent inhibition by this lysine modifier (see the open diamonds in Figure 6), is consistent with a role for lysine residues on nitrite reductase in the productive interaction of the C. reinhardtii enzyme with ferredoxin. Results very similar to those of Figure 6 were obtained when the arginine modifier, phenylglyoxal, replaced N-acetylsuccinimide (data not shown), suggesting that arginine residues on C. reinhardtii nitrite reductase may also be involved in the productive interaction of the C. reinhardtii enzyme with ferredoxin.
Figure 6.
The effect of N-acetylsuccinimide on the activity of C. reinhardtii nitrite reductase. Incubation of the enzyme with N-acetylsuccinimide was carried out as described in Ref. 24. The enzyme concentration during the incubation was approximately 300 µM and the concentration of N-acetylsuccinimide was 3.0 mM. At the indicated times, the incubation mixture was diluted with 250 mM potassium phosphate buffer and activity with either reduced ferredoxin as the electron donor (open squares) or reduced methyl viologen as the electron donor (open circles) was assayed as described in the Methods section. The open diamonds represent an experiment in which the incubation with N-acetylsuccinimide is carried out with the 1:1 complex of the C. reinhardtii nitrite reductase with ferredoxin instead of with the enzyme alone.
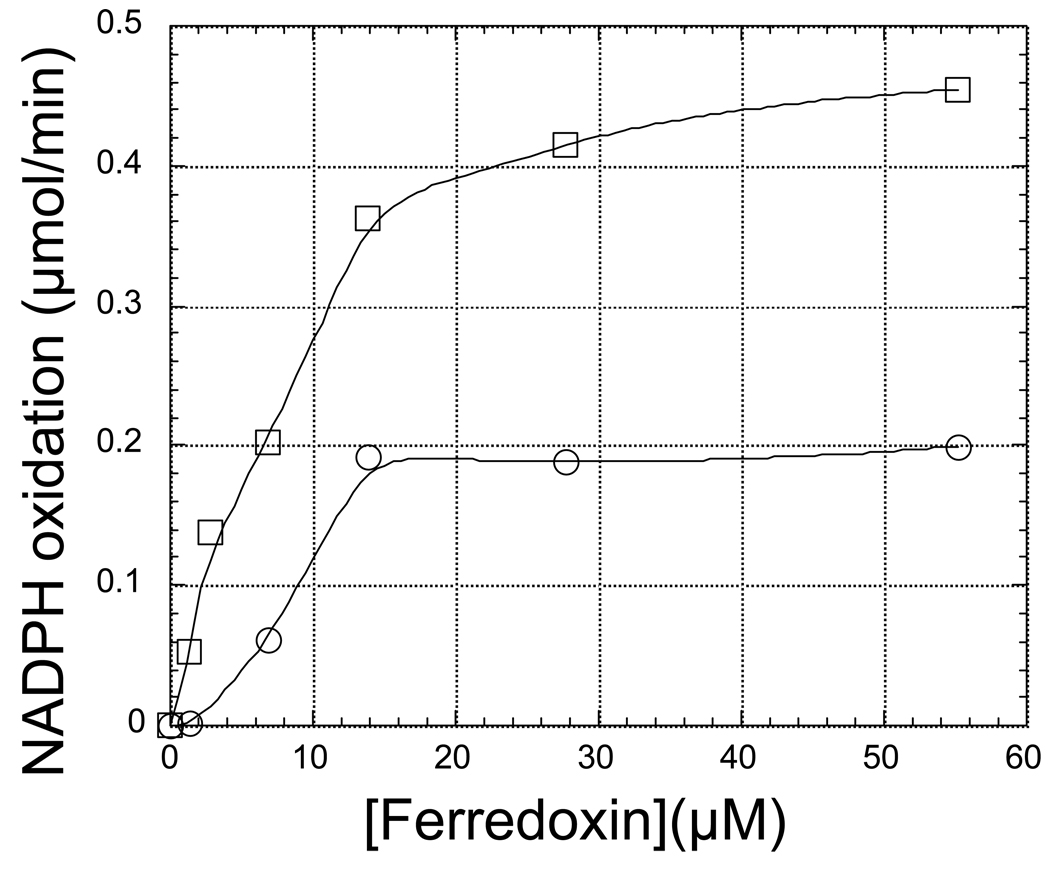
It appears likely that binding of nitrite to oxidized nitrite reductase is too slow to be a mechanistically significant step (Kuznetsova et al. 2004a). In all likelihood, the enzyme is first reduced by ferredoxin, in a one-electron transfer reaction, to the ferrous siroheme state, which then binds nitrite rapidly. A second one-electron reduction by ferredoxin produces the NO adduct of ferrous siroheme (Kuznetsova et al. 2004a), a species that has been shown to behave like a true reaction intermediate (Kuznetsova et al. 2004a). mMuch less is known about subsequent intermediates, but recent flash photolysis experiments (Sétif et al. 2009) indicate that the third electron accepted by the enzyme reduces the iron-sulfur cluster without affecting the redox state of the ferrous siroheme/NO adduct An additional electron must then be transferred to the enzyme from ferredoxin, allowing a two-electron reduction of the NO adduct intermediate to the redox level of hydroxylamine. Although hydroxylamine, which is two electrons less reduced than the final product ammonia, has been proposed as a late intermediate (Vega and Kamin 1977; Lancaster et al. 1979), there is no direct evidence for this proposal (Kuznetsova et al. 2004a; Kuznetsova et al. 2004b). In fact, addition of hydroxylamine to oxidized spinach nitrite reductase produces the ferrous siroheme/NO adduct form of the enzyme (Kuznetsova et al. 2004a; Kuznetsova et al. 2004b) and not, as previously proposed (Vega and Kamin 1977; Lancaster et al. 1979), a hydroxylamine complex of the enzyme. However, as shown in Figure 7 (open circles), hydroxylamine can serve as an electron-accepting substrate for C. reinhardtii nitrite reductase in a coupled assay system where reduced ferredoxin is supplied to the enzyme via the oxidation of NADPH catalyzed by spinach FNR (for comparison, the oxidation of NADPH coupled to the reduction of nitrite under identical conditions is shown with open squares). In parallel experiments (not shown) the product of the hydroxylamine reduction reaction was shown to be ammonia. In control experiments (not shown), no NADPH oxidation (monitored by the decrease in absorbance at 340 nm) was observed in the absence of an electron-accepting substrate for nitrite reductase. The ability of ferredoxin-dependent nitrite reductases to catalyze the reduction of hydroxylamine to ammonia is not confined to the C. reinhardtii enzyme, as results similar to those shown in Figure 7 were obtained with the recombinant spinach enzyme.
Figure 7.
Hydroxylamine reduction catalyzed by C. reinhardtii and spinach nitrite reductase. The 1.0 ml reaction mixture contained, in 62.5 mM potassium phosphate buffer (pH 7.5), 50 µM C. reinhardtii ferredoxin 1, 0.5 mM NADPH, 1.4 µM FNR, 0.3 µM C. reinhardtii nitrite reductase and either 2.0 mM sodium nitrite (open squares) or 2.0 mM hydroxylamine (open circles). The rates were calculated from the absorbance changes at 340 nm, 10 seconds after the reaction was initiated by the addition of either sodium nitrite or hydroxylamine.
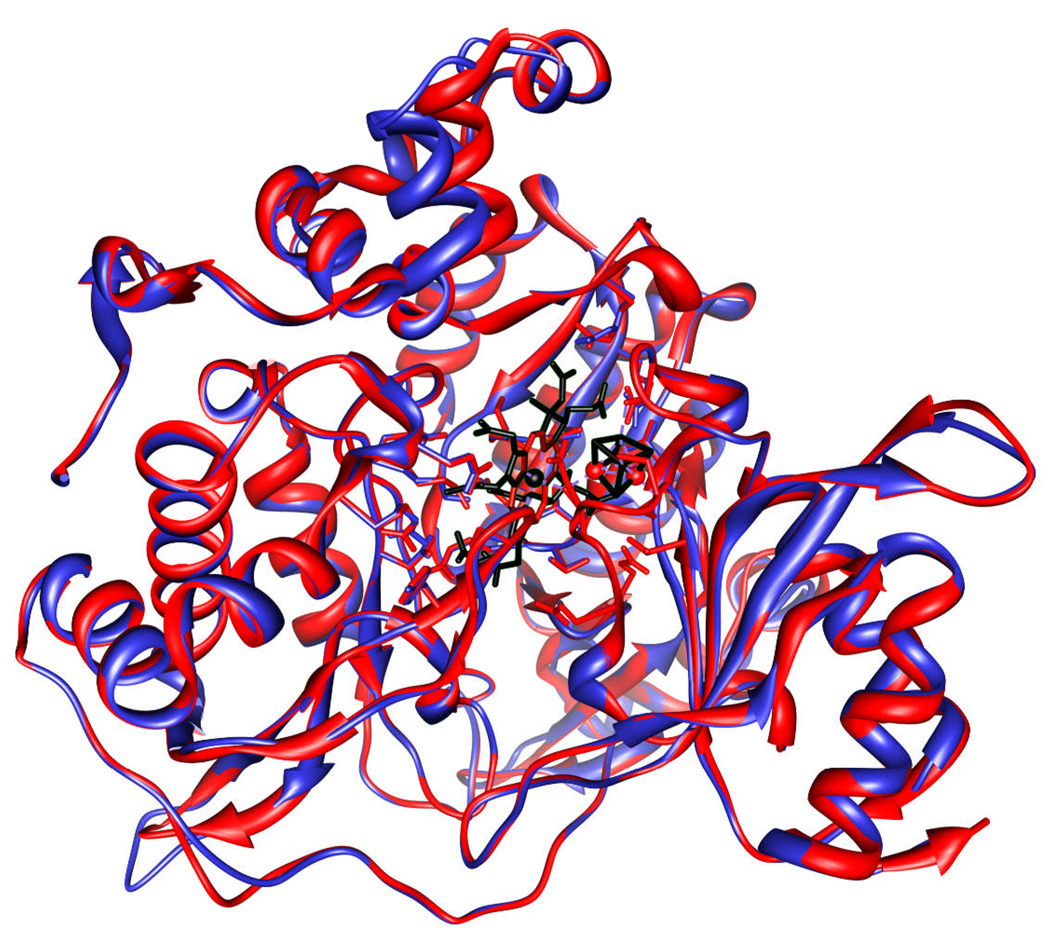
It appeared likely, given the many similarities between the two enzymes, that C. reinhardtii and spinach nitrite reductases might well have similar three-dimensional structures. Figure 8 shows a comparison of the published 2.8 Å resolution structure of the recombinant spinach enzyme with a model for the structure for the C. reinhardtii enzyme that is based on the spinach enzyme structure and the similar amino acid sequences of the two enzymes. The algorithm used (Pettersen 2004) predicts that, with the possible exception of the location of some of the peripheral loops, the two structures will be very similar.
Figure 8.
Superposition of the structural models of the backbones of nitrite reductases from spinach (red) and C. reinhardtii (blue). The siroheme and [4Fe-4S] cofactors are shown in black.
Discussion
A ferredoxin-dependent nitrite reductase from the green alga C. reinhardtii has been expressed in E. coli as a His-tagged recombinant enzyme, purified and extensively characterized. The His-tagged enzyme is relatively easy to purify to homogeneity on a Ni2+ affinity column and, as expression of the C. reinhardtii enzyme in E. coli produces substantially higher yields of pure enzyme than is the case for a His-tagged, recombinant form of the spinach enzyme, the C. reinhardtii nitrite reductase may be particularly useful for biophysical and structural studies. Crystallization trials of the enzyme itself and of the C. reinhardtii nitrite reductase complex with C. reinhardtii ferredoxin are presently underway. Given the predicted similarity between the structures of the spinach and C. reinhardtii nitrite reductases, any information learned about the C. reinhardtii enzyme from a structure at a resolution better than 2.8 Å (e.g., information on the planarity of the siroheme ring, information about the possible location of oriented water molecules near the active site that might function in proton movements that accompany nitrite reduction and information about substrate binding) should be applicable to a better understanding of the mechanisms of the spinach enzyme and other ferredoxin-dependent nitrite reductases as well. As C. reinhardtii chloroplasts contain multiple ferredoxins (Terauchi et al. 2009), the C. reinhardtii nitrite reductase should also prove useful in exploring the question of electron-donor specificity of the different C. reinhardtii chloroplastic ferredoxins for this ferredoxin-dependent enzyme.
Despite recent advances in our understanding of the mechanism of ferredoxin-dependent nitrite reductases (Kuznetsova et al. 2004a; Kuznetsova et al. 2004; Hirasawa et al. 2009; Sétif et al. 2009), considerable uncertainty remains about the complex mechanism involved in a reaction in which electrons enter the enzyme one-at-a-time from ferredoxin and, in an enzyme that only has the capacity to store two electrons; a six-electron reduction of nitrite to ammonia is accomplished. Given the likelihood that this six-electron reduction might be accomplished by a series of three 2-electron steps, it is not surprising that hydroxylamine was proposed as a late intermediate when studies of the enzyme were first initiated, even though one of the earliest studies of a ferredoxin-dependent nitrite reductase (isolated and purified from Cuccurbita pepo) failed to detect any hydroxylamine reductase activity (Hucklesby and Hewitt 1970). While it is not clear why our results with both the C. reinhardtii and spinach enzymes differ from those obtained in 1970 with the C. pepo enzyme, our demonstration that the C. reinhardtii and spinach enzymes can indeed catalyze the conversion of hydroxylamine to ammonia has provided the first concrete evidence for the hypothesis that hydroxylamine might function as a late intermediate in the catalytic cycle and thus represents a significant advance in our understanding of this key enzyme in the photosynthetic nitrate assimilation pathway.
Acknowledgment
This research was supported by contracts from the Chemical Sciences, Geosciences and Biosciences Division, Office of the Basic Energy Sciences, Office of Sciences, U.S. Department of Energy (Contract Numbers DE-FG03-99ER20346 to DBK and DE-FG02-04ER15529 to S.S.M.). F.S. was supported by a fellowship from the DFG (SO706/1-1). The EPR spectroscopy characterizations were funded by a grant from the U.S. National Institutes of Health (GM62524 to MKJ). The authors would like to thank Ms. Shu Fen Lu for her assistance in purifying the C. reinhardtii ferredoxin used in this study, Prof. Toshiharu Hase (Osaka University, Japan) for his suggestion that hydroxylamine be tested as a potential substrate and his advice on the assay system for hydroxylamine reduction, Prof. Emilio Fernández (University of Córdoba, Spain) for his assistance and advice on the cloning of the gene for C. reinhardtii nitrite reductase and Dr. Arthur Grossman (Carnegie Institution) for his gift of the cDNA clone for NII1.
Abbreviations
- EPR
electron paramagnetic resonance
- EST
expressed sequence tag
- FNR
ferredoxin: NADP+ oxidoreductase
- MALDI-ToF
matrix-assisted laser desorption ionization-Time of flight mass spectrometry
- PVDF
polyvinylidene fluoride
- SDS-PAGE
polyacrylamide gel electrophoresis in the presence of sodium dodecylsulfate
References
- Aparicio PJ, Knaff DB, Malkin R. The role of an iron-sulfur center and siroheme in spinach nitrite reductase. Arch Biochem Biophys. 1975;169:102–107. doi: 10.1016/0003-9861(75)90321-5. [DOI] [PubMed] [Google Scholar]
- Bellissimo DB, Privalle LS. Expression of spinach nitrite reductase in Escherichia coli: Site-directed mutagenesis of predicted active site amino acids. Arch Biochem Biophys. 1995;323:155–163. doi: 10.1006/abbi.1995.0021. [DOI] [PubMed] [Google Scholar]
- Cammack R, Hucklesby DP, Hewitt EJ. Electron-paramagnetic-resonance studies of the mechanism of leaf nitrite reductase. Biochem J. 1978;171:519–526. doi: 10.1042/bj1710519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conway EJ. Microdiffusion Analysis and Volumetric Error. Fourth Edition. London: Crosby, Lookwood and Son, Ltd.; 1957. [Google Scholar]
- Curdt I, Singh BB, Jakoby M, Hachtel W, Bohme H. Identification of amino acid residues of nitrite reductase from Anabaena sp. PCC 7120 involved in ferredoxin binding. Biochem Biophys Acta. 2000;1543:60–68. doi: 10.1016/s0167-4838(00)00198-9. [DOI] [PubMed] [Google Scholar]
- Fernández E, Galvan A. Nitrate assimilation in Chlamydomonas. Eukaryot Cell. 2008;7:555–559. doi: 10.1128/EC.00431-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernández E, Llamas A, Galván A. Nitrogen assimilation and its regulation. In: Stern DB, Harris EH, editors. The Chlamydomonas sourcebook. 2nd Edition. Vol 2. New York: Academic Press; 2009. pp. 69–114. Organellar and metabolic processes. [Google Scholar]
- Flores E, Frias JE, Rubio LM, Herrero A. Photosynthetic nitrate assimilation in cyanobacteria. Photosyn Res. 2005;83:117–133. doi: 10.1007/s11120-004-5830-9. [DOI] [PubMed] [Google Scholar]
- Gonzalez-Ballester D, Grossman AR. Sulfur: From acquisition to assimilation. In: Stern DB, Harris EH, editors. The Chlamydomonas sourcebook. 2nd Edition. Vol 2. New York: Academic Press; 2009. pp. 159–188. Organellar and metabolic processes. [Google Scholar]
- Grossman AR, Croft M, Gladyshev VR, Merchant SS, Posewitz MC, Prochnik S, Spalding MH. Novel metabolism of Chlamydomonas through the lens of genomics. Curr Opin Plant Biol. 2007;10:190–198. doi: 10.1016/j.pbi.2007.01.012. [DOI] [PubMed] [Google Scholar]
- Hanikenne M, Merchant SS, Hamel P. Transition metal nutrition: A balance between deficiency and toxicity. In: Stern DB, Harris EH, editors. The Chlamydomonas sourcebook. 2nd Edition. Vol 2. New York: Academic Press; 2009. pp. 333–400. Organellar and metabolic processes. [Google Scholar]
- Harris EH. Chlamydomonas in the laboratory. In: Harris EH, editor. The Chlamydomonas sourcebook. 2nd Edition. Vol 1. New York: Academic Press; 2009. pp. 241–302. [Google Scholar]
- Hase T, Schürmann P, Knaff DB. The Interaction of ferredoxin with ferredoxin-dependent enzymes. In: Golbeck J, editor. Photosystem 1. Dordrecht (The Netherlands): Springer; 2006. pp. 477–498. [Google Scholar]
- Hirasawa M, Tamura G. Ferredoxin-dependent nitrite reductase from spinach leaves. Agric Biol Chem. 1980;44:749–758. [Google Scholar]
- Hirasawa M, Knaff DB. Interaction of ferredoxin-linked nitrite reductase with ferredoxin. Biochim Biophys Acta. 1985;830:173–170. [Google Scholar]
- Hirasawa M, de Best JH, Knaff DB. The effect of lysine- and arginine-modifying regents on spinach ferredoxin:nitrite oxidoreductase. Biochim Biophys Acta. 1993;1140:304–312. [Google Scholar]
- Hirasawa M, Shaw RW, Palmer G, Knaff DB. Prosthetic group content and ligand-binding properties of spinach nitrite reductase. J Biol Chem. 1987;262:12428–12433. [PubMed] [Google Scholar]
- Hirasawa M, Tollin G, Salamon Z, Knaff DB. Transient kinetic and oxidation-reduction studies of spinach ferredoxin:nitrite oxidoreductase. Biochim Biophys Acta. 1994;1185:336–345. doi: 10.1016/0005-2728(94)90249-6. [DOI] [PubMed] [Google Scholar]
- Hirasawa M, Tripathy JN, Somasundaram R, Johnson MK, Bhalla M, Allen JP, Knaff DB. The interaction of spinach nitrite reductase with ferredoxin: A site-directed mutation study. Molec Plant. 2009;2:407–415. doi: 10.1093/mp/ssn098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hucklesby DP, Hewitt EJ. Nitrite and hydroxylamine reduction in higher plants. Biochem J. 1970;119:615–627. doi: 10.1042/bj1190615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hucklesby DP, James DM, Banwell MJ, Hewitt EJ. Properties of nitrite reductase from Cucurbita pepo. Phytochem. 1976;15:599–603. [Google Scholar]
- Ida S, Mikami B. Spinach ferredoxin-nitrite reductase: A purification procedure and characterization of chemical properties. Biochim Biophys Acta. 1986;871:167–176. [Google Scholar]
- Ida S, Iwagami K, Minobe S. Purification and characterization of molecular and immunological properties of rice ferredoxin-nitrite reductase. Agr Biol Chem. 1989;53:2777–2784. [Google Scholar]
- Ip I-M, Kerr J, Ingledew WJ, Wray JL. Purification and characterization of barley leaf nitrite reductase. Plant Sci. 1990;66:155–165. [Google Scholar]
- Jain M, Shrager J, Harris EH, Halbrook R, Grossman AR, Vallon O. EST assembly supported by a draft genome sequence: An analysis of the Chlamydomonas einhardtii transcriptome. Nucleic Acids Res. 2007;35:2074–2083. doi: 10.1093/nar/gkm081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuznetsova S, Knaff DB, Hirasawa M, Lagoutte B, Sétif P. The mechanism of spinach chloroplast ferredoxin-dependent nitrite reductase: spectroscopic evidence for intermediate states. Biochemistry. 2004;43:510–517. doi: 10.1021/bi035662q. [DOI] [PubMed] [Google Scholar]
- Kuznetsova S, Knaff DB, Hirasawa M, Sétif P, Mattioli TA. Reactions of spinach nitrite reductase with its substrate nitrite and a putative intermediate, hydroxylamine. Biochemistry. 2004;43:10765–10774. doi: 10.1021/bi048826r. [DOI] [PubMed] [Google Scholar]
- Lancaster JR, Vega JM, Kamin H, Orme-Johnson NR, Orme-Johnson WH, Krueger RJ, Siegel LM. Identification of the iron-sulfur center of spinach ferredoxin-nitrite reductase as a tetranuclear center and preliminary EPR studies of mechanism. J Biol Chem. 1979;254:1268–1272. [PubMed] [Google Scholar]
- Leustek T, Smith M, Murillo M, Singh DP, Smith AG, Woodcock SC, Awan SG, Warren MJ. Siroheme biosynthesis in higher plants. J Biol Chem. 1997;272:2744–2752. doi: 10.1074/jbc.272.5.2744. [DOI] [PubMed] [Google Scholar]
- Merchant SS, Prochnik SE, Vallon O, Harris EH, Karpowicz SJ, Witman GB, Terry A, Salamov A, Fritz-Laylin LK, Marechal-Drouard L, Marshall WF, Qu LH, Nelson DR, Sanderfoot AA, Spalding MH, Kapitonov VV, Ren Q, Ferris P, Lindquist E, Shapiro H, Grimwood J, Schmutz J, Lucas S, Grigoriev IV, Rokhsar DS, Grossman AR Chlamydomonas community annotation team; JGI annotation team. The Chlamydomonas genome reveals the evolution of key animal and plant functions. Science. 2007;318:245–250. doi: 10.1126/science.1143609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mikami B, Ida S. Spinach ferredoxin-nitrite reductase: Characterization of catalytic activity and interaction of the enzyme with its substrates. J Biochem. 1989;105:47–50. doi: 10.1093/oxfordjournals.jbchem.a122617. [DOI] [PubMed] [Google Scholar]
- Miller RW, Massey V. Dihydroorotic dehydrogenase. I. Some properties of the enzyme. J Biol Chem. 1965;240:1453–1465. [PubMed] [Google Scholar]
- Moseley J, Grossman AR. Phosphate metabolism and responses to phosphorus deficiency. In: Stern DB, Harris EH, editors. The Chlamydomonas sourcebook. 2nd Edition. Vol 2. New York: Academic Press; 2009. pp. 189–216. Organellar and metabolic processes. [Google Scholar]
- Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC, Ferrin TE. UCSF Chimera - A visualization system for exploratory research and analysis. J Comput Chem. 2004;25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- Privalle LS, Privalle CT, Leonardy NJ, Kamin HJ. Interactions between spinach ferredoxin-nitrite reductase and its substrates. Evidence for the specificity of ferredoxin. J Biol Chem. 1985;260:14344–14350. [PubMed] [Google Scholar]
- Quesada A, Gómez I, Fernández E. Clustering of the nitrite reductase gene and a light-regulated gene with nitrate assimilation loci in Chlamydomonas reinhardtii. Planta. 1998;206:259–265. doi: 10.1007/s004250050398. [DOI] [PubMed] [Google Scholar]
- Romero LC, Borrero JA, Galvan F, Vega JM. Prosthetic components and essential groups for activity in ferredoxin-nitrite reductase from Chlamydomonas reinhardtii. J Mol Catal. 1989;57:259–270. [Google Scholar]
- Romero LC, Galván F, Vega JM. Purification and properties of the siroheme-containing ferredoxin-nitrite reductase from Chlmaydomonas reinhardtii. Biochim Biophys Acta. 1987;914:55–63. [Google Scholar]
- Sétif P, Hirasawa M, Cassan N, Lagoutte B, Tripathy JN, Knaff DB. New insights into the catalytic cycle of plant nitrite reductase. Electron transfer kinetics and charge storage. Biochemistry. 2009;48:2828–2838. doi: 10.1021/bi802096f. [DOI] [PubMed] [Google Scholar]
- Shin M, Oshino R. Ferredoxin-Sepharose 4B as a tool for the purification of ferredoxin-NADP+ reductase. J Biochem. 1978;83:357–361. doi: 10.1093/oxfordjournals.jbchem.a131921. [DOI] [PubMed] [Google Scholar]
- Siegel LM, Murphy MJ, Kamin H. Reduced nicotinamide adenine dinucleotide phosphate-sulfite reductase of enterobacteria. I. The Escherichia coli hemoflavoprotein: molecular parameters and prosthetic groups. J Biol Chem. 1973;248:251–264. [PubMed] [Google Scholar]
- Smith JM, Smith WH, Knaff DB. Electrochemical titrations of a ferredoxin-ferreodixn: NADP+ oxidoreductase complex. Biochim Biophys Acta. 1981;635:405–411. doi: 10.1016/0005-2728(81)90038-4. [DOI] [PubMed] [Google Scholar]
- Stoller ML, Malkin R, Knaff DB. Oxidation-reduction properties of photosynthetic nitrite reductase. FEBS Lett. 1977;81:271–274. [Google Scholar]
- Swamy U, Wang M, Tripathy JN, Kim S-K, Hirasawa M, Knaff DB, Allen JP. Structure of spinach nitrite reductase: Implications for multi-electron reactions by the iron-sulfur:siroheme cofactor. Biochemistry. 2005;44:16054–16063. doi: 10.1021/bi050981y. [DOI] [PubMed] [Google Scholar]
- Terauchi AM, Lu S-F, Zaffagnini M, Tappa S, Hirasawa M, Tripathy JN, Knaff DB, Farmer PJ, Lemaire SD, Hase T, Merchant SS. Pattern of expression and substrate specificity of chloroplast ferredoxins from Chlamydomonas reinhardtii. J Biol Chem. 2009;284:25867–25878. doi: 10.1074/jbc.M109.023622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tripathy JN, Hirasawa M, Kim S-K, Setterdahl AT, Allen JP, Knaff DB. The role of tryptophan in the ferredoxin-dependent nitrite reductase of spinach. Photosyn Res. 2007;94:1–12. doi: 10.1007/s11120-007-9198-5. [DOI] [PubMed] [Google Scholar]
- Vega JM, Kamin H. Spinach nitrite reductase. Purification and properties of a siroheme-containing iron-sulfur enzyme. J Biol Chem. 1977;252:896–909. [PubMed] [Google Scholar]
- Vigara J, Garcia-Sánchez MI, Garbayo I, Vilchez C, Vega JM. Purification and characterization of ferredoxin-nitrite reductase from the eukaryotic microalga Monoraphidium braunii. Plant Physiol and Biochem. 2002;40:401–405. [Google Scholar]
- Wilkerson JO, Janick PA, Siegel LM. Electron paramagnetic resonance and optical spectroscopic evidence for interaction between siroheme and tetranuclar iron-sulfur center prosthetic groups in spinach ferredoxin-nitrite reductase. Biochemistry. 1983;22:5048–5954. [Google Scholar]
- Yonekura-Sakakibara K, Onda Y, Ashikari T, Tanaka Y, Kusumi T, Hase T. Analysis of reductant supply systems for ferredoxin-dependent sulfite reductase in photosynthetic and non-photosynthetic organs of maize. Plant Physiol. 2000;122:887–894. doi: 10.1104/pp.122.3.887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zumft WG. Ferredoxin:nitrite oxidoreductase from Chlorella. Purification and properties. Biochim Biophys Acta. 1972;276:363–375. doi: 10.1016/0005-2744(72)90996-5. [DOI] [PubMed] [Google Scholar]