Abstract
Biological membrane fusion is crucial to numerous cellular events, including sexual reproduction and exocytosis. Here, mass spectrometry images demonstrate that the low-curvature lipid phosphatidylcholine is diminished in the membrane regions between fusing Tetrahymena, where a multitude of highly curved fusion pores exist. Additionally, mass spectra and principal component analysis indicate that the fusion region contains elevated amounts of 2-aminoethylphosphonolipid, a high-curvature lipid. This evidence suggests that biological fusion involves and might in fact be driven by a heterogeneous redistribution of lipids at the fusion site.
During membrane fusion, two adjacent lipid bilayers merge and a channel (a fusion pore) forms, which joins the aqueous volumes initially enclosed within the membranes. The protozoan Tetrahymena thermophila (Fig. 1A) is an attractive cell system for membrane fusion studies because it is possible to induce the simultaneous formation of hundreds of fusion pores within a well-defined membrane region of about 8 μm (1, 2). During mating, or conjugation (Fig. 1B), the membranes of two complementary Tetrahymena join at the anterior end and 100- to 200-nm-sized fusion pores form (Fig. 1C) to allow the migration of micronuclei between the cells. Conjugation depends on de novo lipid synthesis (3), and Tetrahymena can readily modify the lipid composition of the cell membrane via intracellular lipid exchange (4). Thus, it seems likely that certain types of lipid are required to allow the mass formation of fusion pores, and that these biophysically relevant lipids may be synthesized or redirected to the fusion site, called the conjugation junction. Additionally, in preparation for conjugation, these cells actively modify their pattern of protein synthesis, and the anterior ends of the cells transform from pointed to blunt in shape and from ciliated and ridged to smooth in texture (2). The dependence of conjugation on lipid synthesis, the membrane morphological changes, and the excess of fusion pores might suggest that there are substantial spatial alterations in the chemistry of the membrane bilayer in the conjugation junction.
Fig. 1.

Hundreds of fusion pores observed during Tetrahymena conjugation result in lipid heterogeneity at membrane fusion sites. Scale bars are 50 μm unless otherwise noted. (A) Brightfield images of Tetrahymena before mating and (B) during full conjugation (arrows). (C) A TEM of a 70-nm cross section through mating Tetrahymena showing eight fusion pores along the conjugation junction in this plane of view. Therefore, an estimated pore density for the entire junction is about 200 fusion pores, in agreement with the literature (2). Scale bar, 2 μm. (D) A schematic of the membrane fusion intermediate structure, the stalk. The wavy lines depict the acyl tail-groups of the membrane phospholipids. The white circles represent the headgroup of PC, a cylinder-shaped lamellar lipid. The black circles represent the headgroup of PE, a cone-shaped nonlamellar lipid. Membrane fusion sites probably contain a large quantity of cone-shaped lipids, because those lipids fit well into contoured intermediate structures. (E) Scanning ion micrograph of mating Tetrahymena thermophila. (F) Brightfield image, ex situ. (G) SIMS image (128 × 128 pixels) for m/z 69 (C5H9+). (H) SIMS image for m/z 184 (PC), demonstrating lipid heterogeneity at the conjugation junction. SIMS images were acquired using a 200-nm beam spot size, resulting in an approximately 250-nm lateral resolution, as in previous work (16). Based on PC packing characteristics in a bilayer, each 1-μm2 pixel contained about 1.5 × 106 PC molecules.
The cellular machinery and thermodynamic driving forces behind biological fusion are a bit of an enigma, although it is widely believed that the machinery involves an intricate cooperation between membrane proteins, the cytoskeletal framework, and lipids (5, 6). The interaction of complementary membrane proteins might dictate the location of fusion and regulate the fusion events (5), and the cell cytoskeleton might play a role by confining fusogenic proteins to domains where fusion occurs in the membrane (6). The existence of biophysically functional lipid domains, or rafts, which are membrane regions concentrated in a particular type of lipid, is well documented (7–10). Lipid movement through the membrane can be restricted and create a heterogeneous distribution of lipids. These structures appear to involve longer-term or semipermanent formations and may drive biological fusion.
During fusion, the lipid bilayer undergoes considerable changes in shape and most likely adopts several intermediate structures with a high radius of curvature (5). One such proposed intermediate structure, the stalk intermediate, has been experimentally observed by x-ray diffraction (11) (Fig. 1D). Thus, if present, lipid domains involved in membrane fusion would form temporarily during the fusion process and would require a driving force and cell-to-cell signaling of some kind. According to the intrinsic curvature hypothesis, cone-shaped nonlamellar lipids, such as phosphatidylethano-lamine (PE), are particularly adept at forming highly contoured structures, such as the stalk intermediate, because they contain hydrophilic headgroups and acyl chain tailgroups of greatly different cross-sectional areas (Fig. 1D) (11–13). This hypothesis suggests that highly fusogenic membrane regions may adjust to contain small amounts of lamellar lipids, including phosphatidylcholine (PC), in comparison to the rest of the cell membrane; and fusion events may preferentially occur at these depleted sites. In Tetrahymena, the formation of the plethora of highly curved fusion pores would require that the entire length of the conjugation junction be concentrated in nonlamellar lipids.
Imaging time-of-flight secondary-ion mass spectrometry (TOF-SIMS) after freeze-fracture was used to examine the lipid distribution along the conjugation junction of mating Tetrahymena. TOF-SIMS has been applied to elemental imaging of the distribution of cancer therapy drugs in single cells (14) as well as molecular imaging of tissue (15), liposomes (16, 17), paramecia (18), and rat pheochromocytoma (PC12) cells (19, 20), but has not yet been demonstrated for subcellular membranes. We incubated conjugating Tetrahymena for 4 hours and then placed a small aliquot of the cell-containing solution between two pieces of silicon for cryogenic freezing. The frozen sample was then freeze-fractured under ultrahigh vacuum (16, 21, 22) and directly transferred into the analysis chamber. After TOF-SIMS imaging, the samples were further analyzed by scanning ion microscopy and brightfield microscopy to ensure that no substantial sample damage occurred during sample preparation and SIMS analysis. In situ scanning ion micrographs provided morphological images of the cell samples and showed no evidence of physical damage to the mating Tetrahymena (Fig. 1E). A brightfield image verified this conclusion (Fig. 1F).
In SIMS studies of cells, a mass-to-charge ratio (m/z) of 69 corresponds to C5H9+, which is most likely an acyl chain fragment from a phospholipid. Because all of the membrane phospholipids contain this species, the distribution of C5H9+ in SIMS images is representative of the total phospholipid distribution. A molecule-specific image of m/z 69 for mating Tetrahymena shows that the phospholipids are homogeneous across the joined cells (Fig. 1G). The uniformity of the entire phospholipid signal over the fusing region of the cells is expected, because the current spatial resolution for this technique is above the size of a fusion pore. Additionally, the homogeneous distribution of m/z 69 indicates that sample topology is not influencing the SIMS images. Any signal variations of other m/z values should be representative of the chemistry of the cell membrane.
The SIMS fragmentation behavior of PC has been well characterized, and through tandem mass spectrometry, m/z 184 has been attributed to the phosphocholine headgroup (15, 23). A substantial decrease in the m/z 184 signal seen at the junction region (Fig. 1H) shows that the phospholipid structure varies considerably in the mating junction because of the abundance of highly curved membrane structures in that region.
Line scans were taken through the molecule-specific images to quantitatively compare the distribution of m/z 184 and m/z 69 across the conjugation junction (Fig. 2). Although there were slight fluctuations that are expected with molecular measurements on the micrometer scale, the m/z 69 signal was constant across the junction (Fig. 2A), whereas the m/z 184 signal clearly dropped at the junction region (Fig. 2B). This heterogeneity was reproduced for the majority of mating Tetrahymena examined (Table 1), with an average decrease in signal of 67 ± 8%. The hydrocarbon signal remained relatively constant across all mating cells, with a slight average increase in signal of 7 ± 11% (24). Principal component analysis verified that PC was decreased in the junction relative to the cell body and also identified a peak at m/z 126 that increased in the junction (Fig. 3A) (25). Compared to the cell body, the membrane junction region of mating Tetrahymena is composed of significantly less PC.
Fig. 2.

Line scans of the molecule-specific images graphically support the idea that PC decreases at the conjugation junction. Data points were collected every 120 nm. (A) Line scan for m/z 69 through the conjugation junction, illustrating that the total lipid content is relatively constant across the mating cells. The inset shows the SIMS image for m/z 69, highlighting the pixels used for the line scan. (B) Line scan for m/z 184 through the junction, demonstrating a sharp decrease in signal at the conjugation junction. The inset shows the SIMS image for m/z 184, highlighting the pixels used for the line scan.
Table 1.
Summary of PC signal intensities (m/z 184) across mating Tetrahymena. The average PC signal intensity was calculated for three regions across mating Tetrahymena membranes: the first cell body (cell body I), the conjugation junction, and the second cell body (cell body II). First, m/z 184 line scans were taken for each of the mating cells, as described in Fig. 2. The line scans were further treated with 15.5% Savitzky-Golay smoothing and then fitted with a chromatography/HVL PeakFit function, resulting in R 2 ≥ 0.98. The signal intensities in the line scan were averaged for each of the three regions indicated in the corresponding columns of Table 1. The regions were selected based on a change of slope in the line scan. All errors are standard deviations. The average signal decrease between the two cell bodies and the conjugation junction was then determined. For the first cell body, the difference between the cell body I intensity and the junction intensity was divided by the cell body I intensity. This was repeated for the second cell, and the two percentages were averaged. The PC signal change for each of cell bodies I and II versus the conjugation junction was used to determine that P < 0.001 (t test). One sample yielded too low a signal to provide conclusive data because of sample charging, and a second sample had extremely high signal variability. Those samples were not used here.
| Trial | Cell body I | Conjugation junction | Cell body II | Signal decrease |
|---|---|---|---|---|
| 1 | 0.591 ± 0.025 | 0.321 ± 0.053 | 0.881 ± 0.057 | 55% ± 10 |
| 2 | 0.926 ± 0.029 | 0.361 ± 0.093 | 0.732 ± 0.041 | 56% ± 12 |
| 3 | 0.619 ± 0.0077 | 0.0751 ± 0.016 | 0.734 ± 0.021 | 89% ± 4 |
| 4 | 0.488 ± 0.020 | 0.240 ± 0.019 | 0.384 ± 0.011 | 44% ± 6 |
| 5 | 0.656 ± 0.0065 | 0.179 ± 0.010 | 0.437 ± 0.0076 | 66% ± 3 |
| 6 | 0.693 ± 0.012 | 0.325 ± 0.021 | 0.806 ± 0.013 | 56% ± 3 |
| 7 | 0.852 ± 0.014 | 0.189 ± 0.045 | 0.560 ± 0.020 | 72% ± 8 |
| 8 | 0.895 ± 0.011 | 0.0622 ± 0.032 | 0.343 ± 0.019 | 87% ± 9 |
| Average | 0.711 ± 0.017 | 0.219 ± 0.044 | 0.610 ± 0.028 | 67% ± 8 |
Fig. 3.
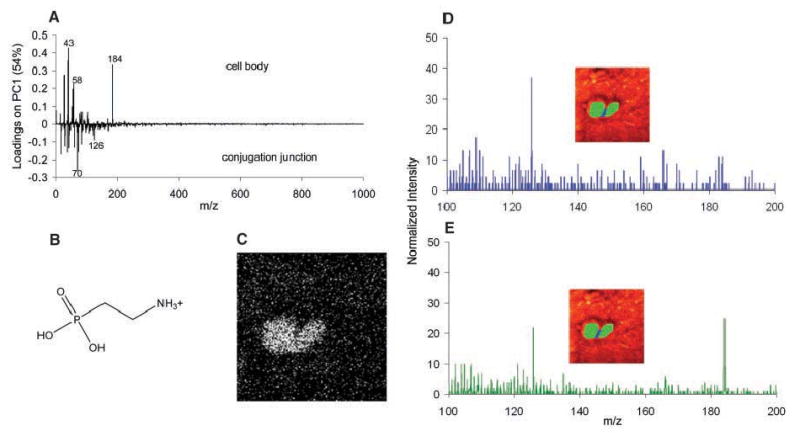
The conjugation junction contains elevated amounts of 2-AEP. Mass spectra from pixels were generated by selecting the pixels of interest using software written in house. (A) Loadings plot from principal component analysis comparing the mass spectra of the cell bodies and the conjugation junction. (B) 2-AEP headgroup fragment corresponding to m/z 126. (C) SIMS image for m/z 126. (D) Mass spectrum from the pixels along the conjugation junction, as indicated in the inset. (E) Mass spectrum from the pixels in the cell bodies, as indicated in the inset.
It is likely that the decreased amount of PC in this junction region correlates with an elevated concentration of 2-aminoethylphos-phonolipid (2-AEP), the phosphonolipid analog of PE. Like PE, 2-AEP is a cone-shaped nonlamellar lipid and should form highly curved membrane fusion structures. 2-AEP makes up about 34% of the Tetrahymena whole-cell membrane phosphorus lipid content, whereas PC represents 26% of the phosphorus lipids (26). 2-AEP represents a class of lipids that contain a variety of fatty acid tailgroups that cannot be detected via the molecular ion with TOF-SIMS. Instead, 2-AEP can be identified with the headgroup fragment ion of m/z 126 (Fig. 3B), similar to the way in which PC is detected with the m/z 184 headgroup ion.
The molecule-specific image of m/z 126 for the mating Tetrahymena appears to be homogeneous across the cells, probably because the entire surface of the Tetrahymena membrane is concentrated in this lipid (Fig. 3C). However, a comparison of the mass spectrum from the pixels of the cell body region with that from the conjugation junction suggests an increase in 2-AEP at the conjugation junction (Fig. 3, D and E). The normalized intensity of m/z 126 and m/z 184 can be directly compared between the two spectra, and not only did the intensity ratio between the two peaks change, but the relative amount of m/z 126 increased in the junction region as m/z 184 decreased. Principal component analysis showed a clear correlation of m/z 126 with the conjugation junction, in comparison to the cell body (Fig. 3A). We note that mammalian cells are not known to contain any substantial amount of 2-AEP (27). Frozen-hydrated rat pheochromocytoma cells and beige mouse mast cells do not localize m/z 126 to the cell structure; thus, its localized increase in the Tetrahymena junction is clearly significant. Tetrahymena can either reorganize or manufacture the necessary lipids and adjust its membrane structure for pore formation during the time necessary for full conjugation. The extent to which lipids in the membrane are involved in these changes and the exact cellular machinery that drives these alterations are unknown, although the changes probably occur during the required period of cell-cell contact before membrane fusion. It is likely that these interactions initiate a biological response that prepares the cells for conjugation by directing a higher concentration of high-curvature lipids, which are adept at fusion pore formation, to the membrane fusion sites.
Supplementary Material
Acknowledgments
Supported in part by a grant from NIH and additional instrumentation support provided by NSF. We thank M. Wagner for sharing his expertise in multivariate statistics and applying principal component analysis to the SIMS data, T. Roddy for helpful discussions and suggestions, M. Hazen and the Pennsylvania State University Electron Microscopy Facility for assistance with the TEM images, and P. Powell and L. Sombers for help with peak fitting and statistical analysis of the data.
Footnotes
Supporting Online Material: www.sciencemag.org/cgi/content/full/305/5680/71/DC1
Materials and Methods
References
References and Notes
- 1.Wolfe J. J Cell Sci. 1985;73:69. doi: 10.1242/jcs.73.1.69. [DOI] [PubMed] [Google Scholar]
- 2.Wolfe J. J Morphol. 1982;172:159. doi: 10.1002/jmor.1051720204. [DOI] [PubMed] [Google Scholar]
- 3.Frisch A, Loyter A, Levy R, Goldberg I. Biochim Biophys Acta. 1978;506:18. doi: 10.1016/0005-2736(78)90431-5. [DOI] [PubMed] [Google Scholar]
- 4.Thompson GA., Jr J Protozool. 1972;19:231. doi: 10.1111/j.1550-7408.1972.tb03442.x. [DOI] [PubMed] [Google Scholar]
- 5.Blumenthal R, Clague MJ, Durell SR, Epand RM. Chem Rev. 2003;103:53. doi: 10.1021/cr000036+. [DOI] [PubMed] [Google Scholar]
- 6.Eitzen G. Biochim Biophys Acta. 2003;1641:175. doi: 10.1016/s0167-4889(03)00087-9. [DOI] [PubMed] [Google Scholar]
- 7.Simons K, Ikonen E. Nature. 1997;387:569. doi: 10.1038/42408. [DOI] [PubMed] [Google Scholar]
- 8.Hao MM, Mukherjee S, Maxfield FR. Proc Natl Acad Sci USA. 2001;98:13072. doi: 10.1073/pnas.231377398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Edidin M. Trends Cell Biol. 2001;11:492. doi: 10.1016/s0962-8924(01)02139-0. [DOI] [PubMed] [Google Scholar]
- 10.D'Onofrio TG, et al. Langmuir. 2003;19:1618. [Google Scholar]
- 11.Yang L, Huang HW. Science. 2002;297:1877. doi: 10.1126/science.1074354. [DOI] [PubMed] [Google Scholar]
- 12.Chernomordik L. Chem Phys Lipids. 1996;81:203. doi: 10.1016/0009-3084(96)02583-2. [DOI] [PubMed] [Google Scholar]
- 13.Gruner SM. Proc Natl Acad Sci USA. 1985;82:3665. doi: 10.1073/pnas.82.11.3665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chandra S, Smith DR, Morrison GH. Anal Chem. 2000;72:104a. doi: 10.1021/ac002716i. [DOI] [PubMed] [Google Scholar]
- 15.McMahon JM, Short RT, McCandlish CA, Brenna JT, Todd PJ. Rapid Commun Mass Spectrom. 1996;10:335. doi: 10.1002/(SICI)1097-0231(199602)10:3<335::AID-RCM516>3.0.CO;2-W. [DOI] [PubMed] [Google Scholar]
- 16.Cannon DM, Pacholski ML, Winograd N, Ewing AG. J Am Chem Soc. 2000;122:603. [Google Scholar]
- 17.Pacholski ML, Cannon DM, Ewing AG, Winograd N. Rapid Commun Mass Spectrom. 1998;12:1232. doi: 10.1002/(SICI)1097-0231(19980930)12:18<1232::AID-RCM319>3.0.CO;2-G. [DOI] [PubMed] [Google Scholar]
- 18.Colliver TL, et al. Anal Chem. 1997;69:2225. doi: 10.1021/ac9701748. [DOI] [PubMed] [Google Scholar]
- 19.Roddy TP, Cannon DM, Ostrowski SG, Winograd N, Ewing AG. Anal Chem. 2002;74:4020. doi: 10.1021/ac025574w. [DOI] [PubMed] [Google Scholar]
- 20.Roddy TP, Cannon DM, Meserole CA, Winograd N, Ewing AG. Anal Chem. 2002;74:4011. doi: 10.1021/ac0255734. [DOI] [PubMed] [Google Scholar]
- 21.Pscheid P, Schudt C, Plattner H. J Microsc. 1981;121:149. doi: 10.1111/j.1365-2818.1981.tb01208.x. [DOI] [PubMed] [Google Scholar]
- 22.Chandra S, Morrison GH. Biol Cell. 1992;74:31. doi: 10.1016/0248-4900(92)90006-m. [DOI] [PubMed] [Google Scholar]
- 23.Roddy TP, Cannon DM, Ostrowski SG, Ewing AG, Winograd N. Anal Chem. 2003;75:4087. doi: 10.1021/ac026353t. [DOI] [PubMed] [Google Scholar]
- 24.S. G. Ostrowski, C. T. Van Bell, N. Winograd, A. G. Ewing, data not shown.
- 25.Jackson JE. A User's Guide to Principal Components. Wiley; Hoboken, NJ: 2003. [Google Scholar]
- 26.Thompson GA, Jr, Nozawa Y. Biochim Biophys Acta. 1977;472:55. doi: 10.1016/0304-4157(77)90014-4. [DOI] [PubMed] [Google Scholar]
- 27.Ansell GB, Hawthorne JN, Dawson RMC. Form and Function of Phospholipids, vol 3, BBA Library. 2. Elsevier Scientific; New York: 1973. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


