Abstract
The understanding of the membrane flow process during autophagosome formation is essential to illuminate the role of autophagy under various disease-causing conditions. Atg9 is the only identified integral membrane protein required for autophagosome formation, and it is thought to cycle between the membrane sources and the phagophore assembly site (PAS). Thus, Atg9 may play an important role as a membrane carrier. We report the self-interaction of Atg9 and generate an Atg9 mutant that is defective in this interaction. This mutation results in abnormal autophagy, due to altered phagophore formation as well as inefficient membrane delivery to the PAS. Based on our analyses, we discuss a model suggesting dual functions for the Atg9 complex: by reversibly binding to another Atg9 molecule, Atg9 can both promote lipid transport from the membrane origins to the PAS, and also help assemble an intact phagophore membrane.
Keywords: membrane biogenesis, mitochondria, protein targeting, stress, vacuole, yeast
Autophagy is a so-called “self-eating” process in eukaryotic cells induced by stress, for example, starvation, hypoxia and infection. In response to these signals, double-membrane autophagosomes enwrap cargos such as bulk cytosol, superfluous organelles, pathogens or protein aggregates, and are subsequently transported to lysosomes (or the vacuole in yeast) for degradation. The resulting macromolecules are released from the vacuole and used for the synthesis of critical proteins and the maintenance of vital cellular functions during starvation. Under other stress conditions, such as those resulting in damaged or dysfunctional organelles, autophagy may protect cells from further damage by sequestering these compartments from the remainder of the cytoplasm. Autophagosome biogenesis is considered a de novo process, in which small membranes are sequentially added onto the phagophore, the initial sequestering structure, leading to expansion and ultimately the formation of a completed autophagosome.1 In budding yeast, this process occurs at a distinct perivacuolar site, known as the phagophore assembly site (PAS).
How the rapid membrane remodeling that is characteristic of autophagosome biogenesis is achieved remains a fundamentally open question. To elucidate the molecular mechanism, we focus on the autophagy-related (Atg) protein Atg9. Atg9 is unique among the current 31 characterized Atg proteins, in that it is the only transmembrane protein required for autophagy, and displays a relatively distinct subcellular localization pattern. Atg9 is present both at the PAS and at multiple peripheral structures; Atg9 partially localizes at mitochondria and possibly at the Golgi complex, both of which may provide membranes to the forming autophagosome.2-4 Thus, we have proposed a cycling model in which Atg9 shuttles between the peripheral sites and the PAS, supported by live imaging studies showing Atg9 movement in the cytoplasm and by the finding of several Atg factors that play a role in Atg9 delivery to and retrieval from the PAS.5-7
During our study of Atg9 trafficking, we noticed that fluorescent-tagged Atg9 is organized in large clusters, suggesting that the protein might self-interact. In addition, two different fusion chimeras, Atg9-GFP and Atg9-DsRed, largely colocalize. We confirm the self-interaction by yeast two-hybrid analysis and coimmunoprecipitation, and further map the interaction site to the C terminus of Atg9. Autophagy is blocked when we mutate this site by deleting five conserved amino acids (Atg9Δ766-770).8
Similar to wild-type Atg9, this loss-of-self-interaction mutant partially localized to mitochondria (Fig. 1), but we found that its delivery to the PAS is less efficient.8 What is the possible cause of this transport defect? On the mitochondrial surface, the wild-type Atg9 clusters undergo highly dynamic movement,2 which is reminiscent of lipid rafts that form in the membrane of the trans-Golgi network (TGN): selected membrane protein cargos accumulate in the rafts, and then bud from the TGN into transport vesicles. Raft-like microdomains have also been suggested to form on the mitochondrial outer-membrane, playing a role in signal transduction.9 Given the recent discovery of mitochondria-derived vesicles,10 we speculate that Atg9 molecules are concentrated into clusters that are organized by self-association at the membrane sources, and this process may be essential for efficient trafficking of the protein (similar to the budding events in the secretory pathway and at the plasma membrane; Fig. 2). Future studies are important to analyze the underlying mechanism that triggers such movement.
Figure 1.
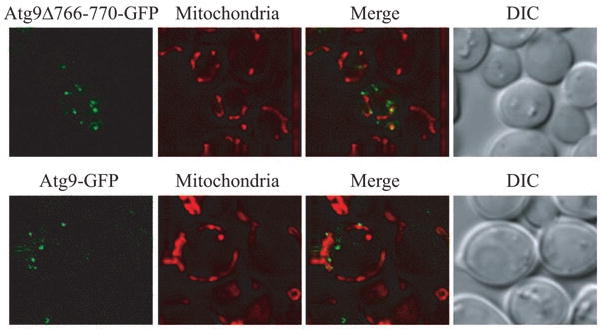
The loss-of-self-interaction mutant partially localizes to mitochondria. atg9Δ cells expressing plasmid-borne CUP1 promoter-driven wild-type Atg9-GFP or Atg9Δ766-770-GFP were cultured to mid-log phase and stained with Mitofluor Red 589 before imaging by fluorescence microscopy. DIC, differential interference contrast.
Figure 2.
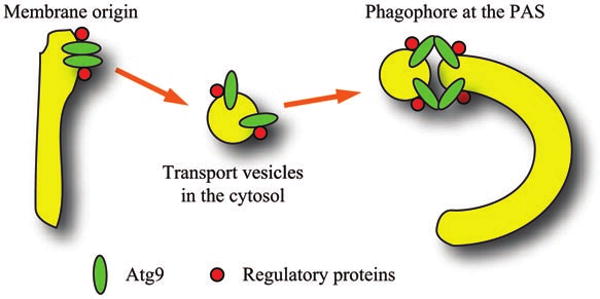
Possible roles of Atg9 self-association during autophagosome formation. Atg9 forms clusters on the surface of membrane sources through self-interaction, which promotes its trafficking to the PAS. Regulation of assembly and disassembly of the Atg9 complex by additional components allows the Atg9-containing vesicles to fuse with the existing phagophore membrane, which leads to phagophore expansion.
Our previous data show that Atg9 delivery to the PAS is mediated by Atg11.7 We find that overexpressing Atg11 enhances the trafficking of the loss-of-self-interaction Atg9 mutant to the PAS. However, autophagy activity is still defective, which suggests that there must be a second function for Atg9 self-interaction at the phagophore assembly site. By fluorescence microscopy, we observe a cup-shaped structure formed by wild-type Atg9 around the cargo RFP-Ape1, in atg1Δ cells. This structure is presumably the growing phagophore, which is unable to complete the autophagosome formation process due to loss of Atg1. Although resolution by fluorescence microscopy is limited, it appeared that the phagophore structure was abnormal in cells expressing the Atg9 self-interaction mutant. To gain additional insight, we examined the phagophore in atg1Δ cells by immunoelectron microscopy. The phagophore membranes associated with Atg9Δ776-770 appear to be more fragmented, and less total membrane is present at the presumed PAS. These results suggest that Atg9 self-interaction has a direct role in phagophore formation. Membrane tethering and fusion usually require the function of SNAREs and Rab family proteins, but no one has yet demonstrated the direct involvement of these factors in autophagosome formation. Here, we propose a membrane-supplying and tethering mechanism involving Atg9 during autophagy, possibly in cooperation with several other autophagy-related proteins that associate with lipids, such as Atg8, and in mammalian cells, Bif-1.11,12 On the other hand, the ability to form cup-shaped phagophores in atg1Δ cells indicates that Atg1 (and its kinase activity) may be dispensable for autophagy induction; together with our finding that Atg1 kinase-defective mutants block the retrieval of Atg9 from the PAS (our unpublished results), the kinase activity of Atg1 seems to have a function at a later step(s) of autophagosome formation.
Interestingly, the loss-of-self-interaction mutant Atg9 did not migrate as a monomer under native conditions, suggesting that other binding proteins may be present in the Atg9 complex. To test this hypothesis, we purified the Atg9 complex by tandem affinity purification and resolved it by SDS-PAGE. Four major protein bands were detected in our system besides Atg9, migrating at the sizes of 94, 50, 28 and 25 kDa. In the future, it would be interesting to characterize these components and their roles in regulating Atg9 self-interaction during autophagosome formation. Notably, from the reported large-scale proteomic studies in yeast, three non-Atg proteins have been identified to copurify with Atg9, Ccr4 (94.7 kD), Cdc39 (240 kD) and Tif5 (45.3 kD).13,14 Among them, Ccr4 and Cdc39 are components of the CCR4-NOT transcriptional complex involved in regulating transcription and destabilizing mRNAs by deadenylation, and Tif5 is the translation initiation factor eIF-5 with a function as a GTPase-activating protein (GAP) for GTP hydrolysis.15 It is essentially unknown in what manner the transcription and translation complexes may function together with Atg9, but the role of Tif5 as a GAP is intriguing because it may provide a hint to study the energy supply for membrane assembly. Therefore, based on these data, we propose dual roles of Atg9 self-interaction: to promote the flow of membrane from peripheral origins to the PAS and to mediate tethering of small membranes to the forming phagophore, possibly with the help of other regulatory factors (Fig. 2). Mass spectrometry and in vitro experiments will be helpful to further solve the conundrum of phagophore assembly.
References
- 1.Mizushima N, Yamamoto A, Hatano M, Kobayashi Y, Kabeya Y, Suzuki K, et al. Dissection of autophagosome formation using Apg5-deficient mouse embryonic stem cells. J Cell Biol. 2001;152:657–68. doi: 10.1083/jcb.152.4.657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Reggiori F, Shintani T, Nair U, Klionsky DJ. Atg9 cycles between mitochondria and the pre-autophagosomal structure in yeasts. Autophagy. 2005;1:101–9. doi: 10.4161/auto.1.2.1840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Juhasz G, Neufeld TP. Autophagy: a forty-year search for a missing membrane source. PLoS Biol. 2006;4:36. doi: 10.1371/journal.pbio.0040036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Young ARJ, Chan EYW, Hu XW, Köchl R, Crawshaw SG, High S, et al. Starvation and ULK1-dependent cycling of mammalian Atg9 between the TGN and endosomes. J Cell Sci. 2006;119:3888–900. doi: 10.1242/jcs.03172. [DOI] [PubMed] [Google Scholar]
- 5.Monastyrska I, He C, Geng J, Hoppe AD, Li Z, Klionsky DJ. Arp2 links autophagic machinery with the actin cytoskeleton. Mol Biol Cell. 2008;19:1962–75. doi: 10.1091/mbc.E07-09-0892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Reggiori F, Tucker KA, Stromhaug PE, Klionsky DJ. The Atg1-Atg13 complex regulates Atg9 and Atg23 retrieval transport from the pre-autophagosomal structure. Dev Cell. 2004;6:79–90. doi: 10.1016/s1534-5807(03)00402-7. [DOI] [PubMed] [Google Scholar]
- 7.He C, Song H, Yorimitsu T, Monastyrska I, Yen WL, Legakis JE, et al. Recruitment of Atg9 to the preautophagosomal structure by Atg11 is essential for selective autophagy in budding yeast. J Cell Biol. 2006;175:925–35. doi: 10.1083/jcb.200606084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.He C, Baba M, Cao Y, Klionsky DJ. Self-interaction is critical for Atg9 transport and function at the phagophore assembly site during autophagy. Mol Biol Cell. 2008;19:5506–16. doi: 10.1091/mbc.E08-05-0544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.McBride HM, Neuspiel M, Wasiak S. Mitochondria: more than just a powerhouse. Curr Biol. 2006;16:551–60. doi: 10.1016/j.cub.2006.06.054. [DOI] [PubMed] [Google Scholar]
- 10.Neuspiel M, Schauss AC, Braschi E, Zunino R, Rippstein P, Rachubinski RA, et al. Cargo-selected transport from the mitochondria to peroxisomes is mediated by vesicular carriers. Curr Biol. 2008;18:102–8. doi: 10.1016/j.cub.2007.12.038. [DOI] [PubMed] [Google Scholar]
- 11.Nakatogawa H, Ichimura Y, Ohsumi Y. Atg8, a ubiquitin-like protein required for autophagosome formation, mediates membrane tethering and hemifusion. Cell. 2007;130:165–78. doi: 10.1016/j.cell.2007.05.021. [DOI] [PubMed] [Google Scholar]
- 12.Takahashi Y, Coppola D, Matsushita N, Cualing HD, Sun M, Sato Y, et al. Bif-1 interacts with Beclin 1 through UVRAG and regulates autophagy and tumorigenesis. Nat Cell Biol. 2007;9:1142–51. doi: 10.1038/ncb1634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gavin AC, Bosche M, Krause R, Grandi P, Marzioch M, Bauer A, et al. Functional organization of the yeast proteome by systematic analysis of protein complexes. Nature. 2002;415:141–7. doi: 10.1038/415141a. [DOI] [PubMed] [Google Scholar]
- 14.Gavin AC, Aloy P, Grandi P, Krause R, Boesche M, Marzioch M, et al. Proteome survey reveals modularity of the yeast cell machinery. Nature. 2006;440:631–6. doi: 10.1038/nature04532. [DOI] [PubMed] [Google Scholar]
- 15.Das S, Ghosh R, Maitra U. Eukaryotic translation initiation factor 5 functions as a GTPase-activating protein. J Biol Chem. 2001;276:6720–6. doi: 10.1074/jbc.M008863200. [DOI] [PubMed] [Google Scholar]


