Main Text
To the Editor: It has long been accepted that productive recombination between X and Y chromosomes in humans is limited to short telomeric portions known as pseudoautosomal regions. It now appears that our picture of X-Y chromosome recombination requires revision because of the observation that gene conversion between X and Y gametologous regions occurs in humans, as reported by Rosser et al.1 in the July 2009 issue of the American Journal of Human Genetics. The data were based on resequencing of X and Y copies of a translocation hotspot (HSA)2 adjacent to the PRKX (MIM 300083) and PRKY (MIM 400008) genes.
In October, our group presented evidence for X-to-Y gene conversion within the VCY (MIM 400012) genes,3 a region that lies within the male-specific portion of the Y chromosome (MSY) and is different from the PRKY region. Thus, both studies indicate that a new form of genetic exchange occurs between the sex-specific portions of X and Y and that this process has been active in recent human evolution.
PRKY and VCY gene conversion hotspots are similar in that both are evolutionarily conserved and lie in regions displaying a high X-Y sequence similarity (∼95%); both have a block of complete X-Y identity (246 bp and 206 bp, respectively). On the other hand, the two hotspots strongly differ from each other with respect to the estimated gene conversion rate per base per generation. A conversion rate per base per generation of 2.5 × 10−6 to 5.4 × 10−6 was estimated for the VCY genes (a generation time of 20 years was assumed).3 By contrast, Rosser et al.1 obtained a range for the PRKY X-to-Y conversion rate per base per generation (∼1.45 × 10−4 to ∼6.82 × 10−3, if a 25-year generation time is assumed) two to three orders of magnitude higher than that estimated for VCY and compared this rate to the rate of Y-Y gene conversion (2.2 × 10−4) between the arms of palindromes, as reported by Rozen et al.4 However, to compare analyses on different sequence lengths meaningfully, it is crucial to divide by the length of the sequence under study to get a per base rate, and Rosser et al.1 don't seem to have done that. In an alternate measure of conversion rate, one can take the total length of the converted tracts and divide by the total length of the tree connecting the sequences in generations, as suggested by Rosser et al.,1 but then divide by the total sequence length, as follows: (where c is the estimated rate of gene conversion per base per generation, n is the number of observed gene conversion events, li is the length in bp of the ith gene conversion event, L is the length in bp of the region under study, and t is the number of generations in the tree). By this formula, one would estimate the probability per generation per site that a site is in a gene conversion event. This recognizes that a single gene conversion event might replace a string of bases, not just one, as a substitution does; it also allows for the dependence on sequence length.
We also analyzed the PRKY HSA hotspot by resequencing a 2,348 bp DNA segment (chrY:7,095,919–7,098,266) (see Web Resources) in 47 Y chromosomes representing a wide coverage of the Y phylogeny and resequenced an 831 bp DNA segment (chrY:7,245,678–7,246,508) encompassing a second translocation hotspot within the PRKY gene (HSB)2 in the same sample set. DNA samples came from collections of the authors, and haplogroup information is as reported.5–8
No SNPs were found in the HSB region. This region has been described as a weaker hotspot of translocation than HSA.2 Our analysis was unable to determine whether the absence of any signal of gene conversion in the HSB region was a consequence of the reduced size of the sample analyzed or a lower frequency of conversion (if any conversion occurs at all).
By contrast, we found eight SNPs by resequencing the HSA region (Figure 1 and Table S1). All the SNPs identified lay in a narrow region of 1.0 kb, encompassing the 246 bp identity block. Three of these SNPs (V139.1, V141, and V145) resulted in the homogenization of pre-existing X-Y sequence differences, most likely as a consequence of X-to-Y gene conversion events. Among these, SNPs V139.1 and V141 resulted from the sequencing of one haplogroup A2 Y chromosome and corresponded to two variant sites identified by Rosser et al.1 in the same haplogroup. A fourth Y chromosome SNP (V144) was found to be shared with the X chromosome (see Table S1) as shown by resequencing 1,213 bp (chrX:3,671,984–3,673,196) (see Web Resources) of the PRKX HSA region in 51 X chromosomes. This suggested an additional gene conversion event between X and Y, although the direction of the genetic change could not be determined. Thus, in total, two or three X-to-Y gene conversion events were identified (Table S2).
Figure 1.
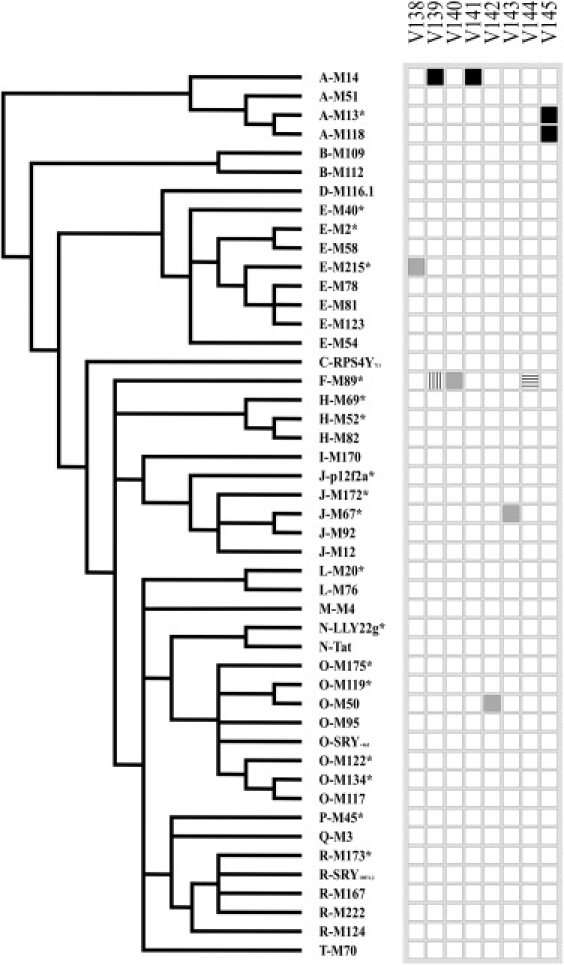
SNPs Identified in the Sequenced Region
To the left, a simplified version of the Y chromosome tree10 showing the phylogenetic relationships among the 47 Y chromosomes analyzed. SNP names are given at the top. Squares represent the allelic state for each SNP: white, ancestral allele; black, the SNP has arisen in an X-Y gametologous sequence variant, and the derived state is equal to the gametologous base on the X; vertical bars, the SNP has arisen in an X-Y gametologous sequence variant, and the derived state is different from the gametologous base on the X; gray, the SNP has arisen in a site identical between X and Y; horizontal bars, the SNP is a shared polymorphism between X and Y.
The 47 Y chromosomes we surveyed included the same haplogroups and paragroups as did the Y chromosomes analyzed by Repping et al.9 Therefore, we estimated the same length for our tree as the tree in the study of Repping et al.,9 i.e., 52,000 generations. Then, as in Rosser et al.,1 we obtained a lower and upper bound on the estimate of the X-to-Y gene conversion rate per base per generation (we used a 25-year generation time) by considering the minimum and maximum number of converted base pairs (Table S2). The formula above yielded a range for the conversion rate per base per generation of 4.1 × 10−8 to 2.4 × 10−6 and of 4.9 × 10−8 to 2.9 × 10−6 for two or three gene conversion events in the HSA, respectively. We used the same approach to recalculate the rate of X-to-Y gene conversion in the VCY genes on the basis of the data reported by Trombetta et al.3 With L = 1,616 bp, t = 35,990 generations (if we assume a 25-year generation time), a total minimum number of converted bases = 93, and a total maximum number of converted bases = 475, this gave a rate of 1.6 × 10−6 to 8.2 × 10−6, similar to the rate we estimated for the PRKY HSA region. Thus, although the present estimates are based on relatively small datasets, analyses on two different regions of the MSY indicate that the rate of X-to-Y gene conversion is one to two orders of magnitude lower than that estimated for Y-Y gene conversion between palindrome arms.4
Defining the per-base X-Y gene conversion rate has widespread significance for understanding the genetic composition and evolution of genes within the MSY. Our results revise and expand previous research1 on X-to-Y gene conversion in the PRKY gene region and offer a revised estimate of the per-base gene conversion rate for this translocation hotspot as compared to that for other regions of the MSY and the Y-Y gene conversion rate4 within the palindromes of the Y chromosome.
Acknowledgments
This work was supported by the Sapienza Università di Roma, Grandi Progetti di Ateneo, and the Italian Ministry of the University, Progetti di Ricerca di Interesse Nazionale 2007 (both to R.S.). B.T. was supported by an “Ateneo della Scienza e della Tecnica” postdoctoral fellowship.
Supplemental Data
Web Resources
The URLs for data presented herein are as follows:
Nucleotide positions according to the February 2009 human reference sequence (GRCh37), http://genome.ucsc.edu/cgi-bin/hgGateway
Online Mendelian Inheritance in Man (OMIM), http://www.ncbi.nlm.nih.gov/Omim
References
- 1.Rosser Z.H., Balaresque P., Jobling M.A. Gene conversion between the X chromosome and the male-specific region of the Y chromosome at a translocation hotspot. Am. J. Hum. Genet. 2009;85:130–134. doi: 10.1016/j.ajhg.2009.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wang I., Weil D., Levilliers J., Affara N.A., De La Chapelle A., Petit C. Prevalence and molecular analysis of two hot spots for ectopic recombination leading to XX maleness. Genomics. 1995;28:52–58. doi: 10.1006/geno.1995.1105. [DOI] [PubMed] [Google Scholar]
- 3.Trombetta B., Cruciani F., Underhill P.A., Sellitto D., Scozzari R. Footprints of X-to-Y gene conversion in recent human evolution. Mol. Biol. Evol. 2010;27:714–725. doi: 10.1093/molbev/msp231. Published online October 6, 2009. [DOI] [PubMed] [Google Scholar]
- 4.Rozen S., Skaletsky H., Marszalek J.D., Minx P.J., Cordum H.S., Waterston R.H., Wilson R.K., Page D.C. Abundant gene conversion between arms of palindromes in human and ape Y chromosomes. Nature. 2003;423:873–876. doi: 10.1038/nature01723. [DOI] [PubMed] [Google Scholar]
- 5.Underhill P.A., Passarino G., Lin A.A., Shen P., Mirazón Lahr M., Foley R.A., Oefner P.J., Cavalli-Sforza L.L. The phylogeography of Y chromosome binary haplotypes and the origins of modern human populations. Ann. Hum. Genet. 2001;65:43–62. doi: 10.1046/j.1469-1809.2001.6510043.x. [DOI] [PubMed] [Google Scholar]
- 6.Cruciani F., Santolamazza P., Shen P., Macaulay V., Moral P., Olckers A., Modiano D., Holmes S., Destro-Bisol G., Coia V. A back migration from Asia to sub-Saharan Africa is supported by high-resolution analysis of human Y-chromosome haplotypes. Am. J. Hum. Genet. 2002;70:1197–1214. doi: 10.1086/340257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cruciani F., La Fratta R., Torroni A., Underhill P.A., Scozzari R. Molecular dissection of the Y chromosome haplogroup E-M78 (E3b1a): A posteriori evaluation of a microsatellite-network-based approach through six new biallelic markers. Hum. Mutat. 2006;27:831–832. doi: 10.1002/humu.9445. [DOI] [PubMed] [Google Scholar]
- 8.Cruciani F., La Fratta R., Trombetta B., Santolamazza P., Sellitto D., Beraud Colomb E., Dugoujon J.M., Crivellaro F., Benincasa T., Pascone R. Tracing past human male movements in northern/eastern Africa and western Eurasia: New clues from Y-chromosomal haplogroups E-M78 and J-M12. Mol. Biol. Evol. 2007;24:1300–1311. doi: 10.1093/molbev/msm049. [DOI] [PubMed] [Google Scholar]
- 9.Repping S., van Daalen S.K.M., Brown L.G., Korver C.M., Lange J., Marszalek J.D., Pyntikova T., van der Veen F., Skaletsky H., Page D.C. High mutation rates have driven extensive structural polymorphism among human Y chromosomes. Nat. Genet. 2006;38:463–467. doi: 10.1038/ng1754. [DOI] [PubMed] [Google Scholar]
- 10.Karafet T.M., Mendez F.L., Meilerman M.B., Underhill P.A., Zegura S.L., Hammer M.F. New binary polymorphisms reshape and increase resolution of the human Y chromosomal haplogroup tree. Genome Res. 2008;18:830–838. doi: 10.1101/gr.7172008. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


