Abstract
Subtelomeres are dynamic structures composed of blocks of homologous DNA sequences. These so-called duplicons are dispersed over many chromosome ends. We studied the human 4q and 10q subtelomeres, which contain the polymorphic macrosatellite repeat D4Z4 and which share high sequence similarity over a region of, on average, >200 kb. Sequence analysis of four polymorphic markers in the African, European, and Asian HAPMAP panels revealed 17 subtelomeric 4q and eight subtelomeric 10qter haplotypes. Haplotypes that are composed of a mixture of 4q and 10q sequences were detected at frequencies >10% in all three populations, seemingly supporting a mechanism of ongoing interchromosomal exchanges between these chromosomes. We constructed an evolutionary network of most haplotypes and identified the 4q haplotype ancestral to all 4q and 10q haplotypes. According to the network, all subtelomeres originate from only four discrete sequence-transfer events during human evolution, and haplotypes with mixtures of 4q- and 10q-specific sequences represent intermediate structures in the transition from 4q to 10q subtelomeres. Haplotype distribution studies on a large number of globally dispersed human DNA samples from the HGDP-CEPH panel supported our findings and show that all haplotypes were present before human migration out of Africa. D4Z4 repeat array contractions on the 4A161 haplotype cause Facioscapulohumeral muscular dystrophy (FSHD), whereas contractions on most other haplotypes are nonpathogenic. We propose that the limited occurrence of interchromosomal sequence transfers results in an accumulation of haplotype-specific polymorphisms that can explain the unique association of FSHD with D4Z4 contractions in a single 4q subtelomere.
Introduction
Subtelomeres are located immediately proximal to the telomeric repeats and are composed of blocks of homologous DNA sequences that are dispersed over different chromosome ends. These domains of sequence homology have arisen through intra- and interchromosomal segmental duplications.1,2 The resulting duplicons can contain tens to hundreds of kilobases of DNA and can be more than 97% identical in sequence between homologous and nonhomologous chromosome ends.3 Some duplicons were shown to be human specific, whereas others have occurred earlier in primate evolution.4 It has been proposed that natural variation in the order and copy number of duplicons contributes to normal phenotypic variation.1,2 Indeed, gene families that require rapid diversification because of their protein function often reside in subtelomeres.5
Large DNA polymorphisms within a single subtelomere were first described for chromosome 16p: three variants that differ by as much as 230 kb in subtelomeric content were reported.6 Of particular interest is the subtelomere of chromosome 4q because of its involvement in the autosomal-dominant myopathy facioscapulohumeral muscular dystrophy (FSHD [MIM 158900]). FSHD is caused by a contraction of the macrosatellite repeat D4Z4 that resides in this chromosome end (reviewed in de Greef et al.7). We previously identified, analogous to chromosome 16p, two 4qter variants, dubbed 4qA and 4qB, on the basis of large sequence variations immediately distal to D4Z4 (Figures 1A and 1B).8,9 A more detailed analysis extended these observations by identifying at least nine different haplotypes of the 4q subtelomere on the basis of subtle, but consistent, sequence variations in and immediately flanking the D4Z4 repeat array. Interestingly, for only one of these haplotypes (4A161) were D4Z4 repeat array contractions shown to be associated with FSHD; contractions in two other common haplotypes (4A166 and 4B163) were not associated with the disease.10
Figure 1.
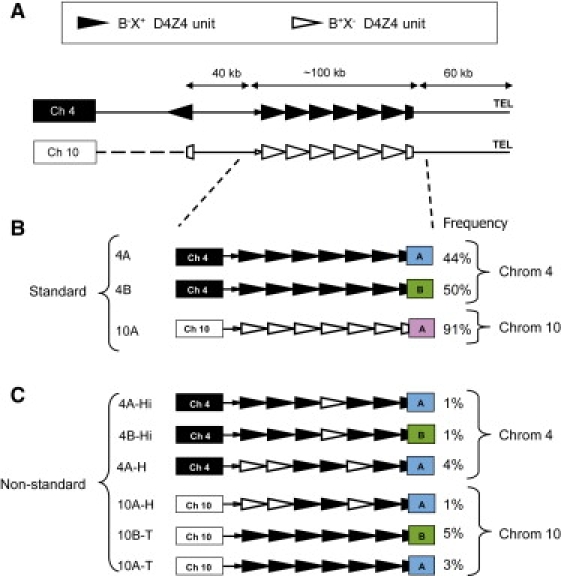
Structure of the D4Z4 Repeat on Standard and Nonstandard Chromosomes 4 and 10
(A) The ends of chromosomes 4q and 10q share a large region of homology that contains the D4Z4 repeat array spanning, on average, 100 kb. Proximal to D4Z4, the homology between chromosomes 4q and 10q extends approximately 40 kb and ends with an inverted D4Z4 repeat unit. Distally, the homology extends 60 kb into the telomere repeats. The standard D4Z4 repeat array on chromosome 4q consists of homogeneous B−X+ repeat units (closed triangles) and on chromosome 10 consists of homogeneous B+X− repeat units (open triangles).
(B) Standard chromosome 4q can end with the distal A or B variation, 4qA or 4qB, respectively, and standard chromosome 10 ends with an A variant.
(C) Nonstandard D4Z4 repeat arrays on chromosome 4q can be grouped in three classes: 4A-Hi, 4B-Hi (Hi means Hybrid internal), or 4A-H (H means Hybrid). In 4A-Hi and 4B-Hi chromosomes, the most proximal unit of the D4Z4 repeat array is B−X+, and in 4A-H chromosomes a B+X− D4Z4 unit can be found on this proximal location. Also, on chromosome 10q three nonstandard chromosomes can be discriminated: 10A-T and 10B-T (T means transferred) and 10A-H. 10A-T and 10B-T chromosomes carry a homogenous D4Z4 array of B−X+ units and end with the distal A or B variation, respectively, whereas 10A-H chromosomes have the same composition as 4A-H chromosomes and end with an A variant. The frequency of the standard and nonstandard chromosomes in the European population is indicated.
The variation in this particular subtelomere is not restricted to chromosome 4q: the D4Z4 repeat array and flanking sequences can also be found in the subtelomere of chromosome 10q (Figure 1).11 Previously, it was suggested that the 4q subtelomere has been transferred onto chromosome 10q.9 The region of homology between both chromosome ends thus contains the D4Z4 repeat array spanning on average 100 kb and extends approximately 40 kb on either side of the D4Z4 repeat array. Proximal to D4Z4, the homology between chromosomes 4q and 10q ends with an inverted D4Z4 repeat unit, whereas distally, the homology extends into the telomere repeats (Figure 1A).9 The D4Z4 repeat units are 99% identical between both chromosomes, but D4Z4 contractions on chromosome 10q are not associated with FSHD.11 The individual repeat units of the 4q and 10q D4Z4 arrays can be discriminated by specific D4Z4 SNPs that affect the recognition sites of the restriction enzymes BlnI and XapI.12,13 Most chromosomes 4q have arrays that are homogeneous for D4Z4 units and have SNP combination B−X+ (D4Z4 units are BlnI resistant and XapI sensitive), whereas chromosome 10q repeat arrays are usually homogeneous for B+X- D4Z4 units (XapI resistant and BlnI sensitive) (Figure 1B).
The sequence similarity between 4q and 10q subtelomeres created an opportunity for further transfers between these nonhomologous chromosome ends. Indeed, aproximately 5% of all chromosomes 4 in the European population carry B+X− D4Z4 repeat units that are normally found on chromosome 10, and the same incidence has been reported for the presence of the opposite configuration: B−X+ D4Z4 repeat units on chromosome 10 (Figure 1C).14,15 In this study, we will refer to these chromosomes as “nonstandard” chromosomes as opposed to “standard” chromosomes that have the normal configuration of B−X+ D4Z4 units on 4q or B+X− D4Z4 units on 10q (Figures 1B and 1C). Nonstandard chromosomes have also been detected in other populations, albeit with different frequencies.16,17 These nonstandard chromosomes are, like standard chromosomes, highly polymorphic for the D4Z4 repeat copy number. This high incidence of nonstandard chromosomes has previously been attributed to recurrent and ongoing interchromosomal exchanges between chromosomes 4 and 10.14,16 However, more recently we have shown that somatic instability of D4Z4 predominantly arises by intrachromosomal rather than by interchromosomal rearrangements.18 Furthermore, we defined a number of haplotypes on chromosomes 4q and 10q on the basis of a specific combination of markers that are distributed over a region of more than 50 kb, which does not support a mechanism of frequent exchanges between these and other chromosome ends.10
To further address this apparent discrepancy and to better understand the dynamic behavior of the subtelomeres of chromosomes 4q and 10q, we initiated a detailed study on the composition and prevalence of D4Z4 haplotypes in European and non-European human populations. We show that most nonstandard chromosomes can be grouped in a small number of nonstandard haplotypes and provide evidence that most nonstandard chromosomes probably originate from only four ancient human-specific translocation events. We were able to construct an evolutionary network of standard and nonstandard 4q and 10q haplotypes. This network allowed us to identify the ancestral 4q and 10q haplotypes and to demonstrate that most nonstandard haplotypes represent ancient key haplotypes in the network, rather than having evolved from recent translocations. Our data thus strongly suggest that only a few sequence transfers have occurred between chromosomes 4q and 10q in human evolution and that subsequent intrachromosomal mutations must explain the large number of standard and nonstandard haplotypes.
Subjects and Methods
DNA Samples for Genotyping
Detailed genotyping of the D4Z4 locus (Figure 2) was performed on DNA samples from European, Chinese, Japanese, and African populations. European samples were collected at the LUMC from unrelated healthy control individuals.15 Almost 70% of this collection consists of Dutch individuals; the other individuals are from different parts of Europe. Blood from these individuals was collected after informed consent was obtained. In addition, we studied DNA isolated from lymphoblastic cell lines collected by the International HapMap Consortium from Yoruban individuals in Ibadan, Nigeria (abbreviation: YRI), Japanese individuals in Tokyo, Japan (abbreviation: JPT), Han Chinese individuals in Beijing, China (abbreviation: CHB) and CEPH (Utah residents with ancestry from Northern and Western Europe) (abbreviation: CEU) (HAPMAPP1, HAPMAPP2, and HAPMAPP3 were obtained from the Coriell Institute).19 The CEU and YRI samples consisted of 30 father-mother-child trios, and the JPT and CHB panels contained samples from 45 unrelated Japanese and 45 unrelated Han Chinese individuals.
Figure 2.
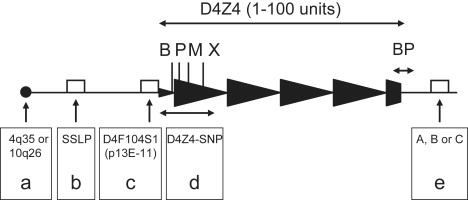
Schematic Overview of the 4q and 10q Subtelomeres with the Key Sequence Variations Used in This Study
The D4Z4 repeat array within the subtelomere of chromosomes 4q and 10q varies in size between 1 and 100 D4Z4 units (3.3–330 kb) and is indicated with closed triangles.
(A) Chromosomal localization of the D4Z4 repeat, chromosome 4q35 or 10q26.
(B) This Simple sequence length polymorphism (SSLP) is a combination of five VNTRs, a 8 bp insertion/deletion, and two SNPs localized 3.5 kb proximal to D4Z4 and varies in length between 157 and 182 bp.
(C) The D4F104S1 (p13E-11) region is localized immediately proximal to D4Z4, is 475 bp in length, and contains 15 SNPs.
(D) The most proximal unit of the D4Z4 repeat array contains several SNPs, of which four result in restriction-site polymorphisms that can be studied by Southern blot analysis (BlnI [B], XapI [X], PvuII [P], and MscI [M] in the first D4Z4 repeat unit of the sequence available with accession number AF117653 at positions 5660, 7512, 6044, and 6694 bp, respectively).
(E) A large sequence variation (A or B) has been detected distal to D4Z49 and can be detected by Southern blot analysis. In addition, we identified 4q chromosomes that fail to hybridize to probes A and B and provisionally called these C.
BP designates the breakpoint between the distal partial D4Z4 unit and the A, B, or C distal variation.
To study the global distribution of the chromosome 4q and 10q SSLP (simple sequence length polymorphism), we genotyped all samples in the Human Genome Diversity Project (HGDP) cell line panel.20 This panel consists of DNA samples from 1051 individuals from 51 globally dispersed human populations. We also used 96 samples from the Y Chromosome Consortium (YCC).21
DNA Isolation
To obtain high-quality DNA for detailed genotyping, we embedded peripheral blood lymphocytes and immortalized lymphoblastic cell lines in agarose plugs (InCert agarose, FMC) at a concentration of approximately 7.5 × 105 cells per plug and treated them with 600 μg/μl pronase and 1% Sarkosyl for 40–48 hr at 37°C. Next, plugs were washed in Tris-EDTA (TE−4) and were stored in 0.5 M EDTA at 4°C. Prior to restriction analysis, plugs were successively equilibrated in TE−4 and the appropriate restriction-enzyme buffer.
Analysis of SSLP and D4F104S1 Sequence
The SSLP sequence is localized approximately 3 kb proximal to the D4Z4 repeat (Figure 2B, nucleotides 1532–1694 [GenBank accession number AF117653]) and was studied by PCR with the use of forward primer 5′-HEX-GGT GGA GTT CTG GTT TCA GC-3′ and reverse primer 5′-CCT GTG CTT CAG AGG CAT TTG-3′. This PCR simultaneously identifies the SSLP size on chromosomes 4q and 10q. The PCR was performed with 5 ng genomic DNA, the Phusion F530-L DNA polymerase (Finzymes), and the supplemented high-fidelity buffer at 98°C for 15 s, 60°C for 30 s, and 72°C for 15 s for 33 cycles. Size differences were determined with the use of an ABI Prism 3100 Genetic Analyzer. The D4F104S1 region (Figure 2C, nucleotides 4384–4858 (GenBank accession number AF117653) of each chromosome was determined with forward primer 5′-CCC AGT TAC TGT TCT GGG TGA-3′ and reverse primer 5′-ATC CCA ATG TCT CCC CAT C-3′. The sequence of the SSLP and the D4F104S1 region as well as the SSLP size of individual chromosomes was identified after the four chromosomes were separated by restriction-enzyme digestion, electrophoresis, and electro-elution of the DNA from the gel slices.10 Primers were designed with Primer3 software.
Analysis of D4Z4 Repeat and Distal Variation
To determine the different SNPs in the proximal D4Z4 repeat unit and the homogeneity of the different D4Z4 units in a repeat array, we applied different methods. The BlnI and XapI polymorphic sites (B and X, Figure 2D) were analyzed by restriction analysis followed by pulsed-field gel electrophoresis (PFGE) and Southern blotting. In three different digestions we distinguish (1) all four complete D4Z4 repeat arrays from chromosomes 4 and 10 (double digestion with EcoRI and HindIII) (2) D4Z4 repeat arrays that are resistant to BlnI, B-X+ and B-X- D4Z4 units (double digestion with EcoRI and BlnI) and (3) D4Z4 repeat arrays that are resistant to XapI, B+X− and B−X− D4Z4 units (digestion with XapI).
The PvuII and the MscI polymorphisms (P and M, Figure 2D) in the proximal unit of the D4Z4 repeat array were analyzed on genomic DNA samples by digestion with either PvuII and BlnI or BglII, BlnI, and MscI. Digestion with PvuII generates chromosome-4-derived fragments of 2849 bp or 4559 bp in size, depending on the presence (P+D4Z4) or absence (P−D4Z4) of the PvuII restriction site, whereas chromosome 10-derived fragments are 2464 bp because of the presence of BlnI.18 Hybridization of probe p13E-11 to Southern blots of genomic DNA digested with MscI reveals chromosome-4-derived fragments of 2900 bp (M+D4Z4) and chromosome-10-derived fragments of 1774 bp because of the presence of a BlnI restriction site in D4Z4 on chromosome 10. Nonstandard 10A176T chromosomes give a fragment of 4061 bp because of the absence of the MscI restriction site (M−D4Z4). For analysis of the A/B variation distal to D4Z4 (Figure 2E), DNA was digested with HindIII only.8 All digestions and PFGE analyses were performed as described previously.10 To determine the chromosomal location of nonstandard D4Z4 repeat arrays (Figure 2A), we digested DNA with NotI, and the PFGE conditions have been described elsewhere.22
The breakpoint between D4Z4 and the region distal to D4Z4 for chromosomes with the distal A or C variation (Figure S2) was determined by a PCR with forward primer 5′-AGC GTT CCA GGC GGG AGG GAA G-3′ and reverse primer 5′-GGT TTG CCT AGA CAG CGT CGG AAG G-3′. The PCR reaction was performed on 100 ng of genomic DNA with 5 μl GC PCR buffer I (Takara), 1.5 μl GC-dNTPs (0.5 mM dATP, 0.5 mM dCTP, 0.5 mM dTTP, 0.3 mM dGTP, and 0.2 mM 7-deaza-dGTP) and 1.0 U of LA-Taq DNA polymerase (Takara) in a total volume of 25 μl. The PCR conditions consisted of an initial denaturation at 94°C for 3 min, followed by 35 cycles of denaturation at 94°C for 30 s, annealing at 68°C for 30 s, and extension at 72°C for 3 min. The breakpoint between D4Z4 and the region distal to D4Z4 for B chromosomes were determined by PCR with primers 5′-AAC GGA GGG AAA GAC AGA GC-3′ and 5′-GCC TGT CCT TAT GTC CAG GAT-3′. The PCR reaction was performed on 100 ng of genomic DNA with 1.5 μl GC-dNTPs (0.5 mM dATP, 0.5 mM dCTP, 0.5 mM dTTP, 0.3 mM dGTP and 0.2 mM 7-deaza-dGTP), 0.4 U of Phusion F530-L DNA polymerase, and supplemented GC buffer in a total volume of 25 μl. The PCR conditions consisted of an initial denaturation at 98°C for 3 min, followed by 35 cycles of denaturation at 98°C for 30 s, annealing at 55°C for 30 s, and extension at 72°C for 30 s. Chromosome-specific sequences were obtained from monochromosomal cell line sources previously described and from GenBank sequences.10 Sequence AF017466 is derived from a chromosome 4B source,9 and AC197422 is a chimpanzee chromosome 3 sequence.23 In addition, to eliminate unwanted chromosomes and specific PCR primers, we selected DNA samples of individuals with a specific haplotype composition that allowed chromosome-specific analysis based on a preceding restriction-enzyme (BlnI) digestion and used specific PCR primers for distal A or B. Primers were designed with Primer3 software.
Hybridization
Southern blots used for D4Z4 sizing and determination of the array homogeneity were successively hybridized with probes p13E-11 (D4F104S1)24 and D4Z4.25 For the determination of the SNPs in the proximal D4Z4 repeat unit, blots were hybridized with probe p13E-11. Southern blots for determination of the distal A/B variation were successively hybridized with probes A and B,8 and blots for chromosomal assignment were hybridized successively with probes p13E-11 and B31.22 All hybridizations, except those with probe D4Z4, were performed in a buffer containing 0.125 M Na2HPO4 (pH 7.2), 10% PEG6000, 0.25 M NaCl, 1 mM EDTA, and 7% SDS for 16–24 hr at 65°C. D4Z4 hybidizations were done in a buffer containing 0.125 M sodium phosphate (pH 7.2), 0.25 M NaCl, 7% SDS, 100 μg/ml of denatured salmon sperm DNA, and 50% deionized formamide. Final washing conditions were 2× SSC-0.1% SDS (p13E-11), 0.1× SSC-0.1% SDS (D4Z4), 1× SSC-0.1% SDS (A), 0.3× SSC-0.1% SDS (B), and 0.3× SSC-0.1% SDS (B31). Southern blots for the analysis of the PvuII and MscI polymorphisms in the most proximal D4Z4 unit were washed in 0.3× SSC-0.1% SDS. All blots were exposed for 16–24 hr to phosphorimager screens and analyzed with the Image Quant software program (Molecular Dynamics).
Statistical Analyses
We used the program Network version 4.5.1.0 to construct a median-joining network of all 4q and 10q haplotypes. For this we used a uniform weight of 10 for all the different polymorphisms that define each distinct haplotype. We used Network Publisher 1.1.0.7 to draw, adjust, and export the final network.
We also used the same dataset to construct a neighbor-joining phylogenetic tree. For this, we used the following subroutines from the Phylip (Phylogeny Inference Package) version 3.68: SeqBoot (in order to create 1000 bootstrap replicates of the original datasets), Pars (to create a single most parsimonious tree for each of the bootstrapped dataset), Neighbor (to make a neighbor joining tree for each of the bootstrapped parsimonious trees, and Consense (to make a single consensus tree). Subsequently, we used Treeview26 to make the neighbor-joining tree with support values of the major branches (Figure S3).
For each population and global region, the allele frequencies, expected heterozygosities, and average number of alleles of the 4q and 10q SSLP loci were calculated with the excel add-in microsatellite toolkit.27 To correlate the observed genetic diversity estimates of each population with geographic distance variation, we used, for each population, the distance from Addis Ababa, Ethiopia via a number of way-points, as previously described by others.28
Results
Genotyping of 4q and 10q D4Z4 Haplotypes
The assignment of the chromosomal origin of the D4Z4 repeat arrays is routinely based on differential sensitivity to the restriction enzymes BlnI and XapI. Standard 4q chromosomes carry D4Z4 repeats with homogeneous arrays of B−X+ D4Z4 units, and standard 10q chromosomes carry D4Z4 repeats with homogeneous arrays of B+X− D4Z4 units. Standard and nonstandard chromosomes 4q and 10q can end with the distal A or B variation (Figure 1).
Previously, we performed detailed genotyping on standard 4qter and 10qter chromosomes (nonstandard chromosomes were not included) in 222 European individuals and showed that all chromosomes could be grouped into nine different 4qter haplotypes and two distinct 10q haplotypes.10 Haplotype differentiation was based on the chromosomal location, the sequence of the SSLP, the sequence variation in D4F104S1 and D4Z4, and the distal variation A and B (Figure 2). In the present study, we concentrate on the question of whether nonstandard chromosomes are the result of numerous and recurrent transfers between chromosomes 4q and 10q, or whether they have arisen from only one or a limited number of founder exchanges. In the latter model, depending on the number of transfers, it should be possible to group nonstandard chromosomes in only a few nonstandard haplotypes, and the intrinsic D4Z4 repeat instability would then be expected to be the underlying cause of the repeat-array copy-number variation within nonstandard chromosomes belonging to the same haplotype. To address this question in more detail, we performed genotyping on standard and nonstandard D4Z4 chromosomes in 444 unrelated European control individuals (Figure 3). The methods used for analyzing the markers in Figure 2 are generally not specific for chromosome 4 or 10. To overcome this problem, we mainly studied family members for whom we could analyze the segregation pattern. This allowed us to determine the exact allele composition of 233 of the 444 controls studied. In addition, the chromosomal origin of the D4Z4 repeat array (standard or nonstandard) was verified in >250 controls by additional PFGE studies with chromosome-4q- and -10q-specific probes.22 Finally, we isolated 145 individual chromosome 4q and 10q D4Z4 fragments that were separated by PFGE for detailed analysis of the SSLP and D4F104S1 sequence. For approximately 100 individuals, we had no DNA available from family members or did not perform chromosome-specific analysis. For these individuals, haplotypes were defined by inference for the most obvious combination of markers on the basis of the most prevalent haplotypes observed in the other approximately 350 controls.
Figure 3.
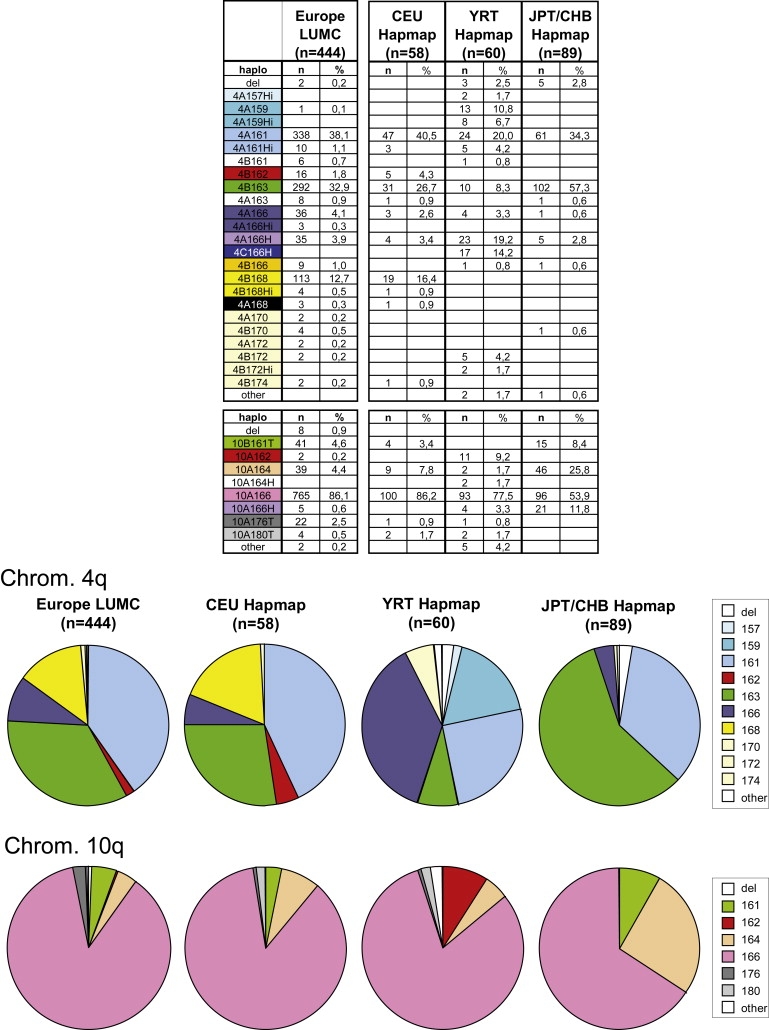
Distribution of 4qter and 10qter Haplotypes in Different Populations
Upper panel: Haplotype distribution of 4q and 10q subtelomeres in 444 European individuals (left column) and three populations from the HAPMAP panels (CEU, YRT, and CHB/JPT; columns on the right) according to SSLP, D4Z4-SNPs, and distal A, B, or C variation. The number of chromosomes per haplotype and frequencies of the different haplotypes are indicated.
Bottom panel: pie charts of the haplotype distributions on chromosomes 4q and 10q in the LUMC-Europe and HAPMAP panels. All chromosomes that have the same SSLP length are indicated by an identical segment color.
The distribution of standard haplotypes is almost identical to that in our previous study; 4A161 and 4B163 are the most prevalent haplotypes on chromosome 4.10 In addition to the previously reported haplotypes, we found evidence for the presence of additional rare haplotypes on chromosomes 4q (4A159, 4A168, 4A170, 4A172, 4B170, 4B172, and 4B174) and 10q (10A162). We found 52 (52/888 = 5.8%) nonstandard chromosomes 4q and 78 (8.7%) nonstandard chromosomes 10q. Consistent with previous reports, most (67/78) nonstandard chromosomes 10q carry homogeneous B−X+ D4Z4 repeat arrays, whereas all nonstandard chromosomes 4q carry hybrid D4Z4 repeat arrays with mixtures of B−X+ and B+X− D4Z4 units.15,29 On the basis of their D4Z4 repeat structure, we could distinguish two hybrid chromosomes; H (hybrid) chromosome ends were detected on chromosomes 4 and 10 and carry a D4Z4 repeat array that begins with B+X− repeat units at the centromeric side, whereas Hi (B−X+ D4Z4 repeat array with some internal B+X− D4Z4 units) chromosome ends were only detected on chromosome 4 and always start with B−X+ D4Z4 units at the proximal end of the repeat (Figure 1C).
Haplotype Distribution in HAPMAP Populations
Previously nonstandard chromosomes have been detected in European and Asian populations,14,16,17 but the prevalence of these chromosomes in the African population was unknown. To gain insight into the distribution of the different standard and nonstandard chromosomes in an African population, we studied the Yoruban (YRI) samples from the HAPMAP project and compared them with the HAPMAP samples from the European (CEU), Japanese (JPT) and Han Chinese (CHB) panels. The inheritance pattern in the father-mother-child trios allowed the chromosomal assignment of the markers studied in the YRI and CEU samples.
As shown in Figure 3, the haplotype distribution among CEU samples (n = 58) is very similar to the distribution among European samples collected by our institute (n = 444). The haplotype distribution in the JPT and CHB panels differs considerably from the European haplotype distributions. Nonstandard hybrid 10q chromosomes are common among the two East Asian population samples, whereas they are almost absent among Europeans. In addition, 4B163 chromosomes are roughly twice as abundant in the JPT and CHB populations (58%) as in the European (32.9% and 26.7% for LUMC and HAPMAP samples, respectively). Not surprisingly, among YRT the haplotype distribution also differs substantially from those of the two east-Asian and the European populations; haplotype 4A159, which is rare in Europeans, is relatively abundant (10.8%) among YRT (Figure 3). In addition, the hybrid 4A166H haplotype is much more abundant among Yorubans (19.2%) than among Europeans (4%), and we discovered many hybrid Hi chromosomes in Yorubans (4A157Hi, 4A159Hi, 4A161Hi, and 4B172Hi). Overall, YRT display an overrepresentation of 4A chromosomes (4A157Hi, 4A159, 4A159Hi, 4A161, 4A161Hi, 4A166, and 4A166H; total 64.9%) compared to 4B chromosomes (4B163, 4B166, 4B172, and 4B172Hi; total 14.9%).
Characterization of Haplotypes
Subsequently, we further characterized and sequenced the SSLP and D4F104S1 regions of individual chromosomes from each haplotype. We sequenced at least one, but often multiple independent chromosomes of almost all haplotypes found in the different populations and composed a haplotype map based on sequence variations (Figure 4). On the basis of sequence similarities in the SSLP and the D4F104S1 region, all haplotypes were categorized in two major groups (major groups 1 [blue] and 2 [orange] in Figure 4). Major group 1 consists mainly of the haplotypes 4A159, 4A161, and 4B163, which are most common in all three HAPMAP populations. Most other standard and nonstandard 4q and 10q haplotypes belong to major group 2.
Figure 4.

Sequence Comparison of Standard and Non-Standard Haplotypes
On the basis of sequence variants in the SSLP, D4F104S1, the proximal D4Z4 unit, the internal D4Z4 unit, and distal sequences, all haplotypes could be grouped into two major haplogroups: major group 1 (blue) and major group 2 (orange). Indicated are the name of the haplotype (column 1), the number of individual chromosomes that were sequenced for each haplotype (column 2), and the specific sequence variants (SNPs, VNTR, and other variables). Column 3 shows the chromosomal location of the D4Z4 haplotype. The top row shows all different possibilities for each sequence variation in the different regions of the D4Z4 locus (SSLP, D4F104S1, proximal D4Z4 unit, internal D4Z4 unit, and distal sequences). The bottom row shows the position of the SNPs in the GenBank sequence AF117653 for all different regions. All SNPs in SSLP and D4F104S1 are depicted as nucleotide changes, and SNPs in D4Z4 are depicted as the presence (1) or absence (2) of a specific restriction site (B = BlnI, P = PvuII, M = MscI, and X = XapI). The last column shows the distal A, B, or C variations indicated by A, AL, B, and C. The distal AL variation has previously been identified and carries a larger (L) partial distal D4Z4 unit but an identical region distal to D4Z4.35
Within the panel of 444 European controls, we sequenced the proximal D4Z4 repeat end of all nonstandard 10q chromosomes with homogeneous B−X+ D4Z4 repeat arrays (n = 67). We discovered that all these nonstandard 10q chromosomes could be grouped into three nonstandard haplotypes (10B161T, 10A176T, and 10A180T; Figure 4). Chromosomes that belong to the largest nonstandard 10q haplotype (10B161T) are all characterized by an SSLP size of 161 bp, the distal variant B, and the P+D4Z4 variant in the proximal D4Z4 unit, which was not detected in 4B161 chromosomes. The remaining nonstandard 10q haplotypes, 10A176T and 10A180T, carry the distal variant A and have an SSLP length of 176 bp and 180 bp, respectively. Further analysis showed that all 10A176T chromosomes carry the unique M−D4Z4 variant in the proximal D4Z4 unit. This combination of a unique SSLP sequence with specific sequence variants, P+D4Z4 or M−D4Z4, implies that all these nonstandard chromosomes originate from only three ancestral translocation events. Similar results were obtained in the HAPMAP panels.
All nonstandard chromosomes with a hybrid repeat structure on chromosomes 4 or 10 were further analyzed in detail for the composition of the repeat array (Figure 5 and Figure S1) and were categorized in either the H or the Hi haplotypes (Figure 1). Most hybrid repeat arrays on chromosomes 4 and 10 belong to the H haplotype, and they are all preceded by an identical SSLP of 166 bp and end with the distal A variant. We name them 4A166H and 10A166H chromosomes. We also discovered a novel chromosome 4q variant with an unknown distal variation that is not recognized by the distal A and B probes, and which we provisionally define as the distal C variation. This distal C variation is only detected in the hybrid chromosomes 4C166H, which are abundant among Yorubans (14.2%). Further analysis showed that 4A166H, 4C166H, and 10A166H chromosomes start with at least one B+X− D4Z4 unit at the proximal site of the repeat, and this is followed by a hybrid repeat structure that consists of B−X+ and B+X− D4Z4 units but also by a novel type of D4Z4 unit that is resistant to both BlnI and XapI (B−X−). These B−X− units were first identified in the most proximal repeat unit of 4A166 chromosomes.10
Figure 5.
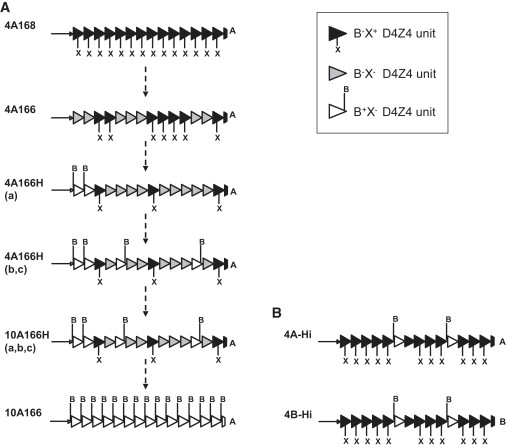
Repeat Array Composition of 4A166 Chromosomes and Typical 4A-H, 10A-H, and 4A-Hi Chromosomes
The composition of the D4Z4 repeat array on the basis of specific SNP combinations for the individual D4Z4 repeat units as determined by Southern blotting with restriction enzymes BlnI (B) and XapI (X). We have identified three different D4Z4 repeat units on the basis of sensitivity to these restriction enzymes: B−X+ (most common in 4q repeat units), B+X- (most common in 10q repeat units), and B-X− D4Z4 units. On the left we depicted the D4Z4 repeat array composition of six different haplotypes (a). The top and bottom haplotypes represent standard chromosomes 4q and 10q with a homogeneous D4Z4 repeat array consisting, respectively, of B−X+ or B+X− D4Z4 units only. A typical 4A166 D4Z4 repeat array consists of arrays of B−X+ units and B−X− units (all units are resistant to BlnI, and most proximal and some internal units are resistant to BlnI and XapI). 4A166H chromosomes have a similar D4Z4 repeat array structure but additionally carry one or more B+X− units at the proximal end of the repeat array, and the D4Z4 repeat array of other 4A166H chromosomes contains one or more B+X− units both at the proximal end and internally. D4Z4 repeat arrays with a similar composition of 4A166H chromosomes have also been identified on chromosome 10: 10A166H. It is conceivable that these four nonstandard hybrid haplotypes represent intermediate haplotypes in the transition from chromosome 4 to chromosome 10 as indicated by the dotted arrows. On the right are depicted two hybrid D4Z4 repeat arrays that are typical for Hi hybrid chromosomes (b). Hi hybrid chromosomes carry a D4Z4 repeat array that mainly consists of B−X+ units and carries some internal B+X− units. Typically, these Hi hybrid chromosomes are identical to different standard 4A and 4qB chromosomes with regard to their proximal and distal sequences except that they carry some B+X− units.
Additional detailed Southern blot analysis of the D4Z4 repeat array structure of 4A166, 4A166H (a, b, and c), and 10A166H (a, b, and c) shows that all repeat arrays are heterogeneous, and each has a specific combination of the three different types of D4Z4 unit (Figure 5 and Figure S1). The sequence similarity of 4A166 and 4A166H chromosomes suggests that they share a common ancestor. Possibly, both for 4A166 chromosomes (loss of XapI site in B−X+ proximal units) and for 4A166Ha chromosomes (gain of BlnI site in proximal B−X− units) the DNA changes in D4Z4 started at the most proximal site of the D4Z4 repeat array. Subsequently, the new D4Z4 units (B−X− on 4A166 and B+X− on 4A166Ha) spread over the entire array (as seen in 4A166Hb and 4A166Hc) by intrachromosomal array rearrangements.
Hi hybrid chromosomes have only been detected in substantial numbers in the African population and are restricted to chromosome 4q. They were found in combination with variable SSLP lengths (157, 159, 161, 166, 168, and 172 bp), corresponding D4F104S1 sequences, and either the distal A or B variation, similar to combinations as detected in the standard 4q haplotypes (4A157Hi, 4A159Hi, 4A161Hi, 4A166Hi, 4B168Hi, and 4B172Hi). As shown in Figure 5B, the D4Z4 repeat array of a typical Hi chromosome consists of B−X+ units, interrupted by a few internal B+X− units.
Evolutionary Network of 4qter and 10qter Haplotypes
Previously, it was suggested that D4Z4 on chromosome 10 evolved from a transfer of the 4q subtelomeric region to 10q.9 Similarly, all 4q and 10q chromosomes that we identified might have evolved from each other, just as 4A166 and 4A166H chromosomes seem to have a common ancestor. To gain more insight into the genetic evolution of all haplotypes, we composed a median-joining network of all distinct haplotype sequences from Figure 4 by using the program Network (Figure 6). In order to reconstruct the evolutionary root of the network, we included the orthologous D4Z4 region in chimpanzees on chromosome 3 (PAN-Ch3); we obtained this sequence from GenBank (accession number AC197422, clone PBT1-134P4).23 As shown in Figure 6, all haplotypes group in six different clusters, (a) and (b) (representing major group 1) and (c), (d), (e), and (f) (representing major group 2). We obtained further independent support for the topology of this median-joining network by analyzing the same dataset by means of PHYLIP. As can be seen in Figure S3, the two unrooted networks are essentially identical, and all major branches are strongly supported by bootstrap values of 95% or higher. The single low support value of 49% is, not surprisingly, positioned at the reticulated (because of SSLP variation) cluster of four haplotypes, including 4A168 (a) and 4A168 (b).
Figure 6.

Evolutionary Network of all 4q and 10q Sequence Haplotypes
A median-joining network based on all distinct 4q and 10q haplotype sequences shown in Figure 4. In order to reconstruct the evolutionary root of the network, we included the orthologous D4Z4 region present on chimpanzee chromosome 3 (chimp). All haplotypes could be grouped in six distinct clusters: (a) and (b) (representing major group 1, left of chimp) and (c), (d), (e), and (f) (representing major group 2, positioned right of chimp). The major difference between haplotypes from clusters (a) and (b) is the distal variation, A and B, respectively. The introduction of the distal B variation between clusters (a) and (b) is indicated with an arrow. Cluster (c) is the only subgroup in major group 2 that contains haplotypes with the distal B variation. Cluster e consists of haplotypes that carry hybrid D4Z4 repeat arrays. This network clearly indicates that only three transfers from 4 to 10 (indicated with a T and a black arrow) and a single transfer of the distal B variation within chromosome 4 (indicated with a gray dotted arrow between clusters b and c) are sufficient to explain all major groups of variants.
Based on the network, the human haplotypes 4A159, 4A161, and 4A161L localize most closely to the chimpanzee haplotype. In combination with the observation that these haplotypes are the most prevalent haplotypes in the YRT HAPMAP samples, this strongly suggests that they represent the oldest human D4Z4 haplotypes. Similarly, the 4A168 haplotype is most probably the oldest haplotype that belongs to major group 2. In cluster (e) all hybrid haplotypes are grouped. According to the network, ancestral D4Z4 repeat units were initially defined by sensitivity to XapI and insensitivity to BlnI (B−X+), and it is possible that B−X− D4Z4 units were first introduced in the 4A166 haplotype by a transversion that disrupted the XapI restriction site. Subsequently, B+X− D4Z4 units likewise evolved by a transition that resulted in a BlnI restriction site giving rise to 4A166H haplotypes (Figure 4A). The African 4C166H haplotype also contains a hybrid D4Z4 repeat array and can also be found in cluster (e). The network shows that the hybrid D4Z4 repeat array on 4A166H was then transferred to 10q, which resulted in the 10A166H haplotypes. Finally, the hybrid D4Z4 repeat array on chromosome 10 homogenized only to B+X− D4Z4 units, which resulted in the most common 10A166 haplotype and gave rise to cluster (f). The 4A168 haplotype most likely also served as translocation ancestor for the nonstandard haplotypes 10A176T and 10A180T (cluster [d]).
It has been shown that 4qA and 4qB chromosomes differ in sequence immediately distal to the D4Z4 repeat.9 To provide further support for a single ancestor for all haplotypes with the distal A variant on chromosomes 4 and 10, we sequenced the breakpoint between D4Z4 and its distal region (BP in Figure 2) in some key haplotypes and analyzed the polymorphisms in this region independently from the evolutionary network reconstruction. As shown in Figure S2a, we observed similar sequences with identical breakpoints in all key 4qA and 10qA haplotypes, except for the 4A161L chromosomes. Interestingly, 4C166H chromosomes showed an identical breakpoint sequence in this region, which provides additional evidence that this haplotype belongs to cluster (e). Finally, a similar breakpoint was also found in the chimp sequence, providing support that all 4qA, 4qC, and 10qA chromosomes are derived from a single ancestral chromosome.
The network contains two clusters, (b) and (c), that carry the distal B variant. This might imply that this distal B variant was transferred twice from an unknown chromosome end (not 4q or 10q, but probably 4p as suggested by van Geel and colleagues9) to chromosome 4q. Alternatively, one of the two clusters could have donated the distal B variant to the other B cluster by a transfer between homologous chromosomes 4q. In the latter model, a single transfer of a distal B end to 4qA is responsible for all different 4qB haplotypes. To gain evidence for this scenario, we sequenced the distal D4Z4 region from several 4B163, 10B161T, and 4B168 chromosomes and show that they all have the same breakpoint between the most distal D4Z4 unit and the distal B sequence (Figure S2b). This suggests that they are all derived from a single transfer event at the distal end of the D4Z4 repeat. Furthermore, the two 10B161T haplotypes in cluster (b) seem to have evolved from a transfer of 4B161 to chromosome 10.
Worldwide Haplotype Distribution
To further estimate the global distribution of 4q and 10q haplotypes, we analyzed the HGDP-CEPH samples supplemented by samples from the YCC panel. Together, these two panels have been used in many global genetic-diversity studies. Individuals originate from all seven major global regions: Africa, Middle East, Europe, South and Central Asia, East Asia, Oceania, and America. Because the amount and quality of the DNA in both panels does not allow PFGE or Southern blot analysis for detailed 4q and 10q studies, these samples were only analyzed by PCR for the SSLP marker on both chromosomes (Figure 7.On the basis of the haplotype analysis of samples in Figure 3, most SSLP sizes (alleles) can mainly be attributed to a single haplotype, at least in those populations studied in detail. SSLP sizes 161 and 163 can be assigned to three or two haplotypes, respectively, but 4A161 and 4B163 are most common. Therefore, we established the SSLP distribution in the seven world populations (Figure 7) and, when necessary, inferred their chromosomal origin from the detailed studies in LUMC and HAPMAP samples. The similarities in the 4q and 10q SSLP distributions between Figures 3 and 7 justifiy this method of inference.
Figure 7.
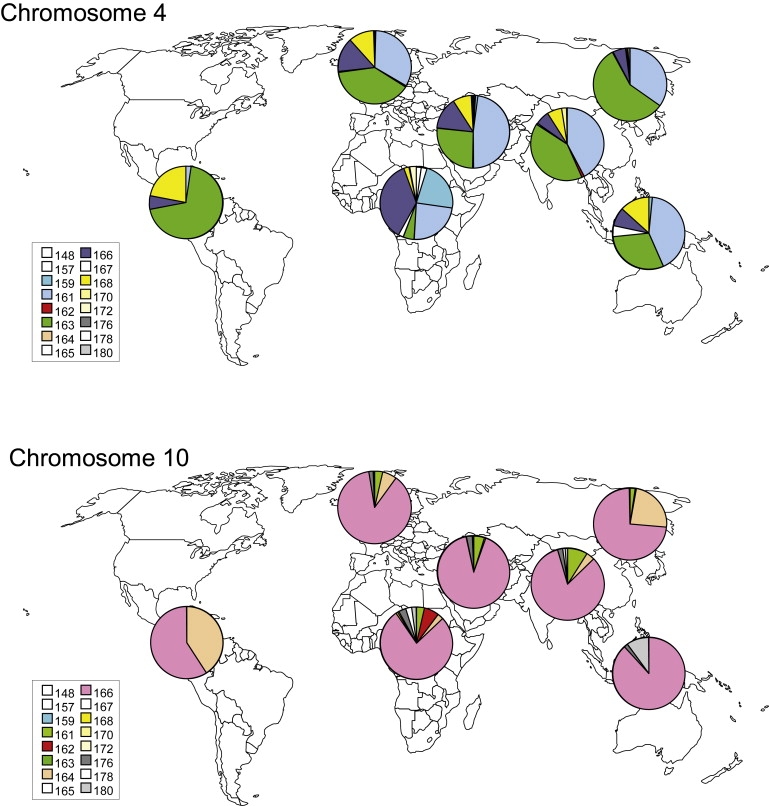
Global Distributions of the 4q and 10q SSLP Alleles
Pie charts showing the allele frequency of the SSLP locus on 4q and 10q chromosomes in seven major global regions (Africa [n = 138], Middle East [n = 163], Europe [n = 175], South and Central Asia [n = 205], East Asia [n = 241], Oceania [n = 30], and America [n = 77]). The color key is indicated on the left. Each population has a unique distribution of 4q and 10q alleles; the oldest (African) populations show the highest variation in the number of alleles, and the youngest (American) populations show the lowest variation.
As shown in Figure 7A, the African region shows the largest allele variation (similar to that of the HAPMAP YRT panel), whereas the allele distribution in other populations is less diverse and represents a good reflection of the two major bottlenecks that have occurred during human evolution. For example, in the East Asia population we identified predominantly SSLP alleles of 161 bp (35%) or 163 bp (58%), which most probably represent haplotypes 4A161 and 4B163 on the basis of a detailed analysis of the Japanese and Han Chinese controls from the HAPMAP panel (Figure 3). In accordance with the occurrence of a second bottleneck during the migration of modern humans into the Americas, Native Americans (n = 77) show an almost complete absence of the 161 bp alleles. We observed that the proportion of 163, 168, and 172 bp alleles representing 4B haplotypes in Figure 3 increased after the migration out of Africa from 10% to 91% in Native Americans (Figure 6A).
Similar to the 4q haplotypes, SSLP sizes that represent all standard and nonstandard 10q haplotypes seem to be present in Africa and therefore are also likely to have arisen before the migration of modern humans out of Africa (Figure 6B). The steady increase in the proportion of 164 bp alleles (10A164 chromosomes) from 3% in Africa to 6% in Europe, 24% in East Asia, and 41% in America provides another example of how the migration of modern humans can be genetically traced with the SSLP marker.
Nonstandard haplotypes 10B161T, 10A176T, and 10A180T are well recognized by SSLP-PCR because of their unique SSLP size on 10q (161, 176, and 180 bp). These alleles seem to be present in all world populations, and their frequency in European and Asian populations is comparable to previous observations.14,16,17 In conclusion, it seems that all standard and nonstandard haplotypes had already evolved before modern humans started their migration out of Africa.
If we correlate the genetic variation of the 4q and 10q SSLP among all globally dispersed populations with their respective geographical distance from east Africa, we see a remarkable difference between 4q and 10q (Figure S4). For the 4q SSLP, we see the classical picture of a decrease in unbiased heterozygosity and average number of alleles as populations become more removed from Africa. For the 10q SSLP, we observe a more complex picture: the average number of alleles decreases with geographical distance, but the average heterozygosity does not.
Discussion
Here we present a study of the recent evolution of two highly homologous subtelomeres at an unprecedented level of detail. Both subtelomeres, 4qter and 10qter, share a region of, on average, 200 kb of sequence homology immediately adjacent to the telomere and include the highly polymorphic macrosatellite repeat D4Z4. Different haplotypes have previously been identified on chromosomes 4 and 10 on the basis of sequence variations proximal, within, and distal to D4Z4.10 These haplotypes seem to have arisen through intrachromosomal sequence changes. On the other hand, subtelomeres are considered dynamic domains of the genome, and the presence of many nonstandard chromosomes were believed to be the result of ongoing rearrangements between chromosomes 4q and 10q.14,16 This apparent conflict triggered us to study the sequence and recent evolution of the human 4q and 10q subtelomeres in more detail.
Recent studies in Old and New World monkeys substantiated earlier observations that the 10q subtelomere originates from an ancestral duplication event from 4qter to the subtelomere of 10q.30 Previously, it was suggested that the distal B variant resulted from an ancestral duplication of 4p to 4q, making the 4qA chromosome ancestral to the 4qB chromosome.9 Our evolutionary network provides additional evidence that the 4qA cluster (a) represents the oldest group of haplotypes with highest sequence similarity to the Chimpanzee sequence. From these haplotypes, it is possible to identify distinct inter- or intrachromosomal rearrangement events that result in all currently known subtelomeric haplotypes of chromosomes 4 and 10. According to the network, on chromosome 4 the distal B variant was first introduced on 4A161, resulting in cluster (b). At a relatively large evolutionary distance from cluster (a), a new 4qA haplotype evolved. This 4A168 haplotype displays many sequence differences in comparison to haplotypes from major group 1 and is probably the ancestor of all other haplotypes that belong to major group 2. Haplotype 4A168 gained the distal B region from cluster (b), creating cluster (c), and although we cannot rule out a low level of meiotic recombination, a more likely scenario is that haplotype 4A168 was involved in a complex reorganization of the D4Z4 sequence by first erasing the XapI restriction site (evolution of B−X− D4Z4 units in 4A166 haplotypes). Subsequently the BlnI restriction site was introduced (evolution of B+X− D4Z4 units in 4A166H haplotypes initially at the most proximal end of the array, afterward at internal D4Z4 units). The hybrid 4q haplotypes, were then transferred to chromosome 10q (10A166H haplotypes), where they served as the ancestors for the standard 10q haplotypes (10A166 and 10A164) (Figure 5). Our study also shows that all chromosome 10q haplotypes carrying homogeneous B−X+ D4Z4 repeat arrays (10A176T, 10A180T, and 10B161T) have evolved from only two major transfer events.
Hi hybrid chromosomes cannot be explained by these ancient translocation events. These haplotypes are mostly found in the African population and are rare outside Africa. The D4Z4 repeat array of these hybrid chromosomes 4 consists of an array of B−X+ D4Z4 units that is interrupted by a few B+X− D4Z4 units. Possibly, B+X− D4Z4 units were transferred from 4A166H (b) and (c) chromosomes onto standard chromosomes and thus gave rise to Hi chromosomes. These transfers might have been provoked by the high frequency of 4A166H chromosomes in the African population.
Previously, the high incidence of nonstandard arrays was attributed to the increased interchromosomal exchange rate between the subtelomeres of both chromosomes, as has been observed for other chromosome ends.14,16 Subtelomeres are shaped by a two-step process. In the first step, a subtelomeric DNA segment is transferred to a nonhomologous chromosome end. Once these transferred DNA segments exist on nonhomologous chromosomes, the probability that they will be subject to subsequent reciprocal or nonreciprocal sequence transfers increases. On the basis of extensive sequence analysis of a 60 kb region that is shared by seven subtelomeres, it was concluded that subtelomeric blocks do not evolve independently but are instead subject to continued interchromosomal interactions.1 Our study of these specific sequence domains of chromosomes 4q and 10q provides detailed insight in the frequency of sequence exchanges between the different chromosome ends during human evolution: each 4q and 10q chromosome end evolves relatively independently from each other, and there is strong linkage disequilibrium between sequences immediately flanking the D4Z4 repeat.
Our current study supports a model in which only four major rearrangements occurred in the subtelomeric D4Z4 locus during human evolution: the B subtelomere was introduced once on chromosome 4A, and the 4q subtelomere was transferred three times to chromosome 10. Although other histories are possible, this represents the most likely evolutionary history on the basis of the median-joining network and the neighbor-joining tree. Most probably, the 4qter-to-10qter transfers from which the 10B161T chromosomes evolved and those from which 10A176T and 10A180T chromosomes evolved were evoked after the initial transfer of 4A166H to chromosome 10q that resulted in 10A166H. This limited rate of exchanges between 4qter and 10qter, rather than a frequent occurrence of repeat exchanges between chromosomes 4q and 10q to explain nonstandard chromosomes, fits well with other observations. Gonosomal mosaicism for D4Z4 repeat-array instability is common.31 When studying mitotic D4Z4 repeat-array instability in detail, the preferred mechanism was consistent with a synthesis-dependent strand-annealing model between sister chromatids rather than homologous chromosomes.18 Additionally, clear population differences can be observed in the frequency of nonstandard haplotypes (high frequency of 4q hybrids in YRT and high frequency of 10A176T in Oceania). Finally, we and others have previously shown a large difference in the average D4Z4 copy number between chromosomes 4 and 10q15,29 and, more recently, among 4A161, 4B163, and 10A166 chromosomes.10 These observations argue against a model of frequent interchromosomal exchanges and support the idea that transfers between 4qter and 10qter and between different haplotypes are infrequent.
Therefore, in this study we unequivocally demonstrate that nonstandard haplotypes have arisen by a few subtelomeric exchanges that already occurred between chromosomes 4q and 10q before the migration of modern humans out of Africa. In addition, this study shows the evolution of the D4Z4 region on chromosome 10q from chromosome 4q via intermediate hybrid haplotypes. This low rate of sequence exchanges can explain why each chromosome end becomes genetically unique and might provide an explanation as to why FSHD is uniquely associated with D4Z4 contractions on 4A161. We postulate that sequence variants specific to the 4A161 haplotype are essential for the development of FSHD. It is interesting to note that this permissive haplotype belongs to the oldest haplogroup and that nonpermissive haplotypes (4B163 and 10A166) are typically younger, suggesting a selection bias toward haplotypes that are nonpermissive for FSHD.
On the basis of the global distribution of alleles of the 4q and 10q SSLP loci, we can further refine the evolutionary model that leads to the present-day distribution and diversity of the 4q and 10q distal regions. It has been demonstrated many times that the genetic variation among globally dispersed human populations is not randomly distributed but follows a remarkably constant decrease as a function of the distance from east Africa.28 Human populations dispersed out of Africa via a complex migration process that included a number of distinct population bottlenecks. When populations undergo a bottleneck, genetic variation rarely remains unaltered.32 In many cases, the first sign of a genetic bottleneck is the rapid reduction of the number of distinct alleles in a population, followed by a reduction of heterozygosity.33 Our observations of a reduction in the average number of alleles and heterozygosity of the 4q SSLP is perfectly in line with other global STR and SNP studies in the same HGDP samples we studied.34 The slightly different results for the 10q SSLP suggests that population bottlenecks only affected global 10q variation in the number of alleles, not in heterozygosity. One plausible explanation is that the 10q variation was still very low at the time of the first out-of-Africa migration and simply could not be influenced further. If this is true, it would mean that most translocation events from 4q to 10q occurred much later in human evolution and relatively shortly before modern humans left Africa for the first time. This corresponds to their relative positions in the median-joining network of all 4q and 10q haplotypes.
This study represents the first in-depth analysis of natural subtelomeric variation in all world populations and provides mechanistic insight into the recent human evolution of the subtelomeric domains of 4q and 10q at an unprecedented level of detail. The evolutionary network and global distribution of all haplotypes it provides will be instrumental to a better understanding of the composition, evolution, and function of these and other chromosome domains that lie adjacent to telomeres.
Acknowledgments
The authors thank Barbara Trask for critical reading and useful comments on the manuscript. This study was financially supported by institutional funds from the Leiden University Medical Center, the Shaw Family Foundation; the Fields Center for FSHD and Neuromuscular Research; the Netherlands Organization for Scientific Research (NWO 917.56.338); a Breakthrough Project Grant from the Netherlands Genomics Initiative (NWO 93.51.8001) to R.J.L.F.L.; and a Muscular Dystrophy Association and Marjorie Bronfman Fellowship grant from the FSH Society to R.J.L.F.L.
Supplemental Data
Web Resources
The URLs for data presented herein are as follows:
Coriell Institute for Medical Research, www.coriell.org
Detailed information on genotyping protocols: http://www.urmc.rochester.edu/fields-center/
GenBank, http://www.ncbi.nlm.nih.gov/Genbank/ (for accession numbers AF117653, AF017466, AP006328, AY028079, AL732375, AL845259, and AC197422)
HGDP-CEPH Human Genome Diversity Cell Line Panel, http://www.cephb.fr/en/hgdp/diversity.php
The International HapMap Project, http://www.hapmap.org/downloads/index.html.en
Network version 4.5.1.0, http://www.fluxus-engineering.com/sharenet.htm
Online Mendelian Inheritance in Man, http://www.ncbi.nlm.nih.gov/Omim/
Phylip version 3.68, http://evolution.genetics.washington.edu/phylip.html
Primer3, http://frodo.wi.mit.edu/cgi-bin/primer3/primer3_www.cgi.
Treeview, http://taxonomy.zoology.gla.ac.uk/rod/treeview.html
Y Chromosome Consortium (YCC), http://ycc.biosci.arizona.edu/
Accession Numbers
All sequences reported in this paper (SSLP, D4F104S1, respectively) have been deposited in GenBank under accession numbers GU480773, GU550600 (4A159); GU480774, GU550601 (4A161); GU480775, GU550602 (4A161L); GU480776, GU550603 (4B163); GU480777, GU550604 (10B161Ta); GU480778, GU550605 (10B161Tb); GU480803, GU550606 (4B161); GU480791, GU550607 (4B162a); GU480792, GU550615 (4B162b); GU480793, GU550608 (4B168a); GU480794, GU550616 (4B168b); GU480795, GU550609 (4B170); GU480796, GU550610 (4B172); GU480797, GU550611 (4A168a); GU480798, GU550612 (4A168b); GU480779, GU550613 (10A176T); GU480780, GU550614 (10A180T); GU480786, GU550617 (4A166); GU480787, GU550618 (4A166Ha); GU480788, GU550629 (4A166Hb); GU480789, GU550620 (4A166Hc); GU480790, GU550619 (4C166H); GU480781, GU550623 (10A166Ha); GU480782, GU550621 (10A166Hb); GU480783, GU550627 (10A166Hc); GU480802, GU550624 (10A166a); GU480784, GU550622 (10A166b); GU480785, GU550628 (10A166c); GU480799, GU550625 (10A164a); GU480800, GU550630 (10A164b); and GU480801, GU550626 (10A162).
References
- 1.Linardopoulou E.V., Williams E.M., Fan Y., Friedman C., Young J.M., Trask B.J. Human subtelomeres are hot spots of interchromosomal recombination and segmental duplication. Nature. 2005;437:94–100. doi: 10.1038/nature04029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Mefford H.C., Trask B.J. The complex structure and dynamic evolution of human subtelomeres. Nat. Rev. Genet. 2002;3:91–102. doi: 10.1038/nrg727. [DOI] [PubMed] [Google Scholar]
- 3.Ambrosini A., Paul S., Hu S., Riethman H. Human subtelomeric duplicon structure and organization. Genome Biol. 2007;8:R151. doi: 10.1186/gb-2007-8-7-r151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bailey J.A., Eichler E.E. Primate segmental duplications: Crucibles of evolution, diversity and disease. Nat. Rev. Genet. 2006;7:552–564. doi: 10.1038/nrg1895. [DOI] [PubMed] [Google Scholar]
- 5.Mefford H.C., Linardopoulou E., Coil D., van den Engh G., Trask B.J. Comparative sequencing of a multicopy subtelomeric region containing olfactory receptor genes reveals multiple interactions between non-homologous chromosomes. Hum. Mol. Genet. 2001;10:2363–2372. doi: 10.1093/hmg/10.21.2363. [DOI] [PubMed] [Google Scholar]
- 6.Wilkie A.O., Higgs D.R., Rack K.A., Buckle V.J., Spurr N.K., Fischel-Ghodsian N., Ceccherini I., Brown W.R., Harris P.C. Stable length polymorphism of up to 260 kb at the tip of the short arm of human chromosome 16. Cell. 1991;64:595–606. doi: 10.1016/0092-8674(91)90243-r. [DOI] [PubMed] [Google Scholar]
- 7.de Greef J.C., Frants R.R., van der Maarel S.M. Epigenetic mechanisms of facioscapulohumeral muscular dystrophy. Mutat. Res. 2008;647:94–102. doi: 10.1016/j.mrfmmm.2008.07.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lemmers R.J., de Kievit P., Sandkuijl L., Padberg G.W., van Ommen G.J., Frants R.R., van der Maarel S.M. Facioscapulohumeral muscular dystrophy is uniquely associated with one of the two variants of the 4q subtelomere. Nat. Genet. 2002;32:235–236. doi: 10.1038/ng999. [DOI] [PubMed] [Google Scholar]
- 9.van Geel M., Dickson M.C., Beck A.F., Bolland D.J., Frants R.R., van der Maarel S.M., de Jong P.J., Hewitt J.E. Genomic analysis of human chromosome 10q and 4q telomeres suggests a common origin. Genomics. 2002;79:210–217. doi: 10.1006/geno.2002.6690. [DOI] [PubMed] [Google Scholar]
- 10.Lemmers R.J., Wohlgemuth M., van der Gaag K.J., van der Vliet P.J., van Teijlingen C.M., de Knijff P., Padberg G.W., Frants R.R., van der Maarel S.M. Specific sequence variations within the 4q35 region are associated with facioscapulohumeral muscular dystrophy. Am. J. Hum. Genet. 2007;81:884–894. doi: 10.1086/521986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bakker E., Wijmenga C., Vossen R.H., Padberg G.W., Hewitt J., van der Wielen M., Rasmussen K., Frants R.R. The FSHD-linked locus D4F104S1 (p13E-11) on 4q35 has a homologue on 10qter. Muscle Nerve. 1995;2:39–44. [PubMed] [Google Scholar]
- 12.Deidda G., Cacurri S., Piazzo N., Felicetti L. Direct detection of 4q35 rearrangements implicated in facioscapulohumeral muscular dystrophy (FSHD) J. Med. Genet. 1996;33:361–365. doi: 10.1136/jmg.33.5.361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lemmers R.J.L., de Kievit P., van Geel M., van der Wielen M.J., Bakker E., Padberg G.W., Frants R.R., van der Maarel S.M. Complete allele information in the diagnosis of facioscapulohumeral muscular dystrophy by triple DNA analysis. Ann. Neurol. 2001;50:816–819. doi: 10.1002/ana.10057. [DOI] [PubMed] [Google Scholar]
- 14.van Deutekom J.C., Bakker E., Lemmers R.J., van der Wielen M.J., Bik E., Hofker M.H., Padberg G.W., Frants R.R. Evidence for subtelomeric exchange of 3.3 kb tandemly repeated units between chromosomes 4q35 and 10q26: implications for genetic counselling and etiology of FSHD1. Hum. Mol. Genet. 1996;5:1997–2003. doi: 10.1093/hmg/5.12.1997. [DOI] [PubMed] [Google Scholar]
- 15.van Overveld P.G., Lemmers R.J., Deidda G., Sandkuijl L., Padberg G.W., Frants R.R., van der Maarel S.M. Interchromosomal repeat array interactions between chromosomes 4 and 10: A model for subtelomeric plasticity. Hum. Mol. Genet. 2000;9:2879–2884. doi: 10.1093/hmg/9.19.2879. [DOI] [PubMed] [Google Scholar]
- 16.Matsumura T., Goto K., Yamanaka G., Lee J., Zhang C., Hayashi Y.K., Arahata K. Chromosome 4q;10q translocations; Comparison with different ethnic populations and FSHD patients. BMC Neurol. 2002;2:7. doi: 10.1186/1471-2377-2-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wu Z.Y., Wang Z.Q., Murong S.X., Wang N. FSHD in Chinese population: Characteristics of translocation and genotype-phenotype correlation. Neurology. 2004;63:581–583. doi: 10.1212/01.wnl.0000133210.93075.81. [DOI] [PubMed] [Google Scholar]
- 18.Lemmers R.J., van Overveld P.G., Sandkuijl L.A., Vrieling H., Padberg G.W., Frants R.R., van der Maarel S.M. Mechanism and timing of mitotic rearrangements in the subtelomeric D4Z4 repeat involved in facioscapulohumeral muscular dystrophy. Am. J. Hum. Genet. 2004;75:44–53. doi: 10.1086/422175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.International HapMap Consortium The International HapMap Project. Nature. 2003;426:789–796. doi: 10.1038/nature02168. [DOI] [PubMed] [Google Scholar]
- 20.Cann H.M., de Toma C., Cazes L., Legrand M.F., Morel V., Piouffre L., Bodmer J., Bodmer W.F., Bonne-Tamir B., Cambon-Thomsen A. A human genome diversity cell line panel. Science. 2002;296:261–262. doi: 10.1126/science.296.5566.261b. [DOI] [PubMed] [Google Scholar]
- 21.Y Chromosome Consortium A nomenclature system for the tree of human Y-chromosomal binary haplogroups. Genome Res. 2002;12:339–348. doi: 10.1101/gr.217602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Buzhov B.T., Lemmers R.J., Tournev I., Dikova C., Kremensky I., Petrova J., Frants R.R., van der Maarel S.M. Genetic confirmation of facioscapulohumeral muscular dystrophy in a case with complex D4Z4 rearrangments. Hum. Genet. 2005;116:262–266. doi: 10.1007/s00439-004-1237-0. [DOI] [PubMed] [Google Scholar]
- 23.Rudd M.K., Endicott R.M., Friedman C., Walker M., Young J.M., Osoegawa K., de Jong P.J., Green E.D., Trask B.J. Comparative sequence analysis of primate subtelomeres originating from a chromosome fission event. Genome Res. 2009;19:33–41. doi: 10.1101/gr.083170.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wijmenga C., Hewitt J.E., Sandkuijl L.A., Clark L.N., Wright T.J., Dauwerse H.G., Gruter A.M., Hofker M.H., Moerer P., Williamson R. Chromosome 4q DNA rearrangements associated with facioscapulohumeral muscular dystrophy. Nat. Genet. 1992;2:26–30. doi: 10.1038/ng0992-26. [DOI] [PubMed] [Google Scholar]
- 25.Ehrlich M., Jackson K., Tsumagari K., Camano P., Lemmers R.J. Hybridization analysis of D4Z4 repeat arrays linked to FSHD. Chromosoma. 2007;116:107–116. doi: 10.1007/s00412-006-0080-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Page R.D. TreeView: An application to display phylogenetic trees on personal computers. Comput. Appl. Biosci. 1996;12:357–358. doi: 10.1093/bioinformatics/12.4.357. [DOI] [PubMed] [Google Scholar]
- 27.Park S. Trinity College, University of Dublin; Dublin: 2001. Trypanotolerance in West African cattle and the population genetic effects of selection. [Google Scholar]
- 28.Handley L.J., Manica A., Goudet J., Balloux F. Going the distance: Hhuman population genetics in a clinal world. Trends Genet. 2007;23:432–439. doi: 10.1016/j.tig.2007.07.002. [DOI] [PubMed] [Google Scholar]
- 29.Rossi M., Ricci E., Colantoni L., Galluzzi G., Frusciante R., Tonali P.A., Felicetti L. The Facioscapulohumeral muscular dystrophy region on 4qter and the homologous locus on 10qter evolved independently under different evolutionary pressure. BMC Med. Genet. 2007;8:8. doi: 10.1186/1471-2350-8-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Clapp J., Mitchell L.M., Bolland D.J., Fantes J., Corcoran A.E., Scotting P.J., Armour J.A.L., Hewitt J.E. Evolutionary conservation of a coding function for D4Z4, the tandem DNA repeat mutated in facioscapulohumeral muscular dystrophy. Am. J. Hum. Genet. 2007;81:264–279. doi: 10.1086/519311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.van der Maarel S.M., Deidda G., Lemmers R.J., van Overveld P.G., van der Wielen M., Hewitt J.E., Sandkuijl L., Bakker B., van Ommen G.J., Padberg G.W. De Novo Facioscapulohumeral Muscular Dystrophy: Frequent Somatic Mosaicism, Sex-Dependent Phenotype, and the Role of Mitotic Transchromosomal Repeat Interaction between Chromosomes 4 and 10. Am. J. Hum. Genet. 2000;66:26–35. doi: 10.1086/302730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nei M. Bottlenecks, genetic polymorphism and speciation. Genetics. 2005;170:1–4. doi: 10.1093/genetics/170.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Luikart G., Cornuet J. Empirical evaluation of a test for identifying recently bottlenecked populations from allele frequency data. Conservation Biology. 1998;12:228–237. [Google Scholar]
- 34.Friedlaender J.S., Friedlaender F.R., Reed F.A., Kidd K.K., Kidd J.R., Chambers G.K., Lea R.A., Loo J.H., Koki G., Hodgson J.A. The genetic structure of Pacific Islanders. PLoS Genet. 2008;4:e19. doi: 10.1371/journal.pgen.0040019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.van Deutekom J.C., Wijmenga C., van Tienhoven E.A., Gruter A.M., Hewitt J.E., Padberg G.W., van Ommen G.J., Hofker M.H., Frants R.R. FSHD associated DNA rearrangements are due to deletions of integral copies of a 3.2 kb tandemly repeated unit. Hum. Mol. Genet. 1993;2:2037–2042. doi: 10.1093/hmg/2.12.2037. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


