Abstract
Obscurin and obscurin-associated kinase are two products of the obscurin transcriptional unit that encodes a recently identified giant muscle-specific protein obscurin. In this study, we characterized the developmental expression and cellular localization of obscurin and obscurin-associated kinase in cardiac muscle cells. We cloned murine obscurin-associated kinase and found that it is abundantly expressed in the heart as two isotypes encoded by 2.2 and 4.9 kb sequences. The 2.2 kb isotype of the kinase was more prominently expressed than the 4.9 kb isotype. Both obscurin and the kinase-like domains were progressively upregulated since the early stages of cardiac development. Obscurin-associated kinase was expressed at higher levels than obscurin at early stages of cardiomyogenesis. Increasing intensity of obscurin expression in the developing heart positively correlated with progressive cell differentiation and was higher in the ventricles compared to the atria. These data were supported by the results of experiments with primary cardiac cell cultures. Obscurin localization changed from a weakly immunopositive diffuse pattern in poorly differentiated cells to an intensely immunolabeled cross-striated distribution at the level of mid-A-bands and Z-disks during the assembly of the myofibrillar contractile apparatus. In dividing myocytes, unlike the interphase cells, obscurin translocated from disassembling myofibrils into a diffuse granulated pattern segregated separately from α-actinini-mmunopositive aggregates. Obscurin-associated kinase was localized mainly to cell nuclei with increasing incorporation into the Z-disks during differentiation. Our results suggest that these two novel proteins are involved in the progression of cardiac myogenesis during the transition to advanced stages of heart development.
Keywords: muscle, myofibrils, heart, obscurin, development, mitosis, cardiac myocytes
Identification and characterization of a several new sarcomeric proteins has significantly expanded our understanding of the structural organization and functional regulation of the contractile system in muscle cells [reviews—Sanger et al., 2005; Ferrari et al., 2006]. A novel giant protein named obscurin (~820 kDa) has been recently found in the contractile apparatus of cardiac and skeletal muscle. Obscurin contains 68 immunoglobulin domains, two fibronectin 3 domains, one calcium/calmodulin binding domain and a RhoGEF domain, all of which can potentially be involved in structural and functional control in muscle cells [Bang et al., 2001; Young et al., 2001; Russell et al., 2002]. We have found that the DNA sequence encoding obscurin is topographically very closely associated with the sequences encoding myosin light chain kinase (MLCK)-like motifs, and that these sequences can be transcribed as a single transcriptional unit [Russell et al., 2002]. Similar data concerning obscurin isoform generation by alternative splicing have been reported recently by Fukuzawa et al. [2005]. Interestingly, MLCK-like protein kinase domains have been observed in several phylogenetic homologs of obscurin in the organisms that belong to different taxonomic groups including Caenorhabditis elegans and Drosophila melanogaster [Small et al., 2004; Sutter et al., 2004]. The structural and functional characterization of the protein products encoded by these domains in different species is important for better understanding of the physiological role of obscurin in striated muscle cells.
The MLCK family plays an essential role in regulation of actin/myosin organization, sarcomere assembly and cytoskeletal dynamics [Aoki et al., 2000; Kamm and Stull, 2001]. Several large muscle-specific proteins such as titin, projectin, twitchin, and stretchin contain protein kinase domains more or less structurally similar to MLCK [for discussion see Sutter et al., 2004]. In addition to their serine/threonine kinase motifs, these proteins contain multiple immunoglobulin (Ig) and fibronectin (Fn) domains that mediate interactions with myosin and other sarcomeric and cytoskeletal components. All these proteins possess both structural and signaling functions. For instance, titin, the largest known member of the family, acts as a molecular ruler, organizing the sarcomeric structure through interprotein interactions. It was found that its kinase domain, known to be required for sarcomere assembly [Mayans et al., 1998], carries some resemblance to MLCK [Seberstyen et al., 1996] and is involved in the control of muscle gene expression and protein turnover [Lange et al., 2005].
Obscurin-associated kinase is the product of the same transcriptional unit that encodes obscurin, and its expression depends on alternative translation initiation sites. In the mammalian heart, the “large” obscurin isoform does not routinely contain the kinase domain, and the kinase-containing sequences can be expressed as a separate entity [Russell et al., 2002]. Therefore, obscurin and obscurin-associated kinase are separate transcripts from a “split” gene, not unlike the giant Drosophila MLCK, stretchin [Champagne et al., 2000]. Earlier we reported that kinase-containing isotypes of obscurin can be expressed autonomously in the overloaded mammalian heart in vivo [Borisov et al., 2003].
The expression of these genes during muscle development and cellular localization of their products has not been comparatively characterized. Assembly of myofibrils and cytoskeletal remodeling are integral components of myocardial differentiation and adaptive responses. For this reason, understanding of the developmental patterns allows us to collect more information concerning the functional role of these proteins. In this study, we examined the developmental expression and cellular localization of obscurin and obscurin-associated kinase.
MATERIALS AND METHODS
Cloning of the Murine Obscurin-Associated Kinase Isoforms
cDNA sequence from the human obscurin-MLCK were compared to the high throughput genome sequence database at the National Center for Biotechnology Information using the BLASTN [Altschul et al., 1990] sequence homology search. The search identified significant homology between the cDNA sequence and sequence from three human chromosome 1q42 BAC clones two mouse chromosome 11 BAC clones (RP23-344L20 and RP23-441I8). The aligned mouse sequence was assembled to create a draft murine obscurin-MLCK cDNA sequence. Three overlapping sets of PCR primers were selected using the Primer Select subroutine of the Lasergene sequence analysis program (5′F: gggcgccggtaccacaggtcactattg, 5′R: ggcgccactagcttcccctcgtag; midF: tggcccggcacctacgag, midR: ggtaccaggcctgccttctttctg; 3′F: gggatccaaccgcacggtggggaaggttacg, 3′R: gtgggcaggaagcgcaagtggtc). The primers were used to amplify the corresponding cDNA sequences from a mouse heart cDNA library (Clontech, Inc.). The PCR products were subcloned and completely sequenced on both strands. This sequence was assembled to create the final murine obscurin-MLCK isoform cDNA sequence. Sequence from the 3′ end of the murine obscurin sequence was identified by BLASTN sequence homology search and used to generate riboprobes for RNA in situ hybridization.
Northern Blot Analysis
Probes were generated from the 5′ and 3′ serine-threonine kinase encoding regions of obscurin MLCK and the 3′UT region of obscurin using random-prime labeling as directed by the manufacturer (Life Technologies, Inc.). Hundred nanograms of each probe was labeled with 5 μCi αP32-dCTP using previously described methods [Russell et al., 2002]. Between hybridizations, the blots were stripped by brief immersion in 1% SDS/0.01% SSC at 100°C.
Reverse Transcription-Quantitative Polymerase Chain Reaction Analysis
Embryos were isolated at 10, 12, 12.5, 14, and 17 dpc and hearts were extracted. Total RNA was isolated from whole heart (10 and 12.5 dpc), left ventricle (12, 14, and 17 dpc), and right ventricle (12, 14, and 17 dpc) using the TRIZOL reagent (Invitrogen). Poly A RNA was prepared from total RNA using Poly A Tract kit (Promega). Primers were designed to amplify terminal kinase, internal kinase, and Rho domains [Heid et al., 1996]. The conditions for RT-PCR were as follows: 30 min at 60°C (reverse transcription), 10 min at 95°C (reverse transcriptase inactivation and AmpliTaq Gold activation), and then 15 s at 95°C, 1 min at 60°C, for 40 cycles (PCR amplification).
Whole Mount In Situ RNA Hybridization
After plasmid linearization with SacI and SacII, respectively, digoxigenin-UTP labeled sense and antisense riboprobes were transcribed from inserts encoding for part of the terminal serine-threonine kinase of obscurin-MLCK (545 bp) and the 3′ untranslated region of obscurin (497 bp; Fig. 1). Hearts were extracted from 12.0, 14.0, and 17.0 dpc mouse embryos. The embryos were fixed overnight at 4°C with 4% paraformaldehyde in phosphate-buffered saline (PBS, pH 7.4). In our experiments we used Dulbecco's modification of phosphate buffered saline (KCl 200 mg/L, KH2PO4 200 mg/L, NaCl 8,000 mg/l, and Na2HPO4 1,150 mg/l). The embryos and hearts were then dehydrated in a graded series of 25, 50, 75, and 100% methanol in PBT (PBS with 0.1% Triton X-100) and stored at −20°C. Following rehydration in graded methanol, embryos, and hearts were processed as described by [Wilkinson, 1992]. The hearts incubated without the specific probe served as a negative control. Omission of the specific probe served as negative control for this technique. After in situ hybridization, specimens were post-fixed in 4% paraformaldehyde in PBS, and digital microscopic images were taken soon after completion of the reaction.
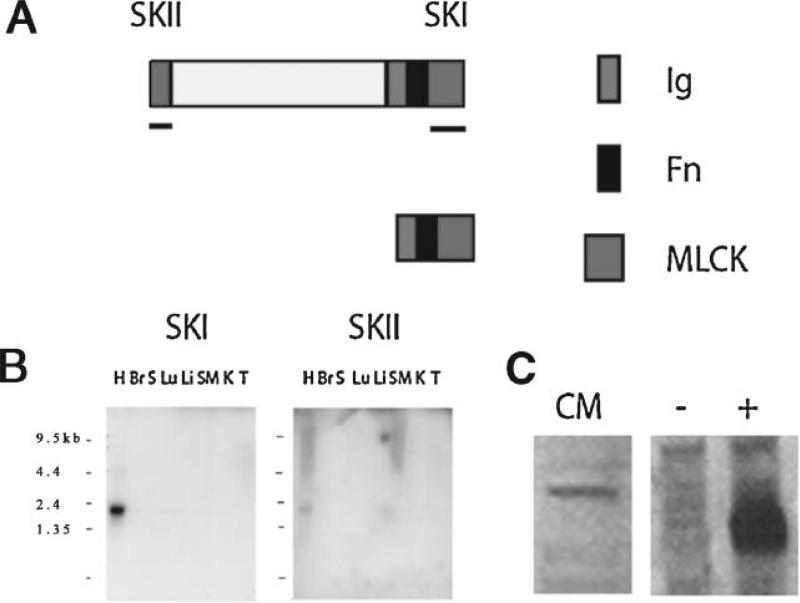
Fig. 1.
A: Schematic representation of the obscurin-associated MLCK-like single and dual kinase isoforms. Putative functional domains identified by PROSITE search include one full and one partial serine threonine kinase (SKI and SKII, respectively), one immunoglobulin-like (Ig), and one fibronectin-like (Fn) domains. Antisense RNA and cDNA probes, for in situ and Northern analysis respectively, were prepared using cDNA sequence from the underlined regions. A polyclonal antibody (link 7) generated to human obscurin-associated MLCK-like kinase recognizes epitopes in the SKI domain (indicated by the pink square and short line on the right). B: Northern blot analysis of obscurin-MLCK mRNA expression in adult murine tissues. The single kinase isoform, detected with the SKI probe, is selectively expressed in the heart (H) and not detectable in other tissues including brain (Br), spleen (S), lung (Lu), liver (Li), skeletal muscle (SM), kidney (K), and testis (T). The dual kinase isoform (top of panel A), detected with the SKII probe, was expressed in the heart and skeletal muscle (SM). Note the 4.6 kb band in the SKII blot. Larger transcripts may reflect inclusion of the kinase domains at the end of the obscurin transcript. C: Western blot analysis using the anti-obscurin-MLCK-like terminal kinase antibody. The antibody detected the native obscurin-MLCK single kinase isoform in lysates from adult rat cardiac myocytes in culture and in lysates of Cos7 cells transfected with an obscurin-MLCK expression construct (a positive control, marked with +) but not with a control plasmid (a negative control, marked with −).
Antibody Preparation and Characterization
A 580 bp cDNA encoding for most of the terminal kinase domain of obscurin-MLCK (Fig. 1) was cloned into the pRSET expression vector and the protein expressed in the E. coli strain BL21(DE3)pLysS. Induction of expression was carried out at 37°C using 0.1 M isopropyl-B-d-thiogalactopyranoside (IPTG). Cells were harvested 5 h after induction of expression, and expressed protein was purified and dialysed according to Invitrogen's Xpress system protein purification protocol. The protein was isolated under denaturing conditions and purified using Invitrogen's ProBond resin. The eluted protein was dialyzed against 10 mM Tris, pH 8.0, 0.1% Triton X-100 overnight at 4°C to remove urea. The purified protein was supplied to Bethyl laboratories (Montgomery, TX) for production of polyclonal rabbit antisera. Antibody purification was performed with protein A column (Pierce Inc., Rockford, IL) following the manufacturer's instructions. Western analysis was performed, using standard procedures, on cell lysates of NIH3T3 cells 48 h after transfection with a plasmid construct expressing the full length single kinase isoform. The work on the antibody development and testing of its specificity was performed by Dr. M. Raeker with the technical assistance of a specialized core at the University of Michigan Medical School.
Primary Cardiac Muscle Cell Cultures
Primary cell cultures of neonatal cardiac myocytes have been isolated from the hearts of 2- to 3-day-old rats by enzymatic dissociation of myocardial tissue with trypsin and collagenase as previously described [Borisov et al., 1985, 1989]. Isolated cells were plated on glass coverslips in 60 mm Petri dishes. The cells were cultivated in a CO2 incubator (5% CO2) in F-12 medium containing 10% of bovine fetal serum and were collected for immunolabeling every 24 h after plating.
Immunocytochemistry and Confocal Microscopy
Cell fixation, processing, and immunolabeling were performed as previously described [Borisov et al., 1989, 2001]. The samples of mouse fetal cardiac tissue were prepared, sagittally cryosectioned, mounted on histological slides and processed according to the protocols that we described earlier [Borisov et al., 2001]. Similar techniques were applied to a set of rat cardiac tissue samples. The development and characterization of the monoclonal antibody to the sarcomeric isoform of α-actinin and its use for studies of myogenic differentiation were previously described [Borisov et al., 1989; Fridlianskaia et al., 1989]. This antibody, clone EA53, is now marketed by Sigma Chemical Co., St. Louis, MO. A polyclonal antibody recognizing the carboxy-terminal region of obscurin was prepared in a rabbit host using standard methods, and its specificity has been examined [Kontrogianni-Konstantopoulos et al., 2003]. This antibody was kindly provided to us by Dr. K. Kontrogianni-Konstantopoulos and Dr. R.J. Bloch, Department of Physiology, the University of Maryland. We used two negative controls in our immunocyto-chemical reactions: (1) omission of the primary antibody to myofibrillar proteins, and (2) the labeling of extracardiac connective tissue known to be immunonegative for both obscurin and α-actinin. Both negative controls demonstrated the specificity of the immunolabeling reactions. Non-specific binding was blocked with 1% bovine serum albumin in PBS. In double-labeling indirect immunofluorescence experiments, we incubated the samples with the monoclonal antibody to α-actinin diluted 1:400 for 2 h, washed the cells three times for 3 min in PBS, then incubated the samples with the polyclonal antibody to obscurin diluted 1:45 for 2 h and washed three times for 3 min in PBS at room temperature. The solution of the secondary anti-mouse and anti-rabbit antibodies conjugated with FITC and rhodamine, respectively, was used to label the cells for 45 min. The secondary antibodies were purchased from Sigma Chemical Co. Immunolabeled samples of cultured cardiac muscle cells were washed three times for 3 min with PBS at room temperature, mounted on histological glass slides using the embedding medium for immunofluorescence (Sigma Chemical Co.) and examined in a Zeiss LSM 510 Meta confocal microscope using the objectives 40× and 63×.
RESULTS AND DISCUSSION
Cloning of the Murine Obscurin-Associated Kinase Isoforms
The amino acid sequences of the human and mouse obscurin-associated kinase isoforms were compared using BLAST sequence similarity search (blastp, pairwise blast format). The assembled mouse and human cDNA sequences would be predicted to encode for 1,585 amino acid and 1,610 amino acid proteins, respectively. Analysis of the predicted proteins, using a PROSITE database search, determined that both the mouse and human obscurin-associated kinase isoforms would have two immunoglobulin domains and two putative serine-threonine kinase domains (Fig. 1). PROSITE search also identified a domain with fibronectin homology between the terminal immunoglobulin and serine-threonine kinase domains that had not been recognized in the human cDNA sequence. This putative fibronectin domain is conserved in the human with 82% amino acid identity and 87% conserved amino acids between the mouse and human amino acid sequence within this 77 amino acid domain.
Both the mouse and human obscurin-associated kinase-like transcripts also share a conserved putative translation initiation site (mouse: ggggccccgtccATGcaggtc; human: gggcccccatccATGcaggta) and predicted amino terminal nine amino acids (MQVTIEDVQ). The search revealed 76% sequence identity between the two predicted proteins with much higher degrees of similarity between the corresponding serine-threonine kinase domains (SK1: 89% identity; SK2: 88% identity; Fig. 1). The most striking divergence between the sequences occurred between the amino terminal serinethreonine kinase domain and the carboxy terminal immunoglobulin domain (65% identity). This amino acid sequence does not have any identified functional domains and has no significant homology to other known proteins. The bulk of this region is encoded for by a single large exon, 1,988 bp in the mouse and 2,063 bp in the human as we demonstrated earlier [Russell et al., 2002]. The putative translation initiation site for the single kinase-containing transcript does not appear to be conserved between the human and the mouse. In both species, a good Kozak consensus start sequence is present at the 3′ end of the large exon, however, the start in the mouse occurs approximately 75 bp upstream of the putative start site in the human.
Expression of Obscurin-Associated Kinase Transcripts
The expression pattern of the murine obscurin-associated kinase isoform was investigated using Northern blotting analysis. As in the human, the obscurin-associated kinase isoforms are predominantly expressed in the heart. In contrast to the human gene, the 2.2 kb, single obscurin-associated kinase-encoding isoform is much more prominently expressed than the 4.9 kb, SK-II-encoding isoform (Fig. 1). The probe corresponding to the carboxy terminal serine-threonine kinase region detected transcripts of 4.9 kb and several larger transcripts as well in both the heart and skeletal muscle. The larger transcripts may represent obscurin isoforms that include this kinase domain, which is supported by Northern analysis with a probe for the murine obscurin RhoGEF encoding region (unpublished results). Although not detectable by Northern analysis, the proteins similar to obscurin-associated kinase do appear to be expressed at low levels in multiple tissues. BLAST sequence homology search revealed murine ESTs from adult inner ear, diaphragm, and neo-natal lung that represented the sequences similar to obscurin-associated kinase transcripts.
The structure of these genes appears to be conserved among mammalian species, and the translation initiation sites for the two genes are approximately 140–170 kb apart in the mouse, rat and human genomes. The transcription initiation site for obscurin-associated kinase isoforms has not been definitively identified as yet, and there is no evidence to suggest that it shares the same initiation site as the large obscurin transcript. Northern analysis with probes from the 5′-untranslated region of obscurin do not detect the obscurin-associated kinase transcript and RT-PCR, using primers from the obscurin 5′-untranslated sequence, have not been able to amplify products when paired with primers from the obscurin-associated kinase region (unpublished results).
The level of expression does suggest that at least some regulatory elements differ between the two sets of transcripts. By Northern analysis, the obscurin-associated kinase 2.2 kb transcript appears to be much more highly expressed than the much larger obscurin transcript in adult mouse tissues. Since Northern analysis can be affected by probe affinity and by low efficiency preparation, separation and transfer of high molecular weight RNAs, the relative amount of expression of obscurin and the kinase was compared using RT-qPCR. The efficiency of amplification for the obscurin and the kinase-encoding products were similar based on serial dilutions of starting RNA. The obscurin-associated kinase single kinase isoform was expressed about 50-fold greater than the obscurin transcript. Again, differences in the efficiency of mRNA isolation between the 25 kb obscurin and the 2.2 kb obscurin-associated kinase transcripts may account for some of the difference. The fold difference is consistent with the findings of Nagase et al. [2000] who isolated fragments of both obscurin and obscurin-associated kinase, which they named KIAA1556 and KIAA1639, respectively. Using reverse-transcription-coupled polymerase chain reaction, the products of which were quantified by enzyme-linked immunosorbent assay, these authors demonstrated that KIAA1556 and KIAA1639 had a similar pattern of distribution in human adult tissues but that, in general, KIAA1639 was expressed approximately 50-fold greater.
The difference in the obscurin-associated kinase isoform expression patterns between the mouse and the human may reflect the differences in the myocardial physiology between the two species. One of the most striking differences between the murine and human heart contractile activity is their resting heart rates. Murine heart rates range from 300 to 400 beats per minute compared to a normal human heart rate of less than 90 beats per minute. This more rapid heart rate shortens the ventricular filling time, necessitating rapid myocardial relaxation. Other important differences include their inotropic and chronotropic responses to adrenergic stimulation. Thus, it remains to be determined whether differences in the kinase isoform expression facilitate some of these physiologic differences.
Developmental Expression of Obscurin and Obscurin-Associated Kinase in Cardiac Tissue
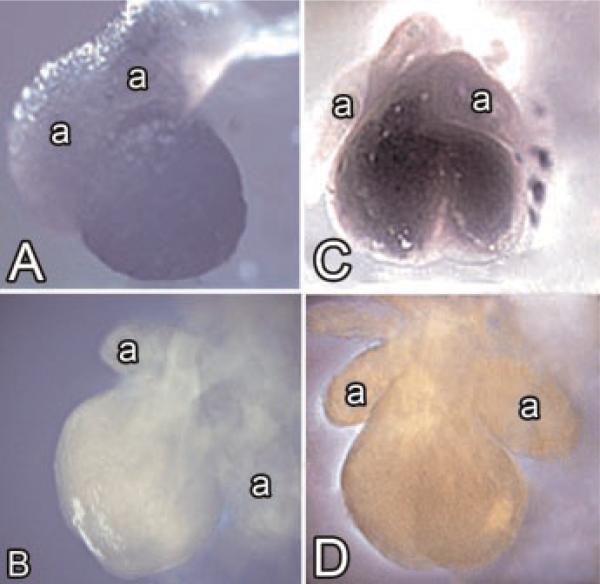
Using RNA in situ hybridization, we found that the expression of obscurin and obscurin-associated kinase transcripts in mouse cardiac tissue was not visually detectable until embryonic day 10, but by day 12 post-conception there was significant expression in both the left and right ventricle (Fig. 2A–D). We used murine hearts in these experiments because our probes were mouse-specific. As noted in the whole mount in situ hybridization analysis of the hearts isolated from 12-day embryos, there was intense reactivity for obscurin-associated kinase in the ventricles and relatively little expression at this point in the developing atria (Fig. 2A,B). Similar differences in compartment-specific expression between the atria and the ventricles have been observed for obscurin (Fig. 2C,D). The intensity of labeling of the ventricular tissue for obscurin-associated kinase was higher than the intensity of labeling for obscurin (compare Fig. 2A,C). The comparison of these two labeling patterns suggests that the intense expression of obscurin during development is activated later than the expression of obscurin-associated kinase.
Fig. 2.
Expression of obscurin and obscurin-associated kinase during mouse cardiac development. Whole mount RNA in situ hybridization of obscurin (A) and obscurin-associated kinase (C). Note weak to moderate level of obscurin expression in the ventricles (A) and the prominent ventricular expression of obscurin-associated kinase (C) 12 days post-conception. Also, note that the atria visible on the upper part of the photos show a considerably lower level of reactivity than the ventricles. A,B show the lateral views of the heart, and C,D show the frontal views of the heart. The letter a marks the atria. B,D show a negative control incubated without the specific probe demonstrating the specificity of the in situ hybridization reaction, 21×. [Color figure can be viewed in the online issue, which is available at www.interscience.wiley.com.]
This localization study supported the results of our Northern blotting experiments that demonstrated low levels of obscurin expression at early stages of cardiomyogenesis. A lower level of obscurin expression in the atrial tissue compared to the ventricles can be explained by the fact that the differentiation of the contrac-tile system in the developing atria occurs at later stages of embryonic development and proceeds less intensely compared to the ventricular myocytes [for literature see Borisov and Rumyantsev, 1991; Rumyantsev, 1991]. Very low levels of expression of obscurin-associated kinase encoding transcripts were also detectable in the developing central nervous system (unpublished data).
The developmental pattern of obscurin expression and localization was further studied using indirect immunofluorescent protein labeling of fetal mouse and rat embryonic tissue sections and in primary cultures of rat cardiac muscle cells. Both obscurin and obscurin-associated kinase were noted to be expressed in the developing heart at the protein level. We found that obscurin expression progressively increased during myocardial development directly correlating with the increasing level of cardiac muscle differentiation. The results of immuno-labeling of the mouse cardiac tissue at different stages of embryonic development are presented in Figure 3A–F. Interestingly, at early stages of cardiac wall morphogenesis the immunolabeling for obscurin in the heart had a tendency to be more intense in the subendocardial developing trabecular areas compared to the forming middle compact layer and the subepicardial layer of the ventricular wall (Fig. 3A). This difference appears to reflect the gradients of cell differentiation in the developing heart with less advanced cell differentiation and increased cell proliferation in the compact layer of the developing ventricles. Several studies demonstrated that as myocytes move away from the compact zone toward the trabecular zone, they become progressively differentiated and their rate of proliferation declines [for literature see Thompson et al., 1990; Tokuyasu, 1990; Rumyantsev, 1991]. In these areas, the cells proliferating at a slower rate have more developed contractile structures. This suggests that the accumulation of obscurin in the subendocardial myocyte populations at this stage is explained by the development of a more mature myofibrillar apparatus. As development proceeded, the intensity of immunolabeling reflecting the level of obscurin expression in the growing heart considerably increased (Fig. 3B–D,F). By day 13 post-conception the presence of obscurin was detected in all myocytes in the developing heart wall, and the transmural gradient observed at earlier developmental stage was not clearly expressed (Fig. 3B). The labeling was specificto muscle cells, and was not found in the non-muscle tissues adjacent to the growing heart (Fig. 3B). An additional negative control that included the omission of the primary antibody (Fig. 3D) also demonstrated the specificity of the immunocytochemical reaction. The intensity of tissue labeling increased, and the development of elements of structured fibrillar pattern rapidly progressed during development (compare Fig. 3A,B). Figure 3E taken at a higher magnification clearly shows that at this stage obscurin was detected both as diffuse labeling and as the cross-striated periodic pattern typical of differentiating myofibrils. This process coincided with the progressive trabeculation in the ventricles. By the time of trabecular fusions and the advanced development of the interventricular septum, the intensity of labeling of myocardial tissue further increased, and obscurin localization acquired more organized granulated, dotted and fibrillar structural patterns (Fig. 3C). After completion of the trabecular fusion and formation of compact ventricular walls, obscurin expression remained upregulated compared to earlier stages, and the structural association of obscurin with the contractile apparatus became the predominant type of localization of this protein (Fig. 3F). The developmental dynamics of obscurin localization in the rat heart was very similar to the structural patterns described above in the mouse heart (unpublished results).
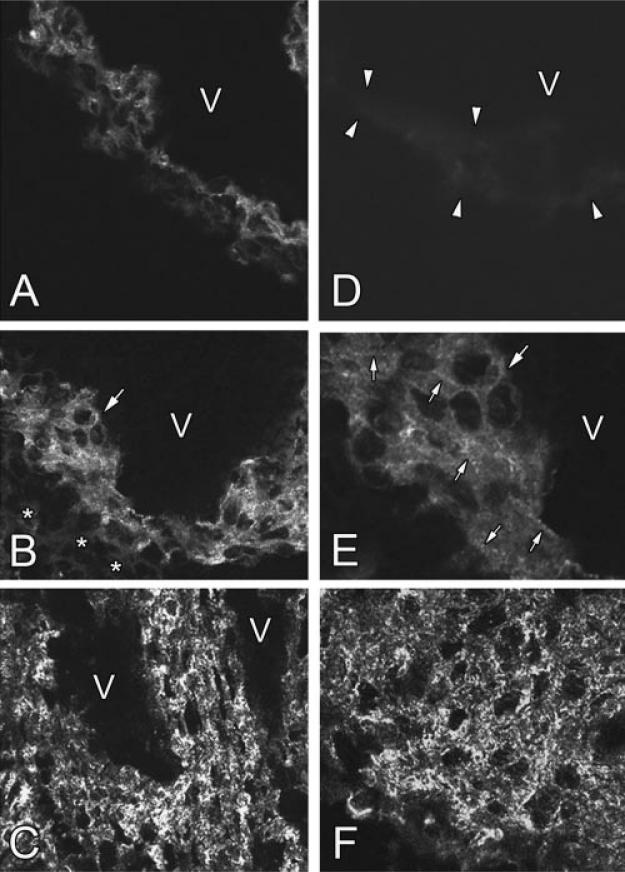
Fig. 3.
Expression of obscurin in the developing myocardium. Immunofluorescent labeling for this protein in the developing free left ventricular wall on days 11, 13, 14.5, and 16-day post-conception is illustrated in A–C,F, respectively. Note the increasing levels of obscurin expression during cardiac development and its shift from the diffused pattern to more intensely labeled structured dotted and periodic fibrillar distribution in the tissue. D: Negative control: the section of the heart on day 13 post-conception incubated only with the secondary antibody (compare to panel B). V marks the ventricular cavity. Asterisks in B indicate the non-muscle tissue immunonegative for obscurin and located close to epicardial surface of the ventricular wall. The arrow in B indicates the area shown in panel E at a high magnification. C: The ventricular wall following trabecular fusion located closely to the apical area of the heart near the interventricular septum. Arrowheads in D show the boundaries of the immunonegative cardiac tissue in a negative control sample. E: The area marked with the large arrow in panel B presented at a higher magnification as a single confocal optical section. The arrows in B and E show the same group of cells. At this stage of obscurin is visualized both as the diffuse labeling pattern and as the developing elements of the striated myofibrillar pattern. Small arrows in E show the elements of sarcomeric cross-striation. A–D, F: 280×; E: 630×.
Obscurin-associated kinase demonstrated a different pattern of intracellular localization. Intense immunopositivity for this protein in vivo was found in mouse and rat cardiac muscle cell nuclei with some moderate presence in the cytoplasm (the insert in Fig. 7B). It was difficult to determine its precise intracellular protein localization in tissue sections because of the heterogeneity of cell populations in vivo, invisibility of clear cell boundaries, very close superimposition of individual cells and neighboring sectioned cell fragments. When we immunolabeled the tissue for sarcomeric proteins as the second label to differentially visualize muscle cells, the fluorescent signal was too intense to identify the details of nuclear and cytoplasmic structure and to determine the intercellular protein localization in individual myocytes. For this reasons, we performed a high resolution microscopic analysis of obscurin and obscurin-associated kinase localization in relation to the contractile apparatus in isolated intact heart cells in culture (see Figs. 7A–D and 8A–D and the text in the next section below).
Fig. 7.
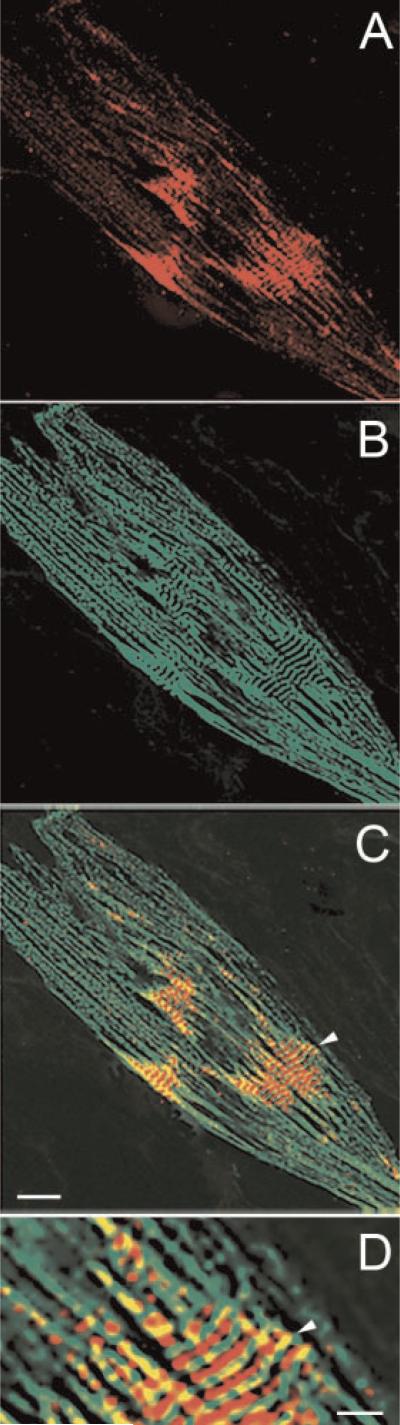
Muscle-specific expression of obscurin-associated kinase in cell cultures of differentiating rat neonatal cardiac muscle cells as demonstrated by double immunolabeling for the sarcomeric isoform of α-actinin (green fluorescence) and obscurin-associated kinase (red fluorescence) on day 4 in culture. A: Immunofluorescent labeling for sarcomeric α-actinin; (B) immunofluorescent labeling for obscurin-associated kinase; the insert shows the intense reactivity of cell nuclei and moderate reactivity of the cytoplasm in the myocardial tissue section of 16-day old rat embryo (C) nuclear staining with Hoechst; (D) combined image of A–C. Note the presence of immunoreactivity for obscurin-associated kinase only in a sarcomeric α-actinin positive muscle cell (the arrowhead in B,C) and the absence of obscurin-associated kinase expression in fibroblasts whose nuclei are seen on the right side of the photo. Bar, 30 μm. [Color figure can be viewed in the online issue, which is available at www.interscience.wiley.com.]
Fig. 8.
Localization of obscurin-associated kinase at advanced stages of cardiomyogenic differentiation in culture. A: Immunofluorescent localization of sarcomeric α-actinin in primary culture of rat neonatal cardiac muscle cells on day 7 in vitro. B: Immunofluorescent localization of obscurin-kinase in the same field. Note the presence of immunolabeling for this protein in myonuclei and in Z-disks of myofibrils. C: A merged image of immunolabeled sarcomeric α-actinin (green fluorescence) and staining for nuclear DNA (blue fluorescence) in the same field shows that obscurin-kinase is localized in the myonuclei. The arrow shows a nucleus of a non-muscle, fibroblastic cell in the upper left corner. This cell is immunonegative for both sarcomeric α-actinin and obscurin-associated kinase; (D) a combined image of the cells immunolabeled for sarcomeric α-actinin and obscurin-associated kinase shows the association of obscurinkinase label with the Z-disks. 400×. [Color figure can be viewed in the online issue, which is available at www.interscience.wiley.com.]
Thus, we found that the tissue specific expression of obscurin and its kinase isotype was upregulated during myogenesis and that the expression of obscurin-associated kinase could be detected at higher level prior to the intense expression of obscurin. This apparent earlier expression of the kinase-like sequence at the transcriptional level appears to allow this protein to attain a higher level of expression in the developing heart sooner than that of obscurin. This interpretation is also supported by our data of reverse transcriptase primed quantitative PCR analysis of obscurin and obscurin-associated kinase mRNA expression under conditions of cardiac hypertrophy in vivo suggesting that the kinase-containing isotype can be upregulated earlier and to a greater extent than obscurin [Borisov et al., 2003].
Differential Cellular Localization of Obscurin and Obscurin-Associated Kinase
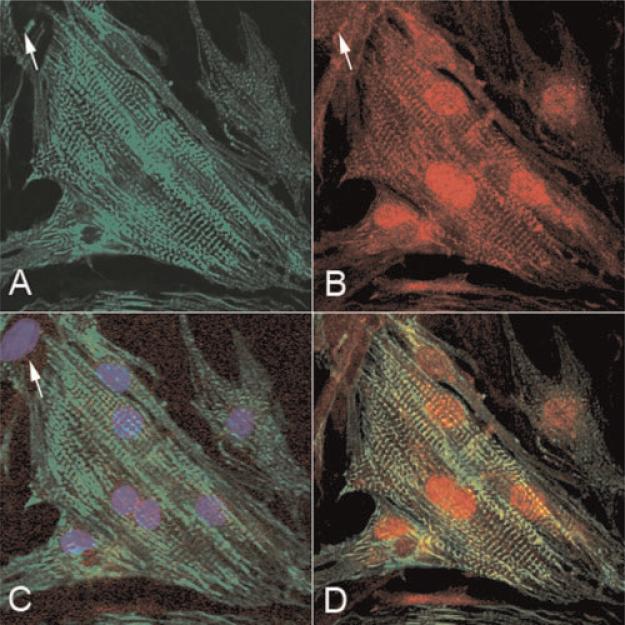
To determine the intracellular localization of obscurin and obscurin-associated MLCK-like kinase more precisely, we studied their topographical distribution in rat cardiac myocytes in culture. It was much more difficult to isolate large numbers of functionally active mouse cardiac muscle cells and to maintain them in culture. For this reason in the present study we used a rat cell model. These experiments allowed us to examine the localization of these proteins in the whole volume of intact, individual myocytes, which was impossible to achieve using tissue sections. Indirect immunolabeling for sarcomeric α-actinin that localizes to the Z-band in mature myofibrils was used to selectively identify myocytes in primary cell cultures and to evaluate the degree of their myofibrillar differentiation [for literature see Borisov, 1991]. Our results showed that in cardiac myocytes obscurin and obscurin-associated kinase were localized differentially at the cellular level. Using double indirect immunofluorescent labeling of cell cultures for sarcomeric α-actinin and obscurin, we found that the localization of obscurin was entirely cytoplasmic and muscle-specific and that it was associated with progressive maturation of the sarcomeric contractile apparatus (Fig. 4A–C). Obscurin was found in mature sarcomeres and was the most abundant in the middle of A-bands at the level of the M-lines, particularly in the myofibrillar clusters. As one can see from Figure 4A–D, double immunolabeling of the sarcomeric isoform of α-actinin and obscurin in the same cells clearly demonstrated that the incorporation of obscurin temporally coincided with the accumulation of laterally aligned mature myofibrils (Fig. 4D). Unlike the interphase cells, during mitotic division obscurin was found in granulated aggregates scattered in the cytoplasm and spatially separated from the granulated aggregates containing α-actinin (Fig. 5). This shows that during mitosis obscurin may form either homophilic protein aggregates or may establish structural complexes with the sarcomeric proteins distinct from α-actinin. Thus, the localization pattern of obscurin is cell cycle-dependent.
Fig. 4.
Cellular localization of obscurin in differentiating rat cardiac muscle cells. A: Localization of obscurin in a poorly differentiated rat early neonatal cardiac myocyte after 4 days in culture; (B) localization of sarcomeric α-actinin in the same field; (C) a merged image showing the localization of obscurin (red fluorescence) and α-actinin (green fluorescence) in the same cell. D: Structural interrelations of obscurin and sarcomeric α-actinin localization in the area marked with the arrowhead in C shown at a higher magnification. Note the topographical association of intense immunolabeling for obscurin with the nascent myofibrillar system and its localization predominantly in the middle of the A-band at the time of formation of mature myofibrillar clusters. Also note that most of premyofibrillar structures immunopositive for sarcomeric α-actinin are only weakly immunopositive for obscurin. Bar, 20 mm (A–C), 6.4 mm (D). [Color figure can be viewed in the online issue, which is available at www.interscience.wiley.com.]
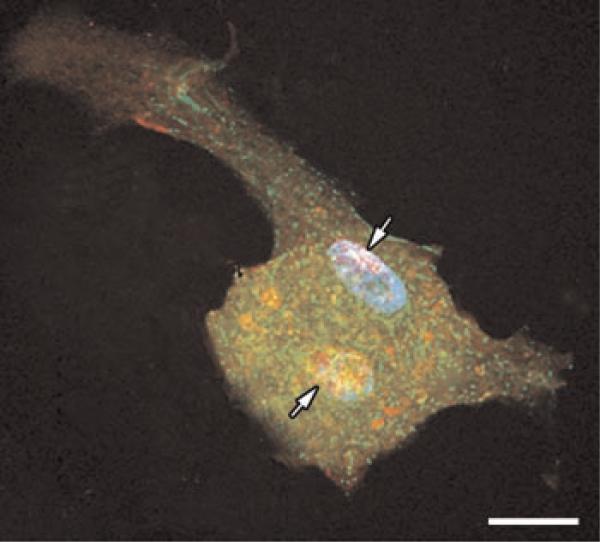
Fig. 5.
Localization of obscurin in cardiac myocytes during mitosis. Double immunofluorescent labeling for obscurin (red fluorescence) and sarcomeric α-actinin (green fluorescence) in a rat neonatal cardiac muscle cell at the stage of early telophase of mitosis. Two daughter nuclei (arrows) are visible in the central area of the cytoplasm. Staining for nuclear DNA with Hoechst dye (blue fluorescence) shows that the nuclei contain highly condensed chromatin typical of the telophase of mitosis. Note that obscurin is segregated in diffusely located granulated bodies that are separated from the protein aggregates containing sarcomeric α-actinin. The fragmented remnants of the contractile system disassembled during mitosis are seen at the cell periphery. Bar, 10 μm. [Color figure can be viewed in the online issue, which is available at www.interscience.wiley.com.]
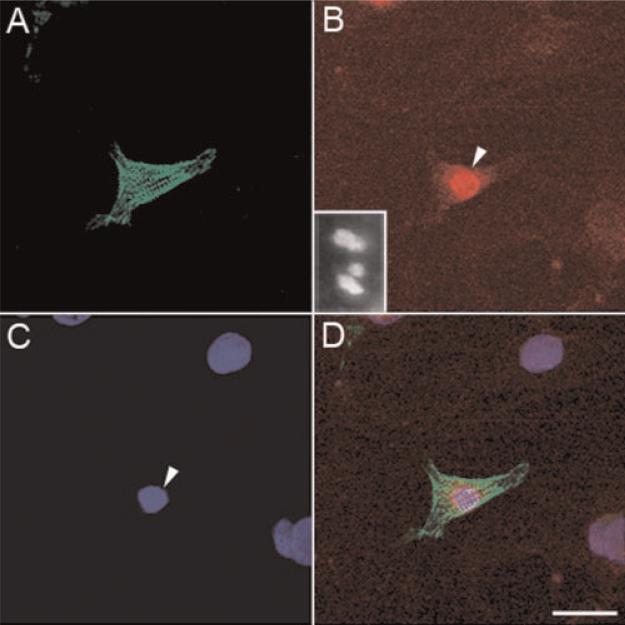
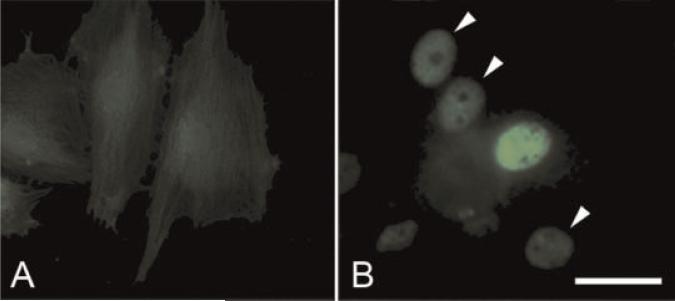
The specificity of the antibody to obscurin used in this experiments has been described earlier [Kontrogianni-Konstantopoulos et al., 2003], and the specificity of the antibody to obscurin-associated kinase called link7 was defined by Western blotting (Fig. 1C). The lysates of Cos7 cells transfected with an obscurin-associated MLCK-like kinase expression construct served as a positive control, and the cells transfected with a non-coding, control plasmid served as a negative control. To further test the specificity of our antibody that we used for the detection of obscurin-associated kinase, we also expressed this protein in a mouse cell line of fibroblast-like undifferentiated cells transfected with a His-tagged full length obscurin-associated kinase (its single kinase isoform) and immunolabeled the cells with an anti-His antibody. The results of these experiments are presented in Figure 6A,B. We found that the antibody intensely reacted to the expressed protein localized in cell nuclei, and no reaction was found in the samples treated with the preimmune serum. This shows that the antibody is specific for obscurin-associated kinase.
Fig. 6.
Immunofluorescent labeling for obscurin-associated protein kinase in the permanent line of H9c2 cells transfected with a non-coding control construct (A) and transfected with His-tagged construct encoding obscurin-associated kinase (B). Note intense fluorescent labeling of anti-His n the nucleus of a transfected cell in the center of the field in panel B. Arrowheads in B show the location of weakly and moderately positive nuclei. Bar, 30 μm. [Color figure can be viewed in the online issue, which is available at www.interscience.wiley.com.]
Our experiments demonstrated that, unlike obscurin, obscurin-associated kinase was localized primarily to myonuclei, and that there was intense expression of this protein in cardiac myocytes with a poorly differentiated contractile system (Fig. 7A–D). During myocyte differentiation, we observed progressive redistribution of obscurin-associated kinase to the cytoplasm, particularly to the Z-bands of myofibrils at advanced stages of differentiation (Fig. 8A–D). There was no immunore-activity in non-myocyte nuclei (Figs. 7 and 8), which clearly indicates that the expression of obscurin-MLCK-like kinase is specifically restricted to muscle cells. Taken together, these data demonstrate that obscurin-associated kinase is expressed early in cardiac muscle development and that its cellular localization is differentiation-dependent.
The differential cellular localization of obscurin and obscurin-associated kinase indicates that these isotypes may have acquired distinct but interrelated roles in vertebrate muscle development during phylogenesis. The close functional relationship of the two is evident from the identification of a D. melanogaster ortholog of obscurin that includes the RhoGEF and multiple Ig domains characteristic of the large obscurin and the kinase domains of obscurin-associated kinase [Sutter et al., 2004]. Obscurin is also expressed in zebrafish muscle [Raeker et al., 2006] where it appears to have similar functions to the orthologs typical of mammalian cardiac myocytes [Borisov et al., 2004, 2006] and skeletal muscle cells [Kontrogianni-Konstantopoulos et al., 2006].
The coordinated temporal expression of obscurin and its derivative obscurin-associated kinase isoform and their possible evolution from a single ancestral gene suggests the possibility of their important functional relationships. The cooperative activity of the kinase-like domains and RhoGTPase signaling to the cytoskeleton has been documented in recent years in other proteins [O'Brien et al., 2000; reviewed by Pfitzer, 2001]. It was shown that activation of the RhoA pathway can inhibit myosin phosphatase activity, thereby enhancing the effects of the kinase-mediated phosphorylation of myosin light chain. There can also be inhibitory interactions between the two signaling pathways since PAK phosphorylation of MLCK by p21-activated kinase, a kinase activated by the Rho family of GTPases, can inhibit MLCK activity [Hoshijima et al., 1998; Sanders et al., 1999; Goeckeler et al., 2000].
Thus, the co-expression of obscurin and obscurin-associated kinase suggests that there may be an important regulatory interaction between the kinase domains associated with obscurin and the RhoGEF domain of this protein. Whether this potential regulatory relationship is active when two are physically separate as occurs in the immature myocyte, or is only present when the two co-localize at the Z-bands of mature myofibrils remains to be determined. However, despite the elements of structural similarity of obscurin-associated kinase to MLCKs, the functional role, the catalytic properties and the substrates of this enzyme-like protein remain to be elucidated.
Interestingly, a recent study by Bowman et al. [2007] suggests that in adult rat skeletal muscle fibers the localization pattern of two alternatively spliced RhoGEF-containing “giant” obscurin isoforms is also differential. These authors reported the development of an antibody to the RhoGEF domain of obscurin that may serve as an important marker for the studies focused on the structural and functional interrelations of RhoGEF and the kinase-regulated signaling pathways.
Obscurin was earlier found in the nascent sarcomeres in the developing chicken and rat myocardium in close association with titin [Young et al., 2001]. At early stages of myofibril formation, obscurin localizes to the Z-disk where it appears to interact with the NH2-terminal Z-disk region of titin and later incorporates into the middle of A-bands [Bang et al., 2001; Young et al., 2001]. These data are consistent with the results of the present study of obscurin expression in developing myocardium in situ and in cardiac muscle cells isolated into differentiating cell culture. This interpretation agrees with our conclusion concerning considerable upregulation of obscurin expression during cardiac development.
As we briefly mentioned above, the hallmark structural motifs of the family of large muscle proteins are the immunoglobulin (Ig), fibronectin 3 (FN3), and serine/threonine kinase MLCK-like domains. Multiple Ig and FN3 domains interact with other proteins anchoring the kinase to specific intracellular sites and may enable the interactions with specific target molecules. They can also provide a physical separation between functional domains of the protein, allowing spatially distinct structural and signaling interactions that are necessary for highly ordered assembly of individual elements of the contractile apparatus in cardiac muscle.
Thus, the potential importance of obscurin, and its associated proteins to striated muscle development and physiology is indicated by their patterns of expression during development and their cellular localization in differentiating cardiac myocytes. The fact that the temporal pattern of their expression coincides with the onset and activation of synthesis and assembly of contractile sarcomeric proteins in developing cardiac and skeletal muscle of vertebrate embryos supports this assertion [for literature on the time course of muscle development see Lyons et al., 1990; Noden et al., 1999; Buckingham et al., 2003]. Taken together, these results suggest that the proteins encoded by the obscurin transcriptional units are directly involved in the progression of myogenic differentiation. The upregulation of both of obscurin and its associated kinase during muscle development suggests that they may have important cooperative roles in cardiac myocyte differentiation. Correct sarcomere assembly in new forming myofibrils depends not only on the integrity of components of the thick and thin filaments but also on the intermediate filament and subsarcolemmal proteins that link sarcomeres to the cytoskeletal apparatus and mediate their interaction with other cytoplasmic organelles. Loss-of-function mutations in many of these cytoskeletal-anchoring proteins can lead to sarcomere dis-array, cardiac dilatation and congestive heart failure. Further definition of the functional role of these proteins during myogenesis and characterization of their structural and regulatory interrelations during myogenic differentiation will be the focus of our future studies.
ACKNOWLEDGMENTS
This work was supported by grants from the National Institutes of Health NIH R01 HL075093-1 and the Muscular Dystrophy Association (MDA3803). The authors are thankful to Dr. Katia Kontrogianni-Konstantopoulos and Dr. Robert Bloch for providing the polyclonal antibody to obscurin used in this study. We also thank Pavel Borisov for help with preparation of the manuscript. We are also thankful to the undergraduate students who provided technical help in the laboratory.
Grant sponsor: NHLBI; Grant number: R01 HL075093-1; Grant sponsor: Muscular Dystrophy Association; Grant number: MDA3803.
REFERENCES
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- Aoki H, Sadoshima J, Izumo S. Myosin light chain kinase mediates sarcomere organization during cardiac hypertrophy in vitro. Nat Med. 2000;6:183–188. doi: 10.1038/72287. [DOI] [PubMed] [Google Scholar]
- Bang ML, Centner T, Fornoff F, Geach AJ, Gotthardt M, McNabb M, Witt CC, Labeit D, Gregorio CC, Granzier H, Labeit S. The complete gene sequence of titin, expression of an unusual ~700-kDa titin isoform, and its interaction with obscurin identify a novel Z-line to I-band linking system. Circ Res. 2001;89:1065–1072. doi: 10.1161/hh2301.100981. [DOI] [PubMed] [Google Scholar]
- Borisov AB. Myofibrillogenesis and reversible disassembly of myofibrils as adaptive reactions of cardiac muscle cells. Acta Physiol Scand. 1991;142(suppl 599):71–80. [PubMed] [Google Scholar]
- Borisov AB, Rumyantsev PP. Atrial myocytes: Myoendocrine cells possessing an enhanced ability to reenter the mitotic cycle in vitro and in vivo. In: Oberpriller JO, Oberpriller JC, Mauro A, editors. The development and regenerative potential of cardiac Muscle. Harwood Academic Publishers; New York: 1991. pp. 115–137. [Google Scholar]
- Borisov AB, Coro Antich RM, Rumyantsev PP. DNA synthesis in cultures of atrial and ventricular rat cardiomyocytes. Tsitologia. 1985;27:990–994. [PubMed] [Google Scholar]
- Borisov AB, Goncharova EI, Pinaev GP, Rumyantsev PP. Changes in a-actinin localization and myofibrillo-genesis in rat cardiomyocytes in culture. Tsitologia. 1989;31:642–646. [PubMed] [Google Scholar]
- Borisov AB, Dedkov EI, Carlson BM. Interrelations of myogenic response, progressive atrophy of muscle fibers, and cell death in denervated skeletal muscle. Anat Rec. 2001;264:203–218. doi: 10.1002/ar.1155. [DOI] [PubMed] [Google Scholar]
- Borisov AB, Raeker MO, Kontrogianni-Konstantopoulos A, Yang K, Kurnit DM, Bloch RJ, Russell MW. Rapid response of cardiac obscurin gene cluster to aortic stenosis: Differential activation of Rho-GEF and MLCK and involvement in hypertrophic growth. Biochem Biophys Res Commun. 2003;310:910–918. doi: 10.1016/j.bbrc.2003.09.035. [DOI] [PubMed] [Google Scholar]
- Borisov AB, Kontrogianni-Konstantopoulos A, Bloch RJ, Westfall MV, Russell MW. Dynamics of obscurin localization during differentiation and remodeling of cardiac myocytes: Obscurin as an integrator of myofibrillar structure. J Histochem Cytochem. 2004;52:1117–1127. doi: 10.1369/jhc.3A6183.2004. [DOI] [PubMed] [Google Scholar]
- Borisov AB, Sutter SB, Kontrogianni-Konstantopoulos A, Bloch RJ, Westfall MV, Russell MW. Essential role of obscurin in cardiac myofibrillogenesis and hypertrophic response: Evidence from small interfering RNA-mediated gene silencing. Histochem Cell Biol. 2006;125:227–238. doi: 10.1007/s00418-005-0069-x. [DOI] [PubMed] [Google Scholar]
- Bowman AL, Kontrogianni-Konstantopoulos A, Hirsch SS, Geisler SB, Gonzales-Serratos H, Russell MW, Bloch RJ. Different obscurin isoforms localize to distinct sites at sarcomeres. FEBS Lett. 2007;581:1549–1554. doi: 10.1016/j.febslet.2007.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buckingham M, Bajaed L, Chang T, Daubas P, Hadchouel J, Meilhac S, Montarras D, Rocancourt D, Relaix F. The formation of skeletal muscle: From somite to limb. J Anat. 2003;202:59–68. doi: 10.1046/j.1469-7580.2003.00139.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Champagne MB, Edwards KA, Erickson HP, Kiehart DP. Drosophila stretchin-MLCK is a novel member of the titin/myosin light chain kinase family. J Mol Biol. 2000;300:759–777. doi: 10.1006/jmbi.2000.3802. [DOI] [PubMed] [Google Scholar]
- Ferrari MB, Podugu S, Eskew JD. Assembling the myofibril: Coordinating contractile cable construction with calcium. Cell Biochem Biophys. 2006;45:317–337. doi: 10.1385/CBB:45:3:317. [DOI] [PubMed] [Google Scholar]
- Fridlianskaia II, Goncharova EI, Borisov AB, Krylova TY, Pinaev GP. Monoclonal antibodies to the muscle isoform of α-actinin–a marker for studies of differentiation of skeletal and cardiac muscle. Tsitologiia. 1989;31:1234–1237. [PubMed] [Google Scholar]
- Fukuzawa A, Idowu S, Gautel M. Complete human gene structure of obscurin: Implications for isoform generation by differential splicing. J Muscle Res Cell Motil. 2005;26:427–434. doi: 10.1007/s10974-005-9025-6. [DOI] [PubMed] [Google Scholar]
- Goeckeler ZM, Masaracchia RA, Zeng Q, Chew TL, Gallagher P, Wysolmerski RB. Phosphorylation of myosin light chain kinase by p21-activated kinase P AK2. J Biol Chem. 2000;275:18366–18374. doi: 10.1074/jbc.M001339200. [DOI] [PubMed] [Google Scholar]
- Heid CA, Stevens J, Livak KJ, Williams PM. Real time quantitative PCR. Genome Res. 1996;6:986–994. doi: 10.1101/gr.6.10.986. [DOI] [PubMed] [Google Scholar]
- Hoshijima M, Sah VP, Wang Y, Chien KR, Brown JH. The low molecular weight GTPase Rho regulates myofibril formation and organization in neonatal rat ventricular myocytes: Involvement of Rho kinase. J Biol Chem. 1998;273:7725–7730. doi: 10.1074/jbc.273.13.7725. [DOI] [PubMed] [Google Scholar]
- Kamm KE, Stull JT. Dedicated myosin light chain kinases with diverse cellular functions. J Biol Chem. 2001;276:4527–4530. doi: 10.1074/jbc.R000028200. [DOI] [PubMed] [Google Scholar]
- Kontrogianni-Konstantopoulos A, Jones EM, van Rossum DB, Bloch RJ. Obscurin is a ligand for small ankyrin 1 in skeletal muscle. Mol Biol Cell. 2003;14:1138–1148. doi: 10.1091/mbc.E02-07-0411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kontrogianni-Konstantopoulos A, Catino DH, Strong JC, Sutter S, Borisov AB, Pumplin DW, Russell MW, Bloch RJ. Obscurin modulates the assembly and organization of sarcomeres and the sarcoplasmic reticulum. FASEB J. 2006;20:2102–2111. doi: 10.1096/fj.06-5761com. [DOI] [PubMed] [Google Scholar]
- Lange S, Xiang F, Yakovenko A, Vihola A, Hackman P, Rostkova E, Kristensen J, Brandmeier B, Franzen G, Hedberg B, Gunnarsson LG, Hughes SM, Marchand S, Sijersen T, Richard I, Edstrom L, Ehler E, Udd B, Gautel M. The kinase domain of titin controls muscle gene expression and protein turnover. Science. 2005;308:1599–1603. doi: 10.1126/science.1110463. [DOI] [PubMed] [Google Scholar]
- Lyons GE, Schiaffino S, Sassoon D, Barton P, Buckingham M. Developmental regulation of myosin gene expression in mouse cardiac muscle. J Cell Biol. 1990;111:2427–2436. doi: 10.1083/jcb.111.6.2427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayans O, van der Ven PF, Wilm M, Mues A, Young P, Furst DO, Wilmanns M, Gautel M. Structural basis for activation of the titin kinase domain during myofibrillogenesis. Nature. 1998;395:863–869. doi: 10.1038/27603. [DOI] [PubMed] [Google Scholar]
- Nagase T, Kikuno R, Hattori A, Kondo Y, Okumura K, Ohara O. Prediction of the coding sequences of unidentified human genes. XIX. The complete sequences of 100 new cDNA clones from brain which code for large proteins in vitro. DNA Res. 2000;7:347–355. doi: 10.1093/dnares/7.6.347. [DOI] [PubMed] [Google Scholar]
- Noden D, Marcucio R, Borycki AG, Emerson CP. Differentiation of avian craniofocal muscles: I. Patterns of early regulatory gene expression and myosin heavy chain synthesis. Dev Dyn. 1999;216:96–112. doi: 10.1002/(SICI)1097-0177(199910)216:2<96::AID-DVDY2>3.0.CO;2-6. [DOI] [PubMed] [Google Scholar]
- O'Brien SP, Seipel K, Medley QG, Bronson R, Segal R, Streuli M. Skeletal muscle deformity and neuronal disorder in Trio exchange factor-deficient mouse embryos. Proc Nat Acad Sci USA. 2000;97:12074–12078. doi: 10.1073/pnas.97.22.12074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfitzer G. Regulation of myosin phosphorylation in smooth muscle. J Appl Physiol. 2001;91:497–503. doi: 10.1152/jappl.2001.91.1.497. [DOI] [PubMed] [Google Scholar]
- Raeker MÖ, Su F, Geisler SB, Borisov AB, Kontrogianni-Konstantopoulos A, Lyons SE, Russell MW. Obscurin is required for the lateral alignment of striated myofibrils in zebrafish. Dev Dyn. 2006;235:1029–2018. doi: 10.1002/dvdy.20812. [DOI] [PubMed] [Google Scholar]
- Rumyantsev PP. Growth and hyperplasia of cardiac muscle cells. Harwood Academic Publishers; New York: 1991. [Google Scholar]
- Russell MW, Raeker MO, Korytkowski KA, Sonneman KJ. Identification, tissue expression and chromosomal localization of human obscurin-MLCK, a member of the titin and Dbl families of myosin light chain kinases. Gene. 2002;282:237–246. doi: 10.1016/s0378-1119(01)00795-8. [DOI] [PubMed] [Google Scholar]
- Sanders LC, Matsumura F, Bokoch GM, de Lanerolle P. Inhibition of myosin light chain kinase by p21-activated kinase. Science. 1999;283:2083–2085. doi: 10.1126/science.283.5410.2083. [DOI] [PubMed] [Google Scholar]
- Sanger JW, Kang S, Siebrands CC, Freeman N, Du A, Wang J, Stout AL, Sanger JM. How to build a myofibril. J Muscle Res Cell Motil. 2005;26:343–354. doi: 10.1007/s10974-005-9016-7. [DOI] [PubMed] [Google Scholar]
- Seberstyen MG, Fritz JD, Wolff JA, Greaser ML. Primary structure of the kinase domain region of rabbit skeletal and cardiac muscle titin. J Muscle Res Cell Motil. 1996;17:343–348. doi: 10.1007/BF00240931. [DOI] [PubMed] [Google Scholar]
- Small TM, Gernert KM, Flaherty DB, Mercer KB, Borodovsky M, Benian GM. Three new isoforms of Caenorhabditis elegans UNC-89 containing MLCK-like protein kinase domains. J Mol Biol. 2004;342:91–108. doi: 10.1016/j.jmb.2004.07.006. [DOI] [PubMed] [Google Scholar]
- Sutter SB, Raeker MO, Borisov AB, Russell MW. Orthologous relationship of obscurin and Unc-89: Phylogeny of a novel family of tandem myosin light chain kinases. Dev Genes Evol. 2004;214:352–359. doi: 10.1007/s00427-004-0413-5. [DOI] [PubMed] [Google Scholar]
- Thompson RP, Lindroth JR, Wong Y- M. Regional differentiation in DNA-synthetic activity in the preseptation myocardium of the chick. In: Clark EB, Takao A, editors. Developmental cardiology: Morphogenesis and function. Mount Kisco-Futura Publishing Company; New York: 1990. pp. 219–234. [Google Scholar]
- Tokuyasu KT. Co-development of embryonic myocardium and myocardial circulation. In: Clark EB, Takao A, editors. Developmental cardiology: Morphogenesis and function. Mount Kisco-Futura Publishing Company; New York: 1990. pp. 205–218. [Google Scholar]
- Wilkinson DG. The theory and practice of in situ hybridization. In: Wilkinson DG, editor. In situ hybridization. IRL Press-Oxford University Press; Oxford-New York: 1992. pp. 7–31. [Google Scholar]
- Young P, Ehler E, Gautel M. Obscurin, a giant sarcomeric Rho guanine nucleotide exchange factor protein involved in sarcomere assembly. J Cell Biol. 2001;154:123–136. doi: 10.1083/jcb.200102110. [DOI] [PMC free article] [PubMed] [Google Scholar]