Abstract
The U3 small nucleolar ribonucleoprotein (snoRNP) is required for three cleavage events that generate the mature 18S rRNA from the pre-rRNA. In Saccharomyces cerevisiae, depletion of Mpp10, a U3 snoRNP-specific protein, halts 18S rRNA production and impairs cleavage at the three U3 snoRNP-dependent sites: A0, A1, and A2. We have identified truncation mutations of Mpp10 that affect 18S rRNA synthesis and confer cold-sensitivity and slow growth. However, distinct from yeast cells depleted of Mpp10, the mutants carrying these truncated Mpp10 proteins accumulate a novel precursor, resulting from cleavage at only A0. The Mpp10 truncations do not alter association of Mpp10 with the U3 snoRNA, nor do they affect snoRNA or protein stability. Thus, the role in processing of the U3 snoRNP can be separated into cleavage at the A0 site, which occurs in the presence of truncated Mpp10, and cleavage at the A1/A2 sites, which occurs only with intact Mpp10. These results strongly argue for a role for Mpp10 in processing at the A1/A2 sites.
Four rRNAs and many proteins comprise the eukaryotic ribosome: the 60S subunit contains 25S (Saccharomyces cerevisiae), 5.8S, and 5S rRNAs, and the 40S subunit houses 18S rRNA. In the nucleolus RNA polymerase I transcribes long, precursor molecules that must be processed to yield the mature 25S, 5.8S, and 18S rRNAs. A schematic of pre-rRNA processing (Fig. 1) shows that the rRNA sequences are flanked by two external transcribed spacers (5′ and 3′) and separated by two internal transcribed spacers (ITS1 and ITS2). Complexes of a small nucleolar RNA and proteins, called snoRNPs, and other nucleolar proteins process this original transcript into the mature rRNAs. The depletion or mutation of many of these processing molecules disrupts pre-rRNA processing (1, 2).
Figure 1.
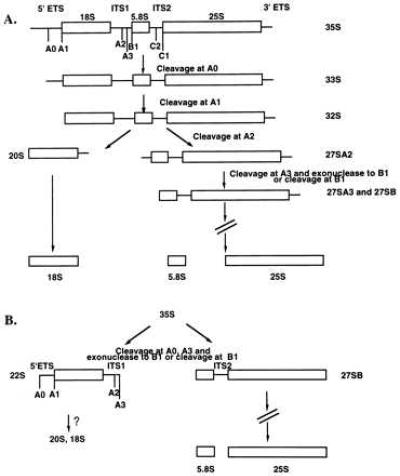
Pre-rRNA processing in S. cerevisiae. (A) The nascent 35S rRNA transcript undergoes successive cleavages at the A0, A1, and A2 sites, dividing it into precursors to the small and large subunit rRNAs. The 20S is processed into the small subunit 18S rRNA. The 27SA2 precursor gives rise to the large subunit rRNAs, 5.8S and 25S. The large subunit RNAs can also be processed from the 27SB precursor, produced by either exonucleolytic degradation to B1 or by direct cleavage at B1. The 27SB precursor undergoes further processing steps to form the mature large subunit rRNAs. (B) Pre-rRNA processing in S. cerevisiae expressing truncated Mpp10 proteins. The levels of rRNA precursors that are generated by cleaving at A1 and A2, the 32S, 20S, and 27SA2 pre-rRNAs, are reduced, whereas the levels of 33S and 23S precursors are similar. A new 22S precursor is visible, resulting from competent cleavage at A0 and deficient cleavage at A1 and A2. It is not known if the 22S precursor is degraded or is slowly processed to form the 18S rRNA.
Many small nucleolar RNAs (snoRNAs) have been identified but U3 is the most abundant in vertebrate cells and exists in a wide evolutionary range of species (from trypanosomes to humans), making it an attractive molecule for studies aimed at elucidation of pre-rRNA processing. In vivo and/or in vitro studies in three different systems, mouse, Xenopus laevis oocytes, and S. cerevisiae, have demonstrated that it is as an essential component for production of 18S rRNA (3–9).
In yeast, processing of the pre-18S rRNA occurs before the cleavage steps that release the mature 5.8S and 25S rRNAs (Fig. 1). The U3 snoRNA has been shown to be necessary for the early processing events that begin at the A0 site and then proceed to the A1 and A2 sites (4, 6, 7, 9). However, other small nucleolar ribonucleoprotein (snoRNPs), including U14 and snR30, are required for formation of 18S rRNA (10, 11). Thus, pre-rRNA processing may be mediated by a multi-snoRNP “processome,” similar to the spliceosome that carries out splicing of pre-mRNAs (12).
Two U3 snoRNA-pre-rRNA base pairing interactions have been identified in yeast (6, 7, 9, 13). One of these interactions, between the U3 snoRNA and a site within the 5′ external transcribed spacers (≈150 nucleotides upstream of the A0 cleavage site), is essential for pre-18S rRNA processing. Base complementarity in this interaction is essential as mutations in the 5′ external transcribed spacer that disrupt the U3-pre-rRNA association impair 18S rRNA production. Processing can be restored with these by making the compensatory mutations in the U3 snoRNA. The sizable distance of this necessary, base paired region from any of the cleavage sites led to a model characterizing the U3 snoRNA as an RNA chaperone. Instead of catalyzing the cleavage, U3 snoRNA may help fold the pre-rRNA and expose cleavage sites to the enzymatically active molecule. Indeed, Rnt1, the yeast homolog of Escherichia coli RNase III (14), is sufficient for cleavage at this site in vitro and cleaves in vivo at this site. The components responsible for cleavage at A1 and A2 have not yet been identified.
Although several U3-specific proteins have been identified in vertebrate cells through immunoprecipitation experiments, only one has been purified and none of their genes have been cloned (15, 16). The genes for two proteins associated with the yeast U3 snoRNP, however, have been cloned: NOP1 (17–19), the yeast homolog of the vertebrate fibrillarin protein, and SOF1 (20). Unlike Nop1, a vertebrate homolog for Sof1 remains unknown. Both proteins are required for yeast viability and production of 18S rRNA (21).
Nop1 is a protein component common to a number of snoRNAs (22), so the phenotypes resulting from mutations in NOP1 may not reflect a function associated with U3. Sof1, on the other hand, is a U3-specific protein that was identified in yeast as an extragenic suppressor of a temperature-sensitive fibrillarin mutant (20). Cells lacking Sof1 mimic the phenotype observed in cells depleted of U3, with impaired cleavage of pre-18S rRNA at the three U3-dependent sites but normal production of 5.8S and 25S rRNAs. Sof1, it appears, plays a necessary role in U3 snoRNP function. Without more detailed studies, however, one cannot acquire further insight into whether Sof1 affects U3 snoRNP assembly and/or the role of the snoRNP in cleavage of the three sites.
Recent studies from our laboratory have characterized a novel protein in yeast and vertebrates, Mpp10, that associates specifically with the U3 snoRNA (J. M. Westendorf, K. N. Konstantinov, S. Wormsley, M. D. Shu, N. Matsumoto-Taniura, F. Pirollet, F. G. Klier, L. Gerace and S.J.B., unpublished results) (23). MPP10 is essential for cell survival in yeast and anti-Mpp10 antibodies specifically immunoprecipitate the U3 snoRNA from yeast cells (23). Depletion of Mpp10 has no effect on 25S rRNA production but halts 18S rRNA production by inhibiting processing at the three U3-dependent sites: A0, A1, and A2.
To better understand its role in pre-rRNA processing, we have made selected mutations in the Mpp10 protein and assayed their functional consequences in yeast. We report here that truncation of only the C terminus of Mpp10 causes the cells to become cold-sensitive, whereas truncation at both ends imparts a slow growth phenotype at all temperatures. Surprisingly, the slow growth results from impaired processing at only the A1 and A2 sites in the pre-rRNA, distinct from the deficiency in pre-rRNA processing that we observe upon depletion of Mpp10. Neither of the truncations affect association of Mpp10 with the U3 snoRNA, nor is U3 snoRNA or protein stability affected. Thus, in characterizing the truncated proteins, we have separated the roles of the U3 snoRNP into cleavage at A0 and cleavage at A1/A2. These results allow preliminary assignment of a role for Mpp10 in processing at the A1/A2 sites.
MATERIALS AND METHODS
Microbiological Medium and Yeast Manipulation.
S. cerevisiae were grown and transformed as described in Dunbar et al. (23). All media used for cell growth, once strains were constructed, was yeast/peptone/dextrose (yeast extract 1%, peptone 2%, glucose 2%).
Yeast Strains.
All constructs were transformed into the haploid yeast strain mpp10∷HIS3 pGAL1∷MPP10. The construction of this strain and its genotype is described in Dunbar et al. (23).
Cloning of the Truncation Mutations.
Oligonucleotides targeted to the desired ends for each of the truncation mutations were used to perform PCR using a full-length MPP10 gene in pET28a as a template. mpp10–1 was generated using ympp10.3 (5′-CCGCGGATCCATGTCAGAACTCTTTGGAGTATTGAAATC-3′) and ympp10.9R (5′-CCCGGAGCTCTCAGACATTGTATATCTCTTGAGG-3′) for the 5′ end and the 3′ end of the mutant gene, respectively. mpp10–2 was generated using ympp10.10 (5′-CCGCGGATCCATGGCAGAACTGGACGAAATC-3′) and ympp10.9R. mpp10–3 required use of ympp10.10 and ympp10.16 (5′-CCCGCTCGAGTCAAAGTTTTCTATTTGTGCT-3′). The fragment corresponding to the terminus, necessary to construct mpp10–4, was generated using ympp10.15 (5′-CCCGCTCGAGTCATGTCGAATGCCTCT-3′) and ympp10.16.
Reactions were carried out under standard conditions using reagents supplied by Perkin–Elmer on a GeneAmp PCR 2400 machine for 20 rounds of amplification using Taq DNA polymerase. Each cycle consisted of 30 sec at 94°C, 30 sec at 55°C, and 30 sec at 72°C.
All PCR fragments were purified using the QIAquick PCR Purification Kit (Qiagen, Chatsworth, CA). The fragments for all the mutants except mpp10–4 were digested with BamHI and AvaI, resolved on a 0.8% gel, and purified using GeneClean (Bio101, Vista, CA). Digested and purified fragments were ligated into p415GPD (AmpR, LEU2, ARS/CEN) at the BamHI and XhoI sites (24). The C-terminus fragment was ligated into the XhoI site of mpp10–2 to make mpp10–4. The fragment was prepared in a similar manner to the other fragments but digested only with XhoI and purified on a 2.0% agarose gel.
Plasmids were extracted from E. coli using QIAprep Spin Miniprep Kits (Qiagen) or Jetstar kits for large scale preparations. Construct identity was verified by restriction digest (BamHI and XhoI) and sequencing of the 5′ and 3′ ends of each insert (automated on an Applied Biosystems 373 Stretch sequencer) using oligos ympp10.13 (5′-GCTACCATCAGAGAGATTGTGAGAAGGC-3′) and ympp10.14 (5′-GAGCTTCAAAAGGCACATTCCG-3′), respectively. Restriction mapping using ClaI and BglII confirmed the correct orientation of the C terminus fragment in mpp10–4. All reagents for digests were obtained from New England Biolabs and reactions were carried out according to their specifications.
Growth Curves.
Yeast were grown in yeast/peptone/dextrose at 30°C until OD600 0.1–0.9. Cells were diluted to OD600 0.050 in yeast/peptone/dextrose and regrown at 30°C. Optical density at 600 nm was measured on a Beckman DU-64 Spectrophotometer at the indicated time points.
Plasmid Retrieval from Yeast.
Plasmids were harvested from yeast using the method shown in ref. 25. To verify the presence of the truncated Mpp10 genes in our mutant strains, the plasmids were restriction mapped and the ends were sequenced on an Applied Biosystems DNA Sequencer at the William Keck facility at Yale.
Western Blot Analysis.
Extracts were made from yeast grown in yeast/peptone/dextrose at 30°C to an OD600 of 0.6–0.8. Cells were washed with water, resuspended in NET-2 (20 mM Tris⋅Cl, pH 7.5/150 mM NaCl/0.05% Nonidet P-40) with protease inhibitors and lysed by vortexing (6 × 45 sec) with 0.45–0.5 mm glass beads. The lysate was cleared by centrifugation for 5 min at 13,200 rpm at room temperature. Total protein (2.5 mg) was separated on 10% SDS/PAGE and transferred onto a nitrocellulose membrane. Expression of Mpp10 in each of the strains was tested by Western blot analysis (enhanced chemluminescence, Amersham) using the anti-yeast Mpp10 antibody characterized in Dunbar et al. (23).
Immunoprecipitations.
2.5 mg of protein A-Sepharose CL-4B (Pharmacia) beads were complexed with anti-yeast Mpp10 (50 μl of rabbit serum) in 0.5 ml of NET-2 (20 mM Tris⋅Cl, pH 7.5/150 mM NaCl/0.05% Nonidet P-40) by nutating overnight at 4°C. Protein A-Sepharose CL-4B was also prepared without antibody for a control (mock). The beads were washed three times with 1 ml NET-2.
Yeast extracts were prepared by the same methods as those used for the Western blot assay except the cells were harvested at OD600 of 5. Extract for 20 OD600 units of cells was complexed with the antibody-bound beads for 1 hr at 4°C. The pellet was washed eight times with 1 ml NET-2. RNA was recovered by PCA acid extraction, ethanol precipitated, and resolved on an 8% denaturing polyacrylamide gel. RNA was transferred to a Zeta-Probe membrane (Bio-Rad) and analyzed by Northern blot with 32P-αUTP labeled anti-sense U3 RNA according to Dunbar et al. (23).
Analysis of Pre-rRNA Processing.
Total RNA preparation, gel conditions, and blotting procedure, were carried out as in Dunbar et al. (23). Growth conditions of the yeast are indicated in the text. Equal amounts of total RNA were loaded in each lane. Blots were probed with several oligonucleotides described in Berges et al. (26). Blots were also probed with oligo z (called oligo c in ref. 27).
RESULTS
Cloning of Mpp10 Truncation Mutations.
Although there is only 30% identity between the yeast and human MPP10 genes, the distribution of charged residues is highly conserved, indicating potential functional significance. We based our yeast MPP10 truncations on these charged regions. Truncated gene fragments of MPP10 were generated by PCR from an intact gene with oligonucleotides directed at the designated truncation sites. All of the Mpp10 gene mutations and the full-length Mpp10 were cloned into a yeast expression vector, p415GPD.
We constructed the following genes: mpp10–1, a C-terminal truncation of 109 amino acids; mpp10–3, an N-terminal truncation of 46 amino acids; and mpp10–2, carrying both truncations. The identity of the mutants was verified by restriction digests and DNA sequencing of the ends from plasmids rescued from yeast. Sequence data revealed several identical amino acid substitutions in the C terminus of mpp10–1 and mpp10–2, both carried in the oligonucleotide used to make the C-terminal truncation. The last five amino acids in these mutants are Arg-Pro-Arg-Val-Met, whereas the corresponding sequences in the wild-type protein are Gln-Pro-Leu-Tyr-Met. To test for the possibility that these substitutions contribute to the observed phenotypes, we cloned a fourth construct, mpp10–4. We constructed this gene by reintroducing the truncated C-terminal sequence onto mpp10–2 rescued from yeast. This mutant represents an N-terminal truncation inclusive of these mutations.
Truncation of Mpp10 Affects Cell Growth.
Strains were constructed by shuffling the plasmids into the mpp10∷HIS3 pGAL1∷MPP10 strain (described in ref. 23). Following selection against the pGAL∷MPP10 plasmid on 5-FOA, these new strains, carrying only full-length or truncated MPP10 genes on the p415GPD plasmid, were streaked onto rich media and grown at 18°C, 22°C, 30°C, and 37°C to test for cold and temperature-sensitivity. The results are shown in Fig. 2. Neither mpp10–3 nor mpp10–4 strains exhibited any cold-sensitive phenotypes and none of the mutants were temperature sensitive (data not shown). Thus, the amino acid substitutions introduced by PCR do not produce a detectable growth phenotype. In contrast, the mpp10–1 strain grows normally at 30°C but shows inhibited growth at 22°C. The mpp10–2 strain, the double truncation, confers slow growth even at 30°C and 37°C, and almost no growth at lower temperatures. Therefore a C-terminal truncation of Mpp10 causes cold-sensitive growth and truncation of both ends causes slow growth even at 30°C.
Figure 2.
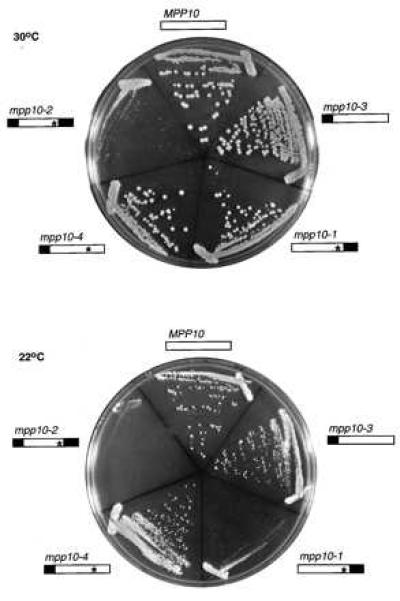
Truncations of the Mpp10 protein result in cold-sensitivity and slow growth of yeast. The Mpp10 protein is represented by □. All four strains bearing Mpp10 truncations, indicated by ▪, were streaked out on rich medium and compared with yeast expressing the full-length Mpp10 protein. Growth was compared at 30°C and 22°C. For the N-terminal truncation, the first 46 amino acids were deleted, and an initiating methionine was added at amino acid 47. For the C-terminal truncation, the last 109 amino acids (amino acids 485–593) were deleted. The asterisk (∗) in mpp10–1, mpp10–2, and mpp10–4 indicate the amino acid substitutions that arose from the oligo used for PCR (refer to Materials and Methods).
To quantify growth differences between MPP10, mpp10–1, and mpp10–2, we recorded growth curves for each strain at 30°C (data not shown). As expected, the strains with MPP10 and mpp10–1 grew at similar rates and the mpp10–2 strain grew at a retarded rate but eventually reached stationary phase. The doubling times were 2, 2.2, and 4 hr for the MPP10, mpp10–1, and mpp10–2 strains, respectively.
To investigate the nature of the growth defect conferred by our truncated proteins, we examined protein levels, U3 snoRNP association, and snoRNA stability, and pre-rRNA processing in our yeast strains.
Truncation of Mpp10 Does Not Affect Levels of Expression.
The expression and stability of the truncated proteins was analyzed by Western blots. Fig. 3 shows that full-length Mpp10, in lane 1, runs anomalously at 110 kDa, even though it has a predicted molecular mass of 67 kDa. This has been observed with other nucleolar proteins that, like Mpp10, also have distinct regions of positive and negative charges (28, 29), and is observed when the Mpp10 protein is expressed in E. coli (23). A band of ≈85 kDa, corresponding to the truncated proteins, is present in both lanes 2 and 3. No size difference between the two proteins is resolvable, even on a lower percentage gel (data not shown). This is not unexpected since even wild-type Mpp10 exhibits anomalous migration. Verification that the yeast strains contain the correct Mpp10 truncations was accomplished by plasmid rescue from the yeast followed by restriction digests and DNA sequencing of the ends of the plasmid inserts (data not shown).
Figure 3.
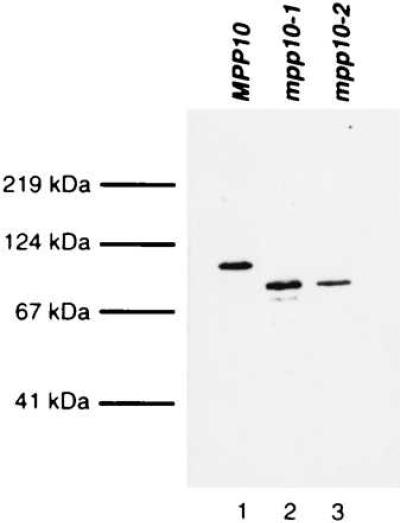
Truncated Mpp10 proteins are stable in yeast and have expression levels similar to that of full-length Mpp10. Western blot analysis of yeast extracts was performed with anti-Mpp10 rabbit serum diluted 1:10,000. Proteins were resolved on 10% SDS/PAGE. The lower, fainter bands in the mpp10–1 lane are degradation products particular to this batch of extract.
Truncation of Mpp10 Does Not Affect U3 snoRNA Association.
To investigate if the truncated Mpp10 proteins were deficient in association with the U3 snoRNA, immunoprecipitations were performed on yeast cell extracts with anti-Mpp10 antibodies. RNA was recovered, from both pellets and total extracts, and resolved on a polyacrylamide gel. For the total extract lanes, we isolated RNA from one-tenth of the volume of extract used for the immunoprecipitation to avoid smearing. Isolated RNA was analyzed via Northern blot by probing with anti-sense U3 snoRNA. Fig. 4 indicates that the mutant Mpp10 proteins associate with the U3 snoRNA to the same extent as wild-type (lanes 5–7). A single band is present in the total extract lanes of all three strains, showing that U3 snoRNA stability is unaffected by truncation of Mpp10 (lanes 1–3). No U3 snoRNA is immunoprecipitable when beads alone are used (lane 4). Therefore the growth defect that we observe in the mpp10–2 strain is not due to a lack of association with the U3 snoRNA nor due to an unstable U3 snoRNA.
Figure 4.
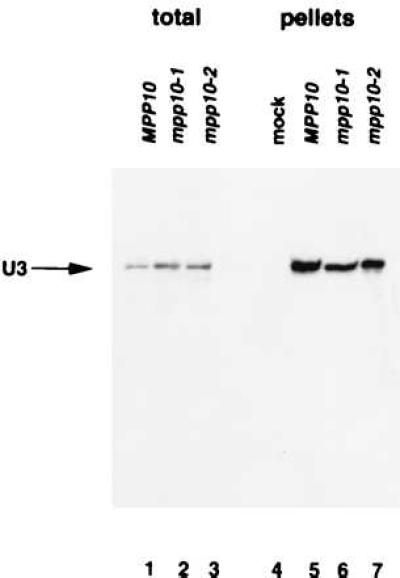
Truncations of Mpp10 do not affect U3 snoRNA association or U3 snoRNA stability. Lanes 1–3 represent RNA isolated from total yeast extract. Lanes 4–7 represent RNA that was isolated from the pellet after immunoprecipitation of yeast extracts with protein A-Sepharose CL 4B beads alone (“mock,” lane 4) or anti-Mpp10 rabbit serum (lanes 5–7). For lanes 1–3, RNA was isolated from 1/10 the volume of extract used for the immunoprecipitations. RNA was resolved on an 8% denaturing polyacrylamide gel. Northern blots were probed with an anti-sense U3 snoRNA.
Truncation of Mpp10 Affects Processing of Pre-18S rRNA Precursors.
The effects of the truncated Mpp10 proteins on pre-rRNA processing was investigated. Total RNA was isolated from yeast grown each to the same density at 30°C (OD600 = 1), resolved on an agarose-formaldehyde gel and analyzed by Northern blot with oligonucleotides that differentiate among mature and precursor rRNAs. The first panel of Fig. 5A shows levels of processed 18S and 25S rRNAs. The steady-state levels of the mature products remain consistent among all three strains (lanes 1–3) when the yeast are grown this way prior to RNA isolation. Refer to Fig. 1 for processing intermediates. The 32S rRNA, resulting from cleavage at A1, is detectable in the wild-type (lane 1), but longer exposures of this panel show that in relation to wild type, levels of 32S are slightly decreased in mpp10–1, and remarkably reduced in mpp10–2 (data not shown). Equal levels of the 33S precursor, across all three strains, is also observable at longer exposures of this panel (data not shown).
Figure 5.
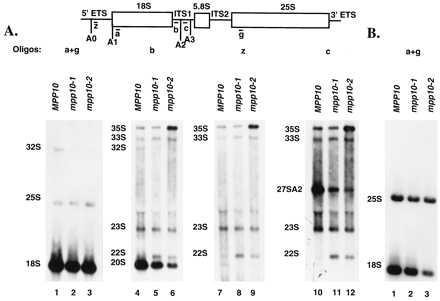
Truncation of the Mpp10 protein impairs processing at sites A1 and A2 in the pre-rRNA. Total RNA from strains bearing full-length MPP10 or the truncations, mpp10–1 and mpp10–2,was resolved on a 1% formaldehyde-agarose gel. Blots were probed with the indicated 32P-labeled oligonucleotides. The relevant cleavage sites are indicated. (A) Northern analysis on RNA that was isolated from strains grown to the same density. (B) Northern blot analysis on RNA isolated from strains grown the same amount of time (10 hr).
Probing with an oligonucleotide that recognizes 18S rRNA precursors demonstrates a deficiency in processing between the two mutants and wild-type (lanes 4–6). Levels of 33S and 23S are consistent among the different strains, but we see a gradual decrease in the accumulation of 20S, the immediate precursor to 18S rRNA, in the mpp10–1 and mpp10–2 strains. Interestingly, the mpp10–1 strain seems to accumulate an intermediate level of 20S, between the levels of wild-type and the mpp10–2 strain. Also significant is the over-accumulation of 35S precursor in the mpp10–2 strain (lane 6) but not in the mpp10–1 strain (lanes 5 and 8). Most importantly, a new 22S precursor is present in the mutants (lanes 5 and 6), but is absent in wild-type (lane 4). This precursor appears in yeast that are deficient in processing at the A1 and A2 sites and is produced by cleavages at the A0 and A3 sites (9, 27). To confirm its identity in our experiments, we reprobed our blots with an oligonucleotide that hybridizes to sequences between the A0 and A1 sites (lanes 7–9). These results clearly indicate that this aberrant precursor is not present in the wild-type (lane 10), but appears in the mutants (lanes 11 and 12). A third oligonucleotide was used to probe for the presence of a precursor to the large subunit RNAs that normally results from cleavage at the A2 site, called 27SA2. The results (lanes 10–12) indicate that the 27SA2 precursor is present in the strain with wild-type Mpp10, but decreased in the two mutant strains. This confirms that cleavage at A2 is impaired in the two mutant strains. Taken together, the results in Fig. 5 suggest that truncation of Mpp10 causes a deficiency in processing primarily at sites A1 and A2 in the pre-rRNA.
We were surprised to detect equivalent levels of 18S and 25S rRNA in our mutant strains when they also exhibit a clear defect in processing of pre-18S rRNA precursors. However, we learned that when we grow the yeast cells, seeded equally, for the same amount of time (10 hr), we observe a graded decrease in 18S levels from wild-type to the mpp10–1 and mpp10–2 mutant strains when compared with 25S rRNA levels (Fig. 5B, lanes 1–3). The effects of the truncation mutations on the pre-rRNA precursors that we observed in Fig. 5B are also visible in Northern blots of yeast grown this way, though they are not as pronounced (data not shown). Growth to equal density prior to RNA analysis, then, aids in determining the nature of the processing defect in cells expressing the Mpp10 truncated proteins.
DISCUSSION
We have made mutations in the Mpp10 protein, a U3 snoRNP component, to better understand its role in pre-rRNA processing in S. cerevisiae. Truncations of the Mpp10 protein cause cold-sensitivity and slow growth. The slow growth is due to a deficiency in processing at only 2 of the 3 U3 snoRNP-dependent sites in the pre-rRNA, visualized by the presence of a new 22S precursor. This is distinct from the deficiency in pre-rRNA processing that we have observed in yeast cells depleted of Mpp10 (23). This processing defect is neither due to instability of the truncated Mpp10 or U3 snoRNA, nor to a lack of association of the truncated proteins with the U3 snoRNA. Thus, our results suggests that an important function of Mpp10 is to enable cleavage at the A1/A2 sites, and these experiments demonstrate a novel separation of the functions of the U3 snoRNP into cleavage at A0 and cleavages at A1 and A2 (Fig. 1).
In yeast, depletion of Sof1, Mpp10, or the RNA component of the U3 snoRNP causes a deficiency in pre-rRNA processing at the A0, A1, and A2 sites in the pre-rRNA. This is indicated by an increase in the 23S and 35S pre-rRNA precursors, and a decrease in the 27SA2 precursor on Northern blots following depletion of any of these components individually (4, 20, 23). This is consistent with the role of the U3 snoRNP in pre-rRNA processing at these three sites. Since the yeast homolog of the E. coli Rnase III protein, Rnt1, cuts at the A0 site in vitro and in vivo (14), the U3 snoRNP may be acting as a chaperone on the pre-rRNA for this reaction.
Although U3 snoRNA depletion causes a deficiency in cleavage at all three sites, specific mutations cause a deficiency at the A1/A2, but not A0, cleavage sites. John Hughes observed the 22S precursor (though referred to as 21S) in yeast that are cold-sensitive and that harbor mutations in the conserved box A sequence of the U3 snoRNA (9). At both 30°C and 16°C, these mutants demonstrate a processing pattern similar to those of our Mpp10 truncation mutants; specifically, a reduction of 32S and 27SA2 and an accumulation of 22S (21S). We do not know if the 22S precursor is a dead-end in pre-rRNA processing, or if it is merely seen when processing at the A1/A2 sites is slowed. Pulse-chase analysis, which might differentiate between these possibilities, has so far been uninformative, since we have been unable to detect aberrant precursors with this methodology even when Mpp10 is depleted (data not shown and ref. 23).
Studies on the U3 snoRNA mutations that confer cold-sensitivity and a deficiency in pre-rRNA processing at the A1 and A2 sites suggests that the U3 snoRNP may play role in the formation of a predicted, conserved pseudoknot at the 5′ end of the 18S rRNA, adjacent to the A1 cleavage site (9, 30). Furthermore, it has been proposed that this pseudoknot-forming interaction is linked to U3-dependent cleavage at sites A1 and A2, since sequences within the highly-conserved box A region of the U3 snoRNA have the potential to base pair with the pseudoknot-forming sequences in the rRNA. One possibility is that the U3 snoRNP, by virtue of base pairing between the snoRNA and the pre-rRNA, brings the pseudoknot-forming sequences proximal to each other, prior to nearby cleavage at A1. This is supported by experimental evidence derived from studying the cold-sensitive U3 snoRNA mutants, since they bear nucleotide changes that would disrupt these interactions. Because our Mpp10 truncation mutants also show deficient A1/A2 processing, perhaps these mutations target the same mechanism. One possibility is that Mpp10 is required for facilitating or maintaining snoRNA-pre-rRNA base pairing, perhaps by contacting both the snoRNP and the pre-rRNA at the same time.
Strikingly, both the Mpp10 truncations and these U3 mutations confer cold-sensitivity, which is a hallmark of molecules involved in macromolecular assembly. Arguments based on thermodynamic principles have asserted that macromolecular assembly mutants are particularly sensitive to cold temperatures. Because these processes are largely driven by hydrophobic interactions, the effects of assembly mutations can be exaggerated at lower temperatures (31). These arguments are also supported by former studies that found that cold-sensitive mutants affecting ribosome assembly were easier to generate than temperature-sensitive ones (32, 33). We do not yet know if the Mpp10 truncations and the cold-sensitive U3 mutants, when coexpressed, cause lethality. If so, this would suggest that the two molecules are functioning in the same pathway.
Our results suggest that slowed growth in the truncation mutant strains is the result of impaired processing of pre-rRNA. Although both mpp10–1 and mpp10–2 strains show inefficient processing at these sites at 30°C, only the double mutant, the mpp10–2 strain, incurs a growth defect. Northern analysis on steady-state levels of rRNA reveals that both the mpp10–1 and mpp10–2 strains have decreased levels of 18S rRNA, mpp10-2 more severely affected than mpp10–1. From wild type to mpp10–1 to mpp10–2, a successive under-accumulation of both 32S and 20S rRNA is observed, indicative that processing efficiency also declines in this graded fashion among the three strains. This suggests that there may be a level of processing deficiency that is tolerable for the cells and allows them to divide normally, as in the mpp10–1 strain. Rates of rRNA production in mpp10–1 seem to fall above this level and the strain grows as quickly as wild-type yeast. However, if the processing efficiency falls below this permissible level, the rate of rRNA production is no longer able to match the demands of a cell dividing at wild-type rates. Therefore, even though processing in the mpp10–2 strain is not drastically different from the mpp10–1 strain, the decrease in efficiency compared with wild-type must exceed the threshold, causing the double mutant to show a sharp two-fold decrease in cell division rate.
Of note is the accumulation of the primary transcript, the 35S rRNA, in the double truncation mutant strain (mpp10–2), but not in the single truncation strain (mpp10–1). It is possible that the appearance of an increase in the 35S precursor occurs when pre-rRNA processing is only severely affected, as an increase in the 35S precursor is also observed for yeast individually depleted of three U3 snoRNP components (9, 20, 23). This is in contrast to results obtained after disrupting pre-rRNA processing in Xenopus oocytes, where levels of the primary transcript remain constant following depletion of the U22 snoRNA, which is also required for pre-18S rRNA processing (34, 35). This may reflect a difference between dividing and nondividing cells, or between yeast and vertebrate cells.
Although our results indicate that part of the Mpp10 protein is required for U3 snoRNA association, it is not known with which part of the U3 snoRNA it is associated. Experiments in vertebrates have indicated that only the 3′ half of U3 is required for fibrillarin binding, and this association requires an intact box C (36). A 55 kDa U3 snoRNP protein identified in hamster cells is also associated with the 3′ half of the U3 snoRNA (16). Determination of the U3 nucleotides required for binding of Mpp10 may strengthen our view that it is involved in the formation of the pseudoknot at the 5′ end of the 18S rRNA, if similar sequences are required for U3 snoRNP association and pre-rRNA processing.
Acknowledgments
We wish to thank these members of the Baserga lab for their help and advice: Steven Wormsley, David Dunbar, and Tina Agentis. We thank Peter Glazer and Chris Yoo for critical reading of the manuscript. S.J.B. is a member of the Yale Cancer Center.
ABBREVIATIONS
- snoRNA
small nucleolar RNA
- snoRNP
small nucleolar ribonucleoprotein
References
- 1.Maxwell E S, Fournier M J. Annu Rev Biochem. 1995;64:897–934. doi: 10.1146/annurev.bi.64.070195.004341. [DOI] [PubMed] [Google Scholar]
- 2.Venema J, Tollervey D. Yeast. 1995;11:1629–1650. doi: 10.1002/yea.320111607. [DOI] [PubMed] [Google Scholar]
- 3.Kass S, Tyc K, Steitz J A, Sollner-Webb B. Cell. 1990;60:897–908. doi: 10.1016/0092-8674(90)90338-f. [DOI] [PubMed] [Google Scholar]
- 4.Hughes J M X, Ares M., Jr EMBO J. 1991;10:4231–4239. doi: 10.1002/j.1460-2075.1991.tb05001.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mougey E B, Pape L K, Sollner-Webb B. Mol Cell Biol. 1993;13:5990–5998. doi: 10.1128/mcb.13.10.5990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Beltrame M, Tollervey D. EMBO J. 1992;11:1531–1542. doi: 10.1002/j.1460-2075.1992.tb05198.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Beltrame M, Henry Y, Tollervey D. Nucleic Acids Res. 1994;22:4057–4065. doi: 10.1093/nar/22.20.4057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Savino R, Gerbi S A. EMBO J. 1990;9:2299–2308. doi: 10.1002/j.1460-2075.1990.tb07401.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hughes J M. J Mol Biol. 1996;259:645–654. doi: 10.1006/jmbi.1996.0346. [DOI] [PubMed] [Google Scholar]
- 10.Li H V, Zagorski J, Fournier M J. Mol Cell Biol. 1990;10:1145–1152. doi: 10.1128/mcb.10.3.1145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Morrissey J P, Tollervey D. Mol Cell Biol. 1993;13:2469–2477. doi: 10.1128/mcb.13.4.2469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fournier M J, Maxwell E S. Trends Biochem Sci. 1993;18:131–135. doi: 10.1016/0968-0004(93)90020-n. [DOI] [PubMed] [Google Scholar]
- 13.Beltrame M, Tollervey D. EMBO J. 1995;14:4350–4356. doi: 10.1002/j.1460-2075.1995.tb00109.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Elela S A, Igel H, Ares M., Jr Cell. 1996;85:115–124. doi: 10.1016/s0092-8674(00)81087-9. [DOI] [PubMed] [Google Scholar]
- 15.Parker K A, Steitz J A. Mol Cell Biol. 1987;7:2899–2913. doi: 10.1128/mcb.7.8.2899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lubben B, Marshallsay C, Rottman N, Luhrmann R. Nucleic Acids Res. 1993;21:5377–5385. doi: 10.1093/nar/21.23.5377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Henriquez R G, Blobel G, Aris J P. J Biol Chem. 1990;265:2209–2215. [PubMed] [Google Scholar]
- 18.Jansen R P, Hurt E C, Kern H, Lehtonen H, Carmo-Fonseca M, Lapeyre B, Tollervey D. J Cell Biol. 1991;113:715–729. doi: 10.1083/jcb.113.4.715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Schimmang T, Tollervey D, Kern H, Frank R, Hurt E C. EMBO J. 1989;8:4015–4024. doi: 10.1002/j.1460-2075.1989.tb08584.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jansen R, Tollervey D, Hurt E C. EMBO J. 1993;12:2549–2558. doi: 10.1002/j.1460-2075.1993.tb05910.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tollervey D, Lehtonen H, Carmo-Fonseca M, Hurt E C. EMBO J. 1991;10:573–583. doi: 10.1002/j.1460-2075.1991.tb07984.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Balakin A G, Smith L, Fournier M J. Cell. 1996;86:823–834. doi: 10.1016/s0092-8674(00)80156-7. [DOI] [PubMed] [Google Scholar]
- 23.Dunbar D A, Wormsley S, Agentis T M, Baserga S J. Mol Cell Biol. 1997;17:5803–5812. doi: 10.1128/mcb.17.10.5803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mumberg D, Muller R, Funk M. Gene. 1995;156:119–122. doi: 10.1016/0378-1119(95)00037-7. [DOI] [PubMed] [Google Scholar]
- 25.Hoffman C-S, Winston F. Gene. 1987;57:267–272. doi: 10.1016/0378-1119(87)90131-4. [DOI] [PubMed] [Google Scholar]
- 26.Berges T, Petfalski E, Tollervey D, Hurt E C. EMBO J. 1994;13:3136–3148. doi: 10.1002/j.1460-2075.1994.tb06612.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lafontaine D, Vandenhaute J, Tollervey D. Genes Dev. 1995;9:2470–2481. doi: 10.1101/gad.9.20.2470. [DOI] [PubMed] [Google Scholar]
- 28.Benton B M, Zang J H, Thorner J. J Cell Biol. 1994;127:623–639. doi: 10.1083/jcb.127.3.623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Meier U T. J Biol Chem. 1996;271:19376–19384. [PubMed] [Google Scholar]
- 30.Dennis P P, Russell A G, Moniz D S M. RNA. 1997;3:337–343. [PMC free article] [PubMed] [Google Scholar]
- 31.Strauss E J, Guthrie C. Genes Dev. 1991;5:629–641. doi: 10.1101/gad.5.4.629. [DOI] [PubMed] [Google Scholar]
- 32.Guthrie C, Nashimoto H, Nomura M. Proc Natl Acad Sci USA. 1969;63:384–391. doi: 10.1073/pnas.63.2.384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tai P C, Kessler D P, Ingraham J. J Bacteriol. 1969;97:1298–1304. doi: 10.1128/jb.97.3.1298-1304.1969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tycowski K T, Shu M D, Steitz J A. Science. 1994;266:1558–1561. doi: 10.1126/science.7985025. [DOI] [PubMed] [Google Scholar]
- 35.Dunbar D A, Ware V C, Baserga S J. RNA. 1996;2:324–333. [PMC free article] [PubMed] [Google Scholar]
- 36.Baserga S J, Yang X W, Steitz J A. EMBO J. 1991;10:2645–2651. doi: 10.1002/j.1460-2075.1991.tb07807.x. [DOI] [PMC free article] [PubMed] [Google Scholar]


