Abstract
Tropical parasitic and infectious diseases, such as leishmaniasis, pose enormous global health threats, but are largely neglected in commercial drug discovery programs. However, the Panama International Cooperative Biodiversity Group (ICBG) has been working to identify novel treatments for malaria, Chagas’ disease, and leishmaniasis through an investigation of plants and microorganisms from Panama. We have pursued activity-guided isolation from an extract of Lyngbya majuscula that was found to be active against leishmaniasis. A new modified linear peptide from the dragonamide series was isolated, dragonamide E (1), along with two known modified linear peptides, dragonamide A (2) and herbamide B (3). Dragonamides A and E, and herbamide B, exhibited antileishmanial activity with IC50 values of 6.5, 5.1, and 5.9 µM, respectively. Spectroscopic and stereochemical data for dragonamide E (1) and herbamide B (3; the spectroscopic and stereochemical data for this substance is incomplete in the literature) are presented as well as comparisons of biological activity within the dragonamide compound family. Biosynthetic differences among marine compounds with a terminal free amide are also discussed.
Tropical diseases such as leishmaniasis, malaria, and Chagas’disease affect millions of people in equatorial countries each year.1 Inhabitants of these developing-world tropical countries are both at high risk from infection and are often those with the least socio-economic ability to obtain proper treatments; therefore, they are the most likely to develop serious and often fatal illnesses.2 Many tropical diseases have developed resistance to existing therapeutics, thus limiting the effectiveness of these treatments. A majority of the existing agents for treating tropical diseases also have serious side effects that reduce patient compliance with treatment regimens, or in some cases, prohibit any treatment at all.1 Tropical diseases are largely overlooked in drug discovery programs by major pharmaceutical companies due to the lack of significant financial return on this expensive and time-consuming process (only 13 of the 1,300 new drugs introduced during the period 1975 through 1999 were for treating tropical diseases).1
The International Cooperative Biodiversity Group (ICBG) program focused in Panama has been searching among marine cyanobacteria and tropical plant endophytes for new, more efficacious, and less expensive treatments for tropical diseases.3–5 It is hoped that these novel agents will show activity against resistant strains of these parasites. During the course of this research, a new modified linear lipopeptide, dragonamide E (1), as well as two related compounds previously reported in the literature, dragonamide A (2), and herbamide B (3), were isolated from a field collected marine cyanobacterium and found to be active against leishmaniasis in an in vitro screening assay.
Results and Discussion
L. majuscula was collected using snorkeling in shallow waters from Bocas del Toro, Panama in 2006, and subsequently extracted and fractionated using normal-phase flash chromatography. One of the fractions (60% EtOAc/hexanes) exhibited strong anti-leishmanial activity (7.2% of control parasite growth at 10 µg/mL, experimental detail in Supporting Information) while a second more polar fraction eluting with 100% EtOAc exhibited less potent anti-leishmanial activity (38.2% of control parasite growth at 10 µg/mL). This more polar (100% EtOAc) fraction was successively purified by RP-SPE column chromatography and RP-HPLC, and two compounds were ultimately isolated, dragonamides E (1) and A (2). Metabolite 2 was the major compound of this fraction and exhibited 1H and 13C NMR signals indicative of phenylalanyl and valinyl residues, along with a fatty acyl chain. Combined with a prominent [M+H]+ peak by APCIMS at m/z 654, these data were fully consistent with literature values for the known metabolite dragonamide A (2).6,7
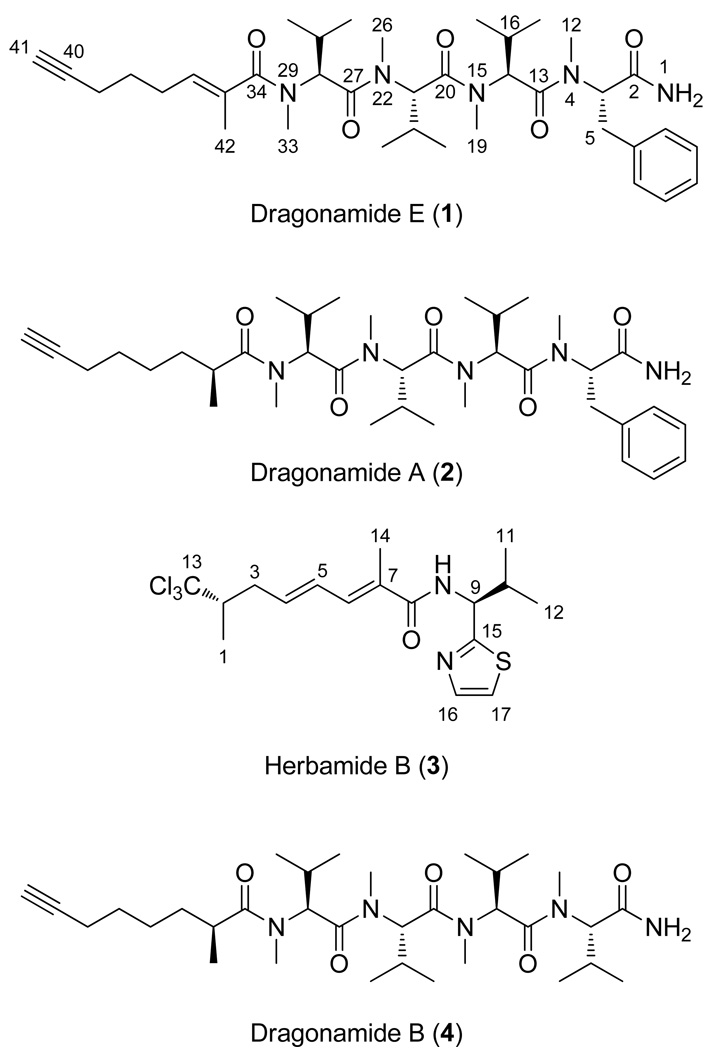
The second eluting and less abundant compound from this fraction, named dragonamide E (1), gave an [M+H]+ peak by HRESI-TOFMS that calculated for C37H58N5O5 (12 degrees of unsaturation) and by database searches was determined to be an unknown substance. The 1H NMR spectra of 1 and dragonamide A (2) were very similar, and both included signals for a terminal NH2 (δ 5.99), an N-methyl phenylalanyl and three N-methyl valinyl residues. While both 1 and 3 existed in a single major conformation in solution, the presence of multiple N-methyl amide functionalities resulted in the presence of several minor conformers as well (see Supporting Information). Compound 1 possessed 13C NMR absorptions at δ 84.0 and 69.0, consistent with a terminal acetylene as in dragonamides A (2) and B (4). However, dragonamide A (2) possesses two more protons and only 11 degrees of unsaturation compared to dragonamide E. In the 1H NMR spectrum of 1, the distinctive signal corresponding to the methine proton on C-35 (δH 2.68; δC 36.2) in 2 was not present. Instead, in 1 the C-35 carbon resonated significantly downfield (δ 133.0) compared to dragonamide A, and by DEPT and HSQC analysis, was identified as a quaternary carbon. These observations suggested that the extra degree of unsaturation in 1 relative to 2 was the result of a double bond between C-35 and C-36. This deduction was confirmed by location of H-36 at δ 5.41 in the 1H NMR and observation of HMBC correlations between H-36 and C-42 (δ 14.4) as well as C-34 (δ 173.9). Correlations observed between the H-42 methyl protons and the H-37 methylene protons by NOESY indicated an E geometry for the C-35–C-36 olefinic bond.
The absolute configurations of the four amino acid residues in dragonamide E (1) were determined using Marfey’s analysis. Following hydrolysis, dragonamide E (1) was derivatized using FDVA Marfey’s reagent and compared with commercially available N-methyl phenylalanine and N-methyl valine standards that were similarly transformed by Marfey’s reagent. All stereogenic centers in dragonamide E (1) were determined to have the l configuration.
The less polar (60% EtOAc/hexanes) yet strongly anti-leishmanial fraction from the primary chromatography was subjected to reversed-phase solid-phase extraction (SPE) and reversed-phase HPLC to yield herbamide B (3), a previously reported but incompletely characterized natural product.6 Several partial structures were determined from NMR analysis of 3 which aided its identification. Specifically, the presence of a modified valine residue was determined based on observation of two methyl doublet signals (δ 0.99 and 0.94), a double-doublet α-proton signal (δ 5.33), a methine (δ 2.36), and an N-H (δ 6.68). The presence of a thiazole ring was determined by the distinctive proton doublet signals at δ 7.27 and 7.75, and was connected to the valine residue through observation of a 3-bond HMBC correlation between C-15 (δ 170.1) and H-10 (δ 2.36). The constellation of a downfield shifted doublet methyl at δ 1.31 with an HMBC correlation to a uniquely downfield shifted quaternary carbon (δ 105.5), and a high field methine proton at δ 2.60 with HMBC correlations to the carbon atom of the aforementioned downfield shifted doublet methyl (δ 16.2), suggested a terminus with methyl and trichloromethyl groups, as in barbamide.8 The remaining downfield singlet methyl (δ 2.02) was assigned to a vinyl position based on its chemical shift, and was placed at C-7 based on a strong HMBC correlation between this resonance and the unsaturated ester carbonyl absorption band at δ 168.5. Piecing these partial structures together identified a molecular framework consistent with that published for herbamide B, and upon detailed comparison, all of the 1H and 13C NMR signals closely matched those published for this structure.6 Our isolate of herbamide B (3) showed an intense [M+H]+ peak by HR APCIMS for C17H24Cl3N2OS, thus confirming the structure assigned previously by NMR analysis alone.6
The stereoconfiguration of herbamide B (3) and the closely related compound, herbamide A, have never been determined. The configuration of C-9 in the modified valine residue of 3 was determined using sequential ozonolysis, hydrolysis, and Marfey’s analysis using the FDVA Marfey’s reagent. This fragment was compared with commercially available valine standards that were also transformed by Marfey’s reagent, and identified this substituent in herbamide B (3) to have an l configuration.
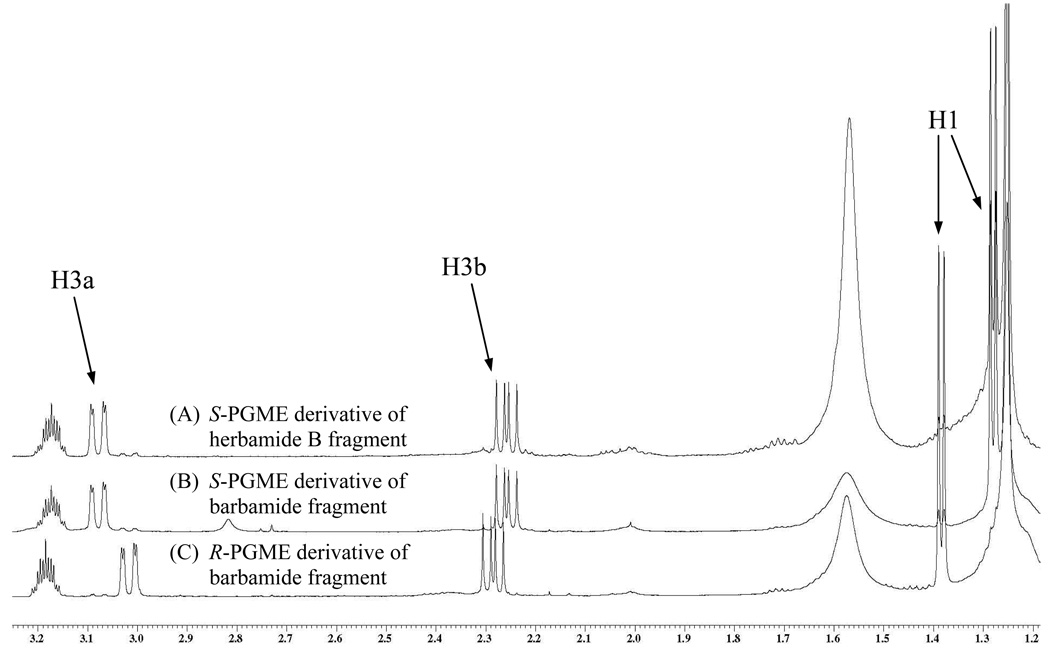
The remaining stereocenter at C-2 of 3 was determined using the semi-synthetic strategy shown in Figure 1. The planar structures of the fragment that results from ozonolysis and oxidative workup of 3 and that resulting from ozonolysis and cleavage of the methyl ester of barbamide are identical. The methine adjacent to the trichloromethyl in barbamide has previously been determined to have the S configuration,9 and an abundant supply of this pure and fully characterized compound was available. After reaction of the barbamide fragment with both enantiomers of 2-phenylglycine methyl ester (PGME), two diastereomeric compounds were obtained with distinctive chemical shifts for the H-1 methyl and H-3 methylene (Figure 2). Herbamide B (3), with unknown configuration at C-2, was subjected to ozonolysis and reaction with S-PGME, and found to have the identical chemical shifts as the barbamide fragment reacted with S-PGME (Figure 2), indicating that herbamide B and barbamide have the same configuration at this methine center. The final, stereochemically complete structure for 3 was therefore determined to have S configuration at C-2 and an l-valine residue at C-9.
Figure 1.
Semi-synthetic strategy utilized to determine stereochemical configuration of herbamide B (3) at C-2.
Figure 2.
1H NMR comparison of chemical shifts for the S-PGME derivative of the herbamide B fragment (A), the S-PGME derivative of the barbamide fragment (B), and the R-PGME derivative of the barbamide fragment (C). The S-PGME derivative of the herbamide B fragment directly overlaps with the S-PGME derivative of the barbamide fragment and has distinctive shift differences from the R-PGME derivative of the barbamide fragment, indicating that herbamide B (3) and barbamide have the same configuration at this center.
The identical configuration of the trichloromethyl for both barbamide and herbamide B is potentially indicative of the conservation of the BarbB1 and BarbB2 enzymes in the herbamide B producer and their chiral preference during trichloromethyl biosynthesis.10,11 However, the subsequent biosynthetic steps for herbamide B likely involve the incorporation of at least one extra PKS module as well as utilization of alternative optional PKS domains (e.g., dehydratase and C-methyltransferase).12 Further studies on the biosynthesis of herbamide B should help illuminate the regions of similarity and divergence from the barbamide pathway.
Compounds 1–3 were tested for activity against three tropical disease parasites, Plasmodium falciparum (malaria), Leishmania donovani (leishmaniasis), and Trypanosoma cruzi (Chagas’ disease). Compound 4 was previously tested in these assays,7 and those results are also reported here to allow for direct comparison with the biological properties of dragonamides A (2), B (3), and E (1). Dragonamide E (1) exhibited moderately strong in vitro activity against leishmaniasis (5.1 µM), with dragonamide A (2) and herbamide B (3) also showing comparable activity against this parasite (6.5 µM and 5.9 µM, respectively). Dragonamide B (4) was inactive against all of the tested parasites, indicating that the aromatic ring-containing residue at the peptide terminus is necessary for activity. Interestingly, dragonamide A (2) previously showed weak activity against malaria parasites;7 however, during the current evaluation, 2 was found to be inactive in this assay (maximum test concentration = 10 µM). The in vitro activity of dragonamide E (1) and herbamide B (3) against the parasite that causes visceral leishmaniasis supports their further examination through in vivo evaluations. This is especially attractive in the case of herbamide B because it was isolated as a major metabolite of this cyanobacterial collection, and hence, analogs could be produced through natural product derivitization strategies.
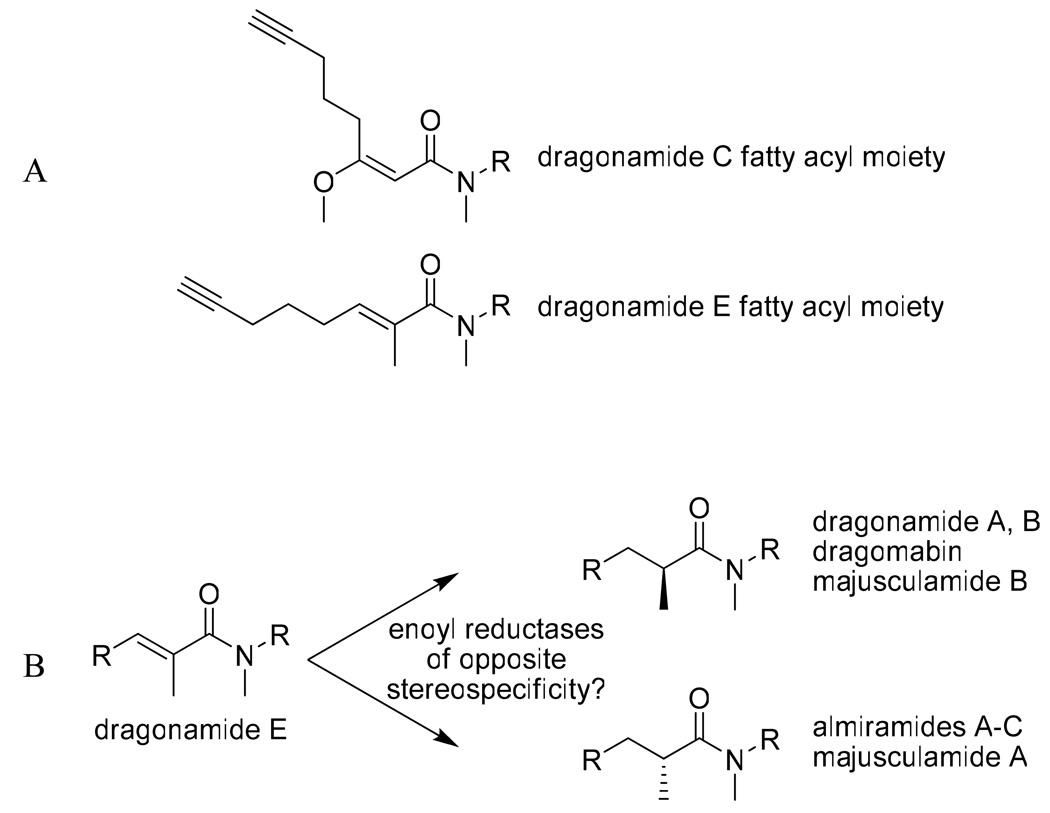
The isolation of dragonamide E (1) represents an interesting addition to the dragonamide family of natural products. This fatty acid moiety has no precedence in the marine literature. All other compounds in the dragonamide series have secondary methyl groups of S configuration at the C-35 or equivalent position, with the exception of dragonamide C which possesses an E configuration olefinic bond between C-31 and C-32 (corresponds to C-35 and C-36 in dragonamide E). However, dragonamide C lacks an α-methyl group (C-42 in compound 1), but rather, possesses a methoxy at C-32 which causes the fatty acyl moiety of dragonamide C to have a distinctively different spatial orientation (Figure 3a). However, it is possible that dragonamide C is an isolation artifact, created during the methanol extraction process from the corresponding β-keto compound; further studies are needed to explore this point. Isolation of dragonamide E with an alkene between C-35 and C-36 of the fatty acyl moiety is potentially important in that it suggests a possible mechanism by which other modified linear peptides are created with both R and S configuration α-methyl groups at this center (Figure 3b), perhaps through enoyl reductases of different facial selectivity, or E versus Z double bonds at this location.
Figure 3.
(A) The distinctive spatial arrangements of the fatty acyl moieties of dragonamide E (1) and dragonamide C, and (B) dragonamide E as a precursor for either the R or S configuration of the fatty acyl moiety, present in dragonamides A and B, dragomabin, and majusculamide B (S moiety) and almiramide A–C and majusculamide A (R moiety).
The isolation of dragonamide E (1) as the fifth compound in the series led us to consider the biosynthetic relationships of these compounds, all isolated from Lyngbya species, as well as for several related compounds with a free amide terminus (Table 2). A total of 18 compounds with a primary amide terminus have been reported from various marine organisms, including cyanobacteria (11 compounds), molluscs (2), sponges (3), and shrimp (2). These have been isolated from collections in both the Pacific Ocean (Japan, Hawaii, and Ecuador) and the Caribbean Sea (Panama, Florida, and Curaçao), indicating no apparent specialization based on geography. A range of biological activities has been reported for these compounds including antiparasitic,6,7 anticancer,13 antifungal,14 diuretic,15 myotropic,15 inhibition of serine protease,16 and cardioexcitatory activity.17
Table 2.
Relationships of Marine Peptides Containing a Terminal Primary Amide Functionality.
| compound | terminus | AA1 | AA2 | AA3 | AA4 | additional AAs | biological activity |
organism | species | location |
|---|---|---|---|---|---|---|---|---|---|---|
| dragonamide A6,7 | NH2 | N-Me l-Phe | N-Me l-Val | N-Me l-Val | N-Me l-Val | leishmaniasis malaria (weak) |
cyanobacteria |
Lyngbya majuscula |
Panama | |
| dragonamide B7 | NH2 | N-Me l-Val | N-Me l-Val | N-Me l-Val | N-Me l-Val | none | cyanobacteria |
Lyngbya majuscula |
Panama | |
| dragonamide C13 | NH2 | N-Me l-Val | N-Me l-Val | N-Me l-Val | N-Me l-Val | cancer (weak) | cyanobacteria |
Lyngbya polychroa |
Florida | |
| dragonamide D13 | NH2 | N-Me l-Val | N-Me l-Val | N-Me l-Val | N-Me l-Val | cancer (weak) | cyanobacteria |
Lyngbya polychroa |
Florida | |
| dragonamide E | NH2 | N-Me l-Phe | N-Me l-Val | N-Me l-Val | N-Me l-Val | leishmaniasis | cyanobacteria |
Lyngbya majuscula |
Panama | |
| carmabin A7,21 | NH2 | N-Me l-Tyr | N-Me l-Ala | l-Ala | N-Me l-Phe | malaria | cyanobacteria |
Lyngbya majuscula |
Panama Curaçao |
|
| carmabin B21,* | NH2 | N-Me Tyr | N-Me Ala | Ala | N-Me Phe | none | cyanobacteria |
Lyngbya majuscula |
Curaçao | |
| dragomabin7 | NH2 | N-Me l-Tyr | N-Me l-Ala | l-Ala | N-Me l-Phe | malaria | cyanobacteria |
Lyngbya majuscula |
Panama | |
| majusculamide A22 | NH2 | N-Me l-Val | N,O-Me2 d-Tyr | none | cyanobacteria |
Lyngbya majuscula |
Hawaii | |||
| majusculamide B22 | NH2 | N-Me l-Val | N,O-Me2 l-Tyr | none | cyanobacteria |
Lyngbya majuscula |
Hawaii | |||
| dysidenamide6 | NH2 | N-Me l-Ala | Cl3 l-Leu | none reported | cyanobacteria |
Lyngbya majuscula |
Panama | |||
| dolastatin C23 | NH2 | l-Phe | l-Pro | N-Me l-Ile | l-Hmp | N,N-Me2 l-Ile | none reported | mollusc |
Dolabella auricularia |
Japan |
| (no name provided)17 | NH2 | l-Phe | d-Trp | d-Asn | cardioexcitatory | mollusc | Aplysia sp. | |||
| halicylindramide E14 | NH2 | d-Phe | N-Me l-Gln | l-Thr | d-Cys(O3H) |
l-Arg-d-Trp-(l-t- Leu)-d-Val-l-Pro-(Br- l-Phe)-d-Ala |
antifungal | sponge |
Halichondria cylindrata |
Japan |
| pseudotheonamide C16 | NH2 | d-Phe | S-v-Tyr | α-formyl l-Dpr | l-Pro | S-k-Arg | serine protease inhibition |
sponge |
Theonella swinhoei |
Japan |
| pseudotheonamide D16 | NH2 | d-Phe | S-v-Tyr | α-formyl l-Dpr | l-Pro | serine protease inhibition |
sponge |
Theonella swinhoei |
Japan | |
| pev-kinin 115,* | NH2 | Gly | Trp | Pro | Ser | Phe-Ser-Ala | diuretic | shrimp |
Penaeus vannamei |
Ecuador |
| pev-kinin 215,* | NH2 | Ala | Trp | Ala | Ser | Phe-Asp | diuretic, myotropic |
shrimp |
Penaeus vannamei |
Ecuador |
Stereochemistry not determined.
One of the more striking differences between the structures of these various primary amide containing natural products lies in the high level of N-methylation of the cyanobacterial compounds; the metabolites of other classes of marine life have little or no N-methylation (Table 2). Compounds from organisms other than cyanobacteria also have a high level of unusual peptidic modifications whereas cyanobacterial compounds with terminal amides have little modification beyond N-methylation. Another distinct aspect of these cyanobacterial compounds lies in their extended polyketide tail, which is largely absent in terminal amide compounds from other organisms.
Within the cyanobacterial compounds, the first two amino acid residues are almost always N-methylated, with the exception of dysidenamide (Table 2). N-methyl valine is the most common first and second residue with N-methyl phenylalanine, tyrosine (also O-methylated in the case of the majusculamides), or alanine also being found in these two residues. The third amino acid residue is either alanine or N-methyl valine (or absent as in the majusculamides and dysidenamide). The fourth residue is always N-methylated and is either valine or phenylalanine. The polyketide tails of these cyanobacterial compounds all end in terminal acetylene functionalities except for carmabin B. A 10-carbon PKS chain is the most common but chain length is variable and ranges from 9 to 13 carbons. Dragonamides A and B, as well as dragomabin, all have the same PKS tail comprising a 2-methyloct-7-ynoyl residue with an S configured secondary methyl group.7 The carmabins are both chain extended, containing two additional carbons in the polyketide chain and an additional methyl substituent. The structural differences in the lipophilic chain likely represent variations in the biosynthetic assembly process, specifically, the incorporation of additional PKS domains in the cluster. As deduced from Table 2, the close structural relationship of these primary amide-containing cyanobacterial peptides suggests an evolutionary relationship in their biosynthetic gene clusters, and further exemplifies a strategy by which these marine prokaryotes diversify their suite of expressed natural products.
Experimental Section
General Experimental Procedures
Optical rotations were obtained with a Jasco P-2000 polarimeter. UV and IR spectra were recorded on a Beckman Coulter DU-800 and Nicolet IR100 FT-IR spectrophotometers, respectively. NMR spectra were recorded on a JEOL Eclipse 400 MHz spectrometer or a Varian Inova 600 MHz spectrometer and referenced to residual solvent signals (δH 7.26, δC 77.2 for CDCl3). Low resolution APCI mass spectra were obtained on a JEOL LC-mate mass spectrometer. High resolution mass spectra were acquired on an Agilent ESI-TOF mass spectrometer. HPLC purifications of natural product isolates were carried out on a Merck Hitachi LaChrom HPLC system with a L-7100 pump, L-7614 degasser, and L-7455 diode array detector using a Prontosil-120 C18 (4.6 × 250 mm, 5µM) RP-HPLC column, with solvent systems as indicated below. HPLC purifications of semi-synthetic compounds were carried out on a Waters HPLC system with a Waters 515 binary pump and a Waters 996 PDA detector using a Phenomenex Synergi Fusion 4µ column (10 × 250 mm), with solvent systems as indicated below.
Collection, Extraction, and Isolation
Lyngbya majuscula was collected in July 2006 by hand using snorkeling methods from mangrove roots in the Bastimentos National Park (9° 14.689 N, 82° 08.366 W) in Bocas del Toro, Panama. After straining through a mesh bag to remove seawater, the sample was stored at 4° C. Voucher specimen number PAB-28-JUL-06-1 is deposited at the INDICASAT sample respository, Panama City, Panama. The sample (63.4 g dry weight) was thawed and repetitively extracted 7 times with 2:1 CH2Cl2/MeOH to afford after solvent evaporation 3.2 g of crude organic extract. The extract was fractionated using flash Si gel column chromatography (Aldrich, Si gel 60, 230–400 mesh, 40 × 180 mm) using 300 mL each of 100% hexanes (A), 9:1 hexanes/EtOAc (B), 4:1 hexanes/EtOAc (C), 3:2 hexanes/EtOAc (D), 2:3 hexanes/EtOAc (E), 1:4 hexanes/EtOAc (F), 100% EtOAc (G), 3:1 EtOAc/MeOH (H), and 100% MeOH (I). Fraction E showed strong antileishmanial activity (only 7.2% of control growth at 10 µg/mL), while fraction G showed good antileishmanial activity (38.2% of control growth at 10 µg/mL). Fraction E was subjected to further fractionation using a Burdick & Jackson C18 RP-SPE cartridge using a MeOH-H2O gradient (1:1, 3:2, 7:3, 4:1 MeOH/EtOAc, 100% MeOH, 100% EtOAc). The 7:3 eluting fraction was subjected to RP-HPLC purification (63% MeOH/37% H2O, 209 nm, 1.0 mL/min) to yield herbamide B (3, 11.2 mg, tR 49.5 min, 0.35% of extract). Fraction G was passed through a Burdick & Jackson C18 RP-SPE cartridge with a 0.45 µm nylon filter using 100% MeOH. The eluent was chromatographed using RP-HPLC (69% MeOH/39%H2O, 209 nm, 1.0 mL/min) yielding dragonamide E (1, 3.0 mg, tR 20.9 min, 0.09% of extract) and dragonamide A (2, 22.3 mg, tR 24.8 min, 0.70% of extract).
Dragonamide E (1)
white amorphous solid; [α]25D −220 (c 0.15, CHCl3); UV (MeOH) λmax (log ε) 203 (5.69), 228 (5.16) nm; IR νmax (film) 3251, 2964, 1688, 1632, 1463, 1393, 1259, 1097 cm−1; 1H NMR and 13C NMR data (CDCl3, 400 MHz), see Table 1; APCIMS m/z (%) 652.5 (80, [M+H]+), 474.3 (100), 361.2 (82), 248 (91); HRESI-TOFMS [M+H]+ m/z 652.4418 (calcd for C37H58N5O5, 652.4437).
Table 1.
NMR Data for Dragonamide E (1) in CDCl3.
| position | δCb, mult. | δHa (J in Hz) | COSY | Selected HMBC (H→C) |
Selected NOESY |
|
|---|---|---|---|---|---|---|
| NH2 | 1 | - | 5.99, br s | - | 1 | |
| N-MePhe | 2 | 171.9, qC | - | - | - | - |
| 3 | 56.6, CH | 5.57, dd (11, 5) | 5 | 2, 5 | ||
| 4 | N | - | - | - | - | |
| 5 | 33.4, CH2 | 3.03, dd (ob) | 3 | 3, 6, 7, 11 | 5 | |
| 3.25, dd (15, 5) | ||||||
| 6 | 136.9, qC | - | - | - | - | |
| 7 | 128.9, CH | 7.16, m | 8 | 5, 9 | 1, 3, 5, 8 | |
| 8 | 128.8, CH | 7.28, m | 7, 9 | 6 | 1, 7 | |
| 9 | 127.2, CH | 7.24, m | 8, 10 | |||
| 10 | 128.8, CH | 7.28, m | 9, 11 | 6 | 1, 11 | |
| 11 | 128.9, CH | 7.16, m | 10 | 5, 9 | 1, 5, 10 | |
| 12 | 32.0, CH3 | 2.90, s | - | 3, 13 | 17, 18 | |
| N-MeVal-1 | 13 | 171.5, qC | - | - | - | - |
| 14 | 58.7, CH | 5.08, d (11) | 16 | 13, 16, 17, 19 | 12, 17 | |
| 15 | - | - | - | - | - | |
| 16 | 27.2, CH | 2.26, m | 17, 18 | 18 | ||
| 17 | 20.2, CH3 | 0.88, d (6) | 16 | 14, 16, 18 | 18 | |
| 18 | 17.6, CH3 | 0.66, d (7) | 16 | 14, 16, 17 | 16, 17 | |
| 19 | 29.9, CH3 | 2.43, s | - | 14, 20 | 21 | |
| N-MeVal-2 | 20 | 169.9, qC | - | - | - | - |
| 21 | 58.2, CH | 4.94, d (11) | 23 | 20, 23, 25, 26 | 19 | |
| 22 | - | - | - | - | - | |
| 23 | 27.3, CH | 2.26, m | 21, 24, 25 | |||
| 24 | 17.9, CH3 | 0.74, d (7) | 23 | 21, 23, 25 | ||
| 25 | 19.9, CH3 | 0.71, d (6) | 23 | 21, 23, 24 | ||
| 26 | 30.5, CH3 | 2.98, s | - | 21, 27 | 28 | |
| N-MeVal-3 | 27 | 170.6, qC | - | - | - | - |
| 28 | 58.0, CH | 5.12, d (11) | 30 | 30, 33, 34 | 26 | |
| 29 | - | - | - | - | - | |
| 30 | 26.9, CH | 2.35, m | 28, 31, 32 | 28, 32 | 31 | |
| 31 | 18.1, CH3 | 0.85, d (ob) | 28 | 28, 30, 32 | 30 | |
| 32 | 19.7, CH3 | 0.84, d (7) | 28 | 28, 30, 31 | ||
| 33 | 30.9, CH3 | 2.93, s | - | 28, 34 | 42 | |
| FA moietyc | 34 | 173.9, qC | - | - | - | - |
| 35 | 133.0, qC | - | - | - | - | |
| 36 | 129.8, CH | 5.41, dt (7, 2) | 37 | 34, 37, 42 | 37 | |
| 37 | 26.6, CH2 | 2.23, m (ob) | 36, 38 | 35, 36, 38, 39 | 36, 38 | |
| 38 | 27.8, CH2 | 1.63, q (7) | 37, 39 | 36, 37, 39, 40 | 37, 39 | |
| 39 | 18.2, CH2 | 2.19, dt (7, 2) | 38 | 37, 40, 41 | 38 | |
| 40 | 84.0, qC | - | - | - | - | |
| 41 | 69.0, CH | 1.97, t (2) | - | |||
| 42 | 14.4, CH3 | 1.82, s | - | 34, 35, 36 | 33 |
Measured at 400 MHz.
Measured at 100 MHz.
FA moiety = (E)-2-methyloct-2-en-7-ynoic acid.
Herbamide B (3)
yellow glass; [α]25D −27 (c 0.20, CHCl3); 1H NMR and 13C NMR data (CDCl3, 400 MHz) were as reported;6 HMBC NMR (H→C) (1→2, 3, 13), (2→1, 3, 4, 13), (3→1, 2, 4, 5, 13), (4→2, 3, 6), (5→3, 6, 7), (6→4, 7, 8, 14), (9→10, 11, 12, 15), (10→9, 11, 12, 15), (11→9, 10, 12), (12→9, 10, 11), (14→6, 7, 8), (16→15, 17), (17→15, 16), (NH→8, 9); APCIMS m/z (%) [M+H]+ 409.5 (41), 375.5 (7), 339.5 (21), 303.4 (69), 265.3 (100); HR APCIMS [M+H]+ m/z 409.0689 (calcd for C17H24Cl3N2OS, 409.0675).
Marfey’s Analysis
A portion of compound 1 (0.2 mg) was hydrolyzed at 120 °C with 6 N HCl for 18 h. A 0.1 M NaHCO3 solution (100 µL) was added to the dried hydrolysate of 1, as well as to standards of N-Me-l-phe, N-Me-d-phe, N-Me-d,l-val, and N-Me-l-val. A solution of 1-fluoro-2,4-dinitrophenyl-5-l-valine-amide (FDVA) in acetone (0.25 mg in 50 µL) was added to each vial. Each vial was sealed and incubated at 90 °C for 5 min. To quench reactions, 50 µL of 2 N HCl were added and then diluted with 100 µL CH3CN. Each of the Marfey’s derivatives of the hydrolysate and standards were analyzed by HPLC using a Phenomenex Jupiter C18 column (4.6 × 250 mm). The reaction mixtures were analyzed starting with 30% CH3CN/70% H2O acidified with 0.02% TFA followed by a gradient elution profile to 70% CH3CN/30% H2O acidified with 0.02% TFA over 60 min at a flow of 0.8 mL/min, monitoring at 340 nm. Retention times for the amino acid standards were N-Me-l-phe 26.3 min; N-Me-d-phe 27.8 min; N-Me-d,l-val 23.8 and 29.6 min; and N-Me-l-val 23.8 min while the hydrolysate gave peaks at 23.8 min and 26.3 min.
Prior to Marfey’s analysis of compound 3, ozonolysis was performed to cleave the thiazole ring. O3 (g) (4% in O2, 1/16 L/min) was bubbled through compound 3 (0.1 mg in 200 µL CH2Cl2) at −78 °C for 5 min. The solvent was removed using N2 (g) and the ozonate was then subjected to hydrolysis at 120 °C with 6 N HCl for 18 h. Marfey’s analysis was undertaken as described above, comparing the ozonolyzed hydrolysate to the d-val and l-val standards. The Marfey’s derivatives of the ozonolyzed hydrolysate and standards were analyzed by HPLC using a Phenomenex Synergi Fusion 4µ column (4.6 × 250 mm). The reaction mixtures were analyzed starting with 15% CH3CN/85% H2O acidified with 0.1% HCO2H followed by a gradient elution profile to 60% CH3CN/40% H2O acidified with 0.1% HCO2H over 45 min at a flow of 0.8 mL/min, monitoring at 340 nm. Retention times for the amino acid standards were d-val 25.3 min and l-val 31.6 min while the ozonolyzed hydrolysate gave a peak at 31.7 min.
Semi-synthesis of Barbamide and Herbamide B Fragments and their PGME Derivatives
Ozonolysis was performed on barbamide to cleave the alkene. O3 (g) (4% in O2, 1/16 L/min) was bubbled through 12.9 mg of barbamide in 200 µL CH2Cl2 at −78 °C for 5 min. This was dried and a mild hydrolysis was performed using 1 mL 1.0N NaOH and 1 mL THF at RT overnight (16 h). To stop the reaction, 2 mL of 1M NaHSO4 was added. This was extracted with diethyl ether (2 × 5 mL). The organic layer was concentrated and was used without purification for the next reaction.
The barbamide fragment was split into two portions and reacted separately with either R-PGME or S-PGME. R-PGME (103.1 mg, 0.51 mmol), Et3N (179 µL, 1.27 mmol), (3-dimethylamino-propyl)-ethyl-carbodiimide hydrochloride (199.4 mg, 1.04 mmol), and 4-dimethylamino pyridine (11.4 mg, 93.29 µmol) were added sequentially to 1.7 mg (8.34 µmol) of the barbamide fragment in CH2Cl2 (10 mL) stirring at 0 °C. This solution was left to warm to room temperature overnight (18 h) and then concentrated under reduced pressure. The residue was re-suspended in EtOAc (75 mL) and filtered. The filtrate was washed sequentially with 0.2M HCl, H2O, 1M NaHCO3, H2O, and saturated NaCl and then dried over NaSO4. This was filtered using Celite and then concentrated under reduced pressure to afford 7.3 mg of impure material. This was subjected to HPLC purification using a Phenomenex Synergi Fusion 4µ column (10 × 250mm) with an isocratic solvent system of 61% CH3CN/39% H2O and a flow of 2 mL/min (retention time 23.7 min) to yield 1.0 mg of the R-PGME derivative of barbamide fragment: (2.84 µmol, 16%); [α]D − 405 (c 0.59, CHCl3); 1H NMR (CDCl3, 600 MHz) δ 7.33–7.39 (m, 5H), 6.49 (d, J = 6.6 Hz, 1H), 5.58 (d, J = 7.1 Hz, 1H), 3.75 (s, 3H), 3.18 (m, 1H), 3.02 (dd, J = 14.9, 2.9 Hz, 1H), 2.29 (dd, J = 15.0, 9.8 Hz, 1H), 1.38 (d, J = 6.5 Hz, 3H); HRESIMS m/z 374.0094 (calcd for C14H16Cl3NO3Na, 374.0088).
The S-PGME derivative of barbamide fragment was synthesized as described for the R-PGME derivative using S-PGME (100.7 mg, 0.50 mmol) to afford 0.6 mg (1.70 µmol, 9%): [α]D + 343 (c 0.35, CHCl3); 1H NMR (CDCl3, 600 MHz) δ 7.33–7.39 (m, 5H), 6.53 (d, J = 6.3 Hz, 1H), 5.59 (d, J = 7.0 Hz, 1H), 3.74 (s, 3H), 3.18 (m, 1H), 3.08 (dd, J = 14.9, 2.5 Hz, 1H), 2.26 (dd, J = 15.0, 9.8 Hz, 1H), 1.28 (d, J = 6.4 Hz, 3H); HRESIMS m/z 374.0091 (calcd for C14H16Cl3NO3Na, 374.0088)..
Ozonolysis was separately performed on herbamide B using O3 (g) (4% in O2, 1/16 L/min) bubbled through 1.0 mg of herbamide B in 200 µL CH2Cl2 at −78 °C for 5 min. This was dried using N2 gas and oxidized using H2O2, then acidified with 1N HCl and partitioned sequentially with ether (2 × 10 mL) and ethyl acetate (2 × 10 mL). The organic layers were combined and concentrated and used without purification for the next reaction.
The herbamide B fragment was reacted only with S-PGME (100.6 mg, 0.50 mmol) as described above to yield 0.7 mg (1.99 µmol, 70%): [α]D + 302 (c 0.24, CHCl3); 1H NMR (CDCl3, 600 MHz) δ 7.33–7.39 (m, 5H), 6.53 (d, J = 6.4 Hz, 1H), 5.59 (d, J = 7.1 Hz, 1H), 3.74 (s, 3H), 3.17 (m, 1H), 3.08 (dd, J = 15.0, 2.6 Hz, 1H), 2.26 (dd, J = 14.9, 9.8 Hz, 1H), 1.28 (d, J = 6.6 Hz, 3H); HRESIMS m/z 374.0093 (calcd for C14H16Cl3NO3Na, 374.0088).
Biological Assays
The three tropical disease bioassays were performed as previously described. Plasmodium falciparum malaria parasites obtained from chloroquine-resistant P. falciparum strain (Indochina W2) were maintained and assayed in human erythrocytes.18 Axenic amastigotes of a WHO reference strain of Leishmania donovani (LD-1S/MHOM/SD/00-strain 1S) were used to assay for anti-leishmanial activity.19 For the Chagas’ assay, a transgenic β-galactosidase-expressing Trypanosoma cruzi strain was used (Tulahuen strain, clone C4).20
Supplementary Material
Figure 4.
Acknowledgment
This research was funded by a Fogarty International Center (FIC) International Cooperative Biodiversity Group (ICBG) grant based in Panama (ICBG U01 TW006634) and by a FIC International Research Scientist Development Award (IRSDA) (K01 TW008002, P.I. MJB). T. L. Suyama provided assistance with the degradation of barbamide and chiral analysis, and J. K. Nunnery assisted with collection of some physical data. We are grateful to Panama’s Autoridad Nacional del Ambiente (ANAM) for providing access to biological samples in Panama.
Footnotes
Supporting Information Available: Table of bioassay results, experimental details of bioassays, 1H, 13C, HSQC, and HMBC NMR spectra of 1 and 3, and gradient HPLC profile of compound 1. This material is available free of charge via the Internet at http://pubs.acs.org.
References and Notes
- 1.Pink R, Hudson A, Mouries MA, Bendig M. Nat. Rev. Drug Discov. 2005;4:727–740. doi: 10.1038/nrd1824. [DOI] [PubMed] [Google Scholar]
- 2.Nwaka S, Hudson A. Nat. Rev. Drug Discov. 2006;5:941–955. doi: 10.1038/nrd2144. [DOI] [PubMed] [Google Scholar]
- 3.Capson TL, Coley PD, Kursar TA. Nat. Biotechnol. 1996;14:1200–1202. doi: 10.1038/nbt1096-1200. [DOI] [PubMed] [Google Scholar]
- 4.Kursar TA, Capson TL, Coley PD, Corley DG, Gupta MB, Harrison LA, Ortega-Barría E, Windsor DM. Pharm. Biol. 1999;37(Suppl):114–126. [Google Scholar]
- 5.Coley PD, Heller MV, Aizprua R, Araúz B, Flores N, Correa M, Gupta M, Solis PN, Ortega-Barría E, Romero LI, Gómez B, Ramos M, Cubilla-Rios L, Capson TL, Kursar TA. Front. Ecol. Environ. 2003;1:421–428. [Google Scholar]
- 6.Jiménez JI, Scheuer PJ. J. Nat. Prod. 2001;64:200–203. doi: 10.1021/np000462q. [DOI] [PubMed] [Google Scholar]
- 7.McPhail KL, Correa J, Linington RG, González J, Ortega-Barría E, Capson TL, Gerwick WH. J. Nat. Prod. 2007;70:984–988. doi: 10.1021/np0700772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Orjala J, Gerwick WH. J. Nat. Prod. 1996;59:427–430. doi: 10.1021/np960085a. [DOI] [PubMed] [Google Scholar]
- 9.Nguyen VA, Willis CL, Gerwick WH. Chem. Commun. (Camb.) 2001:1934–1935. doi: 10.1039/b106087m. [DOI] [PubMed] [Google Scholar]
- 10.Flatt PM, O'Connell SJ, McPhail KL, Zeller G, Willis CL, Sherman DH, Gerwick WH. J. Nat. Prod. 2006;69:938–944. doi: 10.1021/np050523q. [DOI] [PubMed] [Google Scholar]
- 11.Galonic DP, Vaillancourt FH, Walsh CT. J. Am. Chem. Soc. 2006;128:3900–3901. doi: 10.1021/ja060151n. [DOI] [PubMed] [Google Scholar]
- 12.Chang Z, Flatt P, Gerwick WH, Nguyen VA, Willis CL, Sherman DH. Gene. 2002;296:235–247. doi: 10.1016/s0378-1119(02)00860-0. [DOI] [PubMed] [Google Scholar]
- 13.Gunasekera SP, Ross C, Paul VJ, Matthew S, Luesch H. J. Nat. Prod. 2008;71:887–890. doi: 10.1021/np0706769. [DOI] [PubMed] [Google Scholar]
- 14.Li H-y, Matsunaga S, Fusetani N. J. Nat. Prod. 1996;59:163–166. doi: 10.1021/np9600309. [DOI] [PubMed] [Google Scholar]
- 15.Nieto J, Veelaert D, Derua R, Waelkens E, Cerstiaens A, Coast G, Devreese B, Van Beeumen J, Calderon J, De Loof A, Schoofs L. Biochem. Biophys. Res. Commun. 1998;248:406–411. doi: 10.1006/bbrc.1998.8964. [DOI] [PubMed] [Google Scholar]
- 16.Nakao Y, Masuda A, Matsunaga S, Fusetani N. J. Am. Chem. Soc. 1999;121:2425–2431. [Google Scholar]
- 17.Yokotani S, Matsushima A, Nose T, Morishita F, Shimohigashi Y. Pept. Sci. 2005;41:539–540. [Google Scholar]
- 18.Corbett Y, Herrera L, González J, Cubilla L, Capson TL, Coley PD, Kursar TA, Romero LI, Ortega-Barría E. Am. J. Trop. Med. Hyg. 2004;70:119–124. [PubMed] [Google Scholar]
- 19.Williams C, Espinosa OA, Montenegro H, Cubilla L, Capson TL, Ortega-Barría E, Romero LI. J. Microbiol. Methods. 2003;55:813–816. doi: 10.1016/j.mimet.2003.08.013. [DOI] [PubMed] [Google Scholar]
- 20.Molinar-Toribio E, González J, Ortega-Barría E, Capson TL, Coley PD, Kursar TA, McPhail K, Cubilla-Rios L. Pharm. Biol. 2006;44:550–553. [Google Scholar]
- 21.Hooper GJ, Orjala J, Schatzman RC, Gerwick WH. J. Nat. Prod. 1998;61:529–533. doi: 10.1021/np970443p. [DOI] [PubMed] [Google Scholar]
- 22.Marner F-J, Moore RE, Hirotsu K, Clardy J. J. Org. Chem. 1977;42:2815–2819. [Google Scholar]
- 23.Sone H, Nemoto T, Ojika M, Yamada K. Tetrahedron Lett. 1993;34:8445–8448. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.