Abstract
During the development of peripheral ganglia 50 % of the neurons generated undergo apoptosis. How the massive numbers of corpses are removed is unknown. We report that satellite glial cell precursors are the primary phagocytic cells for apoptotic corpse removal in developing mouse dorsal root ganglia (DRG). Confocal and electron microscopic analysis revealed that glial precursors, not macrophages, are responsible for clearing most of the dead DRG neurons. Moreover, we identified Jedi-1, a novel engulfment receptor, and MEGF10, a purported engulfment receptor, as homologs of the invertebrate engulfment receptors Draper and CED-1 expressed in the glial precursor cells. Expression of Jedi-1 or MEGF10 in fibroblasts facilitated binding to dead neurons and knocking down either protein in glial cells, or over expressing truncated forms lacking the intracellular domain, inhibited engulfment of apoptotic neurons. Together, these results reveal the cellular and molecular mechanism by which neuronal corpses are culled during DRG development.
INTRODUCTION
The extensive neuronal cell death that occurs during the ontogenesis of the peripheral ganglia was first described in the developing chick embryo, leading to the discovery of Nerve Growth Factor (NGF)1, 2. An important part of this “tissue sculpting” process is to properly dispose of degenerated cellular components, thereby avoiding any inflammatory response3. Although much progress has been made in understanding the regulation of neuronal cell death4, little is known about how the vast pool of neuronal corpses is eliminated.
In the developing mammalian central nervous system (CNS), glial cells and microglia have been implicated in the clearance of apoptotic neurons. Infiltration of F4/80 positive macrophages from the developing mouse vasculature into the retina and brain is associated with neuronal death. These invading macrophages further differentiate to microglia and engulf and degrade the apoptotic debris5, 6. Early electron microscopy (EM) studies in the developing chick peripheral nervous system (PNS) suggested that macrophages as well as satellite glial cells and their precursors may be involved in clearing neuronal corpses7, 8; nonetheless, the potential function of these glial cells in engulfment and the molecular mechanism involved have since been left unexplored.
The engulfment process utilized by professional phagocytic cells, including macrophages and dendritic cells, is known to involve an array of receptors on the phagocytes able to sense “find-me” and “eat-me” cues exposed by dying cells and “don’t-eat-me” signals by healthy cells9–12. Whether any of these receptors and cues is involved in clearing dead neurons during PNS development is not known. Recently, a Drosophila engulfment receptor, Draper, was identified that is structurally and functionally similar to CED-1, a phagocytic receptor found in Caenorhabditis elegans13, 14. Draper is expressed exclusively in macrophages and glia, and null embryos exhibited defects in the clearance of neuronal corpses and degenerating axons13, 15–17.
Clearance of apoptotic cells is not just for “waste disposal”. Non-ingested apoptotic cells typically undergo secondary necrosis, which not only activates immature dendritic cells to become immunogenic but also exposes the normally sequestered self-antigens18, resulting in an increased risk for auto-immune disease later in life3. To gain insight into how apoptotic neurons are eliminated during dorsal root ganglia (DRG) development, we investigated the cellular and the molecular mechanisms underlying this clearance process. Here, we demonstrate that satellite glial cell (SGC) precursors, are the primary cell type responsible for dead neuron clearance. We also identify two receptors homologous to CED-1 and Draper as mediators of this engulfment process: MEGF10, recently reported to be a CED-1 homolog19, and a novel engulfment receptor, Jedi-1 (also known as PEAR1 or MEGF12).
RESULTS
Apoptotic DRG neurons are engulfed by SGC precursors
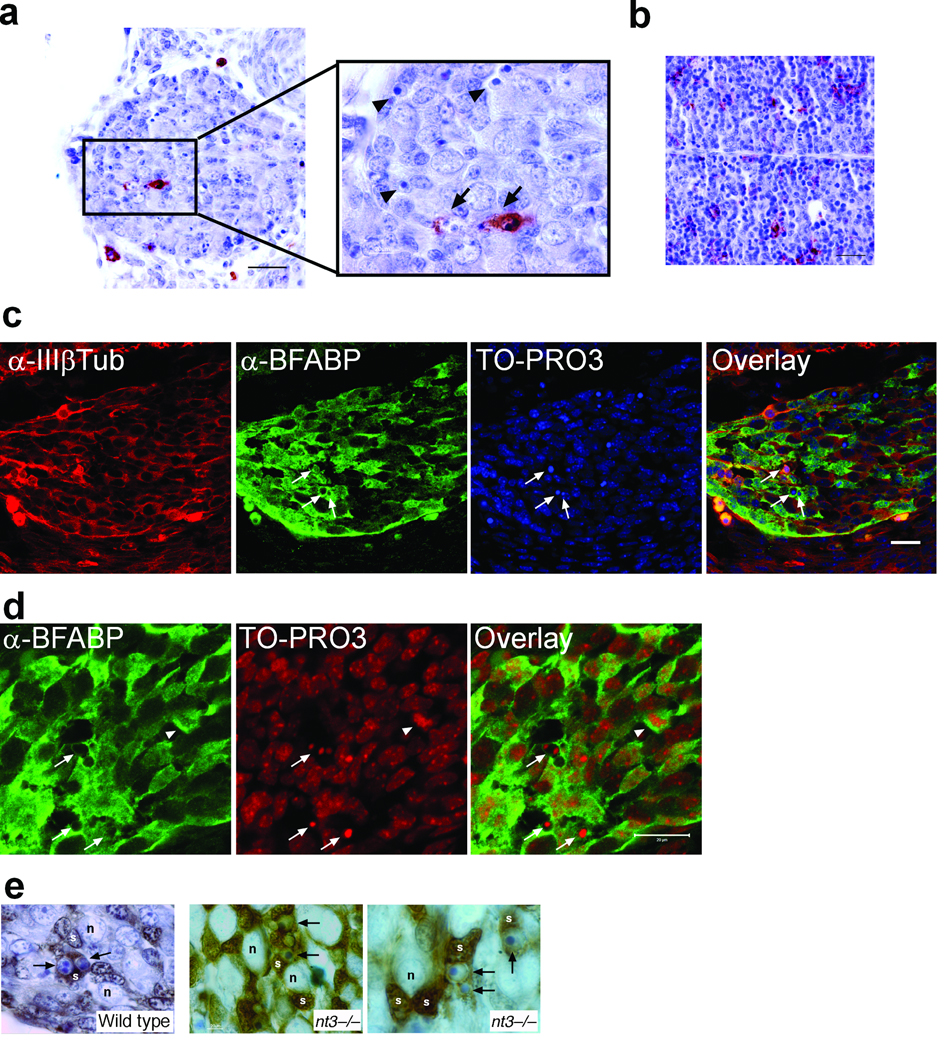
In the mouse embryonic DRG, approximately 50 % of the sensory neurons undergo apoptosis, starting around E11, peaking at E13, then tapering off about E15 20, 21. To determine how these neuron corpses are cleared, we first considered the possibility that macrophages could be responsible. Interestingly, only a few sporadic macrophages, detected by the macrophage-specific antigen F4/80+ 6, were found in the mouse DRG during the period of naturally occurring cell death (Fig. 1a). Specifically, just 0.65 ± 0.58% (n=3) of the total number of cells in the ganglia were F4/80+ at E11 and 0.65 ± 0.65% at E13 (n=3). Moreover, even though F4/80+ cells inside mouse DRG appeared to be encircling dead neurons (Fig. 1a; arrow), most of the apoptotic cells were not associated with macrophages (Fig. 1a; arrow heads). This was not due to a lack of macrophages during this stage or a limitation of detection since many F4/80+ macrophages were present in liver in the same section (Fig. 1b).
Figure 1. Neuron corpses in developing DRG are engulfed by BFABP+ SGC precursors.
(a and b) Immunostaining with F4/80 antibody was used to detect the presence of macrophages in paraffin sections from E12.5 mouse DRG (a) or liver (b). Arrows point to cell corpses engulfed by macrophages, which were rare. Arrowheads point to several cell corpses not engulfed by macrophages. (c and d), Cryosections from E12.5 mouse DRG were immunostained with anti-BFABP to label satellite glial cells, and anti-type III β-Tubulin [α-IIIβTub], to label neurons. Cell nuclei were counterstained with TO-PRO3. (c) Arrows indicate apoptotic cells engulfed by BFABP+ cells. Dorsal is to the right. (d) Enlarged view of nuclear fragments engulfed by BFABP+ cells, indicated by the arrows. The arrowhead indicates a BFABP+ SGC precursor undergoing mitosis. (a–d) Scale bars, 20 µm. (e) Immunohistochemical staining of E12.5 wildtype and nt3−/− DRGs with anti-BFABP [brown] on 5 µm-thick sections counterstained with toluidine blue. S; SGC precursors; n, neurons; arrows point to several apoptotic neurons engulfed/enveloped by BFABP+ SGC precursors.
The primary cell types in the embryonic DRG are neurons and satellite glial cell (SGC) precursors22–25 (Fig. 1c). SGC precursors arise after E10.5 in mice24, 26–28, corresponding to the time of cell death of DRG neurons, and eventually surround the mature neurons, thus positioning them well for engulfment of neuronal corpses. Using an antibody against BFABP (brain fatty acid binding protein), a marker for SGC precursors22–25, we found that most of the apoptotic nuclei appeared to be engulfed by BFABP+ cells (Fig. 1c and d; arrows). We quantified the number of glial precursors surrounding dead cells during the period of normal neuronal apoptosis in the DRG and found 86.5 ± 5.4% of the apoptotic bodies were associated with BFABP+ cells at E11, 73.4 ± 2.8% at E12 and 77.1 ± 3.6% at E13 (n=3). Confocal images with serial optical sections acquired at a higher magnification showed the presence of several apoptotic nuclear remnants inside the BFABP+ cytoplasm (Fig. 1d and data not shown). To be sure that macrophages were not expressing the glial marker, we performed double labeling with BFABP and F4/80 and confirmed that these were separate cell populations (data not shown).
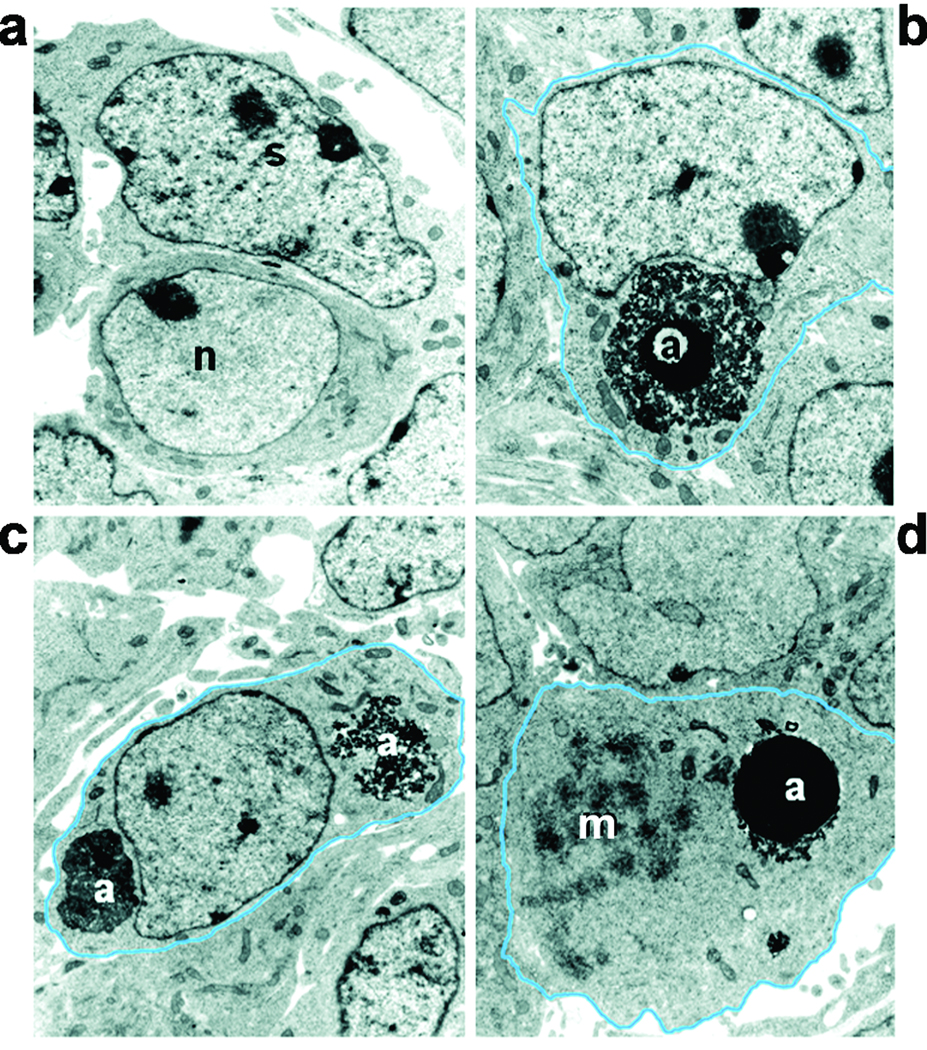
To further demonstrate that SGCs were the primary cell type engulfing apoptotic bodies we examined ganglia of E11 to E13 embryos at the electron microscopy level. Healthy neurons, morphologically characterized by a large nucleus with finely dispersed chromatin and abundant cytoplasm, were typically positioned near elongated cells with a polymorphic nucleus exhibiting a perinuclear chromatin ring and chromatin clumps, typical of satellite glia (Fig. 2a). In 100% of the cases where a cell was surrounding an apoptotic body (53 apoptotic bodies total, in DRG sections from E11, n=15; from E12, n=22 and E13, n=16) the ultrastructure of the engulfing cell resembled that of satellite cells (Fig. 2b, c). We did not find any macrophage-like cells engulfing cell corpses in the electron micrographs. Moreover, we occasionally observed engulfing cells undergoing mitosis (Fig. 2d). Mitotic figures were also observed in BFABP+ cells in the ganglia (Fig. 1d; arrowhead) and 29.3 ± 1.8% of the BFABP+ cells at E12 co-labeled with BrdU 1 h following injection. Since nearly all detectable apoptotic cells in the DRG at this time are neurons 21 and this is the period during which SGC precursors are proliferating, these observations indicate that SGC precursors are responsible for engulfing the dying neurons.
Figure 2. Electron micrographs of apoptotic bodies engulfed and ingested by SGC precursors in embryonic E12.5 DRGs.
(a) A healthy neuron [n] characterized by finely dispersed chromatin and abundant cytoplasm adjacent to a SGC precursor [s] with a characteristic pleomorphic nucleus with chromatin clumps. (b) An apoptotic cell [a] engulfed by a SGC precursor. (c) Debris of two ingested cells [a] inside a SGC precursor. (d) An ingested apoptotic cell present in a cell that is undergoing mitosis (m; mitotic figure, condensing chromosomes).
To further pursue this possibility, we asked whether elevated apoptosis in the developing DRG could be handled by SGC precursors or would lead to macrophage invasion. Using neurotrophin 3 (nt-3) null mice, which lose 70% of their sensory neurons between E11 and E1320, 29, 30, we examined the clearing of dead neurons in the E12 DRG. As in the wild type ganglia, the vast majority of apoptotic cells were surrounded by BFABP+ cells (74.6 ± 6.0%, n=3 embryos) (Fig. 1e), while only 3.1 ± 2.5% were associated with F4/80+ cells (n=3 embryos). This result indicates that SGC precursors are the primary engulfing cell type during DRG development, being sufficient for the job even when the pool of dead neurons is dramatically increased.
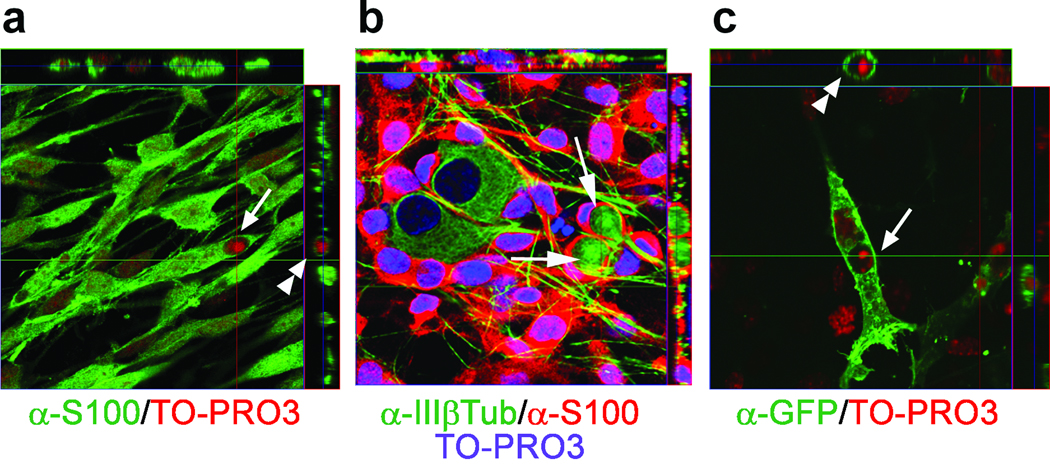
The ability of glial precursors to ingest apoptotic neurons could also be demonstrated in vitro. We established an in vitro engulfment assay using dissociated ganglia from E15 rat or E13 mouse embryos. The cells were first grown in the presence of NGF for 2 to 3 days to keep the sensory neurons alive, followed by removal of NGF to induce their apoptosis. The glial cells were detected with an antibody to BFABP or S100, which labels both SGC and Schwann cells, which most of the glia eventually convert to in culture 31. Virtually all non-neuronal cells in the cultures were BFABP+ and S100+, we did not detect any fibroblasts by Thy 1.1 staining nor did we find any F4/80+ cells (n=3). Following 2 days of NGF withdrawal, 82% of the neurons were apoptotic and the engulfed and ingested dead neurons were detected exclusively inside the glial cells, based on S100 (Fig. 3 a and b, arrows) and BFABP (supplementary Fig. 1) immunostaining and confocal microscopy. We also expressed a membrane bound form of GFP32 (meGFP) in the glial cells to more clearly visualize the internalized corpses. Under these conditions, engulfed nuclear remnants inside phagosomes were clearly observed (Fig. 3c). Taken together with the findings in vivo, these results demonstrate that SGC precursors are the primary cell type responsible for clearing neuronal corpses during the period of naturally occurring cell death in the developing DRG.
Figure 3. Glial cells engulf dead neurons induced by NGF withdrawal in vitro.
Primary mixed cultures of sensory neurons and SGCs were derived from dissociated E15 rat DRG and cultured with NGF for 2 days. The NGF was then removed to induce neuronal apoptosis, and the cultures were fixed and immunostained 2 days later. Images were compiled from orthogonal optical sections showing (a) apoptotic nuclear remnants [TO-PRO3, red] inside the cytoplasm of glial cells [α-S100, green]; (b) neuron corpses [α-IIIβTub, green] enveloped by glial cells [α-S100, red]. Nuclei were stained with TO-PRO3 [blue]. (c) apoptotic nuclear remnants [TO-PRO3, red] present in glial cells expressing membrane bound GFP [α-GFP, green]. The window on the top of each panel shows the Z-axis stacks at the specific Y-axis location [indicated by a green line crossing the panel] and that on the right shows the Z-axis stacks at the specific X-axis location [red line crossing the panel]. Arrows and arrowheads indicated engulfed apoptotic bodies.
Jedi-1 and MEGF10 are expressed in SGC precursors
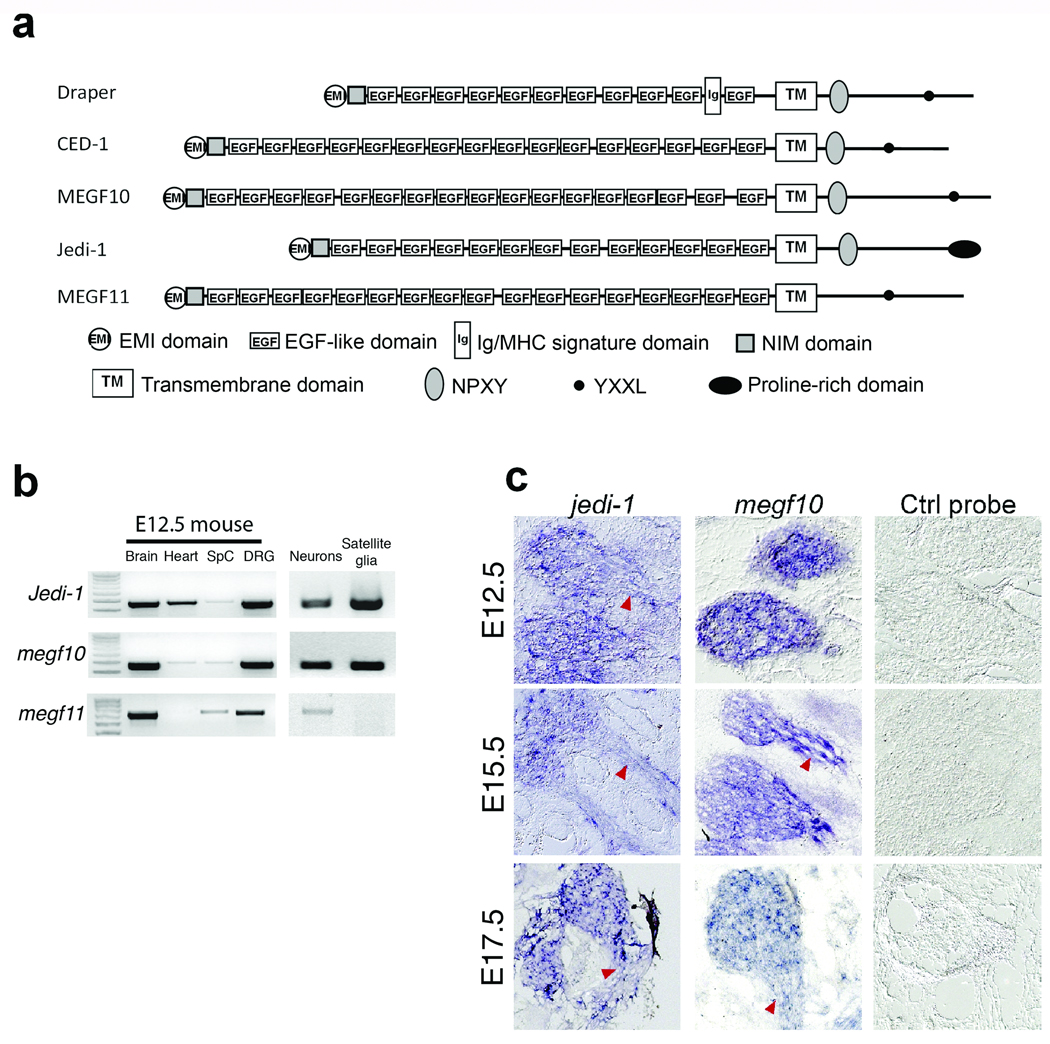
The molecular mechanisms underlying apoptotic cell clearance in the mammalian PNS during development are not known. Draper, a Drosophila protein homologous to the C. elegans CED-1 receptor, was identified as an engulfment receptor expressed on glial cells that was required for clearing degenerating neurons and axons13–17, 33; therefore, we speculated that a Draper/CED-1-like engulfment receptor might exist in SGC precursors to mediate phagocytosis of dead neurons. Three mammalian proteins, MEGF10, MEGF11, and Jedi-1 were identified as highly homologous to Draper and CED-1 using the NCBI blastp program. Two regions in the intracellular domain of CED-1 are required for its engulfment function: an NPXY motif that may serve as a phosphotyrosine binding site and an YXXL motif, a Src Homolog 2 (SH2) domain binding site 14. As shown in Figure 4a (also see supplementary Fig. 2), Draper, and MEGF10 have both NPXY and YXXL motifs, while Jedi-1 has an NPXY sequence and MEGF11 an YXXL, in their putative intracellular regions (Fig. 4a).
Figure 4. Putative Draper and CED-1 homologs, Jedi-1 and MEGF10, are expressed in developing peripheral glial cells.
(a) Schematic representation of the modular architecture of Draper, CED-1 and possible mammalian homologs. A key for the predicted domains and motifs is shown on the bottom. Also see Supplementary Fig. 1 for the sequence alignments of their predicted intracellular domains. (b) RT-PCR detection of Jedi-1, Megf10, or Megf11 mRNA in E13 mouse brain, heart, spinal cord [SpC], whole DRG, and purified DRG neurons or satellite glial cells. 1 Kb DNA markers are on the left. (c) Jedi-1 and Megf10 transcripts were detected in mouse DRG and developing glial cells alongside axons. The developmental stages of the embryos are indicated on the left. Saggital sections; dorsal on the left. Arrowheads indicate the nerves.
To determine if Jedi-1, MEGF10 or MEGF11 could mediate engulfment by SGC precursors, we examined their expression in these cells by RT-PCR. As shown in Fig. 4b, the mRNAs for all of these proteins were present in E12.5 mouse brain and whole DRG; however, only MEGF10 and Jedi-1 were expressed in isolated SGC precursors, indicating that MEGF11 is unlikely to function as an engulfment receptor in DRG development. Interestingly, the mRNA for all three proteins was detected in neurons, although their function there is not known. We then analyzed the expression pattern of Jedi-1 and MEGF10 in the developing mouse DRG at different developmental stages using in situ hybridization (Fig. 4c). At all ages examined (E12.5, E15.5 and E17.5), both Jedi-1 and MEGF10 were observed in the ganglia and in the cells along the nerves, consistent with the location of SGC precursors and immature Schwann cells.
Jedi-1ΔC interferes with apoptotic clearance in C. elegans
Recently, MEGF10 was proposed as a putative CED-1 homolog since it could promote dead thymocyte engulfment when ectopically expressed in HeLa cells19, 34 and expression of an MEGF10::GFP fusion protein under the control of the ced-1 promoter (Pced-1), in ced-1 (e1735) mutant C. elegans partially rescued the engulfment defect of ced-1 (e1735) 19. Whether Jedi-1 exerts a function similar to CED-1 has never been examined. CED-1 and its truncated form lacking the ICD (CED-1DC) similarly cluster around neighboring cell corpses, owing to their ability to recognize an apoptotic cell-surface signal(s) 14. We expressed an intracellular domain truncated Jedi-1 protein, tagged with GFP at its C-terminus (JediΔC::GFP), in engulfing cells under the control of Pced-1 in C. elegans and observed cell surface presentation of a fraction of JediΔC::GFP molecules, although much of it remained inside the cells. Importantly, the portion of JediΔC::GFP present on the surface of gonadal sheath cells, which are engulfing cells for apoptotic germ cells, was clustered around some of the germ cell corpses (Fig. 5). This localized enrichment of JediΔC::GFP around cell corpses suggests that the extracellular domain of Jedi-1 is capable of recognizing a signal displayed on the surface of the dying cell, like that of CED-1. Furthermore, the expression of JediΔC::GFP in wild-type worms resulted in the presence of excessive germ cell corpses (Fig. 5). This result suggests that, among other possibilities, JediΔC::GFP might interfere with the normal engulfment of dead cells by associating with the “eat-me” cue on the surface of cell corpses, thereby preventing endogenous CED-1 or other unknown engulfment receptors from binding these cues and transducing the normal engulfment signal. The expression of CED-1ΔC similarly resulted in the inhibition of cell-corpse engulfment13. We also tested whether ectopic expression of full-length Jedi-1 (Jedi::GFP) in C. elegans engulfing cells could also rescue the cell-corpse removal defects in ced-1 (e1735) mutants. Unfortunately, ectopically expressed Jedi::GFP was retained inside cells, forming protein aggregates and did not result in any rescue of the ced-1 mutant (data not shown).
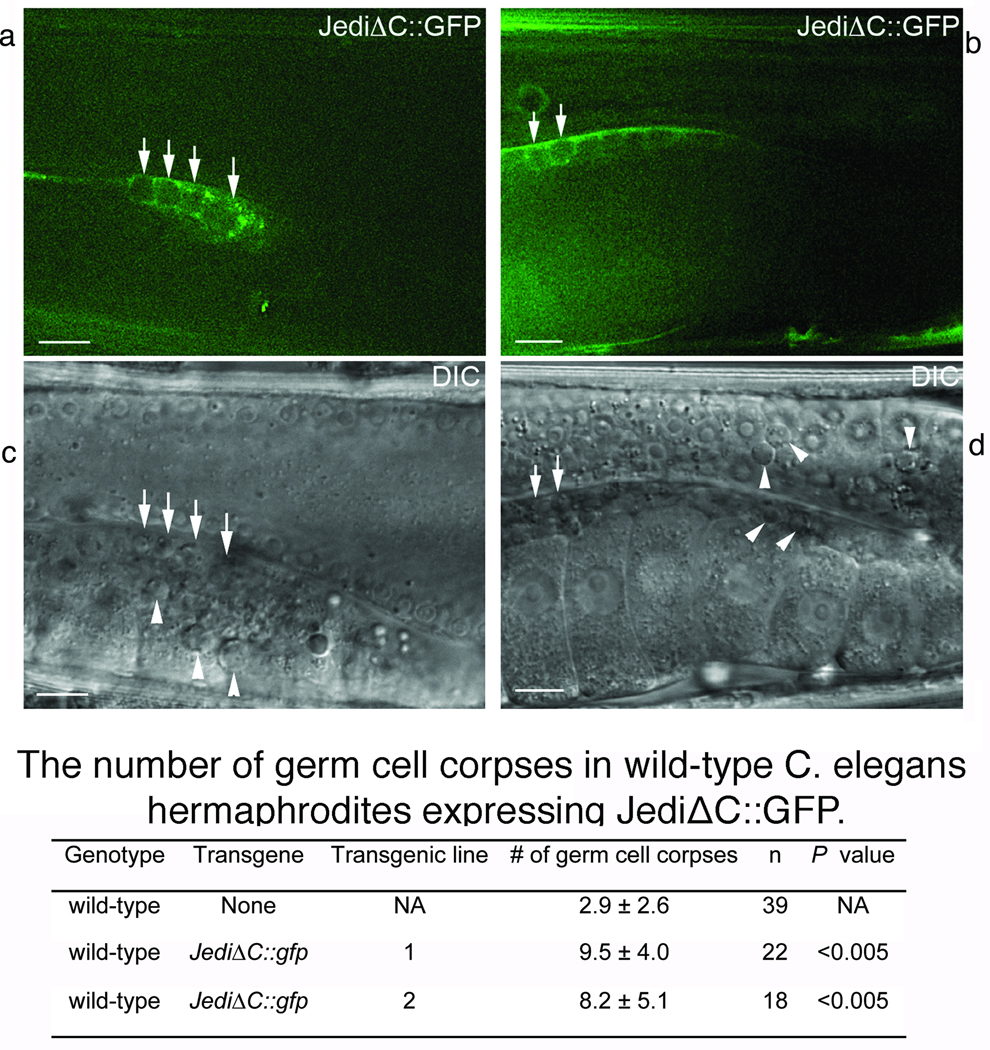
Figure 5. The extracellular domain of Jedi-1 recognizes cell corpses when expressed in C. elegans engulfing cells.
Transgenic animals expressed Jedi-1ΔC::GFP in engulfing cells under the control of Pced-1. All animals analyzed were adult hermaphrodites aged 48 hours post-mid L4 larval stage. GFP (a, b) and corresponding DIC (c, d, respectively) images of part of adult ced-1[e1735] hermaphrodite gonads to indicate the clustering of JediΔC::GFP around germ cell corpses. Arrows indicate a few germ cell corpses labeled with JediΔC::GFP on their surfaces. Arrowheads indicate examples of germ cell corpses not labeled by JediΔC::GFP. Dorsal is to the top. Midbody is to the left. Scale bars: 10 µm. The table shows the number of germ cell corpses in wild type C. elegans hermaphrodites expressing JediΔC::GFP scored under DIC optics in one gonadal arm of each adult hermaphrodite staged 48 hr post the L4 larval stage. Data are presented as mean ± standard deviation. n, the number of animals scored. The data obtained from transgenic and the wild-type control animals are compared and the P values obtained from two-tailed student t-tests.
Jedi-1 and MEGF10 facilitate binding to dead neurons
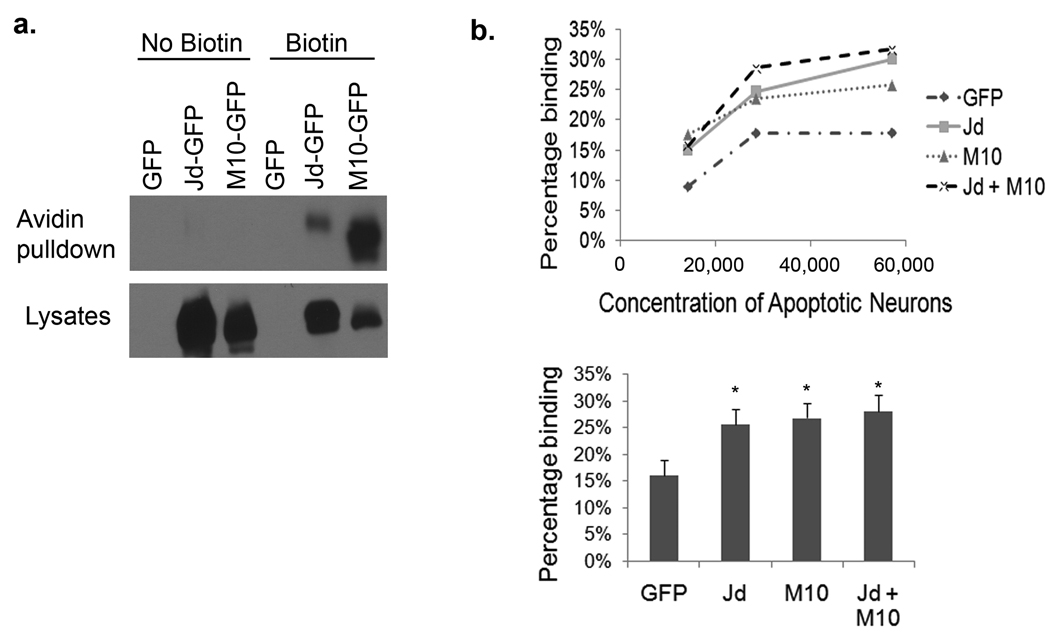
Although CED-1, Draper and MEGF10 are thought to be receptors for apoptotic cells, none of the ligands have been identified nor has it been directly demonstrated that these proteins specifically mediate binding, as opposed to just facilitating the engulfment process. To determine whether Jedi-1 or MEGF10 can act as a receptor for dead neurons, they were transiently expressed in HEK 293 cells and various concentrations of apoptotic neurons, labeled with propidium iodide, were added. We confirmed that Jedi-1 and MEGF10 were expressed and trafficked to the cell surface (Fig. 6a), then the cells expressing these proteins were incubated with neuronal corpses at 4°C to prevent engulfment, since we wanted to specifically assess binding not internalization. After rinsing, the cultures were fixed and the number of dead neurons attached to the transfected cells was scored. Some binding occurred to the control cells, most likely due to other endogenous proteins that can facilitate binding to dead cells, e.g. integrins or PSR 12; however, expression of either Jedi-1 or MEGF10 significantly increased the binding to neuronal corpses (Fig. 6b). This result indicates that these proteins can function as receptors, enhancing binding in the absence of any internalization. Expression of both proteins did not further increase the binding (Figure 6b lower panel), which may indicate they are part of a single binding complex, but the affinity and any cooperativity cannot be accurately determined since the ligand source is an entire dead neuron and not a small, freely diffusible molecule.
Figure 6. Jedi-1 and MEGF10 expressed in HEK 293 cells enable binding to dead neurons.
HEK 293 cells were transfected with GFP-tagged Jedi-1 or MEGF10 and their expression on the cell surface analyzed by biotinylation of the surface proteins followed by precipitation with avidin beads and Western blotting with anti-GFP (a). The biotin reagent was not added to some cells (no biotin) to confirm the specificity of the pull down. The expression of Jedi::GFP and MEGF10::GFP in total cell lysates is shown in the lower panel. (b) Jedi-1 and/or MEGF10 transfected cells were incubated at 4°C with the indicated number of neuronal corpses, induced to undergo apoptosis by NGF withdrawal for 24 hrs and labeled with propidium iodide. After washing off the unbound dead cells, the cultures were fixed with 10% formalin. The percentage of GFP+ cells with at least one PI+ corpse bound is shown (1 representative experiment of 3 is depicted). The lower panel shows the mean ± S.E.M. percent binding at the highest concentration of neurons (*, p < 0.01 relative to GFP transfected cells; n=4).
Over expression of Jedi-1 or MEGF10 increases engulfment
To investigate the involvement of Jedi-1 and/or MEGF10 in neuronal corpse engulfment by SGC precursors, we used the engulfment assay described for figure 3. DRGs from E13.5 mouse embryos were dissociated and grown in the presence of NGF for 2 days. NGF was then removed from the culture and, at the same time, a control (meGFP) or other transgene was transfected into the glial cells. Images of cells expressing these transgenes were acquired with full Z-axis optical sections and the numbers of transfected cells containing at least one fully internalized apoptotic nucleus were determined. Under these conditions, engulfed apoptotic nuclei were observed in about 50 % of meGFP expressing glial cells (Fig. 7c and supplementary Fig. 3a). In contrast, over expression of MEGF10 or Jedi-1 enhanced engulfment of dead neurons by the glial cells: the number of Flag+ or GFP+ cells that had engulfed an apoptotic nucleus increased approximately 50 and 80 % when over expressing either Jedi-1::Flag or MEGF10::GFP, respectively (Fig. 7 and supplementary Fig. 3b–c). Transfection of both receptors into the cells did not further enhance phagocytosis (Fig. 7c), suggesting that there may be an endogenous component in the pathway that is limiting or that the two proteins converge on a common pathway.
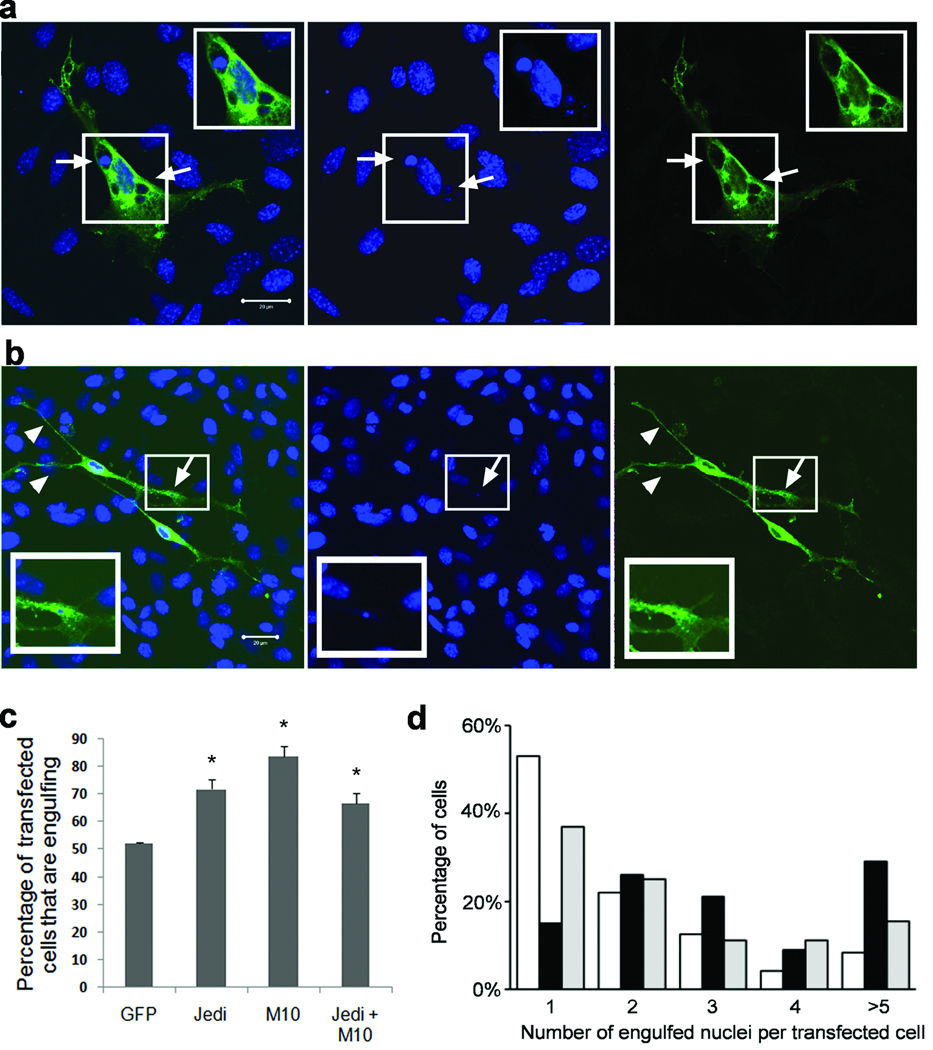
Figure 7. Ectopic expression of Jedi-1 or MEGF10 in glial cells promotes neuronal corpse engulfment.
DRG from E13.5 mice were dissociated and grown in the presence of NGF for 2 days. The glial cells were then transfected with plasmids expressing MEGF10::GFP; Jedi-1::Flag, or meGFP and NGF removed from the cultures. The cells were fixed after 2 days without NGF and immunolabeled with antibodies to GFP or Flag. (a and b) Z-axis optical stacks of confocal images of glial cells expressing MEGF10::GFP (M10::GFP; a, c and d), Jedi-1::Flag (Jd::Flag; b, c and d), or meGFP (c and d) were acquired and the numbers of transfected cells containing at least one ingested nuclear remnant [condensed TO-PRO3 staining] was quantified. Arrows indicate the location of engulfed apoptotic nuclei in these cells. Arrowheads indicate the long processes in Jedi-1::Flag expressing cells. In (c) the error bars = Mean ± s.d. P=0.0002, one-way ANOVA. (d) The number of engulfed nuclei per engulfing cell was also determined and expressed as the percent of transfected cells containing the indicated number of apoptotic nuclei (open bar: meGFP; black bar: MEGF10-GFP; gray bar: Jedi-Flag). Based on a chi-squared analysis, there was a significant difference between all 3 groups of cells (expressing Jedi-1, MEGF10 and meGFP. p<0.0001). Scale bars, 20 µm.
MEGF10::GFP and Jedi-1::Flag accumulated around vacuoles containing apoptotic nuclei (Fig. 7a and b; supplementary Fig. 3, compare optical sections 8–11) and in what appeared to be phagocytic cups (supplementary Fig. 4). The vacuoles were identified as lysosomes or late endosomes by LAMP-1 labeling (supplementary Fig. 5). Interestingly, even though the overall number of cells that were engulfing increased when transfected with Jedi-1 or MEGF10, these cells exhibited some differences: cells expressing MEGF10::GFP contained more vacuoles with apoptotic nuclei than those expressing Jedi-1::Flag, which typically had more long processes and lamellipodia (Fig. 7b, arrowheads). To quantify this difference in engulfment, we scored the number of vesicles with nuclear fragments in each transfected cell. Three or more apoptotic corpses were found in 70% of MEGF10-transfected cells, yet only in ~30% of Jedi-transfected cells, which more often contained one or two engulfed nuclei (Fig. 7d). These observations suggest that over expression of either Jedi-1 or MEGF10 can ultimately increase the engulfment of dead cells, although there appear to be some differences in their mechanisms of action.
The effect of expressing a mutant Jedi-1, lacking the ICD, in C. elegans suggested that this construct could act as an inhibitor of engulfment, preventing the endogenous phagocytic receptor(s) from binding and/or internalizing apoptotic cells (Fig. 5). Indeed, we found that transfection of glial cells with either Jedi-1ΔC::GFP or MEGF10ΔC::GFP in the engulfment assay led to about a 30 % decrease in the number of cells that were engulfing dead neurons when compared with those transfected with meGFP (Fig. 8a and supplementary Fig. 6).
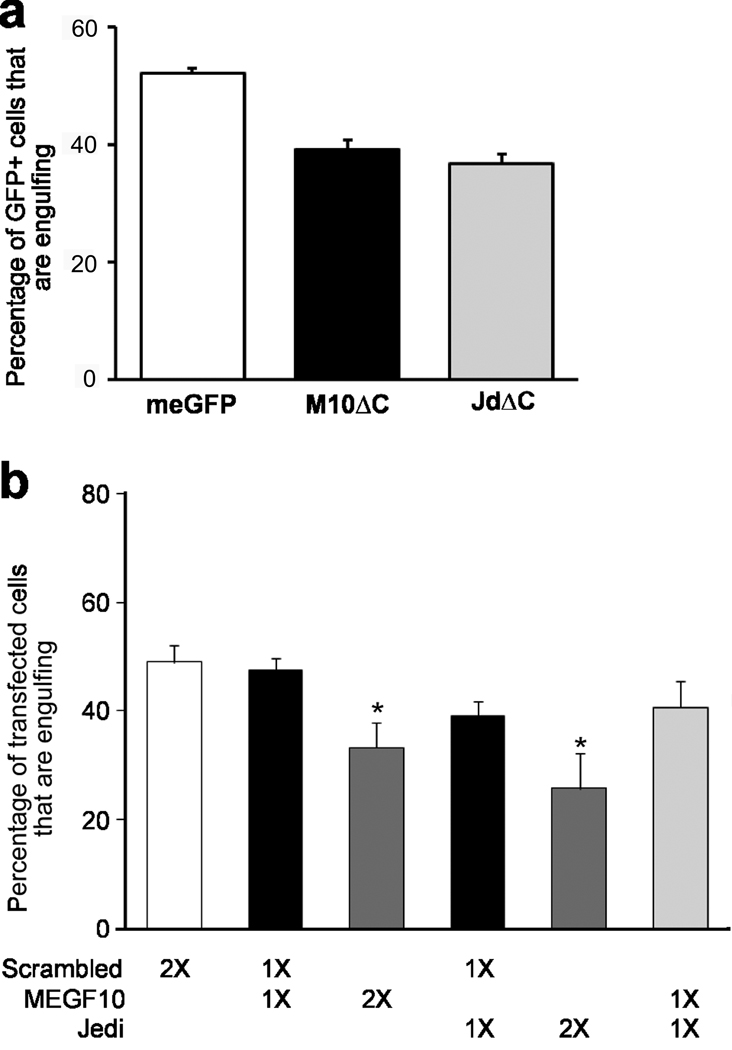
Figure 8. Neuronal corpse engulfment by glial cells requires endogenous Jedi-1 and MEGF10.
The neuronal engulfment assay described in Figure 7a was used to determine whether endogenous MEGF10 and Jedi-1 are required for engulfment. (a) C-terminal truncated forms of MEGF10 or Jedi-1 [M10ΔC or JdΔC respectively] as well as meGFP were transfected into glial cells, and 2 days after NGF withdrawal the number of GFP+ cells with engulfed apoptotic nuclei were counted. Error bar = Mean ± s.d., P=0.0005, one way ANOVA. (b) Plasmids bi-cistronically expressing ZsGreen and shRNAs targeting MEGF10, Jedi-1 or non-targeting shRNA (scrambled) were transfected into the glial cells. A total of 8 µg of DNA was transfected, either in combinations or as a single plasmid and after 2 days the number of ZsGreen+ cells containing engulfed corpses were counted. Error bar = Mean ± s.d., P=0.0008, one way ANOVA.
Knock-down of Jedi-1 or MEGF10 inhibits corpse clearance
The ability of the truncated MEGF10 and Jedi-1 to reduce engulfment suggested that the mutants were interfering with the action of the endogenous protein. Therefore, to directly determine whether endogenous Jedi-1 or MEGF10 is required for neuronal corpse clearance, we used short-hairpin RNA (shRNA) to target these genes (supplementary Fig. 7a and b). Knocking down Jedi-1 or MEGF10 in the glial cells did not alter their morphology (supplementary Fig. 7c); however, it resulted in a 40–50 % decrease in the number of cells with internalized apoptotic neurons relative to those expressing a scrambled shRNA (Fig. 8b). There was no significant difference between the number of engulfing glial cells transfected with the control shRNA (Scr1-1) and those transfected with meGFP (54.3 ± 1.0 vs. 53.4 ± 4.3). Interestingly, knocking down both Jedi-1 and MEGF10 did not further reduce the ability of the transfected cells to engulf apoptotic bodies, further suggesting that the two proteins may function in a common pathway (Fig. 8b). These results indicate that endogenous Jedi-1 and MEGF10 are necessary for neuronal corpse clearance in the embryonic DRG.
DISCUSSION
Although it has long been recognized that there is extensive cell death in the developing peripheral nervous system, the mechanisms responsible for disposing of the cellular “waste” have remained an open question. Our findings demonstrate that SGC precursors in the DRG are the primary cell type responsible for clearing neuronal corpses generated during the period of naturally occurring cell death. In addition, we identified Jedi-1, a novel engulfment receptor, and MEGF10 as two CED-1 homologs expressed in the glial cells and involved in phagocytosing neuronal corpses. Hence, these results reveal the cellular and molecular basis for clearing the neuronal waste generated during the development of sensory ganglia.
Macrophages carry out the removal of cellular debris in many tissues during development or after injury and these cells are known to increase in the DRG after injury to the sciatic nerve35. However, we found few macrophages in the developing DRG, even in animals with unusually high numbers of apoptotic neurons (nt3−/− mice). The phagocytic ability of glial cells in the developing nervous system has been known for some time, although the significance has largely been unrecognized. Axonal fragment ingestion by Schwann cells during Wallerian degeneration after injury was first described some 40 years ago 8. However, macrophages subsequently invade the nerve and clear most of the debris, thereby overshadowing the contribution of the Schwann cells 36. A more recent study demonstrated that Schwann cells play an important phagocytic role in synapse elimination during the development of the neuromuscular junction 37. Early EM studies of the developing chick embryo suggested the presence of degenerated axons and apoptotic neurons in astrocytes, satellite glial cells and Schwann cells7, 8, 38; however, the identity of the engulfing cells was not confirmed due to the absence of immunological markers. Furthermore, in some cases, contradictory observations were reported, suggesting that macrophages were clearing the debris 8. Our findings demonstrate that SGCs, not macrophages, are the primary phagocytes responsible for clearing the dead neurons generated during the normal development of the DRG.
The physiological roles of SGCs are not well understood. They are found in sensory, sympathetic and parasympathetic ganglia, where they cluster around each neuron and regulate the extracellular environment, taking up neurotransmitters similar to astrocytes 39. SGCs also produce numerous neuroactive agents such as neurotrophins and bradykinin, although the functions of these in the ganglia are not clear 39. Following injury the SGCs undergo a morphological change, begin to proliferate and release many of these factors, leading some to suggest that they are involved in neuropathic pain 39, 40. Their phagocytic ability characterized in this study provides a rare glimpse into an important function of SGCs. Whether the glial cells remain the primary cell type responsible for phagocytosing dead neurons in more mature animals, for example, after axotomy or other inducers of neurodegeneration, remains to be determined.
The molecular mechanisms involved in apoptotic neuron clearance in the developing mammalian PNS were previously unknown. In C. elegans, CED-1 was identified as a receptor required for engulfment of apoptotic cells 14. Draper, the Drosophila homolog of CED-1, was shown to mediate dead neuron removal during development13 and in eliminating degenerating axons during metamorphosis and after injury 15, 16, 33. We identified MEGF10 and Jedi-1 as possible homologs based on their predicted structural organization and their expression pattern. MEGF10 was previously suggested to be an engulfment receptor 19, but little is known about Jedi-1. It was shown to be phosphorylated upon platelet activation, although its role in this process was not determined, and its over expression in hematopoietic progenitors reduced the number of cells that committed to a myeloid lineage, suggesting a role in differentiation of these cells. Here, we demonstrate that Jedi-1 functions as a phagocytic receptor, involved in clearing dead sensory neurons.
Interestingly, both Jedi-1 and MEGF10 promoted the engulfment of neuronal corpses by SGCs (Fig. 7 and Fig 8); however, over expression or knock-down of both proteins was no more effective than altering the expression of either alone. We, therefore, suggest that these receptors may converge on a common pathway or even form a complex. However, they also exerted somewhat different actions when over expressed in glial cells, suggesting there are some non-overlapping functions. Almost all the cells transfected with MEGF10 had engulfed more than one apoptotic nuclei (90.6 % ± 1.3) and most had numerous apoptotic nuclear remnants in lysosomes. This could be due to an increase in the number of dead neurons taken up by a single cell or an increase in the number of lysosomes digesting the engulfed material. Increased vacuole formation has been described in HEK293 cells over expressing MEGF10, even when no apoptotic cells were added 34. On the other hand, most of the Jedi-1-over expressing glial cells appeared slimmer with longer processes (Fig. 7b) and contained only one or two visible vacuoles with ingested nuclei, unlike those over expressing MEGF10 (Fig. 7d). It is possible that Jedi has the ability to promote both engulfment and degradation of apoptotic cells, like CED-1 41; whereas mEGF10 only carries the engulfment activity. Of course, we cannot rule out the possibility that over expression of either protein indirectly increases engulfment, for example, by increasing process formation. In any case, these differences suggest that, upon activation by the “eat-me” signals, MEGF10 and Jedi-1 trigger, at least partially, non-overlapping molecular pathways for engulfment.
Such a need for multiple receptors is typical of the phagocytic process. Multiple ligands and receptors are implicated in the recognition and uptake of apoptotic cells by “professional” phagocytes like macrophages 12, 42. Just for phosphatidylserine, the most well studied “eat-me” cue, there are at least four transmembrane receptors, PSR, Tim4, BAI1, and Stabilin-2, that have been shown to bind this phospholipid and transduce an engulfment signal 9. Even in C. elegans, there are two partially redundant pathways that mediate cell corpse removal, with ced-1, ced-6 and ced-7 genes functioning in one pathway and ced-2, ced-5, ced-10 and ced-12 genes acting in the other 43. Recently, a second Drosophila receptor, SIMU was identified and suggested to function as the recognition receptor 44 with Draper acting primarily in the engulfment and degradation process through recruitment of the src family kinases 45.
Programmed cell death and process elimination are essential for the development and maintenance of functional nervous systems. Consequently, considerable effort has been made to understand the molecular mechanisms involved in these events; however, less attention has been given to the resulting byproducts. During such regressive processes, large amounts of degenerated and excess cellular debris are generated that need to be efficiently eliminated. Several reports have provided convincing evidence for a link between defective clearance of apoptotic cells and the development of autoimmunity 3, 46, 47. Hence, it is likely that defects in neural waste clearance during development predisposes an organism to autoimmune attack on the nervous system later in life, although this has yet to be demonstrated.
METHODS
Animals
CD-1 mice, Sprague-Dawley rats or nt3+/+ and −/− mice20 were used for these studies. All experimental procedures using animals were approved by the Institutional Animal Care and Use Committee at Vanderbilt University, the committee on Animal Research at the University of Valencia and conformed to US National Institutes of Health and EU guidelines.
In situ hybridization
For in situ hybridization (ISH), embryos were dissected in cold PBS, fixed in 4% paraformaldehyde overnight at 4°C and transferred to 30% sucrose/PBS prior to cryosection at 20 µm. ISH on sections were performed as described48. Digoxigenin-labeled cRNA probes were: Jedi-1 probe 1and Jedi-1 probe 2, (467–931 bp and 871–1347 bp of GenBank #AF444274); megf10 probe 1 and megf10 probe 2, (1326–1828 bp and 2369–2857 of GenBank # NM_001001979).
Electron microscopy
Isolated E11.5–E13.5 mouse embryos were fixed by immersion in 4% paraformaldehyde (PFA) and 1% glutaraldehyde in PB (0.1M phosphate buffer, pH 7.4) for two hours and thoroughly washed with PB. Body fragments containing lumbar DRGs were postfixed with 1% osmium tetroxide in PB for 90 min at room temperature, dehydrated through a graded ethanol series followed by propylene oxide, and embedded in araldite (Durcupan, Fluka). Samples were sectioned at 1µm and sections stained with 1% toluidine blue for ganglia identification. Ultrathin 60 nm thick sections from lumbar ganglia were collected on formvar-coated slot grids, stained with uranyl acetate and lead citrate, and examined with a Jeol JEM-1010 electron microscope.
RT-PCR
Total RNA from E12.5 CD-1 mouse brain, heart, spinal cord, DRG, and cultured embryonic satellite cells19 or neurons was extracted with TRIzol reagent (Invitrogen) per manufacturer’s recommendation and reverse-transcribed using random primers and Superscript II reverse transcriptase (Invitrogen) following treatment with DNase I (DNA-free kit, Ambion). Resulting cDNAs were analyzed by PCR.
-
Primers: Jedi-1: forward primer, 5'- CCTGCAGCTGCCCACCGGGCTGGA -3'
reverse primer, 5'- CCTGGCAGCCCGGGCCATGCGTGT -3'
Megf10i: forward primer, 5'-GGCGCGCCTGTGTCCCGAGGGGCTTT-3'
reverse primer, 5'-CGGGCGCACAGGTGCAGGTCCCATCC-3'
Megf11: forward primer, 5'-GCGCGCCACGGAAGCAGGCCCCGATG-3'
reverse primer, 5'-CCAGCCTCGGAAGCCCGGGGCGCACA-3'
Plasmids and ISH probes
For information regarding cDNA constructs, truncated fusion constructs, ISH probes, and shRNA containing plasmids please see supplementary methods.
Neuronal engulfment assay
DRG from E13.5 mouse CD1 embryos or E15 rat embryos were dissociated with collagenase A (1 mg/ml, Roche) plus 0.05% Trypsin (1:250; Gibco) and triturated. Then 50,000 cells were plated onto a 25mm round coverslip (#1.5: 0.17 mm; Warner Instruments Inc.) coated with collagen. For mouse cultures, cells were grown in mouse- engulfing medium (MEGM)[1:1 UltraCULTURE serum-free medium (Bio Whittaker): Neural Basal medium (Invitrogen) supplemented with 3% FBS (Hyclone), N2, and B27 (Invitrogen)]. For rat cultures, cells were grown in Basal Medium Eagle (Invitrogen) supplemented with 0.4% glucose, 3% FBS and N2. Cultures were first grown in the presence of 50 ng/ml NGF (Harlan) for 2 days, then the NGF was removed by washing the cultures twice and refeeding with 1:10,000 dilution of monoclonal anti-mouse NGF (Chemicon) to remove any remaining NGF. For mouse cultures, 50 ng/ml of glial growth factor (GGF; R & D Systems) were added to ensure the survival of immature peripheral glial cells. Cells were then transfected with the plasmids indicated using Effectene (Qiagen) according to the manufacturer’s recommendation.
DRG SGCs isolation
DRG SGCs were isolated as previously described25 with the following modifications: E13.5 mouse DRGs were cultured as explants on 35 mm dishes coated with poly-D-Lysine (PDL) and laminin (Invitrogen) in MEGM with 25 ng/ml NGF and 50 ng/ml GGF or 1 µM insulin (Sigma) for 3 days to allow cell migration. Neuron soma and glia still associated with the explants were pinched out using fine forceps and dissecting scissors. The remaining satellite glial cells were dissociated by trypsinization and replated to a 10 cm dish coated with PDL/laminin in MEGM with 1 µM insulin without NGF for 2 days.
Immunostaining and quantification
Embryos, processed as for in situ analysis, were cryosectioned at 12 µm, permeablized and blocked with 0.5% Triton X-100 in 10% bovine calf serum and incubated with primary antibodies: 1:1000 rabbit anti-BFABP (kind gift from Dr. Thomas Muller22), 1:100 F4/80 (Serotec), 1:4 rabbit anti-S100β (ImmunoStar) and/or 1:1000 mouse anti-neuronal class III β-Tubulin (Tuj1; Convance). TO-PRO3 (Invitrogen) was used according to the manufacturer’s recommendation. For immunohistochemistry, nt3+/+ and −/− embryos were processed as described previously20. To quantify neuronal engulfment in vivo, the proportion of apoptotic bodies that appeared surrounded by BFABP+ or F4/80+ cells were counted in sections from at least 6 DRGs from at least 3 embryos at each age.
For immunofluorescence staining of cultured cells, cultures were fixed with 4% paraformaldehyde or 10% Formalin and incubated with primary antibodies: 1:4 anti-S100β; 1:1000 anti-BFABP; 1:500 Chicken anti-GFP (Abcam); 1:500 anti-Flag M2 (Sigma); 1:1000 TUJ1; 1:100 F4/80; 1:25 anti-Thy 1.1 (Serotec); anti-LAMP1 (3.9 mg/ml). (The LAMP1 monoclonal antibody developed by J. T. August was obtained from the Developmental Studies Hybridoma Bank developed under the auspices of the NICHD and maintained by the University of Iowa.) Cultures were then incubated with 200 µg/ml of DNase-free RNase A in PBS for 30 min at RT, rinsed, following which nuclei were visualized with 1 µM TO-PRO3 (Invitrogen). Secondary antibodies used were Alexa488-goat anti-rabbit (1:1000) and Alexa488-goat anti-mouse (1:200) from Invitrogen; Rhodamine X-donkey anti-mouse (1:200–1:400) and Cy2-donkey anti-chicken (1:200) from Jackson ImmunoResearch Laboratories. Photomicrographs of Z-axis series were taken using a Zeiss LSM 510 inverted confocal microscope (Cell Imaging Shared Resource at Vanderbilt University Medical Center) and analyzed with LSM image Browser from Zeiss. For each experiment, at least 50 cells from each condition (done as duplicates or triplicates for each experiment) were counted. The results were obtained from 2–3 independent experiments.
Quantitation of cell proliferation
Pregnant females were injected with 50 µg/kg of body weight bromodeoxyuridine 1 h before sacrifice. Embryos were fixed in Carnoy’s solution and embedded in paraffin. Sections were treated with 2 N HCl for 30 min at 37 °C and neutralized in 0.1 M sodium borate (pH 8.5), for 5–10 min and immunostained using a mouse monoclonal anti-BrdU antibody (1:300; Dako) and anti-BFABP (1:1000).
Apoptotic neuron binding assay
DRG neurons from E13.5 CD1 mouse embryos were isolated as described above, plated on collagen coated coverslips and grown in UltraCULTURE media with 50 ng/ml NGF (Harlan). Non-neuronal cells were eliminated by two 48 h treatments with uridine (10 µM) and fluorodeoxyuridine (10 µM). NGF was removed to induce apoptosis by rinsing and addition of anti-NGF (1:10,000). After 24 hrs, the neurons were harvested by scraping and stained with propidium iodide (PI) for 20 min, rinsed and added to transfected HEK 293 cells.
HEK 293 cells were plated onto collagen coated 8-well glass chamber slides at a density of 20,000 cells per well in DMEM with 10% FBS (Sigma). After 24 h, the cells were transfected using Effectene (Qiagen) and 48 h later, the indicated number of PI-stained apoptotic neurons were added for 1 h at 4C. Unbound neurons were washed away with 3 rinses of PBS and fixed with 10% formalin. To quantify, at least 100 GFP positive HEK 293 cells were counted per condition. The percentage of GFP+ cells with at least one PI+ corpse bound was calculated.
Surface biotinylation
HEK 293 cells were transfected with GFP-tagged Jedi or MEGF10 and 48 hs later the surface expression analyzed by biotinylation of the receptor at 4°C using EZ-Link Sulfo-NHS-Biotin (Pierce) according the manufacturer’s instructions. The biotinylated proteins were precipitated using avidin-agarose (Pierce) and subjected to SDS-PAGE and Western blotting using antiserum to GFP (1:500; Roche).
Analysis of the ectopic expression of JediΔC::GFP in C. elegans
The Pced-1JediΔC::gfp and Pced-1Jedi::gfp fusion constructs were generated from full length Jedi-1::Flag or Jedi-1ΔC::GFP and the ced-1 promoter (Pced-1)14 and were introduced as extrachromosomal arrays into wild-type or ced-1(e1735) backgrounds. Transgenic lines were obtained using standard microinjection techniques49 and the transgenic animals were identified by GFP expression using a fluorescence microscope. Germ cell corpses in hermaphrodite gonads were analyzed using Axioplan 2 compound microscope (Carl Zeiss) with Nomarski DIC accessories, AxiocaCam camera, and the AxioVision v 4.3 imaging software. Germ cell corpses labeled with JediΔC::GFP were identified by fluorescence microscopy under the Delta Vision Deconvolution Microscope (Applied Precision). Fluorescence images were deconvolved using the SoftWoRx software (Applied Precision).
Statistical analysis
The cell corpse data obtained from transgenic and the wild-type control C. elegans were compared and the P values obtained from un-paired, two-tailed student t-tests. The amount of dead neurons binding to transfected HEK 293 cells was compared using P values obtained from paired, two-tailed student t-tests. The data from over expressing Jedi-1 and/or MEGF10 in glial cells shown in Fig. 7c was analyzed using a one-way ANOVA with a Dunnett’s post-test and the data in 7d evaluated using a Mantel-Haenszel Chi squared analysis. To evaluate the effects of over expressing the truncated mutants of Jedi-1 and MEGF10 and knocking them down in the engulfment assay shown in Fig. 8, the data were subjected to a one-way ANOVA with a Dunnett’s post-test.
Supplementary Material
ACKNOWLEDGEMENTS
This work was supported by grants from the NIH NS048249 and NS064278 to B.D.C., Muscular Dystrophy Association Development Grant (MDA4023) to H.H.W., NIH GM067848 to Z.Z., the NIH MARC Predoctoral Fellowship (GM079911) to V.V., and the Ministerios de Ciencia e Innovación (SAF) y de Sanidad (TerCel and Ciberned), Fundación la Caixa y Generalitat Valenciana (Prometeo), Spain, to I.F. The authors thank the Statistics and Methodology Services, Vanderbilt Kennedy Center, for assistance on statistical analysis, and Choya Yoon, Cedric Jones and other members of the Carter lab for technical assistance and helpful suggestions.
Footnotes
Author Contributions
H.H.W. and B.D.C. initiated and developed the overall concept and design of the project. H.H.W. also performed, analyzed, and interpreted most of the experiments and prepared the initial version of the manuscript. E.B. performed the quantitative histological analysis of neuronal corpse engulfment in nt3 +/+ and −/− animals. J.L.S. generated some of the Jedi and MEGF10 constructs, performed the binding experiment and some of the immunostaining analyses. V.V. performed all the experiments with C. elegans. C.B. assisted with the immunostaining on sections and generated the shRNA construct for MEGF10. L.F.R. provided technical expertise for E.M. analysis and critical intellectual input for this study. I.F. performed the E.M. analysis, supervised the quantitative hisotological analysis and provided intellectual input for this study. Z.Z. designed and supervised the C. elegans study and provided intellectual input for this study. B.D.C. directed the overall project and prepared the final version of the manuscript.
REFERENCES
- 1.Bennet MR, Gibson WG, Lemon G. Neuronal cell death, nerve growth factor and neurotrophic models: 50 years on. Auton Neurosci. 2002;95:1–23. doi: 10.1016/s1566-0702(01)00358-7. [DOI] [PubMed] [Google Scholar]
- 2.Hamburger V, Levi-Montalcini R. Proliferation, differentiation and degeneration in the spinal ganglia of the chick embryo under normal and experimental conditions. J Exp Zool. 1949;111:457–501. doi: 10.1002/jez.1401110308. [DOI] [PubMed] [Google Scholar]
- 3.Savill J, Dransfield I, Gregory C, Haslett C. A blast from the past: clearance of apoptotic cells regulates immune responses. Nat Rev Immunol. 2002;2:965–975. doi: 10.1038/nri957. [DOI] [PubMed] [Google Scholar]
- 4.Yuan J, Lipinski M, Degterev A. Diversity in the mechanisms of neuronal cell death. Neuron. 2003;40:401–413. doi: 10.1016/s0896-6273(03)00601-9. [DOI] [PubMed] [Google Scholar]
- 5.Hume DA, Perry VH, Gordon S. Immunohistochemical localization of a macrophage-specific antigen in developing mouse retina: phagocytosis of dying neurons and differentiation of microglial cells to form a regular array in the plexiform layers. J Cell Biol. 1983;97:253–257. doi: 10.1083/jcb.97.1.253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Perry VH, Hume DA, Gordon S. Immunohistochemical localization of macrophages and microglia in the adult and developing mouse brain. Neuroscience. 1985;15:313–326. doi: 10.1016/0306-4522(85)90215-5. [DOI] [PubMed] [Google Scholar]
- 7.O'Connor TM, Wyttenbach CR. Cell death in the embryonic chick spinal cord. J Cell Biol. 1974;60:448–459. doi: 10.1083/jcb.60.2.448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pannese E. The response of the satellite and other non-neuronal cells to the degeneration of neuroblasts in chick embryo spinal ganglia. Cell Tissue Res. 1978;190:1–14. doi: 10.1007/BF00210032. [DOI] [PubMed] [Google Scholar]
- 9.Bratton DL, Henson PM. Apoptotic cell recognition: will the real phosphatidylserine receptor(s) please stand up? Curr Biol. 2008;18:R76–R79. doi: 10.1016/j.cub.2007.11.024. [DOI] [PubMed] [Google Scholar]
- 10.Gregory CD, Brown SB. Apoptosis: eating sensibly. Nat Cell Biol. 2005;7:1161–1163. doi: 10.1038/ncb1205-1061. [DOI] [PubMed] [Google Scholar]
- 11.Grimsley C, Ravichandran KS. Cues for apoptotic cell engulfment: eat-me, don't eat-me and come-get-me signals. Trends Cell Biol. 2003;13:648–656. doi: 10.1016/j.tcb.2003.10.004. [DOI] [PubMed] [Google Scholar]
- 12.Henson PM, Hume DA. Apoptotic cell removal in development and tissue homeostasis. Trends Immunol. 2006;27:244–250. doi: 10.1016/j.it.2006.03.005. [DOI] [PubMed] [Google Scholar]
- 13.Freeman MR, Delrow J, Kim J, Johnson E, Doe CQ. Unwrapping glial biology: Gcm target genes regulating glial development, diversification, and function. Neuron. 2003;38:567–580. doi: 10.1016/s0896-6273(03)00289-7. [DOI] [PubMed] [Google Scholar]
- 14.Zhou Z, Hartwieg E, Horvitz HR. CED-1 is a transmembrane receptor that mediates cell corpse engulfment in C. elegans. Cell. 2001;104:43–56. doi: 10.1016/s0092-8674(01)00190-8. [DOI] [PubMed] [Google Scholar]
- 15.Awasaki T, et al. Essential role of the apoptotic cell engulfment genes draper and ced-6 in programmed axon pruning during Drosophila metamorphosis. Neuron. 2006;50:855–867. doi: 10.1016/j.neuron.2006.04.027. [DOI] [PubMed] [Google Scholar]
- 16.MacDonald JM, et al. The Drosophila cell corpse engulfment receptor Draper mediates glial clearance of severed axons. Neuron. 2006;50:869–881. doi: 10.1016/j.neuron.2006.04.028. [DOI] [PubMed] [Google Scholar]
- 17.Manaka J, et al. Draper-mediated and phosphatidylserine-independent phagocytosis of apoptotic cells by Drosophila hemocytes/macrophages. J Biol Chem. 2004;279:48466–48476. doi: 10.1074/jbc.M408597200. [DOI] [PubMed] [Google Scholar]
- 18.Savill J, Fadok V. Corpse clearance defines the meaning of cell death. Nature. 2000;407:784–788. doi: 10.1038/35037722. [DOI] [PubMed] [Google Scholar]
- 19.Hamon Y, et al. Cooperation between engulfment receptors: the case of ABCA1 and MEGF10. PLoS ONE. 2006;1:e120. doi: 10.1371/journal.pone.0000120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Farinas I, Yoshida CK, Backus C, Reichardt LF. Lack of neurotrophin-3 results in death of spinal sensory neurons and premature differentiation of their precursors. Neuron. 1996;17:1065–1078. doi: 10.1016/s0896-6273(00)80240-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.White FA, et al. Synchronous onset of NGF and TrkA survival dependence in developing dorsal root ganglia. J Neurosci. 1996;16:4662–4672. doi: 10.1523/JNEUROSCI.16-15-04662.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kurtz A, et al. The expression pattern of a novel gene encoding brain-fatty acid binding protein correlates with neuronal and glial cell development. Development. 1994;120:2637–2649. doi: 10.1242/dev.120.9.2637. [DOI] [PubMed] [Google Scholar]
- 23.Schreiner S, et al. Hypomorphic Sox10 alleles reveal novel protein functions and unravel developmental differences in glial lineages. Development. 2007;134:3271–3281. doi: 10.1242/dev.003350. [DOI] [PubMed] [Google Scholar]
- 24.Taylor MK, Yeager K, Morrison SJ. Physiological Notch signaling promotes gliogenesis in the developing peripheral and central nervous systems. Development. 2007;134:2435–2447. doi: 10.1242/dev.005520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Woodhoo A, Dean CH, Droggiti A, Mirsky R, Jessen KR. The trunk neural crest and its early glial derivatives: a study of survival responses, developmental schedules and autocrine mechanisms. Mol Cell Neurosci. 2004;25:30–41. doi: 10.1016/j.mcn.2003.09.006. [DOI] [PubMed] [Google Scholar]
- 26.Britsch S, et al. The transcription factor Sox10 is a key regulator of peripheral glial development. Genes Dev. 2001;15:66–78. doi: 10.1101/gad.186601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Farinas I, Cano-Jaimez M, Bellmunt E, Soriano M. Regulation of neurogenesis by neurotrophins in developing spinal sensory ganglia. Brain Res Bull. 2002;57:809–816. doi: 10.1016/s0361-9230(01)00767-5. [DOI] [PubMed] [Google Scholar]
- 28.Maro GS, et al. Neural crest boundary cap cells constitute a source of neuronal and glial cells of the PNS. Nat Neurosci. 2004;7:930–938. doi: 10.1038/nn1299. [DOI] [PubMed] [Google Scholar]
- 29.Ernfors P, Lee KF, Kucera J, Jaenisch R. Lack of neurotrophin-3 leads to deficiencies in the peripheral nervous system and loss of limb proprioceptive afferents. Cell. 1994;77:503–512. doi: 10.1016/0092-8674(94)90213-5. [DOI] [PubMed] [Google Scholar]
- 30.Tessarollo L, Vogel KS, Palko ME, Reid SW, Parada LF. Targeted mutation in the neurotrophin-3 gene results in loss of muscle sensory neurons. Proc Natl Acad Sci U S A. 1994;91:11844–11848. doi: 10.1073/pnas.91.25.11844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Murphy P, et al. The regulation of Krox-20 expression reveals important steps in the control of peripheral glial cell development. Development. 1996;122:2847–2857. doi: 10.1242/dev.122.9.2847. [DOI] [PubMed] [Google Scholar]
- 32.Okada A, Lansford R, Weimann JM, Fraser SE, McConnell SK. Imaging cells in the developing nervous system with retrovirus expressing modified green fluorescent protein. Exp Neurol. 1999;156:394–406. doi: 10.1006/exnr.1999.7033. [DOI] [PubMed] [Google Scholar]
- 33.Hoopfer ED, et al. Wlds protection distinguishes axon degeneration following injury from naturally occurring developmental pruning. Neuron. 2006;50:883–895. doi: 10.1016/j.neuron.2006.05.013. [DOI] [PubMed] [Google Scholar]
- 34.Suzuki E, Nakayama M. MEGF10 is a mammalian ortholog of CED-1 that interacts with clathrin assembly protein complex 2 medium chain and induces large vacuole formation. Exp Cell Res. 2007;313:3729–3742. doi: 10.1016/j.yexcr.2007.06.015. [DOI] [PubMed] [Google Scholar]
- 35.Griffin JW, George R, Ho T. Macrophage systems in peripheral nerves. A review. J Neuropathol Exp Neurol. 1993;52:553–560. doi: 10.1097/00005072-199311000-00001. [DOI] [PubMed] [Google Scholar]
- 36.Hirata K, Kawabuchi M. Myelin phagocytosis by macrophages and nonmacrophages during Wallerian degeneration. Microsc Res Tech. 2002;57:541–547. doi: 10.1002/jemt.10108. [DOI] [PubMed] [Google Scholar]
- 37.Bishop DL, Misgeld T, Walsh MK, Gan WB, Lichtman JW. Axon branch removal at developing synapses by axosome shedding. Neuron. 2004;44:651–661. doi: 10.1016/j.neuron.2004.10.026. [DOI] [PubMed] [Google Scholar]
- 38.Aldskogius H, Arvidsson J. Nerve cell degeneration and death in the trigeminal ganglion of the adult rat following peripheral nerve transection. J Neurocytol. 1978;7:229–250. doi: 10.1007/BF01217921. [DOI] [PubMed] [Google Scholar]
- 39.Hanani M. Satellite glial cells in sensory ganglia: from form to function. Brain Res Brain Res Rev. 2005;48:457–476. doi: 10.1016/j.brainresrev.2004.09.001. [DOI] [PubMed] [Google Scholar]
- 40.Fenzi F, Benedetti MD, Moretto G, Rizzuto N. Glial cell and macrophage reactions in rat spinal ganglion after peripheral nerve lesions: an immunocytochemical and morphometric study. Arch Ital Biol. 2001;139:357–365. [PubMed] [Google Scholar]
- 41.Yu X, Lu N, Zhou Z. Phagocytic receptor CED-1 initiates a signaling pathway for degrading engulfed apoptotic cells. PLoS Biol. 2008;6:e61. doi: 10.1371/journal.pbio.0060061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ravichandran KS, Lorenz U. Engulfment of apoptotic cells: signals for a good meal. Nat Rev Immunol. 2007;7:964–974. doi: 10.1038/nri2214. [DOI] [PubMed] [Google Scholar]
- 43.Reddien PW, Cameron S, Horvitz HR. Phagocytosis promotes programmed cell death in C. elegans. Nature. 2001;412:198–202. doi: 10.1038/35084096. [DOI] [PubMed] [Google Scholar]
- 44.Kurant E, Axelrod S, Leaman D, Gaul U. Six-microns-under acts upstream of Draper in the glial phagocytosis of apoptotic neurons. Cell. 2008;133:498–509. doi: 10.1016/j.cell.2008.02.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ziegenfuss JS, et al. Draper-dependent glial phagocytic activity is mediated by Src and Syk family kinase signalling. Nature. 2008 doi: 10.1038/nature06901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Nagata S. Autoimmune diseases caused by defects in clearing dead cells and nuclei expelled from erythroid precursors. Immunol Rev. 2007;220:237–250. doi: 10.1111/j.1600-065X.2007.00571.x. [DOI] [PubMed] [Google Scholar]
- 47.Silva MT, do Vale A, Dos Santos NM. Secondary necrosis in multicellular animals: an outcome of apoptosis with pathogenic implications. Apoptosis. 2008;13:463–482. doi: 10.1007/s10495-008-0187-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wu HH, et al. Autoregulation of neurogenesis by GDF11. Neuron. 2003;37:197–207. doi: 10.1016/s0896-6273(02)01172-8. [DOI] [PubMed] [Google Scholar]
- 49.Jin Y, Jorgensen E, Hartwieg E, Horvitz HR. The Caenorhabditis elegans gene unc-25 encodes glutamic acid decarboxylase and is required for synaptic transmission but not synaptic development. J Neurosci. 1999;19:539–548. doi: 10.1523/JNEUROSCI.19-02-00539.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.