Abstract
Rationale
Ca2+ control of troponin-tropomyosin position on actin regulates cardiac muscle contraction. The inhibitory subunit of troponin, cardiac troponin I (cTnI) is primarily responsible for maintaining a tropomyosin conformation that prevents crossbridge cycling. Despite extensive characterization of cTnI, the precise role of its C-terminal domain (residues 193–210) is unclear. Mutations within this region are associated with restrictive cardiomyopathy, and C-terminal deletion of cTnI, in some species, has been associated with myocardial stunning.
Objective
We sought to investigate the effect of a cTnI deletion -removal of 17 amino acids from the C-terminus- on the structure of troponin-regulated tropomyosin bound to actin.
Methods and Results
A truncated form of human cTnI (cTnI1–192) was expressed and reconstituted with Troponin C and Troponin T to form a mutant troponin. Using electron microscopy and 3D-image reconstruction, we show that the mutant troponin perturbs the positional equilibrium dynamics of tropomyosin in the presence of Ca2+. Specifically, it biases tropomyosin position toward an “enhanced Cstate” that exposes more of the myosin-binding site on actin than found with wild-type troponin.
Conclusions
In addition to its well-established role of promoting the so-called “blocked-state” or “B-state”, cTnI participates in proper stabilization of tropomyosin in the “Ca2+-activated state” or “C-state”. The last 17 amino acids fulfill this stabilizing role. The data are consistent with a “fly-casting” model in which the mobile C-terminus of cTnI ensures proper conformational switching of troponin-tropomyosin. Loss of actin-sensing function within this domain, by pathological proteolysis or cardiomyopathic mutation, may be sufficient to perturb tropomyosin conformation.
Keywords: troponin, thin filament, myocardial stunning, cardiomyopathy
Introduction
Lesions of the myofilament proteins are a common cause of inherited and acquired forms of heart disease1, 2. Such defects in thin filament protein, cardiac troponin I (cTnI), have been implicated in both hypertrophic and restrictive cardiomyopathy as well myocardial stunning. For example, mutations within the C-terminal domain of cTnI gene which cause amino acid substitutions R192H, G203S, and K206Q lead to hypertrophic and/or restrictive cardiomyopathy3, 4, whereas removal of the C-terminal 17 amino acids from cTnI by Ca2+-dependent proteolysis has been implicated in models of myocardial stunning5–9, a condition that arises from brief ischemia that substantially depresses contractile function without causing cell death2, 10. In fact, expression of truncated cTnI1–193, at levels <20% relative to endogenous cTnI, is sufficient to substantially compromise systolic and diastolic function11 in mice. Moreover, C-terminal degradation of cTnI has been observed in patients undergoing coronary artery bypass graft surgery12. Given the role of this domain of cTnI in genetic and acquired heart disease, efforts are underway to fully understand its function.
TnI is one of the three subunits that comprise the troponin complex that regulates muscle contraction by controlling the position of tropomyosin on actin filaments in response to Ca2+ 13, 14. Known as the inhibitory subunit of troponin, it prevents myosin binding to actin, in diastole, by maintaining tropomyosin over the outer edge of actin filaments. In systole, Ca2+ binds to the Ca2+-receptor subunit of troponin, troponin C (TnC), which causes a conformational change that promotes its interaction with cTnI and coincident release of TnI inhibitor regions from actin. This causes the average position of tropomyosin to shift across the face of actin and thereby expose myosin-binding sites that are then accessible for myosin to begin crossbridge cycling15.
The domains of TnI, which contain multiple binding sites for TnC, TnT, actin and tropomyosin have been extensively characterized16 and their organization clarified by the low and high Ca2+ crystal structures of cardiac and skeletal muscle troponin17, 18. However, only an incomplete picture of regulatory switching of tropomyosin on actin can be garnered from the troponin structures, since the C-terminus of TnI is unresolved, owing to its high flexibility19. Given the pathological significance of lesions with the C-terminus of cTnI, we sought to determine how truncation of TnI affects the prime function of troponin, viz, its ability to modulate tropomyosin position on actin filaments. We discuss newly acquired structural data in the context of recent biochemical and biophysical studies of cTnI1–192 and a newly-proposed model of cTnI function20
Methods
Protein preparation
F-actin and bovine cardiac tropomyosin were purified by standard methods21. Methods describing the expression, purification and reconstitution of troponin subunits are described in the supplemental material.
Electron microscopy
Thin filaments were reconstituted by mixing actin, tropomyosin and wild-type or mutant troponin in a ratio of 7:2:2 (F-actin: 10–20 µmol/L) in both low- and high-calcium buffers (low Ca2+: 5 mmol/L PIPES/5 mmol/L sodium phosphate buffer (pH 7.1), 100 mmol/L NaCl, 3 mmol/L MgCl2, 0.2 mmol/L EGTA, 1mmol/L NaN3, 1mmol/L DTT; high-Ca2+: same buffer supplemented with 2 mmol/L CaCl2). Uranyl acetate staining is described in the supplemental material. Electron microscopy was carried out on a Philips CM120 transmission electron microscope using low-dose methods (12e−/Å), the details of which are described in references 22–24.
3D-Image reconstruction from electron micrographs
Electron micrographs were digitized and analyzed by two distinct yet complementary methods of image reconstruction22, 23. First, data were analyzed by helical reconstruction, a Fourier-space filtering and averaging method, using the Brandeis Helical Package essentially as detailed in 23. Given the subtle, yet statistically and biologically significant changes that we observed in thin filament structure, the results were cross-validated by further analysis of micrographs from an independent protein preparation using the real-space single-particle averaging method of Egelman25, as described by Pirani et al.22 (see Online Figure III for a comparison of results obtained from both reconstruction methods).
Results
Electron microscopy and 3D reconstruction of wild-type and cTnI1–192-containing thin filaments
Thin filaments were formed from F-actin, cardiac tropomyosin and troponin complexes under conditions known to saturate the filaments with regulatory proteins22–24 “Wild-type” and mutant troponin complexes, reconstituted from subunits expressed in E. coli, were used for comparison. The mutant troponin contained a truncated form of cTnI (TnI1–192) but included otherwise normal troponin subunits, human cTnC and cTnT. Filaments were negatively stained in uranyl acetate and recorded by low-dose electron microscopy23. EM of the thin filaments showed characteristic double-helical arrays of actin monomers, tropomyosin strands, and troponin densities repeating with a 40 nm periodicity (Fig. 1). EM images of reconstituted filaments prepared from separately expressed and purified proteins were analyzed independently by the first and last authors; the raw images and 3D reconstructions generated from the two data sets were indistinguishable from each other and thus combined for analysis here. Filaments were studied by both helical reconstruction of relatively long filament stretches (~200 to 400 nm)26 and by single particle methods on filament segments (~40 nm)25; results from the two methods were completely consistent and reproducible.
Figure 1. Electron micrographs of reconstituted thin filaments.
(a) Bare actin filaments. (b) Actin filaments reconstituted with wild-type troponin and tropomyosin in Ca2+-free buffer. (c) Actin filaments reconstituted with wild-type troponin and tropomyosin in the presence of Ca2+. (d) Actin filaments reconstituted with mutant troponin and tropomyosin in Ca2+-free buffer. (e) Actin filaments reconstituted with mutant troponin and tropomyosin in the presence of Ca2+. Arrowheads point to the globular heads of the troponin complex. The white arrows highlight tropomyosin strands. Scale bar = 50 nm.
Reconstructions of thin filaments showed actin subunits and densities that were attributable to tropomyosin (Fig. 2). The longitudinally continuous tropomyosin strands were well defined in both control filaments containing “wild-type” troponin-tropomyosin and in filaments containing the mutant cTnI1–192. Inspection of the reconstructions showed that the mutation did not interfere with the ability of tropomyosin to undergo a Ca2+-induced shift from the outer domain (Ao) to the inner domain (Ai) of actin; thus, the impact of both the wild-type troponin and mutant troponin on directed tropomyosin movement is normal in both sets of filaments. In fact, in high Ca2+ conditions, tropomyosin localized further onto Ai in filaments containing mutant cTnI (Fig. 2c and Fig. 3c) than it did in filaments with the wild-type TnI (Fig. 2b and Fig. 3b). Thus, while the direction of the tropomyosin movement was the same in both samples, the magnitude of the movement was greater in the mutant (superimposed in Figure 2A/B-d). As a consequence, less lingering tropomyosin density touched Ao at high Ca2+ in the mutant than in the wildtype. In contrast, no obvious differences in tropomyosin position on F-actin were found for the low Ca2+ data (Fig. 2g and Fig. 3g).
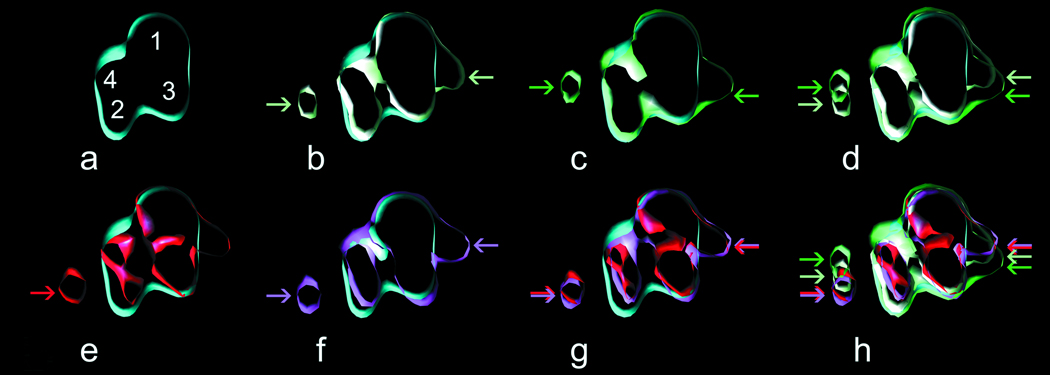
Figure 2. Mutant troponin affects the average position of tropomyosin on actin.
3D reconstructions of filaments are depicted in longitudinal view. (b–d) show 3D-structures of filaments incubated in Ca2+ whereas (e–g) depict reconstructions of Ca2+-free filaments. The bare actin filament is depicted in (a). Subdomains 1 and 2 comprise the outer domain of actin (Ao) whereas the inner domain (Ai) consists of subdomains 3 and 4. Panel (b) depicts actin-tropomyosin with wild-type troponin in Ca2+. Panel (c) depicts of mutant-controlled tropomyosin on actin in Ca2+. In (d) the positions of tropomyosin from (b) and (c) are superimposed. Panels (e) and (f) show the average position of tropomyosin conferred by wild-type and mutant troponin, respectively, in the absence of Ca2+. The results of (e) and (f) are superimposed in (g). All structures are superimposed in (h).
Figure 3. Cross-sections of wild-type- and mutant-controlled thin filaments.
Panels correspond to those described in figure 2. Structures in Ca2+ (b–d); structures in EGTA (e–h); wild-type + Ca2+ (b); mutant + Ca2+ (c); panels (b) and (c) are superimposed in (d). wild-type in EGTA (e); mutant in EGTA (f); panels (e) and (f) are superimposed in (g). All structures are superimposed in (h)
The effect of TnI1–192 at high Ca2+ is statistically significant
Helical projection, i.e. projection of densities down the helical axis of F-actin and tropomyosin, provides a means of defining the average position of tropomyosin relative to actin in reconstructions. Comparison of helical projections confirmed that tropomyosin is localized differently in wild-type and mutant thin filaments, but again such a distinction was only detected for the high Ca2+-treated sample. The distinction was subtle, but became obvious following difference density analysis that isolated the respective tropomyosin densities from actin. Here maps of F-actin (no tropomyosin) were simply subtracted from those of thin filaments. The resulting tropomyosin densities then were superimposed on reference maps of bare F-actin and compared (Fig. 4e). In the presence of Ca2+, tropomyosin controlled by mutant troponin, containing cTnI1–192 was shifted azimuthally by ~9° more than it was by wild-type troponin (Figure 4e). Point by point analysis of the maps using Student’s t-test methodology27, 28, showed that this difference in tropomyosin position was statistically significant at 95% confidence levels. (also, see Online Figure IV, which demonstrates further that the distinctions noted are statistically significant). As the average position of tropomyosin in the mutant is further from the low Ca2+, blocking state than it is in control filaments, we call it the “enhanced-on state position”. Differences in tropomyosin positions for low Ca2+ filaments were not obvious or statistically significant.
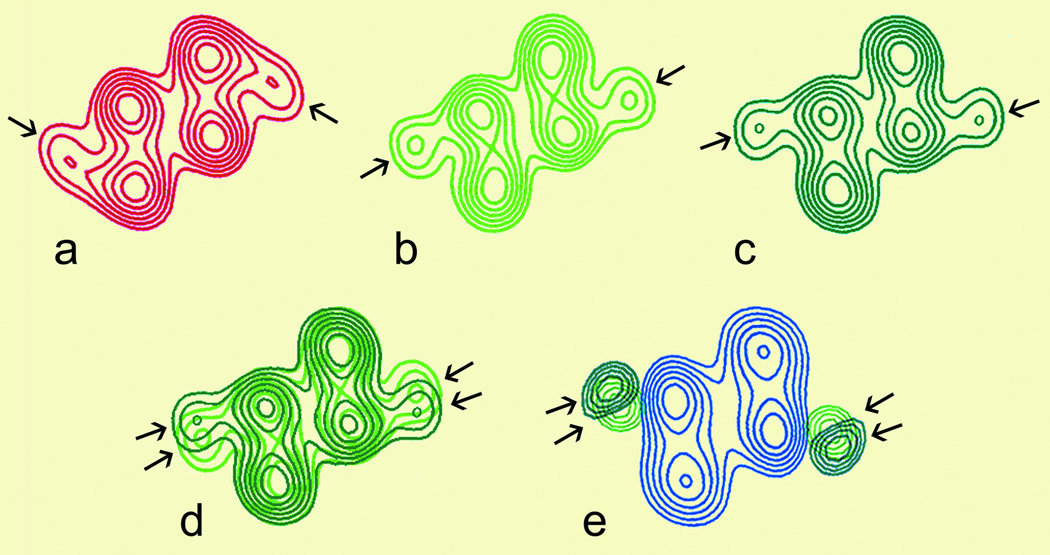
Figure 4. Helical projections illustrate the impact of mutant troponin on the tropomyosin position.
(a) Actin-tropomyosin-troponin filaments in Ca2+-free solution. Tropomyosin sits on the outer domain of actin. (b) In Ca2+, tropomyosin adopts an average position over the inner domain of actin. (c) Mutant Troponin containing cTnI1–192, is also responsive to Ca2+ and tropomyosin again adopts and average position over the inner domain of actin. (d) Superimposing results from (b) and (c) shows that tropomyosin, controlled by mutant troponin, has shifted azimuthally to adopt an average position further from the outer actin domain by about 9°. (e) To better visualize the image densities arising from the tropomyosin strands, the image density of bare actin filaments was subtracted from (b) and (c). The densities of tropomyosin controlled by wild-type (light green) or mutant troponin (dark green) were superimposed over the helical projection of bare actin (blue). The difference in the centroid positions of tropomyosin density were statistically significant at >95%.
Tropomyosin equilibrium position on thin filaments is altered by cTnI1–192
Tropomyosin is thought to oscillate laterally over a narrow region of the flat surface of actin22, 29; however, in the presence of troponin, its equilibrium balance becomes more biased towards specific regulatory positions on actin24, 30, 31 viz. those of the low Ca2+ B-state or the high Ca2+ C-state. The results above suggest that the mutant caused a re-balancing between positional states or possible development of a new equilibrium position for tropomyosin. Cross-correlation tools32, 33 comparing the experimental data to thin filament models with different tropomyosin locations, were used to sort and classify short filament segments into positional categories. An analysis of high Ca2+ filaments indicated that ~3.5 times more mutant filament data fitted better to the “enhanced C-state” than to the wild-type C-state position, whereas the reverse was true for wild-type data, where more of the data belonged to the C-state category (Table 1).
Table 1.
Distribution of filament segments sorting to different regulatory states (%).
| Sample | B-state | C-state | enhanced-C-state |
|---|---|---|---|
| Ca2+-treated filaments with wild type troponin |
29 | 43 | 28 |
| Ca2+-treated filaments with mutant troponin |
26 | 17 | 57 |
Discussion
Control of tropomyosin conformation by TnI1–192
The C-terminal half of cTnI harbors three well characterized domains 1) an actin-binding region that inhibits actomyosin ATPase activity (inhibitory peptide; residues 128–147), 2) a region that binds to the N-terminal domain of troponin C in the presence of Ca2+ (switch peptide; residues 148–163) and 3) a second actin-binding site (residues 168–188). The function of the remaining C-terminal residues (residues 189–210) is largely unknown. In the absence of Ca2+ this highly flexible domain19 adopts a more definedstructure as it binds to actin. Image reconstruction of thin filaments saturated with the C-terminal half of TnI show that the inhibitory region binds to actin at its N-terminus (subdomain 1). Residues downstream of the inhibitory peptide span the cleft of the long-pitch helical actin strands, much like the smooth muscle inhibitory protein caldesmon34, and drape over subdomains 3 and 4 of the adjacent actin, where they abut tropomyosin and stabilize it in the blocked state (B-state)31.
The human cTnI1–192 construct, like the form that recapitulates the phenotype of myocardial stunning in mice11, lacks the last 17 amino acids. Previous biochemical studies35 showed that cTnI1–192, alone, bound both actin and actin-tropomyosin with the same affinity as full length cTnI. Yet when cTnI1–192 was reconstituted into troponin, the complex could not fully inhibit ATPase activity in the absence of Ca2+. This suggested that either cTnI could not maintain tropomyosin in a fully competent B-state or that equilibrium dynamics between the B- and C-states of the thin filament of might be altered. As shown in Fig. 2g/Fig. 3g, mutant troponin caused no statistically discernible difference in the average position of tropomyosin in the presence of EGTA. Thus residues 193–210 of cTnI, downstream of its major actin-binding regions, are not required to generate the B-state position of tropomyosin.
In the presence of Ca2+, the mutant troponin displays higher maximal Ca2+-activated actin-tropomyosin-S1 ATPase than does the wild-type Tn35, 36. Similar observations of both higher basal and Ca2+-activated ATPase activity35 were noted in studies of the murine variant of restrictive cardiomyopathy mutant, R193H37. Wild-type troponin could confer comparable maximal activity, provided that thin filaments were fully activated by non-cycling NEM-S1 heads. The data could be best explained by a shift in the tropomyosin equilibrium from the inactive to the active state37 or, in the context of a 3-state structural model, a shift that would favor transition to the myosin-induced state (M-state). Here, incorporation of cTnI1–192 into troponin, in the presence of Ca2+, evinces a tropomyosin position shifted further over the inner domain of actin (i.e. subdomains 3 and 4) than typically observed with wt troponin in Ca2+. The 9° azimuthal shift of tropomyosin, though subtle, is most discernible in the cross section of the filament (figure 3d) and the helical projection (figure 4e). The tropomyosin displacement is about half of tropomyosin's width and just shy of the fully activated M-state observed in the presence of Ca2+ and docked myosin-S1 heads.
Note that the average position of tropomyosin is a function of the frequency with which one regulatory configuration or another is adopted, i.e. the “enhanced C-state” is not a fixed position on actin but rather is associated with a readjusted distribution of positional states (Table 1). Tropomyosin regulated by wild-type troponin can also adopt the enhanced C-state, albeit less frequently, i.e. in 28% of wild-type filaments vs. 57% among mutant filaments. Therefore, the position of tropomyosin defined by mutant troponin is not a new structural state, per se, but rather a perturbation of the natural equilibrium distribution of tropomyosin on actin. This increases the propensity for tropomyosin to be found further over the inner domain of actin that is comprised of subdomains 3 and 4. Hence actin more easily can bind myosin crossbridges.
The data in Fig 2d/3d and Fig 4e are the first to depict alterations of thin filament structure by a pathological lesion of troponin, and they provide insight into how cTnI1–192, alters the Ca2+-sensitivity of myofilaments. Specifically, we and others have shown that cTnI1–192 increases the Ca2+-sensitivity of the ATPase reaction35, 36, and cTnI1–192 incorporation into rat trabeculae and human myofibrils increases the Ca2+-sensitivity of steady-state isometric tension. To determine the mechanism, Tachampa et al.36 measured the mutant’s effect on Ca2+-affinity for TnC within the troponin complex. Though isolated troponin showed no difference, Ca2+ affinity was increased when mutant troponin was bound to thin filaments36. However, myofilament Ca2+-sensitivity also reflects the degree to which Ca2+-binding would ultimately affect tropomyosin movement. The primary novel finding of this study is that Ca2+-binding to mutant troponin shifts the average position of tropomyosin not to the normal C-state, but to a state that more closely resembles myosin-induced M-state over subdomain 3 and 4 of actin, thereby exposing more of the myosin binding site on actin. Thus, the integrity of the C-terminus of TnI appears to mediate proper equilibrium transitions between the B-, C- and near-M-states and its effect can only be observed properly in the context of the entire thin filament.
Intact cTnI is therefore necessary for stabilization of the natural C-state of tropomyosin on actin, in addition to its established role as stabilizer the B-state in the absence of Ca2+. Ca2+-activation of the thin filament is widely held to involve removal of cTnI from actin as the switch peptide of cTnI (residues 148–163) binds to the N-terminal domain of TnC. However, simple removal of cTnI inhibition in Ca2+, and release of the B-state, is inconsistent with recent biochemical studies showing that cardiomyopathy mutations within distinct C-terminal domains of cTnI perturb tropomyosin equilibrium differently37, 38. While some mutations, notably within the inhibitory peptide, appear to exhibit B-state defects, others closer to the C-terminus are more consistent with defects of the C-state. Moreover, in vitro motility analysis has shown that the sliding velocity of thin filaments regulated by TnIG203S and TnIK206Q was Ca2+-sensitized and ultimately higher in maximum Ca2+, indicative of greater thin filament activation39. Therefore, lesions within the last 17 amino acids of cTnI may cause aberrant thin filament activation by destabilizing the C-state of tropomyosin in favor of a conformation that more closely resembles the fully active M-state.
Proposed mechanism of C-state stabilization by the C-terminus of TnI
If cTnI is an active participant in C-state stabilization, the salient question is, how? Recently, an innovative “fly-casting” hypothesis has been proposed20 to describe the manner by which the highly-disordered C-terminal region of cTnI might contribute to muscle regulation. When Ca2+ binds to the N-terminal of TnC, and the TnC-binding switch peptide of cTnI binds the hydrophobic pocket of TnC, the TnI inhibitory peptide and second actin-binding domain are removed from actin in the process. However, the fly-casting hypothesis posits that residues that lie C-terminal to the TnI switch peptide (the mobile domain) would continue to participate in long-range sampling, or sensing, of the thin filament via ionic interactions. This would effectively catalyze TnI binding to actin when Ca2+ dissociates from TnC19, 20. If the weak transient ionic interactions between the mobile domain of cTnI and actin destabilize binding of the TnI switch peptide to the N-terminal domain of TnC20, then C-terminal deletions of thin filament-bound cTnI would, conversely, confer higher affinity for TnC. Indeed, we noted this previously, as cTnI1–192-mediated inhibition of actin-tropomyosin activated ATPase was more easily reversed by TnC in the presence of Ca2+ (see 35 and Fig 3B therein).
From the perspective of the fly-casting model, cTnI1–192 is deficient in two ways. It lacks the last 17 amino acids and therefore casts a shorter “fishing line” with which to “sense” actin. It also lacks four basic residues, 3 lysines and 1 arginine. Since Lys and Arg are critical determinants of actin-binding affinity in the inhibitory region40 and second actin-binding sites of TnI41, these residues in the C-terminus of TnI may also interact weakly/transiently with Asp and Glu residues on the actin filament. These weak TnI-actin interactions could well be sufficient to stabilize tropomyosin in a wild-type C-state, given the correspondingly low local affinity of tropomyosin for F-actin42. We suspect that pathophysiological changes to the C-terminus of TnI that abrogate its transient ionic interactions with actin, or impinge upon its flexibility, may destabilize the C-state and thereby favor movement of troponin-tropomyosin to an enhanced-C-state. Biochemical studies have shown that cardiomyopathies arising from cTnI mutations alter the equilibrium positions of tropomyosin37, 38 and lesions within the C-terminus, D190H R192H, G203S and R206Q likewise exert Ca2+-sensitizing effects on contractility38, 39. These mutants would also be worth investigating structurally.
Role of the C-terminus of cTnI in heart function
Though difficult to extrapolate complex functional sequelae from static structures, the conformation of tropomyosin conferred by the mutant troponin suggests possible mechanisms by which deletion of the C-terminus of cTnI might influence heart function. The exposure of myosin-binding sites on actin is a key determinant of the rate of crossbridge attachment (so-called fapp in the nomenclature of 2-state crossbridge models)43. Our results with the mutant troponin would therefore favor a higher rate of crossbridge attachment at any given Ca2+ concentration, which is consistent with recent work on stunned rat myocardium44. However, work with both models of stunned myocardium, and the mutant troponin in rat trabeculae, show that force production is substantially compromised11, 36, 44, 45, and at a higher energetic cost36, 44. This is indicative of increased crossbridge turnover that stems from an offsetting increase in the rate of crossbridge detachment (gapp)44, 45. It is possible that destabilization of tropomyosin from the C-state toward the M-state by the mutant may decrease an energetic barrier to crossbridge detachment. Finally, functional studies of the mutant troponin have shown lower cooperativity of Ca2+-activated force production in rat trabeculae36, human cardiomyocytes46, and in a transgenic mouse model of myocardial stunning11. We submit that stunning characterized by low levels of cTnI truncation, could well cause destabilization of tropomyosin as seen here, and thereby dampen propagation of Ca2+ activation along the thin filament.
Indeed, previous work in a transgenic mouse model of stunning indicates that systolic and diastolic heart function is compromised when only 9–17% of the troponins contain truncated cTnI11. Destabilization of tropomyosin position would be expected to have large functional consequences in muscle, even with such low levels of cTnI proteolysis. Given the semi-rigid nature of tropomyosin, local destabilization would be propagated beyond a single troponin-tropomyosin regulatory unit. In other words, since thin filament regulatory switching is cooperative, an effect at one site, in this case due to mutant cTnI, will have a delocalized effect on neighboring sites even if those sites contain wild-type troponin.
Summary
In conclusion, a key issue regarding thin filament activation in striated muscle is: what are the precise molecular determinants that govern the movement of tropomyosin on actin, viz. what protein interactions influence its equilibrium position in the presence and absence of Ca2+? Increased Ca2+-sensitivity observed in multiple studies of cTnI1–192 involves both higher affinity of Ca2+ for troponin36, and alteration of tropomyosin conformation on actin. This change in conformation reveals that cTnI actively stabilizes the natural C-state tropomyosin position in the presence of Ca2+. In diastole, residues 128–192 of cTnI are sufficient to generate the B-state, whereas determinants within the last 17 amino acids are critical to the C-state in systole. This work informs our understanding of myocardial stunning models characterized by cTnI proteolysis at the C-terminus, and suggests a framework for the consideration of restrictive and/or hypertrophic cardiomyopathy mutations within the same domain.
Novelty and Significance
What is known:
Mutations or proteolysis within the C-terminus of cTnI cause heart dysfunction.
The function of this intrinsically-disordered domain is ill-defined.
There is currently no structural framework that helps us understand why mutations in this domain would be harmful.
What new information does this article contribute:
The C-terminus of cTnI stabilizes the Ca2+-activated state of tropomyosin on actin, likely through transient ionic interactions with actin.
This tropomyosin-stabilizing function means that cTnI actively participates in proper myofilament activation in systole in addition to its established role of promoting muscle relaxation in diastole
Cardiomyopathy mutations within the C-terminus of cTnI that affect charge or flexibility may mimic its deletion, altering the natural movements of tropomyosin and, in turn, influencing myofilament crossbridge interactions.
Lesions within the C-terminus of cTnI have severe consequences for heart function, yet this region has no assigned molecular function that would help refine models of contraction or explain its pathophysiology. Here, we have shown that a critical function of this unstructured domain is to stabilize the Ca2+-activated state of the thin filaments. Removal of the C-terminus of TnI perturbs the structure and equilibrium movements of tropomyosin on actin in the presence of Ca2+. This is first documented change in thin filament structure caused by a pathological modification of cTnI. The data suggest an expanded role for cTnI in muscle regulation. It is more than a simple inhibitory protein that promotes muscle relaxation in diastole; it is an active participant in proper myofilament activation in systole. Because defects in the C-terminus of cTnI can be propagated along tropomyosin strands, a small degree of proteolysis, as might occur in myocardial stunning, can have a disproportionate effect on heart function.
Supplementary Material
Acknowledgements
We would like to thank Larry Tobacman and Wei-Dong Gao for their feedback on this manuscript.
Sources of Funding
This study was funded by NIH grants RO1 HL36153 (WL), RO1 AR34711 (RC), RO1 HL63038 (AMM & JVE), P01 AR41637 (CLAW), and by an American Heart Association Postdoctoral Fellowship (New England affiliate) to DBF. DBF would also like to acknowledge early support from the BBRI Scholar Program (Watertown, MA).
Disclosures
None.
Non-standard Abbreviations and Acronyms
- TnI
troponin I
- cTnI
the cardiac troponin I
- cTnI1–192
truncated cTnI lacking 17 amino acids at the C-terminus
- TnC
troponin C
- cTnC
cardiac troponin C
- TnT
troponin T
- cTnT
cardiac troponin T
- B-state
the blocked state of the thin filament
- C-state
the Ca2+-induced closed state of the thin filament
- M-state
the myosin-induced fully-active open state of the thin filament
- EM
electron microscopy
- PIPES
piperazine-N,N’-bis(ethanesulfonic acid)
- EGTA
ethylene glycol tetraacetic acid.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Subject Codes: Myocardial Biology [104; Structure], Myocardial Biology [105; Contractile Function]
References
- 1.Morita H, Seidman J, Seidman CE. Genetic causes of human heart failure. J Clin Invest. 2005;115:518–526. doi: 10.1172/JCI200524351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bolli R, Marban E. Molecular and Cellular Mechanisms of Myocardial Stunning. Physiol Rev. 1999;79:609–634. doi: 10.1152/physrev.1999.79.2.609. [DOI] [PubMed] [Google Scholar]
- 3.Kimura A, Harada H, Park J-E, Nishi H, Satoh M, Takahashi M, Hiroi S, Sasaoka T, Ohbuchi N, Nakamura T, Koyanagi T, Hwang T-H, Choo J-A, Chung K-S, Hasegawa A, Nagai R, Okazaki O, Nakamura H, Matsuzaki M, Sakamoto T, Toshima H, Koga Y, Imaizumi T, Sasazuki T. Mutations in the cardiac troponin I gene associated with hypertrophic cardiomyopathy. Nat Gen. 1997;16:379–382. doi: 10.1038/ng0897-379. [DOI] [PubMed] [Google Scholar]
- 4.Mogensen J, Kubo T, Duque M, Uribe W, Shaw A, Murphy R, Gimeno JR, Elliott P, McKenna WJ. Idiopathic restrictive cardiomyopathy is part of the clinical expression of cardiac troponin I mutations. J Clin Invest. 2003;111:209–216. doi: 10.1172/JCI16336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gao WD, Atar D, Liu Y, Perez NG, Murphy AM, Marban E. Role of troponin I proteolysis in the pathogenesis of stunned myocardium. Circ Res. 1997;80:393–399. [PubMed] [Google Scholar]
- 6.Gao WD, Liu Y, Mellgren R, Marban E. Intrinsic Myofilament Alterations Underlying the Decreased Contractility of Stunned Myocardium : A Consequence of Ca2+-Dependent Proteolysis? Circ Res. 1996;78:455–465. doi: 10.1161/01.res.78.3.455. [DOI] [PubMed] [Google Scholar]
- 7.McDonough JL, Arrell DK, Van Eyk JE. Troponin I Degradation and Covalent Complex Formation Accompanies Myocardial Ischemia/Reperfusion Injury. Circ Res. 1999;84:9–20. doi: 10.1161/01.res.84.1.9. [DOI] [PubMed] [Google Scholar]
- 8.Van Eyk JE, Powers F, Law W, Larue C, Hodges RS, Solaro RJ. Breakdown and Release of Myofilament Proteins During Ischemia and Ischemia/Reperfusion in Rat Hearts : Identification of Degradation Products and Effects on the pCa-Force Relation. Circ Res. 1998;82:261–271. doi: 10.1161/01.res.82.2.261. [DOI] [PubMed] [Google Scholar]
- 9.Westfall MV, Solaro RJ. Alterations in myofibrillar function and protein profiles after complete global ischemia in rat hearts. Circ Res. 1992;70:302–313. doi: 10.1161/01.res.70.2.302. [DOI] [PubMed] [Google Scholar]
- 10.Kloner RA, Jennings RB. Consequences of Brief Ischemia: Stunning, Preconditioning, and Their Clinical Implications: Part 1. Circulation. 2001;104:2981–2989. doi: 10.1161/hc4801.100038. [DOI] [PubMed] [Google Scholar]
- 11.Murphy AM, Kogler H, Georgakopoulos D, McDonough JL, Kass DA, Van Eyk JE, Marban E. Transgenic Mouse Model of Stunned Myocardium. Science. 2000;287:488–491. doi: 10.1126/science.287.5452.488. [DOI] [PubMed] [Google Scholar]
- 12.McDonough JL, Labugger R, Pickett W, Tse MY, MacKenzie S, Pang SC, Atar D, Ropchan G, Van Eyk JE. Cardiac Troponin I Is Modified in the Myocardium of Bypass Patients. Circulation. 2001;103:58–64. doi: 10.1161/01.cir.103.1.58. [DOI] [PubMed] [Google Scholar]
- 13.Kobayashi T, Jin L, de Tombe P. Cardiac thin filament regulation. Pflüg Arch Eur J Phy. 2008;457:37–46. doi: 10.1007/s00424-008-0511-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kobayashi T, Solaro RJ. Calcium, Thin Filaments, and the Integrative Biology of Cardiac Contractility. Annu Rev Physiol. 2005;67:39. doi: 10.1146/annurev.physiol.67.040403.114025. [DOI] [PubMed] [Google Scholar]
- 15.Lehman W, Craig R. Tropomyosin and the steric mechanism of muscle regulation. Adv Exp Med Biol. 2008;644:95–109. doi: 10.1007/978-0-387-85766-4_8. [DOI] [PubMed] [Google Scholar]
- 16.Solaro RJ, Rosevear P, Kobayashi T. The unique functions of cardiac troponin I in the control of cardiac muscle contraction and relaxation. Biochem Biophys Res Commun. 2008;369:82–87. doi: 10.1016/j.bbrc.2007.12.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Vinogradova MV, Stone DB, Malanina GG, Karatzaferi C, Cooke R, Mendelson RA, Fletterick RJ. Ca2+-regulated structural changes in troponin. Proc Nat’l Acad Sci USA. 2005;102:5038–5043. doi: 10.1073/pnas.0408882102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Takeda S, Yamashita A, Maeda K, Maeda Y. Structure of the core domain of human cardiac troponin in the Ca2+-saturated form. Nature. 2003;424:35–41. doi: 10.1038/nature01780. [DOI] [PubMed] [Google Scholar]
- 19.Blumenschein TMA, Stone DB, Fletterick RJ, Mendelson RA, Sykes BD. Dynamics of the C-Terminal Region of TnI in the Troponin Complex in Solution. Biophys J. 2006;90:2436–2444. doi: 10.1529/biophysj.105.076216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hoffman RMB, Blumenschein TMA, Sykes BD. An Interplay between Protein Disorder and Structure Confers the Ca2+ Regulation of Striated Muscle. J Mol Biol. 2006;361:625–633. doi: 10.1016/j.jmb.2006.06.031. [DOI] [PubMed] [Google Scholar]
- 21.Pirani A, Vinogradova MV, Curmi PMG, King WA, Fletterick RJ, Craig R, Tobacman LS, Xu C, Hatch V, Lehman W. An Atomic Model of the Thin Filament in the Relaxed and Ca2+-Activated States. J Mol Biol. 2006;357:707–717. doi: 10.1016/j.jmb.2005.12.050. [DOI] [PubMed] [Google Scholar]
- 22.Pirani A, Xu C, Hatch V, Craig R, Tobacman LS, Lehman W. Single Particle Analysis of Relaxed and Activated Muscle Thin Filaments. J Mol Biol. 2005;346:761–772. doi: 10.1016/j.jmb.2004.12.013. [DOI] [PubMed] [Google Scholar]
- 23.Moody C, Lehman W, Craig R. Caldesmon and the structure of smooth muscle thin filaments: electron microscopy of isolated thin filaments. J Mus Res Cell Motil. 1990;11:176–185. doi: 10.1007/BF01766496. [DOI] [PubMed] [Google Scholar]
- 24.Lehman W, Hatch V, Korman V, Rosol M, Thomas L, Maytum R, Geeves MA, Van Eyk JE, Tobacman LS, Craig R. Tropomyosin and actin isoforms modulate the localization of tropomyosin strands on actin filaments. J Mol Biol. 2000;302:593–606. doi: 10.1006/jmbi.2000.4080. [DOI] [PubMed] [Google Scholar]
- 25.Egelman EH. A robust algorithm for the reconstruction of helical filaments using single-particle methods. Ultramicroscopy. 2000;85:225–234. doi: 10.1016/s0304-3991(00)00062-0. [DOI] [PubMed] [Google Scholar]
- 26.Owen C, DeRosier D. A 13-A map of the actin-scruin filament from the limulus acrosomal process. J Cell Biol. 1993;123:337–344. doi: 10.1083/jcb.123.2.337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Milligan RA, Flicker PF. Structural relationships of actin, myosin, and tropomyosin revealed by cryo-electron microscopy. J Cell Biol. 1987;105:29–39. doi: 10.1083/jcb.105.1.29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Trachtenberg S, DeRosier DJ. Three-dimensional structure of the frozen-hydrated flagellar filament : The left-handed filament of Salmonella typhimurium. J Mol Biol. 1987;195:581–601. doi: 10.1016/0022-2836(87)90184-7. [DOI] [PubMed] [Google Scholar]
- 29.McKillop DF, Geeves MA. Regulation of the interaction between actin and myosin subfragment 1: evidence for three states of the thin filament. Biophysical Journal. 1993;65:693–701. doi: 10.1016/S0006-3495(93)81110-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lehman W, Galinska-Rakoczy A, Hatch V, Tobacman LS, Craig R. Structural Basis for the Activation of Muscle Contraction by Troponin and Tropomyosin. J Mol Biol. 2009;388:673–681. doi: 10.1016/j.jmb.2009.03.060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Galinska-Rakoczy A, Engel P, Xu C, Jung H, Craig R, Tobacman LS, Lehman W. Structural Basis for the Regulation of Muscle Contraction by Troponin and Tropomyosin. J of Mol Biol. 2008;379:929–935. doi: 10.1016/j.jmb.2008.04.062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Frank J, Radermacher M, Penczek P, Zhu J, Li Y, Ladjadj M, Leith A. SPIDER and WEB: Processing and Visualization of Images in 3D Electron Microscopy and Related Fields. J Struct Biol. 1996;116:190–199. doi: 10.1006/jsbi.1996.0030. [DOI] [PubMed] [Google Scholar]
- 33.Galkin VE, Orlova A, Lukoyanova N, Wriggers W, Egelman EH. Actin Depolymerizing Factor Stabilizes an Existing State of F-Actin and Can Change the Tilt of F-Actin Subunits. J Cell Biol. 2001;153:75–86. doi: 10.1083/jcb.153.1.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Foster DB, Huang R, Hatch V, Craig R, Graceffa P, Lehman W, Wang CLA. Modes of Caldesmon Binding to Actin: Sites of Caldesmon Contact and Modulation of Interactions by Caldesmon Phosphorylation. J Biol Chem. 2004;279:53387–53394. doi: 10.1074/jbc.M410109200. [DOI] [PubMed] [Google Scholar]
- 35.Foster DB, Noguchi T, VanBuren P, Murphy AM, Van Eyk JE. C-Terminal Truncation of Cardiac Troponin I Causes Divergent Effects on ATPase and Force: Implications for the Pathophysiology of Myocardial Stunning. Circ Res. 2003;93:917–924. doi: 10.1161/01.RES.0000099889.35340.6F. [DOI] [PubMed] [Google Scholar]
- 36.Tachampa K, Kobayashi T, Wang H, Martin AF, Biesiadecki BJ, Solaro RJ, de Tombe PP. Increased Cross-bridge Cycling Kinetics after Exchange of C-terminal Truncated Troponin I in Skinned Rat Cardiac Muscle. J Biol Chem. 2008;283:15114–15121. doi: 10.1074/jbc.M801636200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mathur MC, Kobayashi T, Chalovich JM. Some Cardiomyopathy-Causing Troponin I Mutations Stabilize a Functional Intermediate Actin State. Biophys J. 2009;96:2237–2244. doi: 10.1016/j.bpj.2008.12.3909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kobayashi T, Solaro RJ. Increased Ca2+ Affinity of Cardiac Thin Filaments Reconstituted with Cardiomyopathy-related Mutant Cardiac Troponin I. J of Biol Chem. 2006;281:13471–13477. doi: 10.1074/jbc.M509561200. [DOI] [PubMed] [Google Scholar]
- 39.Kohler J, Chen Y, Brenner B, Gordon AM, Kraft T, Martyn DA, Regnier M, Rivera AJ, Wang CK, Chase PB. Familial hypertrophic cardiomyopathy mutations in troponin I (K183Δ, G203S, K206Q) enhance filament sliding. Physiol Genom. 2003;14:117–128. doi: 10.1152/physiolgenomics.00101.2002. [DOI] [PubMed] [Google Scholar]
- 40.Van Eyk JE, Hodges RS. The biological importance of each amino acid residue of the troponin I inhibitory sequence 104–115 in the interaction with troponin C and tropomyosin-actin. J Biol Chem. 1988;263:1726–1732. [PubMed] [Google Scholar]
- 41.Tripet B, Van Eyk JE, Hodges RS. Mapping of a second actin-tropomyosin and a second troponin C binding site within the C terminus of troponin I, and their importance in the Ca2+-dependent regulation of muscle contraction. J Mol Biol. 1997;271:728–750. doi: 10.1006/jmbi.1997.1200. [DOI] [PubMed] [Google Scholar]
- 42.Holmes K, Lehman W. Gestalt-binding of tropomyosin to actin filaments. J Mus Res Cell Motil. 2008;29:213–219. doi: 10.1007/s10974-008-9157-6. [DOI] [PubMed] [Google Scholar]
- 43.Gordon AM, Homsher E, Regnier M. Regulation of Contraction in Striated Muscle. Physiol Rev. 2000;80:853–924. doi: 10.1152/physrev.2000.80.2.853. [DOI] [PubMed] [Google Scholar]
- 44.Gao WD, Dai T, Nyhan D. Increased cross-bridge cycling rate in stunned myocardium. Am J Physiol Heart Circ Physiol. 2006;290:H886–H893. doi: 10.1152/ajpheart.00493.2005. [DOI] [PubMed] [Google Scholar]
- 45.Gao WD, Atar D, Backx PH, Marban E. Relationship Between Intracellular Calcium and Contractile Force in Stunned Myocardium : Direct Evidence for Decreased Myofilament Ca2+ Responsiveness and Altered Diastolic Function in Intact Ventricular Muscle. Circ Res. 1995;76:1036–1048. doi: 10.1161/01.res.76.6.1036. [DOI] [PubMed] [Google Scholar]
- 46.Narolska NA, Piroddi N, Belus A, Boontje NM, Scellini B, Deppermann S, Zaremba R, Musters RJ, dos Remedios C, Jaquet K, Foster DB, Murphy AM, van Eyk JE, Tesi C, Poggesi C, van der Velden J, Stienen GJM. Impaired Diastolic Function After Exchange of Endogenous Troponin I with C-Terminal Truncated Troponin I in Human Cardiac Muscle. Circ Res. 2006;99:1012–1020. doi: 10.1161/01.RES.0000248753.30340.af. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.