Abstract
Many biological processes are regulated by gradients of bioactive chemicals. Thus, the generation of materials with embedded chemical gradients may be beneficial for understanding biological phenomena and generating tissue-mimetic constructs. Here we describe a simple and versatile method to rapidly generate materials containing centimeter-long gradients of chemical properties in a microfluidic channel. The formation of chemical gradient was initiated by a passive-pump-induced forward flow and further developed during an evaporation-induced backward flow. The gradient was spatially controlled by the backward flow time and the hydrogel material containing the gradient was synthesized via photopolymerization. Gradients of a cell-adhesion ligand, Arg-Gly-Asp-Ser (RGDS), was incorporated in the poly(ethylene glycol)-diacrylate (PEG-DA) hydrogels to test the response of endothelial cells. The cells attached and spread along the hydrogel material in a manner consistent with the RGDS gradient profile. A hydrogel containing PEG-DA concentration gradient and constant RGDS concentration was also generated. The morphology of cells cultured on such hydrogel changed from round in the lower PEG-DA concentration regions to well-spread in the higher PEG-DA concentration regions. This approach is expected to be a valuable tool to investigate the cell-material interactions in a simple and high-throughput manner and to design graded biomimetic materials for tissue engineering applications.
Keywords: hydrogel gradient, microfluidics, passive pump
1. Introduction
Interactions between cells and materials have been shown to profoundly regulate the cellular behaviors, such as morphology, adhesion, locomotion and gene expression.[1-6] Thus, the ability to recreate and study such interactions may facilitate the development of bioactive and biomimetic materials for various biological and biomedical applications.[1, 7] Numerous studies have shown that both chemical and physical properties of the matrix materials are important to determine the cellular behaviors in their surrounding natural or artificial microenvironments.[8] It should be noted that most previous investigations of the cell-material interactions have used individual material samples with uniform chemical or physical properties, which limit the numbers of samples for testing and its application in studies that require continuous variations in the material properties (chemical or physical), such as haptotaxis[9, 10] or durotaxis.[11, 12] Most of the biomaterials designed for tissue engineering applications also lack the spatially and structurally defined anisotropic properties that exists in native tissues.[13, 14] Biomaterials with continuous variance in properties are therefore of great interests to create biomimetic cellular niches for biological investigation and tissue engineering applications.[15, 16]
Numerous approaches have been adopted to engineer material gradient with chemical variance, which includes manipulating the diffusion pattern or duration of exposure of the photocrosslinkable materials to UV or laser irradiation,[17-19] electrochemistry,[20, 21] plasma polymerization,[22, 23] fluidic gradient mixers[24, 25] and microfluidic gradient generator.[2, 5, 26, 27] However, most of the abovementioned approaches required long and sophisticated fabrication procedures involving expertise and expensive equipments. In addition, it is normally difficult to generate long-range material gradients, which will increase the number of conditions and accuracy for the quantitative investigation of cell-material interactions, and be important to recreate the graded materials for tissue engineering applications.[28] For example, to generate long-range hydrogel gradients by pure diffusion, it may take several days to establish a gradient of the desired molecules from the source to the sink before polymerization. In the case of microfluidic gradient generators, the length of the gradient is determined by the combined length of all the input streams. Thus, in order to generate a centimeter-long gradient, a complicated microfluidic network containing a large number of input streams will be required. Such technical difficulties may be the reason that previous microfluidic-based techniques are limited to generate the gradients that were a few hundred microns in size.
Here we develop a rapid and simple microfluidic-based approach to generate gradient of chemical properties within biologically-relevant hydrogels. With the integration of portable microfluidic manipulation and photo-polymerization, the hydrogel gradient can be rapidly achieved with minimal requirements in costs or expertise. The size of such gradients is in centimeter length scale, which is difficult to achieve by most of the existing methods. Our approach for making materials with embedded chemical gradient is based on using a portable microfluidic device for generating molecular concentration gradient, which is initiated by a passive-pump-induced forward flow and further developed during an evaporation-induced backward flow.[29] Here we engineered spatially controlled centimeter long hydrogels by initially generating a concentration gradient of photo-crosslinkable hydrogel precursors, which contain a mixture of PEG-DA, photo-initiator and/or PEG derivative of cell-adhesive peptide RGDS (Acr-PEG-RGDS) in the microfluidic device. The concentration gradient (either Acr-PEG-RGDS gradient or PEG-DA gradient) was photopolymerized later to form the material gradient with variance in either RGDS concentration or PEG-DA concentration. This microfluidic-based material gradient approach enables rapid and easy generation of biomaterials with a continuous variance in concentration, cell adhesiveness and porosity, which is envisioned to be a powerful tool for a wide range of biological and tissue engineering applications.
2. Results
The gradient generation and stabilization processes are illustrated in Figure 1. The gradient materials were generated inside a fluidic channel that was formed by reversibly sealing a PDMS mold containing the impression of the channel to a glass slide (Figure 1A). The channel was first pre-filled with PEG-DA solution from the outlet. Then, 6 μL of PEG-DA solution containing chemicals of interest were introduced from the inlet (Figure 1B). The forward flow rate was measured to be 1mm/s, which did not cause leaking from the reversibly sealed fluidic channel (Figure 1C). The gradient was generated during the period of evaporation-induced backward flow (Figure 1D). The backward flow was stopped by placing the fluidic system into a humidified chamber, and subsequently the hydrogel precursors were photopolymerized to form the gradient hydrogel (Figure 1E). As shown in Figure 1F, the resultant hydrogel is comprised of three different regions: 1) a positive control region with bioactive chemicals of interest or the highest PEG-DA concentration, 2) a centimeter-long gradient region, and 3) a negative control region with no bioactive chemicals of interest or the lowest PEG-DA concentration.
Figure 1.
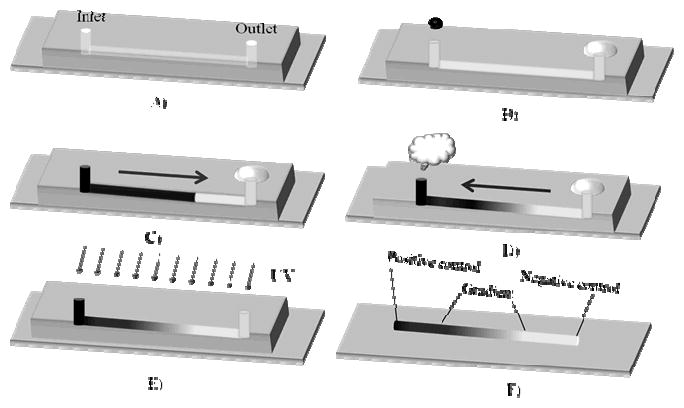
Schematic diagram of the gradient generation and stabilization by photopolymerization. A) the gradient generation device consists of a PDMS mold reversibly sealed to a TMS-PMA treated glass slide, B) the channel was pre-filled with low concentration of PEG-DA precursors from the outlet and a high concentration of PEG-DA precursors containing chemicals of interest was introduced from the inlet, C) the solution of interest flowed spontaneously into the channel by passive-pumping-induced forward flow, D) the gradient was generated by the combined effect of evaporation-induced backward flow and molecular diffusion, E) the gradient was stabilized by photopolymerization, F) the hydrogel gradient was obtained after demolding.
Several factors were essential to create the centimeter-long gradient hydrogel. First, The PDMS mold should be hydrophobic to maintain the spherical shape of the droplets, which are critical to enable the passive pumping process. Therefore, the PDMS mold should not be plasma treated. Second, the length of the gradient could be tuned by controlling the backward flow time. As shown in Figure 2A, when the solution was fully introduced into the channel, the flow profile was parabolic and the length of gradient was about 4 mm. The parabolic profile was gradually flattened in the first 5 min due to the evaporation-induced backward flow. As the backward flow continued, a centimeter-long gradient could be generated within 30 min. Furthermore, the concentration formed an inverted parabolic profile. As shown in Figure 2B, when the backward flow time is longer than 30 min, there was little change in the gradient length. This was probably due to the low water evaporation efficiency with the increasing solution concentration at the inlet. Therefore, the backward flow time was selected as 30 min for the subsequent experiments.
Figure 2.
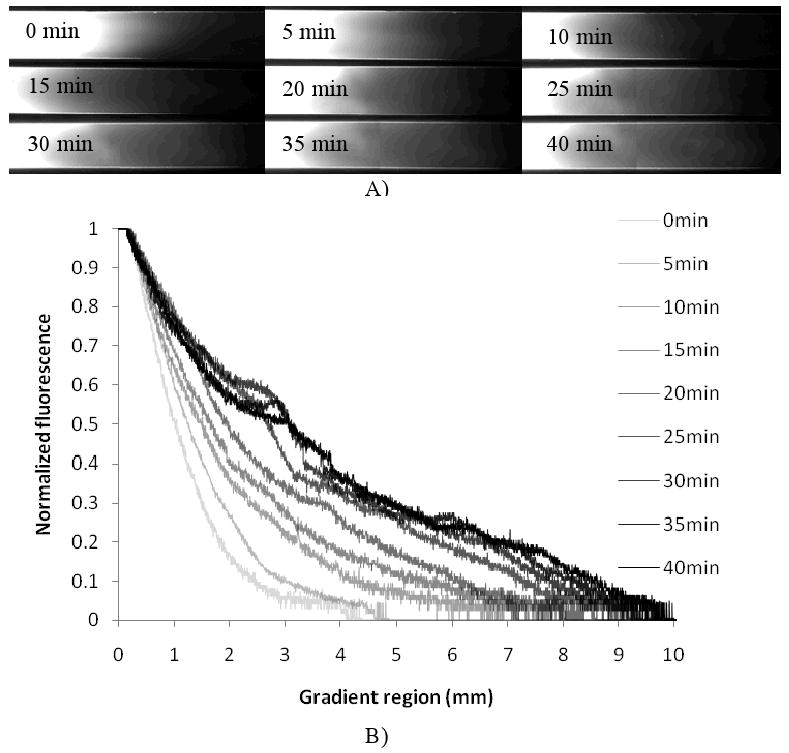
Investigation of the backward flow time to regulate gradient formation. A) Fluorescent images to visualize the gradient at different backward flow times, B) Normalized fluorescence distribution along the channel.
To form the hydrogel gradient, the hydrogel precursor solution was photopolymerized. The concentration of pre-filled solution is a key factor to ensure the integrity of the gradient in the final hydrogel after peeling off the PDMS mold. We pre-filled the channel with different concentrations of PEG-DA solution (0 wt%, 2 wt% and 5 wt%), and then introduced 20 wt% PEG-DA containing 0.05 wt% rhodamine from the inlet to visualize the gradient formation. As shown in Figure 3A-C, the rhodamine gradient disappeared immediately after demolding for the 0 wt% and 2 wt% solutions, but for the 5 wt% PEG-DA solution, an intact hydrogel gradient could be obtained. Figure 3D-F show the phase images of the gradient regions generated in the three conditions. As it can be seen, a hydrogel with a positive control region, a gradient region and a negative control region could be generated only in the case of 5 wt%. The normalized fluorescence distribution along the hydrogel, before and after demolding, were quantified using ImageJ software (Figure 3G and 3H). As it can be seen, the three regions (positive control, gradient and negative control) were created for all pre-filled solutions before demolding, and there was no significant difference in gradient profile. However, after demolding, a large portion of the gradient regions as well as the negative control regions were lost in conditions that were pre-filled PEG-DA concentrations of 0 wt% and 2 wt%.
Figure 3.
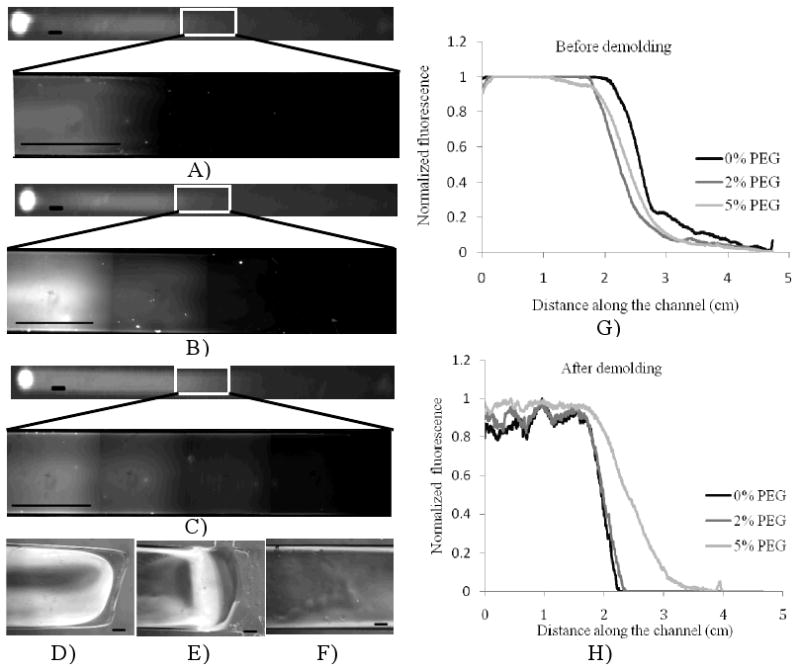
The effect of pre-filled precursor concentration on hydrogel gradient formation (backward flow time: 30 min). A-C) fluorescent images of hydrogel with 0%, 2% and 5% PEG-DA respectively as pre-filled solution after demolding; D-F) phase image of gradient with 0%, 2% and 5% PEG-DA solution as pre-filled solution respectively; G-H) normalized fluorescence distribution before and after demolding. Scale of figure 3a-c is 2mm and that of figure 3d-f is 300μm
To demonstrate the graded properties of the hydrogels generated from 5 wt% PEG-DA solution as pre-filled solution and 40 wt% PEG-DA solution, the hydrogels were either air-dried or freeze-dried and subsequently visualized using scanning electron microscope (SEM). For air-drying, the water in the hydrogel gradually evaporated at room temperature and a film was created. For freeze-drying, the hydrogel was frozen and then lyophilized to generate a porous material upon the sublimation of ice crystals. Figure 4A shows the representative SEM images of the three regions in the same air-dried hydrogel sample. A thinner layer was formed close to the outlet side with a graded intermediate layer in the middle and a thicker layer formed close to the inlet side. The thickness of air-dried hydrogel sample at the 5 wt% PEG-DA side was about 5 μm and that at the 40 wt% PEG-DA side was about 40 μm. Fig. 4B shows the porous structures of the freeze-dried hydrogel sample. As it can be seen, the porosity was gradually increased from the inlet to outlet. It should be noted that the viscosity of the hydrogel precursors may influence the forward and backward flow and the resultant hydrogel gradient. Therefore, this may be a limitation of this technique as handling hydrogel precursors with high viscosity is difficult.
Figure 4.
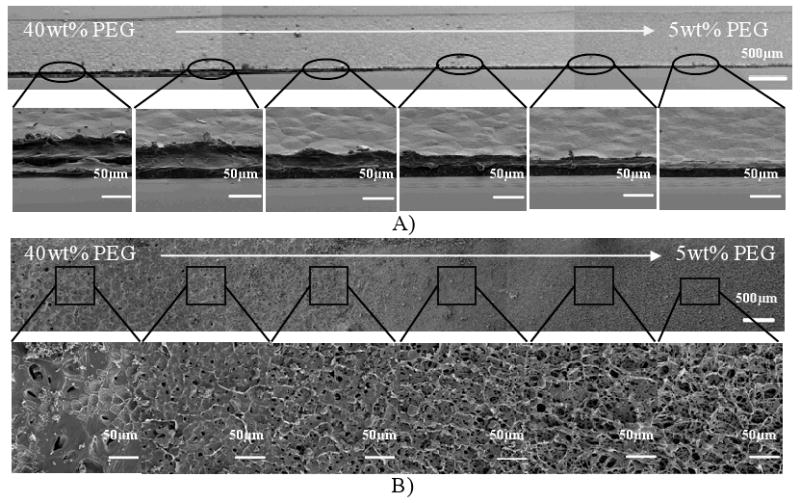
Characterization of air-dried and freeze-dried PEG-DA concentration gradient hydrogels. A) SEM images of the cross-sections of the air-dried hydrogels in the gradient region, B) SEM images of the freeze-dried hydrogel in the porosity gradient region.
To demonstrate the bioactivity of the hydrogel gradient, we generated a gradient of hydrogel conjugated with adhesive RGDS ligands, and investigated the attachment of human umbilical vein endothelial cells (HUVECs). As shown in Figure 5A-C, the cell number and morphology were significantly different in the three regions of the hydrogel. At the positive control region containing 8.0 mM RGDS, many HUVECs attached and spread well on the hydrogel surface, whereas, few cells attached to the negative control region due to the lack of protein adsorption on the hydrophilic PEG-DA surfaces. At the gradient region, the cell numbers followed a similar decreasing trend as the concentration of RGDS ligands. A closer look at the cell morphology in the gradient region (Figure 5D) shows that in the lower RGDS concentration region, the cells remained round in shape with little spreading, whereas they spread towards the higher RGDS concentration region.
Figure 5.
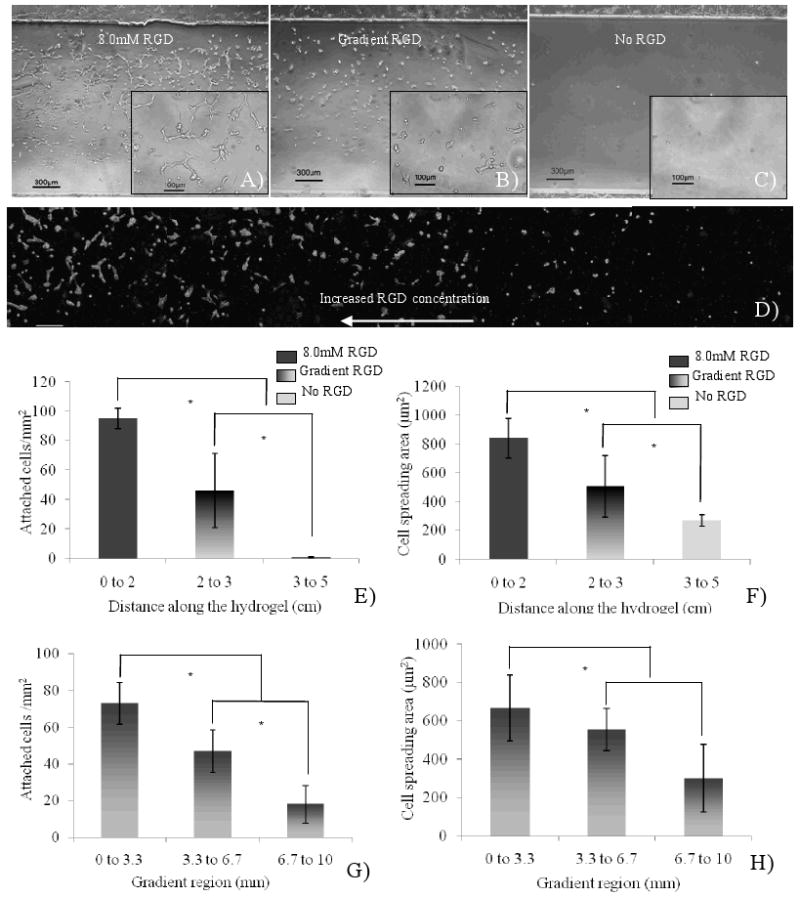
Generation of RGDS gradient hydrogel to guide HUVECs attachment and spreading. A) positive control regions with 8.0mM RGDS, B) RGDS gradient regions, C) negative control regions without RGDS, D) phase images of endothelial cells on RGDS-gradient hydrogel (scale bar: 200 μm), E- F) quantification of cell number and spreading area along the hydrogel, G-H) quantification of cell number and spreading area in gradient region (*, p<0.05).
The cell number and spreading area along the hydrogel were quantified as shown in Figure 5E and 5F. The same trend was observed as before: a higher cell attachment can be seen at the positive control region, a gradually decreasing cell attachment is found at the gradient region, and a lower cell attachment is apparent at the negative control region. The cell density at the positive control region was 93±7 cells/mm2, and the cell spreading area was 842±136 μm2, while at the negative control region, there were few cells and the cell spreading area was 271±41 μm2. For the gradient region, the larger error bars indicate that the HUVECs are distributed in a gradient manner, which is further validated via the quantification of cell number in the gradient region (Fig. 5G). Meanwhile, the concentration of RGDS tethered to the hydrogel also affects cell spreading area as shown in Figure 5H.
To investigate the effect of the PEG-DA concentration gradient on cell behavior, we generated gradient hydrogels ranging from 30 wt% PEG-DA to 5 wt% PEG-DA with a uniform concentration (5.0 mM) of Acr-PEG-RGDS. As shown in Figure 6A, the cell morphology on different regions of the hydrogel was significantly different. At the negative control region (5 wt% PEG-DA), the cells were round in shape and did not spread. In contrast, at the positive control region (30 wt% PEG-DA), the cells were well-spread and appeared to be well-adhered to the underlying substrate. Most notably, the morphology of the cells located on the gradient regions was gradually changed from round to well-spread. We also quantified the cell spreading area at the three regions of the hydrogel. The results (Figure 6B) showed that the average cell spreading area in the gradient region was 705±273 μm2 and that for positive control and negative control were 662±71 μm2 and 316±19 μm2, respectively. In the gradient region, the cell spreading area gradually increased in a gradient manner (Figure 6C). However, it should be noted that the cell number attached on the hydrogel increased with the decrease of PEG-DA concentration, which was also observed when the cells were cultured on the hydrogels with constant PEG-DA concentrations (5 wt%, 15 wt% and 30 wt%) in the same condition (see supplementary).
Figure 6.
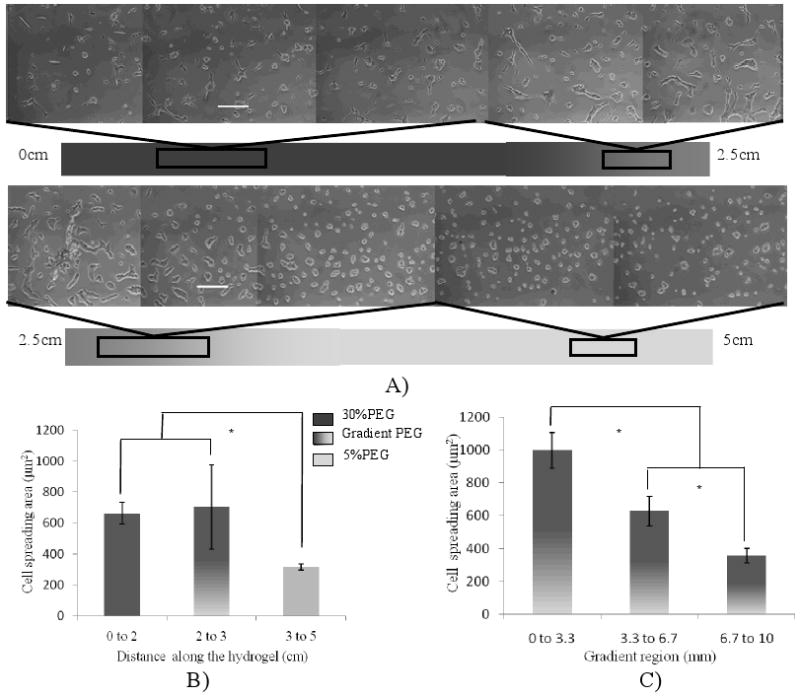
Hydrogels with PEG-DA concentration gradient and constant RGDS concentration to guide HUVECs spreading. A) representative phase contrast images to show the cell morphologies at different regions of a 5cm-long hydrogel (scale bar: 200 μm), B) quantification of the cell spreading area along the PEG-DA concentration gradient hydrogel, C) quantification of the cell spreading area in the gradient region (*, p<0.05).
3. Discussion and Conclusions
The goal of the present study is to develop a simple, rapid and versatile approach for the generation of stable long-range gradient of hydrogels using microfluidic method. Several features of the current approach for gradient generation distinguish it from the existing methods: 1) the resultant hydrogel gradient has three different regions: positive control, negative control and gradient region, which provides a high-throughput platform for biological studies with little experimental error; 2) the size of the hydrogel gradient is in centimeter length scale; 3) the generation of the gradient is rapid (within 30 min), highly dynamic (throughout the backward flow stage) and spatially/temporally controllable (by controlling the evaporation-induced backflow); 4) the gradient can be formed by consuming low amounts of the molecules of interest (from 2–6 μL for the current microfluidic channel design); and 5) the approach is simple and highly reproducible in a portable microfluidic device, requiring only a pipette for implementation.
To maintain the integrity of the hydrogel gradient, the concentration of pre-filled PEG-DA solution should be at least 5 wt% for photopolymerization. On the other hand, to generate the gradient profile in the final hydrogel as predicted using rhodamine, the concentration of PEG-DA solution should be less than 40 wt%. Higher concentrations of PEG-DA solution were too viscous to fill the microfluidic channel by our surface tension driven process. Therefore, it is possible to use higher concentrations by simple modifications to the experimental procedures, such as by using a syringe pump. In this study, we have successfully created a gradient hydrogel with 40 wt% and 5 wt% PEG-DA solutions. When the hydrogel was freeze dried, a porous scaffold was generated with gradient porosity. This may be of particular importance for engineering tissues with gradient porous structures and material distribution such as cartilage[30]. It can be envisioned that a functional scaffold with different biomaterial distribution and gradient porous structures could be fabricated by combining the presented gradient generation method with freeze drying method. Meanwhile, it is possible to incorporate bioactive molecules or proteins (e.g., growth factors) into the porous scaffolds in a controlled manner.
Cellular behaviors, such as adhesion and spreading, are largely regulated by the presence of adhesive molecules. To address these issues, we first generated a RGDS gradient in the cell-repellent PEG-DA hydrogels. The HUVECs cultured on the chemical gradient hydrogel show dramatically different responses dependent on whether they are located on the positive control, the gradient region and the negative control in the same hydrogel. Especially in the gradient region, the cell number gradually increased with the addition of RGDS peptide.
For the hydrogel with the gradients of PEG-DA concentration, we found that the morphology of the cells located on the gradient regions was gradually changed from round to well-spread, which corresponds well with what has been reported previously for vascular smooth muscle cells cultured on the hydrogels with concentration gradient of polyacrylamide[5]. However, the cell number attached on the hydrogels showed an opposite trend in comparison with cell spreading area. The negative effect of PEG-DA concentration on cell attachment might be attributed to the blocking effect of PEG-DA on the RGDS ligand exposed to cells cultured on the hydrogel. Since PEG is well-known for its blocking effect on bioactive surfaces, higher PEG-DA concentration may lead to relatively lower RGDS ligand density exposure to cells on the hydrogel surface.
In conclusion, we have presented a simple and versatile approach to rapidly generate biologically-relevant hydrogels with chemical gradient by combining passive-pump induced forward flow, evaporation-induced backward flow and photopolymerization techniques in a microfluidic channel. The parameters affecting the generation of gradient hydrogel were investigated such as backward flow time and the lowest concentration of pre-filled PEG-DA solution. The resultant hydrogel contains a positive control region, a gradient region and a negative control region, which makes the experimental results more accurate and comparable. Porous tissue engineering scaffolds with gradient porosity variation could be generated by freeze drying the hydrogel with PEG-DA concentration gradient. The response of endothelial cells (adhesion and spreading) to the resultant chemical gradient hydrogels further validated that the presented gradient hydrogel generation method is a promising platform to study biological processes such as cell migration and cell-material interaction.
4. Experimental
Materials
PEG-DA (Molecular Weight 4000) was purchased from Monomer-Polymer & Dajac Labs and Acr-PEG-RGDS were synthesized as previously described[31, 32]. Briefly, Gly-Arg-Gly-Asp-Ser (GRGDS, 1mg/ml, Bachem) was reacted with an equimolar amount of acrylate-PEG-N-hydroxysuccinimide(3500Da, Jenkem Technology) in sodium bicarbonate buffer (50 mM, pH 8.2) for 2 h at room temperature. The product was dialyzed, freeze dried and stored at -20°C until use. The photo-initiator was 2-hydroxy-1-[4-(hydroxyethoxy)phenyl]-2-methyl-l-propanone (D2959, Ciba Geigy, at a concentration of 0.5 wt%). All other reagents were purchased from Sigma-Aldrich (St. Louis, MO) unless specifically mentioned.
Microfluidic device
The microfluidic device consists of a PDMS mold with a straight fluidic channel (50mm×2.0mm×100μm) and a bottom glass slide[29]. The PDMS mold was fabricated using standard soft-lithography methods and reversibly sealed to the glass coverslip. An inlet and outlet of the microchannel were created by a sharp punch (hole radius: 0.4 mm). The glass slide was pre-treated with 3-(trimethoxysilyl) propyl methacrylate (TMS-PMA) to create free methacrylate groups on the glass surface, which would promote the adhesion of PEG-DA hydrogels after UV exposure.
Generation of chemical gradient hydrogels
The microchannel was initially introduced with the pre-filling solution, namely Dulbecco's Phosphate Buffered Saline (DPBS, Gibco, Carlsbad, CA) or a low-concentrated PEG-DA solution. Pre-filling solution (200 μL) was pipetted onto the outlet opening and PEG-DA solution (2 μL) with chemical molecules of interest or higher concentration of PEG-DA was dropped onto the inlet opening. The difference of the surface tension between the two drops induced the automatic flow from inlet to outlet. Three drops of PEG-DA solution (2 μL) were introduced from the inlet. If the inlet was not refilled, the forward flow would stop and a backward flow would occur induced by the evaporation of the solution from the inlet at the room humidity (30%). The pre-polymer concentration gradient was mainly developed during the backward flow process. To visualize the dynamic process of the gradient formation, 0.05wt% rhodamine was mixed with the PEG-DA solution in the inlet and a series of fluorescent images were captured every 5 min after backflow occurred using a fluorescent microscope (Nikon Eclipase TE2000-U, AVON, MA). The normalized fluorescence intensity in the gradient region was quantified using ImageJ software.
The specific hydrogel precursor with concentration gradient was stabilized in the final hydrogels upon photopolymerization (UV exposure: 10mW/cm2 for 70s). To ensure the integrity of gradient in the hydrogel, different concentrated PEG-DA solutions (0 wt%, 2 wt% and 5 wt%) were used as the pre-filled solutions. The final gradient hydrogel was characterized by a Kodak Gel Logic 100 Imaging System, phase-contrast and fluorescent microscope. The normalized fluorescence intensity distribution in the hydrogel before and after demolding was quantified using ImageJ software.
Characterization of PEG-DA concentration gradient hydrogel
To fabricate PEG-DA concentration gradient hydrogel, PEG-DA solution (40 wt%) was used as the drop solution (2 μL) and PEG-DA solution (5 wt%) was selected as pre-filled solution. The resultant hydrogels were air-dried, cut with a scalpel blade to obtain a cross section, sputter-coated with gold and imaged using SEM (ULTRA 55, ZEISS). If the gradient hydrogel was freeze dried, porous scaffold with porosity gradient would be created.
Fabrication of RGDS gradient hydrogel to guide cell attachment
To create Acr-PEG-RGD gradient hydrogel for cell attachment study, PEG-DA solution (20 wt%) was first filled in the microchannel and three drops of PEG-DA solution (20 wt%, 2μL) containing Acr-PEG-RGDS (8.0 mM) were introduced in the inlet. HUVECs were cultured with endothelial cell basal medium (EBM-2, Clonetics) supplemented with vascular endothelial growth factor (VEGF, 0.5 mL), hydrocortisone (0.2 mL), epidermal growth factor (rhEGF, 0.5 mL), ascorbic acid (0.5 mL), r-human fibroblast growth factor-B (rhFGF-B, 2.0 mL), heparin (0.5 mL), recombinant long R insulin-like growth factor (R3-IGF-, 0.5mL) and gentamicin sulfate amphotericin-B (GA-10000, 5 mL) at 37°C in a humidified incubator. Upon trypsinization, the cells were seeded on the surface of hydrogels with Acr-PEG-RGDS gradient, which were pre-washed five times in DPBS and two times in EBM-2. The cell seeding density was 4×104 cells/cm2. After 6 h of incubation, the hydrogels were rinsed with sterile PBS for three times to wash away unattached cells and visualized with phase-contrast microscope. Cells were quantified by counting attached cell number in a minimum of eight images from three individual hydrogels.
Cell study on PEG-DA concentration gradient hydrogel
To create the PEG-DA concentration gradient hydrogel for cell study, PEG-DA solution (5 wt%) was first filled in the microchannel and three drops of PEG-DA solution (30 wt%, 2 μL) were introduced in the inlet. To promote the adhesion of HUVECs onto the PEG-DA concentration gradient hydrogel, Acr-PEG-RGDS (5.0 mM) were incorporated into both solutions. HUVECs were seeded onto the hydrogels at a density of 1×104 cells/cm2. After 24 h of incubation, the cells were fixed for 15 min in a glutaraldehyde solution (0.75% in PBS), and then observed under phase-contrast microscope. The effect of PEG-DA concentration on the cell morphology was investigated.
Statistical analysis
All quantitative Data is expressed as mean±standard deviation. Statistical analysis was performed with one-way analysis of variance (ANOVA) and Tukey HSD tests for post-hoc comparison using SPSS 14.0 statistical package (SPSS Inc. Chicago, USA). Values of p< 0.05 were considered statistically significant.
Supplementary Material
Acknowledgments
This research was funded by the US Army Engineer Research and Development Center, the Institute for Soldier Nanotechnology, and the NIH (HL092836 and DE019024). J. H. was partially sponsored by the China Scholarship Council (CSC), the Program for Changjiang Scholars and Innovative Research Team in University (IRT0646), China. We would like to thank Lifeng Kang, Yi Dong, Adam Hacking, Behnam Zamanian and Shahriar Hojjati Emami for the scientific and technical support.
Footnotes
Supporting Information is available online from Wiley InterScience or from the author.
Contributor Information
Jiankang He, Center for Biomedical Engineering, Department of Medicine, Brigham and Women's Hospital, Harvard Medical School, Boston, MA, 02115, USA; Harvard-MIT Division of Health Sciences and Technology, Massachusetts Institute of Technology, Cambridge, MA, 02139, USA; State Key Laboratory of Manufacturing Systems Engineering, Xi'an Jiaotong University, Xi'an, Shaanxi, 710049, China.
Yanan Du, Center for Biomedical Engineering, Department of Medicine, Brigham and Women's Hospital, Harvard Medical School, Boston, MA, 02115, USA; Harvard-MIT Division of Health Sciences and Technology, Massachusetts Institute of Technology, Cambridge, MA, 02139, USA.
Jose L Villa-Uribe, Center for Biomedical Engineering, Department of Medicine, Brigham and Women's Hospital, Harvard Medical School, Boston, MA, 02115, USA; Harvard-MIT Division of Health Sciences and Technology, Massachusetts Institute of Technology, Cambridge, MA, 02139, USA.
Changmo Hwang, Center for Biomedical Engineering, Department of Medicine, Brigham and Women's Hospital, Harvard Medical School, Boston, MA, 02115, USA; Harvard-MIT Division of Health Sciences and Technology, Massachusetts Institute of Technology, Cambridge, MA, 02139, USA.
Dichen Li, State Key Laboratory of Manufacturing Systems Engineering, Xi'an Jiaotong University, Xi'an, Shaanxi, 710049, China
Ali Khademhosseini, Email: alik@rics.bwh.harvard.edu, Center for Biomedical Engineering, Department of Medicine, Brigham and Women's Hospital, Harvard Medical School, Boston, MA, 02115, USA; Harvard-MIT Division of Health Sciences and Technology, Massachusetts Institute of Technology, Cambridge, MA, 02139, USA.
References
- 1.Khademhosseini A, Langer R, Borenstein J, Vacanti JP. Proceedings of the National Academy of Sciences of the United States of America. 2006;103:2480. doi: 10.1073/pnas.0507681102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Burdick JA, Khademhosseini A, Langer R. Langmuir. 2004;20:5153. doi: 10.1021/la049298n. [DOI] [PubMed] [Google Scholar]
- 3.Engler AJ, Sen S, Sweeney HL, Discher DE. Cell. 2006;126:677. doi: 10.1016/j.cell.2006.06.044. [DOI] [PubMed] [Google Scholar]
- 4.Lutolf MP, Hubbell JA. Nature biotechnology. 2005;23:47. doi: 10.1038/nbt1055. [DOI] [PubMed] [Google Scholar]
- 5.Zaarl N, Rajagopalan P, Kim SK, Engler AJ, Wong JY. Advanced Materials. 2004;16:2133. [Google Scholar]
- 6.Peppas NA, Hilt JZ, Khademhosseini A, Langer R. Advanced Materials. 2006;18:1345. [Google Scholar]
- 7.Liu WF, Chen CS. Materials Today. 2005;8:28. [Google Scholar]
- 8.Sniadecki NJ, Anguelouch A, Yang MT, Lamb CM, Liu Z, Kirschner SB, Liu Y, Reich DH, Chen CS. Proceedings of the National Academy of Sciences of the United States of America. 2007;104:14553. doi: 10.1073/pnas.0611613104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Harris A. Experimental cell research. 1973;77:285. doi: 10.1016/0014-4827(73)90579-x. [DOI] [PubMed] [Google Scholar]
- 10.Brandley BK, Schnaar RL. Developmental biology. 1989;135:74. doi: 10.1016/0012-1606(89)90159-0. [DOI] [PubMed] [Google Scholar]
- 11.Lo CM, Wang HB, Dembo M, Wang Y. Biophysical Journal. 2000;79:144. doi: 10.1016/S0006-3495(00)76279-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wong JY, Velasco A, Rajagopalan P, Pham Q. Langmuir. 2003;19:1908. [Google Scholar]
- 13.Wang M. Biomaterials. 2003;24:2133. doi: 10.1016/s0142-9612(03)00037-1. [DOI] [PubMed] [Google Scholar]
- 14.Langer RS, Vacanti JP. Scientific American. 1999;280:86. doi: 10.1038/scientificamerican0499-86. [DOI] [PubMed] [Google Scholar]
- 15.DeLong SA, Gobin AS, West JL. J Control Release. 2005;109:139. doi: 10.1016/j.jconrel.2005.09.020. [DOI] [PubMed] [Google Scholar]
- 16.DeLong SA, Moon JJ, West JL. Biomaterials. 2005;26:3227. doi: 10.1016/j.biomaterials.2004.09.021. [DOI] [PubMed] [Google Scholar]
- 17.Lo CT, Throckmorton DJ, Singh AK, Herr AE. Lab on a chip. 2008;8:1273. doi: 10.1039/b804485f. [DOI] [PubMed] [Google Scholar]
- 18.Hypolite CL, McLernon TL, Adams DN, Chapman KE, Herbert CB, Huang CC, Distefano MD, Hu WS. Bioconjugate Chemistry. 1997;8:658. doi: 10.1021/bc9701252. [DOI] [PubMed] [Google Scholar]
- 19.Herbert CB, McLernon TL, Hypolite CL, Adams DN, Pikus L, Huang CC, Fields GB, Letourneau PC, Distefano MD, Hu WS. Chemistry & biology. 1997;4:731. doi: 10.1016/s1074-5521(97)90311-2. [DOI] [PubMed] [Google Scholar]
- 20.Plummer ST, Bohn PW. Langmuir. 2002;18:4142. [Google Scholar]
- 21.Ratcliff EL, Hillier AC. Langmuir. 2007;23:9905. doi: 10.1021/la700827w. [DOI] [PubMed] [Google Scholar]
- 22.Whittle JD, Barton D, Alexander MR, Short RD. Chemical Communications. 2003:1766. [Google Scholar]
- 23.Alexander MR, Whittle JD, Barton D, Short RD. Journal of Materials Chemistry. 2004;14:408. [Google Scholar]
- 24.Chu B, Liang D. Electrophoresis. 2002;23:2602. doi: 10.1002/1522-2683(200208)23:16<2602::AID-ELPS2602>3.0.CO;2-X. [DOI] [PubMed] [Google Scholar]
- 25.Liang D, Song L, Quesada MA, Tian Z, Studier FW, Chu B. Electrophoresis. 2000;21:3600. doi: 10.1002/1522-2683(200011)21:17<3600::AID-ELPS3600>3.0.CO;2-H. [DOI] [PubMed] [Google Scholar]
- 26.Jeon NL, Dertinger SKW, Chiu DT, Choi IS, Stroock AD, Whitesides GM. Langmuir. 2000;16:8311. [Google Scholar]
- 27.Dertinger SK, Jiang X, Li Z, Murthy VN, Whitesides GM. Proceedings of the National Academy of Sciences of the United States of America. 2002;99:12542. doi: 10.1073/pnas.192457199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Weiner S, Wagner HD. Annual Review of Materials Science. 1998;28:28. [Google Scholar]
- 29.Du Y, Shim J, Vidula M, Hancock MJ, Lo E, Chung BG, BJ T, Khabiry M, Cropek DM, Khademhosseini A. Lab on a chip. 2009;9:761. doi: 10.1039/b815990d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Woodfield TB, Van Blitterswijk CA, De Wijn J, Sims TJ, Hollander AP, Riesle J. Tissue engineering. 2005;11:1297. doi: 10.1089/ten.2005.11.1297. [DOI] [PubMed] [Google Scholar]
- 31.Burdick JA, Anseth KS. Biomaterials. 2002;23:4315. doi: 10.1016/s0142-9612(02)00176-x. [DOI] [PubMed] [Google Scholar]
- 32.Hern DL, Hubbell JA. Journal of biomedical materials research. 1998;39:266. doi: 10.1002/(sici)1097-4636(199802)39:2<266::aid-jbm14>3.0.co;2-b. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


