Abstract
We report that the activity of a rationally engineered protein tyrosine phosphatase (PTP) mutant can be fully rescued by the addition of the biarsenical fluorescein derivative FlAsH, a compound that does not affect the activity of wild-type PTPs.
Small molecules that specifically control protein function are invaluable tools for chemical biology. While many known compounds that act on intracellular proteins are enzyme inhibitors, direct chemical activation of enzymes is also a potentially powerful tool for studying complex processes.1 However, few small-molecule enzyme activators have been identified and most potential target enzymes do not contain known natural allosteric-activation sites.2
Targeted chemical rescue, in which the activity of a mutationally inactivated target is specifically increased by the addition of a small molecule, presents an attractive strategy for circumventing the lack of known enzyme-activation sites. Unfortunately, examples of biologically useful chemical rescue are rare, as the prerequisites for rescue that works in a cellular environment—an inactivating mutation that can be overcome by a small molecule, in addition to an activating compound that is cell permeable and not generally toxic at the active concentrations—are difficult to obtain. Nevertheless, a few demonstrations of the utility of enzyme-activity rescue in chemical biology are known.1 In one important example, Cole and co-workers have shown that an inactivated form of Src tyrosine kinase can be turned on in living cells by the addition of imidazole, which complements an active-site “hole” in the mutated target kinase.3
Protein tyrosine phosphatases (PTPs) are a large and important class of signaling enzymes that catalyze the dephosphorylation of phosphotyrosine in protein substrates.4 Mammalian cells ubiquitously use tyrosine phosphorylation as a key regulatory element, and improperly regulated PTP activity has been implicated in a range of human diseases including leukemia, solid-tumor cancers, diabetes, and several autoimmune disorders.5 Moreover, a number of PTPs are tumor-suppressor proteins,5 making this superfamily particularly attractive for targeted enzyme-activation strategies: compounds that can activate specific PTPs in a cellular context could represent important tools for investigating the mechanisms of tumor suppression in engineered models of oncogenesis. However, no stratgies for specifically “turning on” a target PTP are known.
We have recently shown that a catalytically critical loop of the PTP domain (the “WPD loop”) can be engineered to contain cysteine-rich elements that bind the biarsenical reagent FlAsH6 (Fig. 1A).7 This approach has lead to the identification of several engineered PTPs that are strongly inhibited by FlAsH, a cell-permeable small molecule that does not affect the activity of wild-type PTPs.8 There is, however, nothing inherently inhibitory about the action of FlAsH on engineered enzymes—it could, in principle, be used to stabilize either an active or inactive conformation, and it has been previously shown that FlAsH can activate an engineered antibiotic-resistance enzyme.9 Here we show that targeting of FlAsH to the WPD-loop can be used to potently and specifically turn on the activity of a biologically important tyrosine phosphatase, PTP-PEST.
Fig. 1.
Engineering a FlAsH-activatable mutant of PTP-PEST. (A) Chemical structure of FlAsH. (B) Amino acid sequences of the WPD-loop regions of wild-type (WT) PEST and the FlAsH-activatable PEST mutant PEST-CC-204.
PEST is a wide-ranging and ubiquitously expressed signaling molecule, which is involved in the regulation of osteoclast activation,10 cell motility and adhesion,11 the immune response,12 and apoptosis.13 To engineer a potentially FlAsH-activatable form of PEST, we identified the PEST WPD loop from PTP primary sequences alignments4 (no crystal structure of PEST has been reported), and we introduced a series of cysteine mutations and small cysteine-containing insertions at various points in the loop (Supplementary Fig. 1). (Previous data from our lab14 and others15 have established that the presence of an entire canonical CCXXCC sequence is not always required for a protein to bind FlAsH.) Most of the cysteine-enriched WPD-loop mutants demonstrated no novel sensitivity to FlAsH, the exception being PEST-CC-204, which differs from the wild-type enzyme only by the insertion of a two-cysteine unit immediately following the second conserved proline of the WPD loop (Fig. 1B).
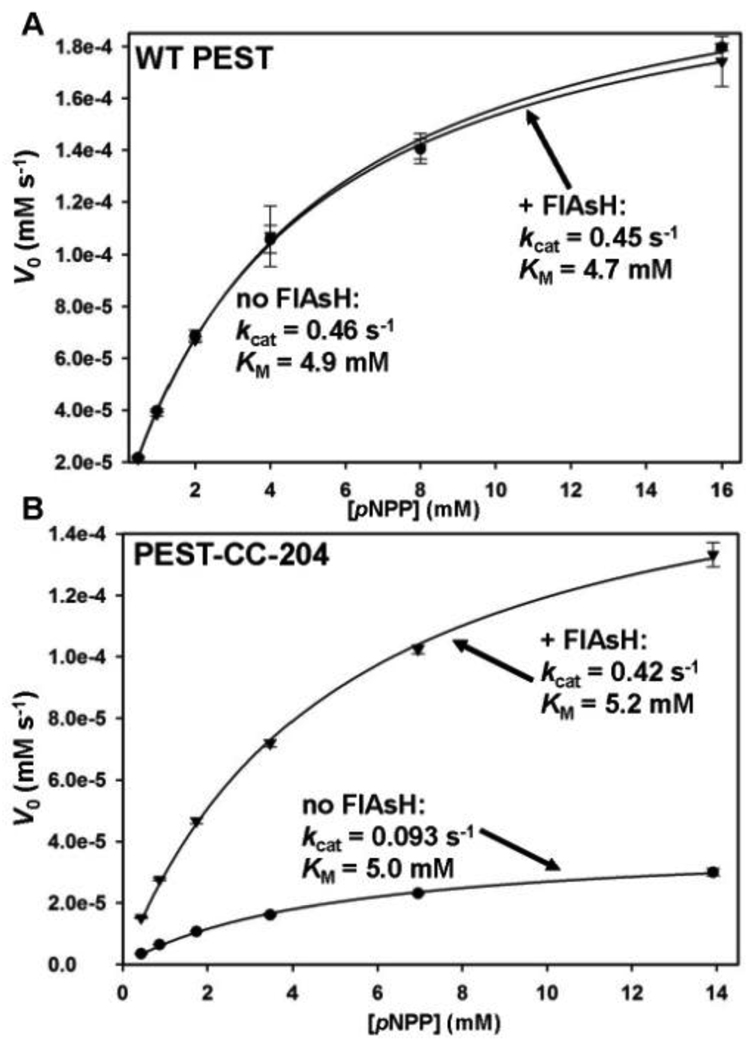
We expressed and purified the six-histidine tagged catalytic domain of PEST-CC-204 and evaluated its in vitro activity using the PTP substrate para-nitrophenyl phosphate (pNPP). Not surprisingly, the inherent activity of PEST-CC-204 is substantially diminished with respect to the wild-type enzyme: in the absence of FlAsH ligand, the catalytic-rate-constant value (kcat) of the mutant enzyme is roughly five-fold lower than the corresponding wild-type value (Fig. 2).
Fig. 2.
FlAsH-induced activation of PEST-CC-204. (A) WT PEST (2.5 µM) or (B) PEST-CC-204 (2.5 µM) was incubated in the absence or presence of FlAsH (10 µM), diluted, and assayed for activity with the PTP substrate pNPP at the indicated concentrations. The initial rates of the resulting reactions were fit to the Michaelis-Menten equation to derive the indicated kinetic constants. Error bars represent the standard deviations from three independent measurements at a particular pNPP concentration.
FlAsH is neither an inhibitor nor an activator of wild-type PTPs and, in agreement with previous results on PEST and other PTPs,8 pre-incubation of wild-type PEST with FlAsH had no significant effect on its activity (Fig. 2A). By contrast, pre-incubation of PEST-CC-204 with a four-fold excess of FlAsH gave rise to a strong increase in PTP activity, to a level (kcat = 0.42 s−1, Fig. 2B) that is comparable with that of wild-type PEST in the absence of FlAsH (kcat = 0.46 s−1, Fig. 2A).
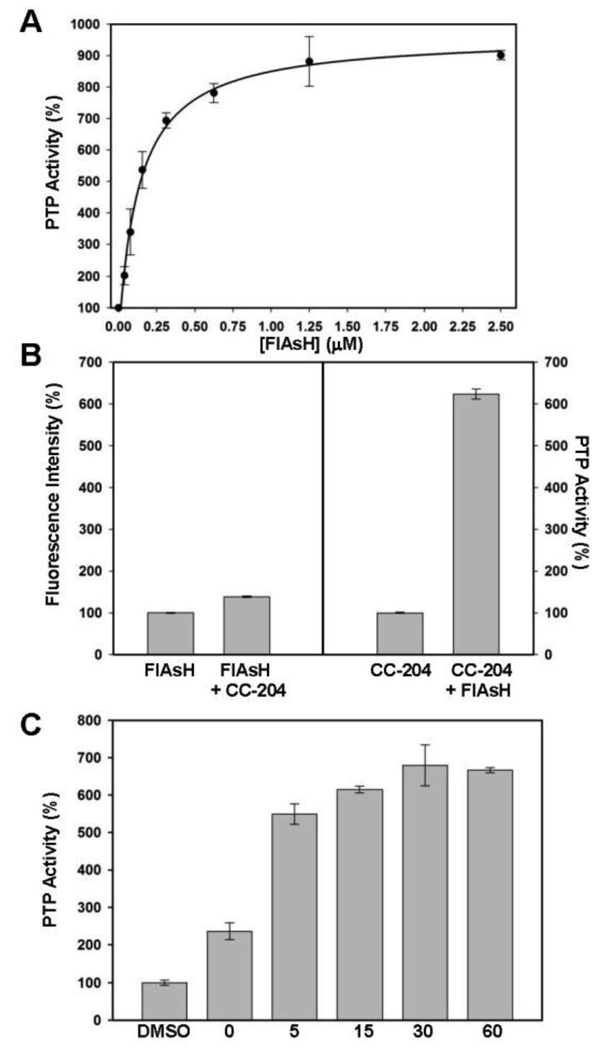
To test whether the degree of PEST-CC-204 rescue was dependent on the FlAsH dosage, we performed a concentration-dependence activation assay (Figure 3A). We observed that the activation level of FlAsH is highly concentration-dependent and saturable, with half-maximal activation achieved at roughly 200 nM FlAsH and full activation (an approximately 9-fold increase in activity) acheieved at 1–2 µM FlAsH. It should be noted that FlAsH so potently activates PEST-CC-204 that the half-maximal activation value approaches the concentration of enzyme in the assay (125 nM PEST-CC-204, the lowest concentration that can be reasonably used in the pNPP-based assay). Under conditions of such potent activation, the measured half-maximum and saturation values are dependent on the enzyme concentration used: only approximately two equivalents of FlAsH were necessary to activate the enzyme to wild-type levels (five-fold activation).
Fig. 3.
Potent, dose-dependent, and rapid activation of PEST-CC-204. (A) Concentration-dependence of FlAsH-mediated activation: PEST-CC-204 (125 nM) was incubated in the absence or presence (indicated concentrations) of FlAsH. After 2.5 hours, PTP reactions were started by the addition of pNPP (4 mM). (B) FlAsH fluorescence of activated PEST-CC-204: Solutions containing either an absence or presence of PEST-CC-204 (125 nM) in the absence or presence of FlAsH (1.25 µM) were incubated at room temperature. After 2.5 hours, the FlAsH fluorescence values (left panel) and the PTP activities (with 5 mM pNPP, right panel) of the solutions were measured. Data were normalized to the FlAsH-only (for fluorescence) and enzyme-only (for PTP activity) controls. (C) Time-dependence of FlAsH-mediated activation: PEST-CC-204 (2.5 µM) was pre-incubated in the absence or presence of FlAsH (10 µM) for the indicated number of minutes, diluted and assayed for activity with pNPP (16 mM). (A–C) In all panels, “PTP Activity (%)” is defined as the initial velocity in the presence of FlAsH divided by the initial velocity of the vehicle-only (100%) control and error bars represent the standard deviations from three independent measurements.
FlAsH was developed by Tsien and co-workers as a fluorescence-based protein-visualization tool,6 and it is generally assumed in the biarsenical literature that FlAsH/protein binding and increases in FlAsH fluorescence go hand-in-hand.15 Interestingly, FlAsH/PEST-CC-204 preincubation conditions that induced strong activation of PTP activity gave rise to only marginal increases in FlAsH fluorescence (Fig. 3B), presumably due to the fact that PEST-CC-204 contains only half (CC) of the classical FlAsH target site (CCXXCC). (Free FlAsH undergoes excited-state quenching, and it is thought that constrained rotation about the aryl-arsenic bonds is the key to induced fluorescence in the full tetracysteine/FlAsH complex.6) To our knowledge, PEST-CC-204 activation provides the first demonstration that FlAsH can serve as a potent modulator of protein function even in cases in which its utility as a protein-visualization tool is largely absent.
Because FlAsH is not a rapid, reversible protein binder—biarsenicals make covalent sulfur-arsenic bonds with their peptide targets6, 16—the timescale of FlAsH action can be a significant issue when using the compound for controlling protein function. In order to determine the timescale of FlAsH-induced PEST-CC-204 activation we performed a time-course activation experiment. We compared the activity of an enzyme-FlAsH incubation with that of an enzyme-vehicle incubation over the course of one hour. The data shown in Figure 3C indicate that activation of 2C-204 PTP-PEST by FlAsH is rapid: within five minutes activation was approximately 80% complete, and within thirty minutes it was maximized. Even at “0 minutes” of pre-incubation (FlAsH and pNPP added at the same time), two-fold activation of PEST-CC-204 was observed, showing that within the course of the activity assay itself (three minutes), the mutant bound to and was activated by FlAsH.
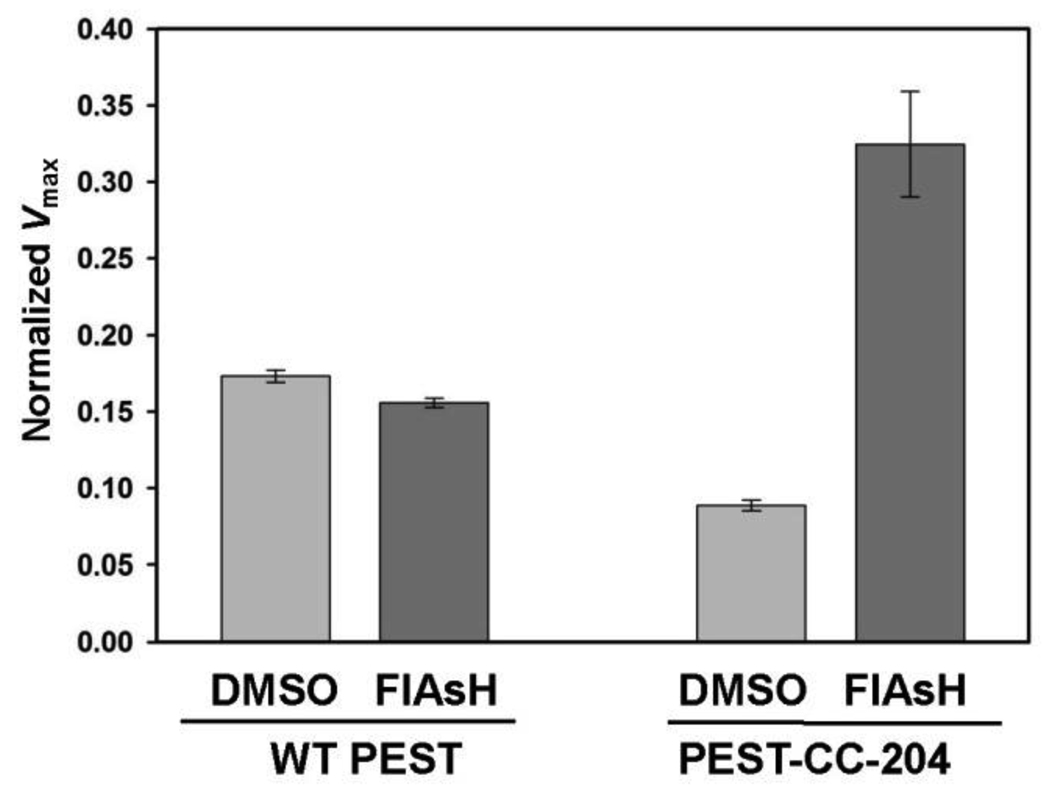
Having established that FlAsH activates purified PEST-CC-204, we then sought to determine whether FlAsH could act in the context of a complex proteomic mixture. Since bacterial genomes do not encode PTPs, total PTP activity in crude lysates from E. coli strains that exogenously express either wild-type PEST or PEST-CC-204 can be used for a direct readout of PEST activity. Consistent with the kinetic constants measured on purified proteins (Fig. 2), the specific activity of PEST-CC-204-expressing lysate is lower than that of wild-type expressing lysate in the absence of FlAsH (Fig. 4). Addition of FlAsH, however, strongly and specifically increased the PTP activity of the PEST-CC-204-containing lysate. Future work in engineered cell lines will be necessary to determine whethter PEST-CC-204 is specifically activatable in the context of a mammalian cell.
Fig. 4.
Activation of PEST-CC-204 in a complex proteome. Crude cell lysates from E. coli strains expressing either WT PEST or PEST-CC-204 were incubated in the absence or presence (10 µM) of FlAsH. After 2.5 hours, the PTP activities of the lysates were assayed as in Figure 1. To normalize the data from different lysates, the measured maximum velocity (µM s−1) of each lysate divided by its respective total protein concentration (mg mL−1). Error bars represent the standard deviations from two (WT) and four (PEST-CC-204) independent lysis experiments.
In conclusion, we have demonstrated that the biarsenical reagent FlAsH is useful as an enzyme-activating small molecule for a member of one of the largest and most important families of cell-signaling enzymes, the PTPs. Our results show that a short peptide insertion can be used to engineer a novel activation site into PTP-PEST despite a lack of three-dimensional structural information for the wild-type enzyme. More generally, we have shown that FlAsH-binding “half-sites” can be used to confer dramatic small-molecule sensitivity on a target protein, even in the absence of a strong fluoresence change upon formation of the FlAsH/protein complex. Since the mammalian-cell permeability of biarsenical reagents is well established,17 it is likely that FlAsH-induced PEST-CC-204 activation can be used to delineate the precise functions of PEST in cultured cells and that FlAsH can be added to the short list of biologically useful chemical-rescue reagents.
Supplementary Material
Acknowledgments
This research was supported by the National Institutes of Health (1 R15 GM071388-01A1) and Amherst College. The authors thank Dr. Xin-Yu Zhang for helpful conversations.
Footnotes
Electronic Supplementary Information (ESI) available: Supplementary Figure 1 and complete experimental protocols. See DOI: 10.1039/b000000x
Notes and References
- 1.Bishop AC, Chen VL. J. Chem. Biol. 2009;2:1. doi: 10.1007/s12154-008-0012-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hardy JA, Wells JA. Curr. Opin. Struct Biol. 2004;14:706. doi: 10.1016/j.sbi.2004.10.009. [DOI] [PubMed] [Google Scholar]
- 3.Qiao YF, Molina H, Pandey A, Zhang J, Cole PA. Science. 2006;311:1293. doi: 10.1126/science.1122224. [DOI] [PubMed] [Google Scholar]
- 4.Andersen JN, Mortensen OH, Peters GH, Drake PG, Iversen LF, Olsen OH, Jansen PG, Andersen HS, Tonks NK, Moller NP. Mol. Cell. Biol. 2001;21:7117. doi: 10.1128/MCB.21.21.7117-7136.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tonks NK. Nat. Rev. Mol. Cell Biol. 2006;7:833. doi: 10.1038/nrm2039. [DOI] [PubMed] [Google Scholar]
- 6.Griffin BA, Adams SR, Tsien RY. Science. 1998;281:269. doi: 10.1126/science.281.5374.269. [DOI] [PubMed] [Google Scholar]
- 7.Zhang XY, Bishop AC. J. Am. Chem. Soc. 2007;129:3812. doi: 10.1021/ja069098t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhang XY, Chen VL, Rosen MS, Blair ER, Lone AM, Bishop AC. Bioorg. Med. Chem. 2008;16:8090. doi: 10.1016/j.bmc.2008.07.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Erster O, Eisenstein M, Liscovitch M. Nat. Methods. 2007;4:393. doi: 10.1038/nmeth1046. [DOI] [PubMed] [Google Scholar]
- 10.Granot-Attas S, Elson A. Eur. J. Cell Biol. 2008;87:479. doi: 10.1016/j.ejcb.2008.01.006. [DOI] [PubMed] [Google Scholar]
- 11.Garton AJ, Tonks NK. J. Biol. Chem. 1999;274:3811. doi: 10.1074/jbc.274.6.3811. [DOI] [PubMed] [Google Scholar]
- 12.Veillette A, Rhee I, Souza CM, Davidson D. Immunol. Rev. 2009;228:312. doi: 10.1111/j.1600-065X.2008.00747.x. [DOI] [PubMed] [Google Scholar]
- 13.Hallé M, Tremblay ML, Meng TC. Cell Cycle. 2007;6:2773. doi: 10.4161/cc.6.22.4926. [DOI] [PubMed] [Google Scholar]
- 14.Zhang X-Y, Bishop AC. Biochemistry. 2008;47:4491. doi: 10.1021/bi800014c. [DOI] [PubMed] [Google Scholar]
- 15.Stroffekova K, Proenza C, Beam KG. Pflugers Arch. 2001;442:859. doi: 10.1007/s004240100619. [DOI] [PubMed] [Google Scholar]; Wang T, Yan P, Squier TC, Mayer MU. Chembiochem. 2007;8:1937. doi: 10.1002/cbic.200700209. [DOI] [PubMed] [Google Scholar]; Luedtke NW, Dexter RJ, Fried DB, Schepartz A. Nat. Chem. Biol. 2007;3:779. doi: 10.1038/nchembio.2007.49. [DOI] [PMC free article] [PubMed] [Google Scholar]; Krishnan B, Gierasch LM. Chem. Biol. 2008;15:1104. doi: 10.1016/j.chembiol.2008.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Madani F, Lind J, Damberg P, Adams SR, Tsien RY, Graslund AO. J. Am. Chem. Soc. 2009;131:4613. doi: 10.1021/ja809315x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Griffin BA, Adams SR, Jones J, Tsien RY. Meth. Enzymol. 2000;327:565. doi: 10.1016/s0076-6879(00)27302-3. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.