Abstract
MicroRNAs (miRNAs) regulate gene expression by guiding Argonaute proteins to specific target mRNA sequences. Identification of bona fide miRNA target sites in animals is made challenging by uncertainties regarding the base-pairing requirements between miRNA and target as well as the location of functional binding sites within mRNAs. Here we present the results of a comprehensive strategy aimed at isolating endogenous mRNA target sequences bound by the Argonaute protein ALG-1 in C. elegans. Using cross-linking and ALG-1 immunoprecipitation coupled with high-throughput sequencing (CLIP-seq), we identified extensive ALG-1 interactions with specific 3′ untranslated region (UTR) and coding exon sequences and discovered features that distinguish miRNA complex binding sites in 3′ UTRs from those in other genic regions. Furthermore, our analyses revealed a striking enrichment of Argonaute binding sites in genes important for miRNA function, suggesting an autoregulatory role that may confer robustness to the miRNA pathway.
miRNAs function as ~22-nucleotide (nt) RNAs that target messenger RNAs (mRNAs) for degradation or translational repression1,2. A single miRNA can potentially repress hundreds of genes by binding with partial sequence complementarity to mRNAs3,4. By combinatorial regulation of thousands of genes, the miRNA pathway critically influences many developmental programs as well as cellular homeostasis, the disruption of which leads to human disease. Thus, an outstanding challenge has been to distinguish biologically relevant miRNA-target interactions. To date, identification of miRNA target sites has been dependent largely on computational methods that have limited capability for predicting specific and physiologically relevant targets. Addressing this need, several studies have reported biochemical approaches to isolate targets by immunoprecipitation of miRNA effector complexes containing miRNA–mRNA duplexes5–10. Despite reduction of the search space for miRNA target sites from within all transcribed genes to a subset of immuno-precipitated RNAs, the identification of miRNA binding sequences is still not directly obtained and usually relies on subsequent computational searches for complementary sites within the precipitated transcripts. Here we have narrowed the regions recognized by miRNA effector complexes to approximately 100-nt sequences. The identification and analysis of sequences directly associated with Argonaute protein in vivo in C. elegans has enabled the discovery of distinct features related to this core component of the miRNA-induced silencing complex (miRISC) as well as its interaction with mRNA target sites.
RESULTS
ALG-1 CLIP-seq in C. elegans identifies known miRNA targets
As miRNAs guide Argonaute proteins to specific complementary sequences in mRNAs, we applied the CLIP-seq (also referred to as HITS-CLIP) method11–13 to capture and identify the miRNA and target-site sequences bound by the miRNA complex (miRISC) in developing worms. A recent application of this approach in mouse brain resulted in a map of Argonaute binding sites in this tissue14. C. elegans offers several advantages for applying the CLIP-seq procedure to detect global Argonaute protein–RNA interactions. A single Argonaute protein, ALG-1, is largely responsible for miRNA function, and viable alg-1 genetic mutants exist15. A short but well-established list of miRNA targets expected to be bound by ALG-1 at discrete positions is available16–31. Of these targets, extensive studies have confirmed that lin-41 is regulated by let-7 miRNA during the fourth larval (L4) stage via two clustered sequences, let-7 complementary sites 1 and 2 (LCS1 and LCS2)26–28. We used this example to optimize the CLIP-seq method to detect bona fide ALG-1 binding sites (Supplementary Fig. 1). Synchronized L4-stage wild-type (WT) worms and alg-1(gk214) mutants (hereafter referred to as alg-1(−)), which lack the anti–ALG-1 antibody epitope sequence, were treated with UV irradiation to stabilize in vivo protein-RNA interactions (Supplementary Fig. 1a). A custom antibody specific for the C. elegans ALG-1 protein (Supplementary Fig. 1b) was used to enrich for ALG-1 complexes expected to include miRNA and target RNA species. Immunoprecipitated complexes were processed for isolation of sequences protected by ALG-1 protein from nuclease digestion.
We obtained 3,864,848 and 5,127,241 reads from WT and alg-1(−) CLIP-seq libraries, respectively, out of which 1,651,523 (42.7%) and 695,895 (13.6%) mapped uniquely to the repeat-masked C. elegans genome (Supplementary Fig. 2a). Using MIResque, a microRNA prediction algorithm designed to analyze small-RNA reads obtained from high-throughput sequencing (S. Aigner and G.W.Y., unpublished data), 136 previously reported miRNAs, 37 of which represent the ‘star’ strand, and 1 novel miRNA gene were identified in the WT library (Supplementary Table 1). Heterogeneity in the terminal sequences, which primarily consisted of lost nucleotides from the 3′ ends, could be due to the cloning method, but in two cases we found base additions to the 5′ ends that altered the seed sequence. Identification of the distinct pool of miRNAs bound to ALG-1, which included a large number of star sequences, enabled selective analysis of pairing capacity between miRNAs and mRNA sequences associated with ALG-1 at the stage of sample collection.
To correctly assign CLIP-seq reads to authentic transcribed regions, we reannotated the 5′ and 3′ untranslated regions (UTRs) of gene loci using publicly available 36-bp reads obtained from high-throughput sequencing of poly(A)-selected cDNA libraries from the L3 and L4 stages of C. elegans larval development32. Reads that mapped upstream and downstream of currently annotated genes were used to redefine the 5′ and 3′ UTRs. In total, the 5′ and/or 3′ ends of 8,231 genes (40% of genes in the genome) were reannotated by our analysis. The median (average ± s.d.) lengths of bases extended for 5′ and 3′ UTRs are 56 (391 ± 621) and 215 (543 ± 700) nt, respectively. This substantial change in the landscape of C. elegans gene predictions was important for defining the genic location of ALG-1 binding sites and for choosing control sequences for computational analyses (see below).
To distinguish important and specific ALG-1 binding sites, we developed a new version of our CLIP-cluster identification algorithm12. Briefly, for each of the three biological replicates of the ALG-1 CLIP-seq experiments (WT or alg-1(−)), we first defined ‘regions’ in each gene by extending the sequencing reads to account for the length of the RNA fragments in our CLIP libraries (Supplementary Fig. 2a). To retain biologically reproducible regions while accounting for the different number of sequenced reads in each replicate library, we weighted regions that overlapped across replicate experiments by the fraction of reads in the region relative to all the reads in that experiment mapping within the gene. Regions that passed our stringent threshold corresponded to being reproducible in at least two of rheww replicate experiments (see Online Methods). Reads within accepted regions were further integrated from replicates to form a ‘cluster’, and clusters containing more reads than statistically expected were kept for further analyses. Finally, clusters that overlapped by at least 25% between WT and alg-1(−) were removed as potential sources of false positives, such as reads from highly abundant rRNA and protein-coding genes (see act-5 gene in Supplementary Fig. 2b).
In total, 5,310 WT and 826 alg-1(−) clusters were identified, 4,806 of which were unique to WT (Supplementary Fig. 2a), representing 3,093 genes, approximately one-fifth of the annotated C. elegans protein-coding genes expressed at this stage in development. Over half of these genes contained a single cluster (Supplementary Fig. 2c). The CLIP-seq results provided a significantly refined and biologically based dataset for identifying miRNA target sites and studying ALG-1 binding properties. Compared to the entire transcriptome, 3′ UTRs only, or 3′ UTRs of mRNAs from miRISC immunoprecipitates, we greatly reduced the search space for functional regions, by a factor of 47, 20 or 5, respectively (see Online Methods for calculation). The tracks for the reannotated gene regions, WT and alg-1(−) reads and clusters are available at the UCSC genome browser (http://genome.ucsc.edu) under ‘ALG1 CLIP-seq’ within the ‘Regulation’ section in the ce6 genome.
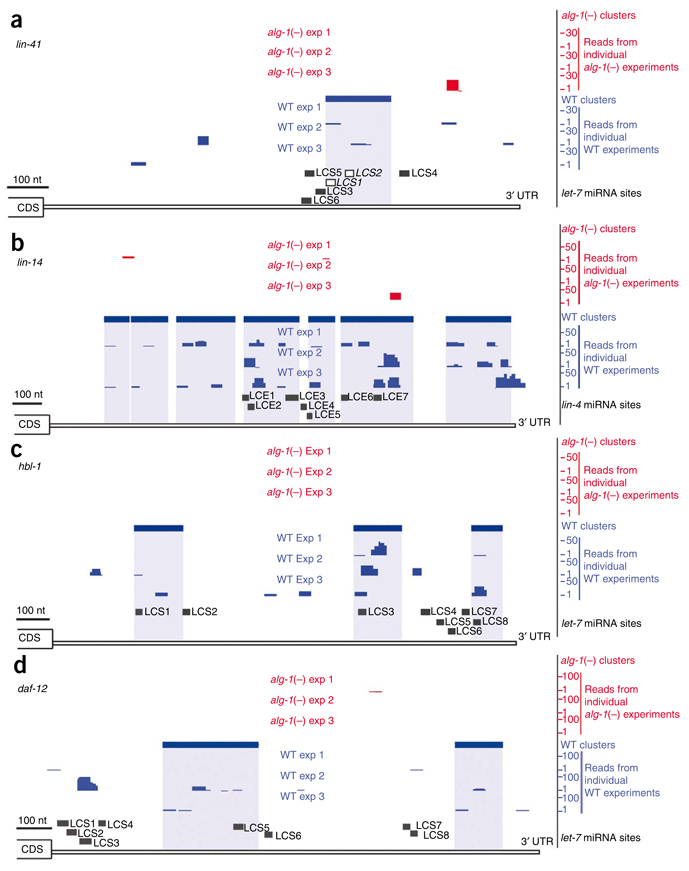
Isolation of sequences containing well-established miRNA target sites demonstrates the sensitivity of the ALG-1 CLIP-seq method. Extensive genetic and reporter gene experiments have pointed to LCS1 and LCS2 in the lin-41 3′ UTR as critical sequences for miRNA regulation of this gene26–28. Our ALG-1 CLIP-seq results identified a series of reads forming a significant cluster that maps directly on top of the closely spaced LCS1-LCS2 region (Fig. 1a). Notably, regulation of lin-41 by let-7 miRNA results in substantial mRNA degradation33. Thus, the detection of lin-41 by ALG-1 CLIP-seq demonstrates the sensitivity of this method for detecting miRNA targets regardless of regulatory mechanism. The first discovered miRNA target, lin-14, is regulated by lin-4 miRNA via multiple 3′ UTR complementary elements (LCEs)23,29. We identified three significant clusters that encompass the proposed LCEs 1–3, 5, and 6–7, respectively (Fig. 1b). Another cluster, toward the end of the lin-14 3′ UTR, is consistent with evidence that this gene is also regulated by other miRNAs26,34. Multiple let-7 binding sites have been predicted to mediate regulation of hbl-1 and daf-12 (refs. 16,18,24), and clusters cover a select few of the LCSs in the 3′ UTRs of these genes (Fig. 1c,d). Thus, ALG-1 CLIP-seq provides direct biochemical evidence for predicted miRNA target sites and reveals regions of greater relative occupancy by miRISC within a regulated 3′ UTR.
Figure 1.
MicroRNA targets identified by ALG-1 CLIP-seq in L4-stage worms. Graphical depictions of the number and location of reads from alg-1(−) (red upper tracks) and WT (blue lower tracks) from three biological replicates, CLIP-derived clusters (solid rectangular boxes over the reads) and putative miRNA binding sites in the 3′ UTR of mRNA transcripts (LCS, let-7 complementary sequence; LCE, lin-4 complementary element). (a) lin-41 3′ UTR. Of the six predicted LCSs, LCS1 and LCS2 (open boxes) have experimentally validated let-7 sites26–28. (b) lin-14 3′ UTR. Deletion of all the predicted LCE sites or of LCEs 1–5 results in misregulation of lin-14 expression23,29. (c) hbl-1 3′ UTR16,24,31. The sites for let-7 miRNA (LCSs 1–8) binding have been predicted but not experimentally tested. (d) daf-12 3′ UTR. Deletion of the predicted LCSs 1–4 or 5–8 in reporter constructs leads to misregulation of reporter gene expression18.
Of 13 well-established miRNA target genes in C. elegans16–21,23–30, all but 3 were found to contain at least one significant 3′-UTR cluster (Table 1). Moreover, the clusters include the cognate miRNA target site for 9 of these 10 genes. The majority of these miRNA target genes were also found to be enriched in ALG-1 interacting proteins 1 and 2 (AIN-1 and AIN-2, members of the GW182 family of proteins) immunoprecipitation experiments6. Beyond showing miRISC association with specific endogenous mRNAs, the ALG-1 CLIP-seq dataset contributes nucleotide-level resolution of the actual target region (Table 1 and Supplementary Table 2).
Table 1.
ALG-1 CLIP-seq Identifies sequences that mediate regulation by miRNAs
| miRNA::mRNA target | AIN-IPa | ALG-1 CLIP-seq | mRNAbalg-1(−)/WT | Evidence for miRNA targetingc |
Reference(s) | |
|---|---|---|---|---|---|---|
| Cluster in 3′ UTR | Cognate miRNA site | |||||
| lin-4::lin-14 | • | • | • | UP | G/R-3 | 23,29 |
| lin-4::lin-28 | • | UP | G/R-3 | 25 | ||
| let-7/mir-48/84/241::hbl-1 | • | • | • | UP | G/R-2 | 16,24,31 |
| let-7::lin-41 | •e | • | • | UP | G/R-3 | 26–28 |
| let-7/mir-84::let-60 | • | • | • | NC | G/R-2 | 20 |
| let-7::daf-12 | • | • | • | UP | G/R-2d | 18 |
| let-7::pha-4 | • | •f | •f | UP | G/R-2 | 18 |
| let-7::lss-4 | • | • | • | UP | G/R-2 | 18 |
| let-7 or mir-273::die-1 | • | • | • | UP | G/R-3 | 17,18 |
| let-7::nhr-25 | • | UP | G | 19 | ||
| let-7::T14B1.1 | • | • | • | UP | R-1 | 22 |
| lsy-6::cog-1 | • | NC | G/R-3 | 21 | ||
| mir-61::vav-1 | • | • | NC | G/R-3 | 30 | |
A list of individual miRNA::target pairs that are supported by genetic and reporter assays was adapted from previous work44. Bullets denote detection of the target mRNA in AIN-1 or AIN-2 immunoprecipitation experiments6, identification of 3′-UTR sequence by ALG-1 CLIP-seq and whether the region includes the cognate miRNA regulatory site. The relative expression of target mRNA levels in alg-1(−) compared to WT worms (UP, upregulation; NC, no change) is shown. Evidence for miRNA targeting is detailed in footnote c, and references are provided in the final column.
mRNA transcripts enriched (>0.5 average percentile rank) in AIN-1 or AIN-2 immunoprecipitations (P < 0.01).
t-statistic difference of more than 2.5 with P < 0.01.
Genetic suppression (G) or GFP/lacZ reporter (R) evidence of posttranscriptional regulation. R-1, 3′ UTR mediates post-transcriptional regulation, and mutation of target site(s) alters regulation. R-2, 3′ UTR regulation is lost in animals bearing mutations in the miRNA. R-3, miRNA and target site(s) are required for post-transcriptional regulation.
Regulation disrupted upon deletion of LCSs 1–4 or LCSs 5–8.
Detected by qRT-PCR in the AIN-2 immunoprecipitation.
A region that includes the cognate miRNA site is covered by sequencing tags but only in one experiment, thus not achieving statistical significance.
Genomic and sequence properties of ALG-1 binding sites
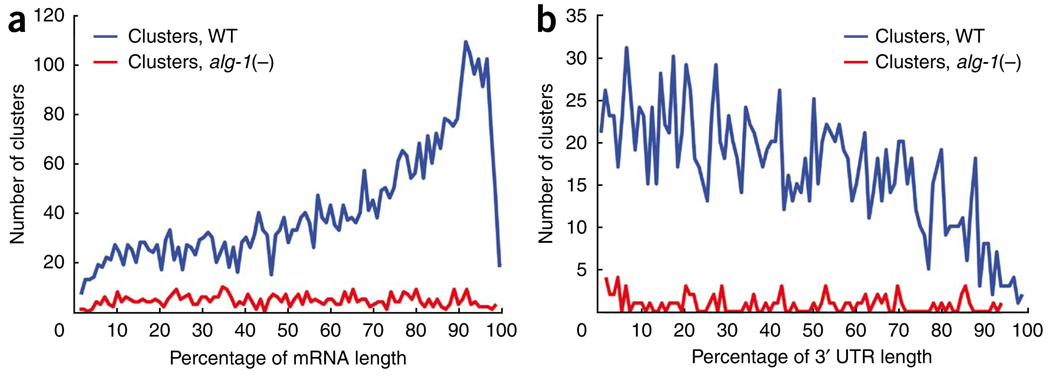
Although most genetic and computational studies support a bias for the location of miRNA target sites in 3′ UTRs, functional interaction of miRISC at other genic positions has also been demonstrated1,35–39. To study the global distribution of ALG-1 binding in C. elegans protein-coding genes, we mapped the positions of clusters relative to the length of targeted mRNAs. We observed a distinct profile of CLIP-derived cluster (CDC) occupancy proximal to the 3′ ends of spliced mRNAs from WT but not alg-1(−) worms (Fig. 2a). Notably, the frequency of clusters throughout the composite gene model was higher in WT than in alg-1(−) worms, showing that ALG-1 binding extends to other genic regions (Fig. 2a). Furthermore, the CDC distribution, as a percentage of 3′-UTR length, was not enriched proximal to the stop codon or poly(A) sites, in contrast to the bias for predicted miRNA target sites residing near either end of mammalian 3′ UTRs1 (Fig. 2b). In fact, the fraction of clusters that mapped a given distance from the stop codon largely mirrored the distribution of 3′-UTR lengths in C. elegans (Supplementary Fig. 3). In total, 1,656 (34.5%) of CDCs were located in 3′ UTRs, 2,473 (51.5%) in coding exons, 602 (12.5%) in introns and 75 (1.6%) in 5′ UTRs.
Figure 2.
Relative position of ALG-1 binding sites across protein-coding genes. (a) Distribution of WT (blue) and alg-1(−) (red) clusters across a composite mRNA length. Cluster position is depicted as a percentage of the gene region, from the beginning to the end of spliced transcripts. (b) Distribution of WT (blue) and alg-1(−) (red) clusters across the 3′ UTR region. Cluster position is depicted as a percentage of the region from the annotated translational stop codon to the end of transcripts, as defined by our reannotation of C. elegans genes.
To characterize the sequence properties of ALG-1 binding sites, we subjected CDCs and a control set of randomly derived clusters (RDCs) to a battery of computational analyses. In order to perform as equitable a comparison as possible, we minimized biases due to GC content, evolutionary conservation, genic region and length of the bound region when selecting RDCs (Supplementary Fig. 4a,b). Furthermore, RDCs were selected from genes depleted of ALG-1 binding sites. Caveats of this approach are that a chosen RDC may actually be bound by ALG-1 at a different developmental stage or that the target mRNA may be present at such low abundance that it is not detected. Our ability to detect the lin-41 LCS1-LCS2 region (Fig. 1a and Supplementary Fig. 1c) despite strong downregulation of this mRNA at the L4 stage33 suggests that this second point is a minor issue. In spite of the potential limitations for assigning RDCs, our results corroborate expected properties and identify new ones associated with miRISC binding to endogenous sequences (CDCs) on a global scale.
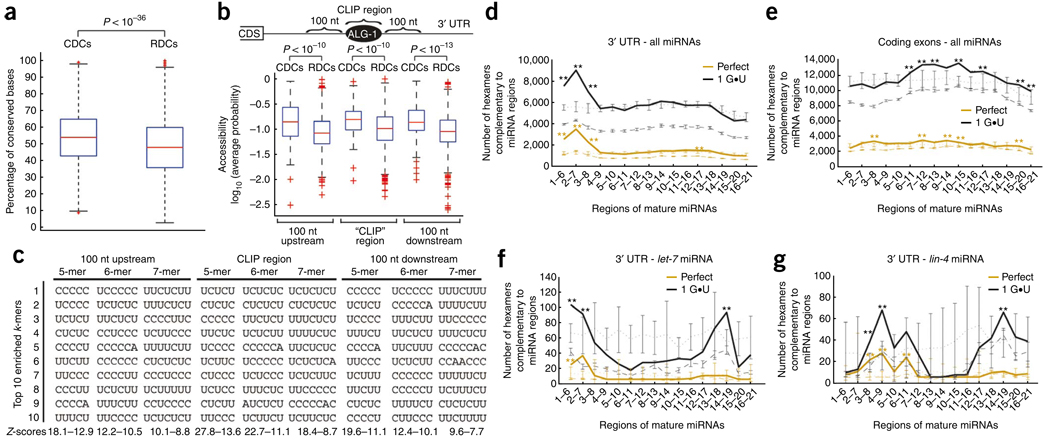
Preferential evolutionary conservation is a common feature used to predict miRNA target sites40–42. Indeed, we observed substantially higher conservation levels within CDCs compared to RDCs in 3′ UTRs (Fig. 3a) and a similar trend for coding exon and intron regions (Supplementary Fig. 5a). Also, consistent with the observation that functional miRNA target sites are frequently located in RNA sequences of higher accessibility (in other words, less secondary structure)43–47, the ALG-1–bound regions (CDCs), as well as the 100-nt upstream and downstream flanking sequences, were significantly more accessible than RDCs in the 3′ UTRs (P < 10−10) (Fig. 3b). However, this was not true for CDCs in the other genic regions (Supplementary Fig. 5b). It has also been suggested that a high local AU content is responsible for the more accessible 3′ UTR sites targeted by miRNAs48,49. Thus, we analyzed the nucleotide composition within and 100 nt upstream and downstream of 3′ UTR CDCs to search for motifs statistically enriched relative to RDCs. Unexpectedly, the ten most enriched 5- to 7-mers in 3′ UTR CDCs and their flanking regions are almost exclusively composed of CU nucleotides (P < 10−4), revealing alternative sequence elements that may mediate miRNA–ALG-1 target recognition and regulation in C. elegans (Fig. 3c). Moreover, this striking pattern was not associated with CDCs from 5′ UTR, coding exon or intron regions (Supplementary Fig. 5c) or with clusters from alg-1(−) animals, indicating that the CU-rich motifs are a specific characteristic of ALG-1–bound regions in 3′ UTRs.
Figure 3.
Attributes enriched in ALG-1 binding sites within 3′ UTRs. (a) Box plots of the conservation levels measured as the fraction of perfectly conserved nucleotides between genome-wide alignments of C. elegans and HG004659 and GM084317 brenneri in CLIP-derived clusters (CDCs) and randomly derived clusters (RDCs). CDCs are significantly more conserved than RDCs as assessed by the Kolgomorov-Smirnov two-sample test (P < 10−36). (b) Box plots of RNA accessibility, measured as the average probability of being unpaired, of CDC and RDC and their corresponding flanking sequences (100 nt upstream or downstream). CDCs and flanking sequences are significantly more accessible than RDCs in the same locations as assessed by the Kolgomorov-Smirnov two-sample test (P < 10−10). (c) The ten most enriched k-mers (k = 5, 6, 7) within or 100 nt upstream or downstream of CDCs, compared to RDCs, are shown along with the range of Z-scores for the specific categories. (d–g). The number of conserved hexamers within CDCs (solid line) and RDCs (dashed line) that base-pair to miRNA or scrambled miRNA regions (dotted line), allowing for zero (orange) or only one G•U base pair (black). Error bars in dashed and dotted lines represent the s.d. among ten independent sets of RDCs and scrambled miRNAs, respectively. Hexamers within 3′-UTR CDCs and RDCs (d) or coding-exon CDCs and RDCs (e) that base-pair to cloned miRNAs or shuffled versions of cloned miRNAs. Hexamers within 3′-UTR CDCs and RDCs that base-pair to the let-7 or shuffled let-7 miRNA (f) and lin-4 or shuffled lin-4 miRNA (g). Regions of the miRNA(s) that have statistically enriched numbers of complementary hexamers within CDCs when compared to RDCs or shuffled miRNAs are denoted by * (P < 0.01) and ** (P < 10−6) as measured by a Z-test.
Multiple computational prediction methods and extensive reporter validation assays point to the miRNA ‘seed’ (defined as perfect pairing between miRNA bases 2–7 and the target site) as a primary determinant for specific target recognition40–42,50. Indeed, the top ten most highly cloned miRNAs in our immunoprecipitations have significantly more frequent seed pairs within the 3′-UTR CDCs than do the least-cloned miRNAs (P < 0.0045; Supplementary Fig. 6a), and this general trend was also observed when all cloned miRNAs were analyzed (Supplementary Fig. 6b). To globally assess whether the seed or any other mature miRNA region showed enriched pairing capacity to CDCs, we used the complete set of miRNAs associated with ALG-1 in our experiments (Supplementary Table 1). We calculated the number of conserved hexamers present in CDCs that have perfect complementarity to regions within cloned mature miRNAs or have one conserved G•U wobble pair (Fig. 3d–g). Our analysis shows that a statistically significant (P < 10−6) number of conserved hexamers from 3′-UTR CDCs were complementary to bases 1–6, 2–7 and 3–8 of miRNAs compared to controls: conserved RDC hexamers paired to miRNAs or CDC hexamers paired to scrambled miRNAs. We observed the strongest signal for bases 2–7 (seed), with the allowance of one G•U pair generating the highest number of sites within the 3′-UTR clusters (Fig. 3d). This trend was not observed for CDCs in the other genic regions (Fig. 3e and Supplementary Fig. 7). Unexpectedly, CDCs within coding exons showed statistically significant pairing to the central region of miRNAs (P < 10−6) (Fig. 3e). We extended our analysis to include not only perfect conservation with or without G•U pairs but also cases in which there would be a G•U pair in one of the two Caenorhabditis species but a perfect match in the other species (G•U and G-C, respectively, ‘semiconserved hexamers’) (Supplementary Fig. 7). This enabled us to assess the specific contributions of conservation of sequence versus pairing capacity to the patterns of miRNA complementarity to sites in CDCs. Overall, perfect conservation coupled with one G•U contributed most substantially to the pairing capacity of miRNAs to 3′-UTR and coding-exon CDCs. The percentage of 3′-UTR clusters containing a perfectly conserved seed match allowing zero and only one G•U base pair was 55% and 63%, respectively, in comparison to 30% and 41% in RDCs (P < 10−4). These results indicate that, although seed pairing is an important determinant of miRNA–mRNA interaction in the 3′ UTRs, other pairing conformations may contribute significantly to ALG-1 binding in vivo. We also analyzed the pairing capacity of the archetypical miRNAs let-7 and lin-4 within 3′-UTR CDCs (Fig. 3f,g). Despite the caveat that a number of these CDCs may not be targeted only by let-7 or lin-4, we observed a strong pairing capacity for the let-7 seed at positions 2–7. Unexpectedly, we observed a significant enrichment for pairing at positions 14–19 when allowing a single G•U base pair at the 3′ end of the let-7 miRNA (Fig. 3f). Notably, lin-4 also showed significant 3′ base-pairing at the same positions, but the strongest signal at the 5′ end of the miRNA was at positions 4–9 (Fig. 3g). The ability of lin-4 to base-pair with potential target sites at positions other than the canonical 2–7 may indicate that individual miRNAs show specific pairing preferences with different outcomes for gene regulation.
Expression and functional biases of ALG-1 mRNA targets
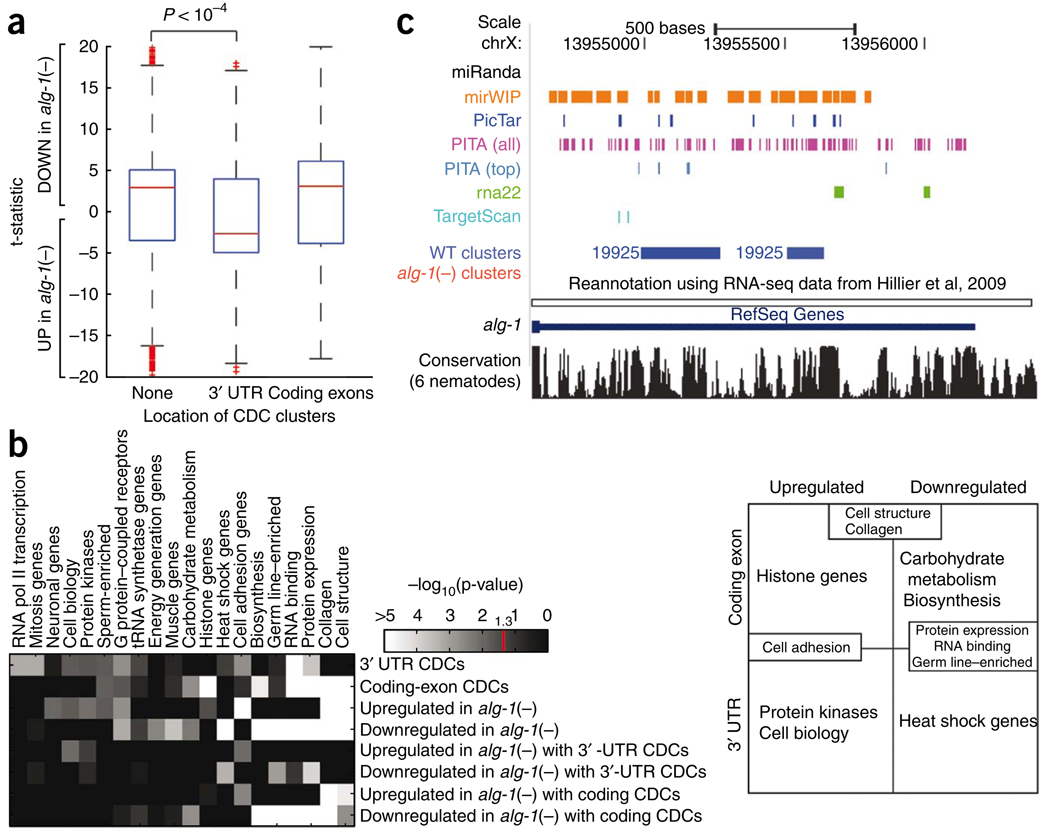
Regulation by miRNAs can result in substantial degradation of target mRNA levels or translational repression with little, if any, mRNA destabilization2. Given that the overwhelming majority of clusters reside in the 3′ UTR and coding exons, we sought to investigate whether the location of clusters affects mRNA levels. To test whether genes bound by ALG-1 at the 3′ UTR and coding exons were subject to regulation at the mRNA level, we performed microarray experiments comparing WT to alg-1(−) L4-stage worms. Consistent with previous reports that miRNA regulation can result in substantial target-mRNA degradation in C. elegans33,51, lin-41, lin-14, lin-28 and many other established miRNA targets were upregulated in the alg-1(−) mutant worms (Table 1 and Supplementary Table 2). Notably, genes containing 3′-UTR clusters were strongly upregulated in alg-1(−) mutants compared to genes that had no ALG-1–bound sites (Fig. 4a). In contrast, no relationship was detected between mRNA expression levels and genes with clusters in coding exons (Fig. 4a). These findings suggest that the mechanism of target regulation may be different for genes with ALG-1 binding sites in 3′ UTRs versus coding exons.
Figure 4.
Relationship between ALG-1 binding and mRNA expression levels. (a) Effects of ALG-1 binding on mRNA levels. Box plots representing the differential expression (as a t-statistic) of genes from biological replicate microarray experiments comparing alg-1(−) to WT L4-stage worms. Genes are divided into those that contained no CDCs and those that contained CDCs only within 3′ UTRs or coding exons. Compared to genes with no CDCs or coding-exon CDCs, genes with 3′-UTR CDCs are significantly more upregulated in alg-1(−) relative to WT as assayed by the Wilcoxon rank-sum test (P < 10−4). (b) Functional enrichment of genes that have CDCs only within 3′ UTR or coding exons that are up- or downregulated in alg-1(−) worms using significantly enriched (P < 0.05 in at least one row; Holm-Bonferroni corrected) functional categories defined by the C. elegans Topomap algorithm52. The intensity on the heat-map denotes −log10(p value). Genes represented by these functional categories can be divided in a matrix (right) depending on the location of the CDCs (3′ UTRs or coding exons), and whether the genes are up- or downregulated in the alg-1(−) mutants relative to WT worms. Several categories occupy multiple cells in the matrix, for example “Cell structure,” “Collagen,” “Cell adhesion,” “Protein expression,” “RNA binding” and “Germ line–enriched.” (c) UCSC Genome Browser view depicting clusters in the 3′ UTR of the alg-1 gene (blue, WT clusters; red, alg-1(−) clusters, none present) and the predicted miRNA binding sites by the various algorithms.
We next asked if genes with ALG-1 binding sites or expression changes in alg-1(−) compared to WT worms were enriched (P < 0.05) in particular functional classes based on the “Topomap” categories, which group co-regulated genes from extensive microarray datasets52. Notably, several functional categories were distinctly associated with genes that contained CDCs in 3′ UTRs versus coding exons and were up- or downregulated in alg-1(−) mutants (Fig. 4b). For example, genes belonging to the functional classes “Protein kinases” and “Cell biology” are enriched for containing 3′-UTR CDCs and being upregulated in alg-1(−) worms. Genes in the “Histone” category are also associated with upregulation but tend to have CDCs in their coding exons. This difference in locality of miRNA binding may be related to the typically short and nonpolyadenylated status of histone mRNAs53. Some functional categories included genes with CDCs in the 3′ UTR and coding exons and/or up- and downregulated genes. The overlap in categories is not surprising given the large fraction of genes with ALG-1–bound regions and the likely widespread direct and indirect effects on mRNA expression by the miRNA pathway. Our results reveal biological pathways targeted in vivo by ALG-1 in developing worms and indicate that some gene categories tend to be differentially bound and regulated by ALG-1.
miRNA pathway genes are enriched in ALG-1 targets
During our analyses of categories of genes bound by ALG-1, we discovered a strong enrichment for genes implicated in the miRNA pathway. CDCs in the 3′ UTR in the alg-1 gene indicate autoregulation of this core miRNA factor (Fig. 4c). Additionally, significant clusters were identified in the 3′ UTRs of ain-1 and ain-2, and mRNA levels of these genes and of the alg-1 homolog alg-2 were found to be upregulated in alg-1(−) worms (Supplementary Table 3). The potential cross-regulation of these miRNA effector genes may explain the nonlethal phenotype associated with loss of any single one of these genes6,15. To investigate the extent of ALG-1 regulation of miRNA pathway genes, we analyzed two published lists of genes specifically connected to miRNA function by proteomic and genetic evidence6,54. We observed that this network of miRNA pathway genes showed statistically significant enrichment in ALG-1 CDCs (30 out of 39 genes identified by proteomics and 15 out of 44 identified by genetics), compared to an expectation of ~16% (P < 10−4; Supplementary Table 3). We speculate that cross-regulation of these genes may confer robustness to the miRNA pathway by relaxing repression of miRNA cofactors to compensate for insufficiencies in major components such as ALG-1.
ALG-1–bound regions provide a resource for miRNA target predictions
A number of different algorithms (mirWIP, rna22, PicTar, TargetScan, PITA and miRanda) are available for predicting miRNA target sites in C. elegans genes22,35,44,45,53,55. Most of these prediction methods use a common set of criteria (seed, conservation and energy requirements), except for PITA, which does not require conservation, and rna22, which uses a different set of parameters. Because predictions are typically available for 3′-UTR sequences, we assessed the ability of these methods to detect predicted miRNA target sites within the ALG-1–bound 3′-UTR CDCs (Supplementary Fig. 8, tracks for the predicted sites from these algorithms are available under ‘ALG 1 CLIP-seq’ within the ‘Regulation’ section in the ce6 genome). Although 93% of the 3′-UTR CDCs contained a miRNA target site predicted by at least one of the algorithms (1,539 CDCs), only 3% of the CDCs had at least one site predicted by all 6 programs (52 CDCs). As an example, five of the six target prediction programs list potential miRNA target sites, largely disparate in both location and number, in the alg-1 3′ UTR (Fig. 4c); our results narrow the regions recognized and bound by ALG-1 in vivo at the specific developmental stage tested. The prominent disparity among prediction methods has been previously noted44 and emphasizes the value of the ALG-1 CLIP-seq as a tool to improve miRNA target identification.
DISCUSSION
We present a global snapshot of an endogenous miRISC RNA binding profile in whole animals. We demonstrate that binding of the core miRNA effector protein Argonaute is strongly enriched at the 3′ ends of transcripts, although substantial numbers of CDCs also reside within the 5′ UTR, coding exonic and intronic regions of genes as well. A striking signature of the ALG-1–bound 3′-UTR CDCs emerged: the regions showed greater sequence conservation and accessibility, they contained and were flanked by CU-rich motifs, they were enriched for sequences complementary to the 5′-end seed regions of miRNAs and they were associated with upregulation of mRNA expression in the alg-1(−) mutant background. Although some of these characteristics were shared with clusters in other genic regions, the marked overall differences in 3′ UTR versus other regions suggests that separate rules may regulate ALG-1 binding to distinct positions within an mRNA. The importance of context could underlie the conflicting conclusions that have been drawn about the ability of miRNAs to target different regions in mRNAs56–58 and, in some cases, the failure of reporter assays to demonstrate miRNA regulation of genes bound and regulated by alg-1 (see Supplementary Table 2). In addition to providing a map of ALG-1 interaction sites for the C. elegans protein-coding genes potentially under miRNA regulation in late larval development (see Supplementary Fig. 9 and Supplementary Data), compared to previously available methods our strategy substantially reduced the search space by factors of 5, 20 and 47 for identifying direct miRNA target sites. Although we detected a strong signal for pairing to the miRNA seed region in 3′-UTR ALG-1–bound sites, ~40% of the ALG-1 clusters lacked conserved seed pairing capacity, indicating that more flexible base-pairing rules may guide a large fraction of miRNA target recognition in vivo. Furthermore, the observation of different patterns for let-7 or lin-4 miRNA paired to sites within ALG-1–bound sequences raises the possibility of individual miRNA pairing rules. The discovery of miRNA pathway genes as an exceptional class of genes bound and regulated by endogenous alg-1 suggests that cross-regulation of miRNA cofactors contributes substantially to this essential post-transcriptional control mechanism. In conclusion, our analyses and data provide a framework and a rich resource for understanding in vivo miRNA–mRNA interactions in a context-specific manner.
METHODS
Methods and any associated references are available in the online version of the paper at http://www.nature.com/nsmb/.
Supplementary Material
ACKNOWLEDGMENTS
The authors thank G. Ruvkun, X. Fu, W. McGinnis and members of our laboratories for critical reading of the manuscript. We thank B. Hehli, S. Hunter and S. Bagga for technical assistance, V. Ambros and M. Hammell for providing the list of mirWIP predictions and D. Bartel for helpful advice. MLW is supported by the Genetics Training Program at the University of California, San Diego and a graduate fellowship from Genentech. This work was supported by grants from the US National Institutes of Health (GM071654-01 to A.E.P. and HG004659 and GM084317 to G.W.Y.), the Keck, Searle, V., Emerald and Peter Gruber Foundations (A.E.P.) and the Stem Cell Program at the University of California, San Diego (G.W.Y.).
Footnotes
Accession codes. Microarray CEL files have been deposited at the Gene Expression Omnibus database repository under accession number GSE19138.
Note: Supplementary information is available on the Nature Structural & Molecular Biology website.
AUTHOR CONTRIBUTIONS
A.E.P. and G.W.Y. designed and directed the project; A.E.P., D.G.Z., G.W.Y. and M.T.L. wrote the paper; D.G.Z. and T.Y.L. performed the experiments; M.T.L., M.L.W., K.R.H, T.Y.L. and G.W.Y. performed the bioinformatics analyses.
COMPETING INTERESTS STATEMENT
The authors declare no competing financial interests.
Reprints and permissions information is available online at http://npg.nature.com/reprintsandpermissions/.
References
- 1.Bartel DP. MicroRNAs: target recognition and regulatory functions. Cell. 2009;136:215–233. doi: 10.1016/j.cell.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chekulaeva M, Filipowicz W. Mechanisms of miRNA-mediated post-transcriptional regulation in animal cells. Curr. Opin. Cell Biol. 2009;21:452–460. doi: 10.1016/j.ceb.2009.04.009. [DOI] [PubMed] [Google Scholar]
- 3.Lim LP, et al. Microarray analysis shows that some microRNAs downregulate large numbers of target mRNAs. Nature. 2005;433:769–773. doi: 10.1038/nature03315. [DOI] [PubMed] [Google Scholar]
- 4.Sood P, Krek A, Zavolan M, Macino G, Rajewsky N. Cell-type-specific signatures of microRNAs on target mRNA expression. Proc. Natl. Acad. Sci. USA. 2006;103:2746–2751. doi: 10.1073/pnas.0511045103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zhang L, Hammell M, Kudlow BA, Ambros V, Han M. Systematic analysis of dynamic miRNA-target interactions during C. elegans development. Development. 2009;136:3043–3055. doi: 10.1242/dev.039008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zhang L, et al. Systematic identification of C. elegans miRISC proteins, miRNAs, and mRNA targets by their interactions with GW182 proteins AIN-1 and AIN-2. Mol. Cell. 2007;28:598–613. doi: 10.1016/j.molcel.2007.09.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hendrickson DG, Hogan DJ, Herschlag D, Ferrell JE, Brown PO. Systematic identification of mRNAs recruited to argonaute 2 by specific microRNAs and corresponding changes in transcript abundance. PLoS One. 2008;3:e2126. doi: 10.1371/journal.pone.0002126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Beitzinger M, Peters L, Zhu JY, Kremmer E, Meister G. Identification of human microRNA targets from isolated argonaute protein complexes. RNA Biol. 2007;4:76–84. doi: 10.4161/rna.4.2.4640. [DOI] [PubMed] [Google Scholar]
- 9.Karginov FV, et al. A biochemical approach to identifying microRNA targets. Proc. Natl. Acad. Sci. USA. 2007;104:19291–19296. doi: 10.1073/pnas.0709971104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Easow G, Teleman AA, Cohen SM. Isolation of microRNA targets by miRNP immunopurification. RNA. 2007;13:1198–1204. doi: 10.1261/rna.563707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Licatalosi DD, et al. HITS-CLIP yields genome-wide insights into brain alternative RNA processing. Nature. 2008;456:464–469. doi: 10.1038/nature07488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yeo GW, et al. An RNA code for the FOX2 splicing regulator revealed by mapping RNA-protein interactions in stem cells. Nat. Struct. Mol. Biol. 2009;16:130–137. doi: 10.1038/nsmb.1545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sanford JR, et al. Splicing factor SFRS1 recognizes a functionally diverse landscape of RNA transcripts. Genome Res. 2009;19:381–394. doi: 10.1101/gr.082503.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chi SW, Zang JB, Mele A, Darnell RB. Argonaute HITS-CLIP decodes microRNA-mRNA interaction maps. Nature. 2009;460:479–486. doi: 10.1038/nature08170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Grishok A, et al. Genes and mechanisms related to RNA interference regulate expression of the small temporal RNAs that control C. elegans developmental timing. Cell. 2001;106:23–34. doi: 10.1016/s0092-8674(01)00431-7. [DOI] [PubMed] [Google Scholar]
- 16.Abrahante JE, et al. The Caenorhabditis elegans hunchback-like gene lin-57/hbl-1 controls developmental time and is regulated by microRNAs. Dev. Cell. 2003;4:625–637. doi: 10.1016/s1534-5807(03)00127-8. [DOI] [PubMed] [Google Scholar]
- 17.Chang S, Johnston RJ, Jr., Frokjaer-Jensen C, Lockery S, Hobert O. MicroRNAs act sequentially and asymmetrically to control chemosensory laterality in the nematode. Nature. 2004;430:785–789. doi: 10.1038/nature02752. [DOI] [PubMed] [Google Scholar]
- 18.Grosshans H, Johnson T, Reinert KL, Gerstein M, Slack FJ. The temporal patterning microRNA let-7 regulates several transcription factors at the larval to adult transition in C. elegans. Dev. Cell. 2005;8:321–330. doi: 10.1016/j.devcel.2004.12.019. [DOI] [PubMed] [Google Scholar]
- 19.Hayes GD, Frand AR, Ruvkun G. The mir-84 and let-7 paralogous microRNA genes of Caenorhabditis elegans direct the cessation of molting via the conserved nuclear hormone receptors NHR-23 and NHR-25. Development. 2006;133:4631–4641. doi: 10.1242/dev.02655. [DOI] [PubMed] [Google Scholar]
- 20.Johnson SM, et al. RAS is regulated by the let-7 microRNA family. Cell. 2005;120:635–647. doi: 10.1016/j.cell.2005.01.014. [DOI] [PubMed] [Google Scholar]
- 21.Johnston RJ, Hobert O. A microRNA controlling left/right neuronal asymmetry in Caenorhabditis elegans. Nature. 2003;426:845–849. doi: 10.1038/nature02255. [DOI] [PubMed] [Google Scholar]
- 22.Lall S, et al. A genome-wide map of conserved microRNA targets in C. elegans. Curr. Biol. 2006;16:460–471. doi: 10.1016/j.cub.2006.01.050. [DOI] [PubMed] [Google Scholar]
- 23.Lee RC, Feinbaum RL, Ambros V. The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell. 1993;75:843–854. doi: 10.1016/0092-8674(93)90529-y. [DOI] [PubMed] [Google Scholar]
- 24.Lin SY, et al. The C. elegans hunchback homolog, hbl-1, controls temporal patterning and is a probable microRNA target. Dev. Cell. 2003;4:639–650. doi: 10.1016/s1534-5807(03)00124-2. [DOI] [PubMed] [Google Scholar]
- 25.Moss EG, Lee RC, Ambros V. The cold shock domain protein LIN-28 controls developmental timing in C. elegans and is regulated by the lin-4 RNA. Cell. 1997;88:637–646. doi: 10.1016/s0092-8674(00)81906-6. [DOI] [PubMed] [Google Scholar]
- 26.Reinhart BJ, et al. The 21-nucleotide let-7 RNA regulates developmental timing in Caenorhabditis elegans. Nature. 2000;403:901–906. doi: 10.1038/35002607. [DOI] [PubMed] [Google Scholar]
- 27.Slack FJ, et al. The lin-41 RBCC gene acts in the C. elegans heterochronic pathway between the let-7 regulatory RNA and the LIN-29 transcription factor. Mol. Cell. 2000;5:659–669. doi: 10.1016/s1097-2765(00)80245-2. [DOI] [PubMed] [Google Scholar]
- 28.Vella MC, Choi EY, Lin SY, Reinert K, Slack FJ. The C. elegans microRNA let-7 binds to imperfect let-7 complementary sites from the lin-41 3′UTR. Genes Dev. 2004;18:132–137. doi: 10.1101/gad.1165404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wightman B, Ha I, Ruvkun G. Posttranscriptional regulation of the heterochronic gene lin-14 by lin-4 mediates temporal pattern formation in C. elegans. Cell. 1993;75:855–862. doi: 10.1016/0092-8674(93)90530-4. [DOI] [PubMed] [Google Scholar]
- 30.Yoo AS, Greenwald I. LIN-12/Notch activation leads to microRNA-mediated down-regulation of Vav in C. elegans. Science. 2005;310:1330–1333. doi: 10.1126/science.1119481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Abbott AL, et al. The let-7 MicroRNA family members mir-48, mir-84, and mir-241 function together to regulate developmental timing in Caenorhabditis elegans. Dev. Cell. 2005;9:403–414. doi: 10.1016/j.devcel.2005.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hillier LW, et al. Massively parallel sequencing of the polyadenylated transcriptome of C. elegans. Genome Res. 2009;19:657–666. doi: 10.1101/gr.088112.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bagga S, et al. Regulation by let-7 and lin-4 miRNAs results in target mRNA degradation. Cell. 2005;122:553–563. doi: 10.1016/j.cell.2005.07.031. [DOI] [PubMed] [Google Scholar]
- 34.Chendrimada TP, et al. MicroRNA silencing through RISC recruitment of eIF6. Nature. 2007;447:823–828. doi: 10.1038/nature05841. [DOI] [PubMed] [Google Scholar]
- 35.Miranda KC, et al. A pattern-based method for the identification of MicroRNA binding sites and their corresponding heteroduplexes. Cell. 2006;126:1203–1217. doi: 10.1016/j.cell.2006.07.031. [DOI] [PubMed] [Google Scholar]
- 36.Shen WF, Hu YL, Uttarwar L, Passegue E, Largman C. MicroRNA-126 regulates HOXA9 by binding to the homeobox. Mol. Cell. Biol. 2008;28:4609–4619. doi: 10.1128/MCB.01652-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Duursma AM, Kedde M, Schrier M, le Sage C, Agami R. miR-148 targets human DNMT3b protein coding region. RNA. 2008;14:872–877. doi: 10.1261/rna.972008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Forman JJ, Legesse-Miller A, Coller HA. A search for conserved sequences in coding regions reveals that the let-7 microRNA targets Dicer within its coding sequence. Proc. Natl. Acad. Sci. USA. 2008;105:14879–14884. doi: 10.1073/pnas.0803230105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tay Y, Zhang J, Thomson AM, Lim B, Rigoutsos I. MicroRNAs to Nanog, Oct4 and Sox2 coding regions modulate embryonic stem cell differentiation. Nature. 2008;455:1124–1128. doi: 10.1038/nature07299. [DOI] [PubMed] [Google Scholar]
- 40.Lewis BP, Burge CB, Bartel DP. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell. 2005;120:15–20. doi: 10.1016/j.cell.2004.12.035. [DOI] [PubMed] [Google Scholar]
- 41.Brennecke J, Stark A, Russell RB, Cohen SM. Principles of microRNA-target recognition. PLoS Biol. 2005;3:e85. doi: 10.1371/journal.pbio.0030085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Krek A, et al. Combinatorial microRNA target predictions. Nat. Genet. 2005;37:495–500. doi: 10.1038/ng1536. [DOI] [PubMed] [Google Scholar]
- 43.Robins H, Li Y, Padgett RW. Incorporating structure to predict microRNA targets. Proc. Natl. Acad. Sci. USA. 2005;102:4006–4009. doi: 10.1073/pnas.0500775102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hammell M, et al. mirWIP: microRNA target prediction based on microRNA-containing ribonucleoprotein-enriched transcripts. Nat. Methods. 2008;5:813–819. doi: 10.1038/nmeth.1247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kertesz M, Iovino N, Unnerstall U, Gaul U, Segal E. The role of site accessibility in microRNA target recognition. Nat. Genet. 2007;39:1278–1284. doi: 10.1038/ng2135. [DOI] [PubMed] [Google Scholar]
- 46.Long D, et al. Potent effect of target structure on microRNA function. Nat. Struct. Mol. Biol. 2007;14:287–294. doi: 10.1038/nsmb1226. [DOI] [PubMed] [Google Scholar]
- 47.Zhao Y, Samal E, Srivastava D. Serum response factor regulates a muscle-specific microRNA that targets Hand2 during cardiogenesis. Nature. 2005;436:214–220. doi: 10.1038/nature03817. [DOI] [PubMed] [Google Scholar]
- 48.Baek D, et al. The impact of microRNAs on protein output. Nature. 2008;455:64–71. doi: 10.1038/nature07242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Grimson A, et al. MicroRNA targeting specificity in mammals: determinants beyond seed pairing. Mol. Cell. 2007;27:91–105. doi: 10.1016/j.molcel.2007.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lewis BP, Shih IH, Jones-Rhoades MW, Bartel DP, Burge CB. Prediction of mammalian microRNA targets. Cell. 2003;115:787–798. doi: 10.1016/s0092-8674(03)01018-3. [DOI] [PubMed] [Google Scholar]
- 51.Ding XC, Grosshans H. Repression of C. elegans microRNA targets at the initiation level of translation requires GW182 proteins. EMBO J. 2009;28:213–222. doi: 10.1038/emboj.2008.275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kim SK, et al. A gene expression map for Caenorhabditis elegans. Science. 2001;293:2087–2092. doi: 10.1126/science.1061603. [DOI] [PubMed] [Google Scholar]
- 53.Dominski Z, Marzluff WF. Formation of the 3′ end of histone mRNA: getting closer to the end. Gene. 2007;396:373–390. doi: 10.1016/j.gene.2007.04.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Parry DH, Xu J, Ruvkun G. A whole-genome RNAi screen for C. elegans miRNA pathway genes. Curr. Biol. 2007;17:2013–2022. doi: 10.1016/j.cub.2007.10.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ruby JG, et al. Large-scale sequencing reveals 21U-RNAs and additional microRNAs and endogenous siRNAs in C. elegans. Cell. 2006;127:1193–1207. doi: 10.1016/j.cell.2006.10.040. [DOI] [PubMed] [Google Scholar]
- 56.Kloosterman WP, Wienholds E, Ketting RF, Plasterk RH. Substrate requirements for let-7 function in the developing zebrafish embryo. Nucleic Acids Res. 2004;32:6284–6291. doi: 10.1093/nar/gkh968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Lytle JR, Yario TA, Steitz JA. Target mRNAs are repressed as efficiently by microRNA-binding sites in the 5′ UTR as in the 3′ UTR. Proc. Natl. Acad. Sci. USA. 2007;104:9667–9672. doi: 10.1073/pnas.0703820104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Gu S, Jin L, Zhang F, Sarnow P, Kay MA. Biological basis for restriction of microRNA targets to the 3′ untranslated region in mammalian mRNAs. Nat. Struct. Mol. Biol. 2009;16:144–150. doi: 10.1038/nsmb.1552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Yeo GW, et al. Alternative splicing events identified in human embryonic stem cells and neural progenitors. PLOS Comput. Biol. 2007;3:1951–1967. doi: 10.1371/journal.pcbi.0030196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Siepel A, et al. Evolutionarily conserved elements in vertebrate, insect, worm, and yeast genomes. Genome Res. 2005;15:1034–1050. doi: 10.1101/gr.3715005. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.