Abstract
Systemic lupus erythematosus (SLE) is a prototypic autoimmune disease with complex genetic inheritance. Recently, single nucleotide polymorphisms (SNPs) in BANK1 and TNFSF4 have been shown to be associated with SLE in Caucasian populations, but it is not known whether they are also involved in the disease in other ethnic groups. Recent data from our genome-wide association study (GWAS) for 314 SLE cases and 920 controls collected in Hong Kong identified SNPs in and around BANK1 and TNFSF4 to be associated with SLE risk. On the basis of the results of the reported studies and our GWAS, SNPs were selected for further genotyping in 949 SLE patients (overlapping with the 314 cases in our GWAS) and non-overlapping 1042 healthy controls. We confirmed the associations of BANK1 and TNFSF4 with SLE in Chinese (BANK1, rs3733197, odds ratio (OR)=0.84, P=0.021; BANK1, rs17266594, OR=0.61, P=4.67 × 10−9; TNFSF4, rs844648, OR=1.22, P=2.47 × 10−3; TNFSF4, rs2205960, OR=1.30, P=2.41 × 10−4). Another SNP located in intron 1 of BANK1, rs4522865, was separately replicated by Sequenom in 360 cases and 360 controls and was also confirmed to be associated with SLE (OR=0.725, P=2.93 × 10−3). Logistic regression analysis showed that rs3733197 (A383T in ankyrin domain) and rs17266594 (a branch point-site SNP) from BANK1 had independent contributions towards the disease association (P=0.037 and 6.63 × 10−8, respectively). In TNFSF4, rs2205960 was associated with SLE independently from the effect of rs844648 (P=6.26 × 10−3), but not vice versa (P=0.55). These findings suggest that multiple independent genetic variants may be present within the gene locus, which exert their effects on SLE pathogenesis through different mechanisms.
Keywords: SLE, BANK1, TNFSF4, Chinese, genetic association
Introduction
Systemic lupus erythematosus (SLE) (OMIM 152700) is a prototypic autoimmune disease characterized by heterogeneous systemic involvement, production of a wide-spectrum of autoantibodies and involvement of complex genetic and environmental components.1 Prevalence of the disease and severity of its manifestations vary among different populations, and SLE is more common in Asians (46.7/100 000) than in Caucasians (20.7/100 000).2 In Hong Kong, the estimated point of prevalence is 0.06% (0.1% among women).3
Recently, several studies including association studies with genome-wide approach and candidate-gene approach have shown new susceptibility genes for SLE, including BANK1 and TNFSF4.4, 5 BANK1 encodes a B-cell-specific scaffold protein and its activation can affect B cell-receptor-induced calcium mobilization from intracellular calcium stores.6 In the populations of European ancestry, a non-synonymous substitution (rs10516487, R16H), a branch point-site single nucleotide polymorphism (SNP) (rs17266594) and a SNP in the ankyrin domain (rs3733197, A383T) have been shown to be associated with SLE, and may contribute to sustained B-cell receptor signaling and subsequent B-cell hyperactivity characterized in the disease.4 TNFSF4 encodes a cytokine that is expressed on CD40-stimulated B cells and antigen-presenting cells to provide CD28-independent costimulatory signals to T cells.7 TNFSF4-mediated signal was found to inhibit IL-10-producing CD4+ type 1 regulatory T-cell function8 and IL-17 production in vitro.9 In two cohorts from UK and Minnesota, a haplotype in the upstream region of TNFSF4, marked by SNPs rs844644 and rs2205960, has been shown to correlate with increased cell surface TNFSF4 expression and TNFSF4 transcript and to be associated with SLE.5
Despite the convincing evidence of disease association, these reported studies only involved samples from populations of European ancestry. However, the importance of replication in a much different population, in terms of validation of an association and discovery of population differences, should not be overlooked.10, 11 The Chinese has a much higher SLE prevalence and more severe disease manifestations than the Europeans, and thus a heavier genetic load is suspected.2 Population differences in susceptibility genes were also reported recently in our population.12 Besides, showing population differences may enlighten some genetic risks that are specific toward certain ethnic groups, which may also help elucidate the ethnic differences in terms of disease prevalence and severity.
Results
Genome-wide association study
We extracted genotype data on SNPs spanning from 100 kb both upstream and downstream of BANK1 and TNFSF4 in our genome-wide association study (GWAS), which was conducted on 314 SLE cases and 920 controls by Illumina Human 610-Quad arrays (Illumina San Diego, CA, USA). Twenty-one SNPs in and around BANK1 showed significant association with the disease. These included rs10516487, the non-synonymous SNP that have been reported in the study of Kozyrev et al.4 In TNFSF4, 11 SNPs were found to confer disease risk by GWAS, including rs2205960, rs844644, rs844648 and a few others that have been reported to be associated with SLE in the study of Graham et al.5 The allelic association results were shown in Table 1.
Table 1.
Results of genome-wide association study in 314 cases and 920 controls
| Gene | No. | SNP | BP | Allele | MAF−case | MAF−control | OR | 95% (CI) | P-values |
|---|---|---|---|---|---|---|---|---|---|
| BANK1 | rs12648641 | 102 878 773 | A<C | 0.286 | 0.238 | 1.29 | (1.05–1.58) | 0.0161 | |
| rs7695738 | 102 920 255 | A<G | 0.175 | 0.134 | 1.38 | (1.08–1.76) | 0.0107 | ||
| 1 | rs4522865 | 102 934 911 | G<A | 0.381 | 0.457 | 0.73 | (0.61–0.88) | 8.49 × 10−4 | |
| rs11944613 | 102 937 818 | G<A | 0.247 | 0.297 | 0.78 | (0.63–0.96) | 0.0166 | ||
| 2 | rs10028805 | 102 956 273 | A<G | 0.344 | 0.407 | 0.77 | (0.63–0.92) | 5.52 × 10−3 | |
| 3 | rs4276281 | 102 965 803 | C<A | 0.522 | 0.449 | 1.34 | (1.12–1.61) | 1.46 × 10−3 | |
| 4* | rs10516487 | 102 970 099 | T<C | 0.139 | 0.183 | 0.72 | (0.56–0.93) | 0.0119 | |
| 5 | rs10516486 | 102 970 299 | G<A | 0.522 | 0.450 | 1.34 | (1.12–1.61) | 1.59 × 10−3 | |
| rs13129744 | 102 975 754 | C<T | 0.150 | 0.189 | 0.75 | (0.59–0.97) | 0.0253 | ||
| rs12498977 | 102 988 700 | A<G | 0.201 | 0.250 | 0.76 | (0.60–0.94) | 0.01299 | ||
| 6 | rs1421624 | 102 993 987 | A<G | 0.148 | 0.198 | 0.70 | (0.55–0.90) | 5.60 × 10−3 | |
| rs10516484 | 103 011 108 | A<G | 0.148 | 0.197 | 0.71 | (0.55–0.91) | 6.09 × 10−3 | ||
| rs17031755 | 103 011 377 | A<G | 0.217 | 0.266 | 0.76 | (0.62–0.95) | 0.0149 | ||
| rs13137047 | 103 023 059 | A<G | 0.148 | 0.197 | 0.71 | (0.55–0.91) | 6.09 × 10−3 | ||
| rs7694392 | 103 042 449 | C<T | 0.350 | 0.401 | 0.81 | (0.67–0.97) | 0.0242 | ||
| rs6532979 | 103 053 410 | A<G | 0.295 | 0.338 | 0.82 | (0.67–1.00) | 0.0477 | ||
| rs2112139 | 103 102 448 | T<C | 0.357 | 0.405 | 0.82 | (0.68–0.98) | 0.0331 | ||
| rs2850374 | 103 112 346 | G<T | 0.311 | 0.357 | 0.81 | (0.67–0.99) | 0.0362 | ||
| rs2631267 | 103 167 495 | G<A | 0.162 | 0.202 | 0.77 | (0.60–0.97) | 0.0290 | ||
| rs12641017 | 103 235 884 | T<G | 0.108 | 0.081 | 1.38 | (1.02–1.86) | 0.0377 | ||
| rs17032126 | 103 307 514 | G<A | 0.202 | 0.251 | 0.76 | (0.61–0.95) | 0.0141 | ||
| TNFSF4 | rs1234302 | 171 388 187 | G<T | 0.149 | 0.105 | 1.49 | (1.14–1.95) | 3.11 × 10−3 | |
| rs1234313 | 171 432 870 | G<A | 0.411 | 0.364 | 1.22 | (1.01–1.47) | 0.0370 | ||
| 1* | rs1234315 | 171 445 086 | T<C | 0.538 | 0.445 | 1.46 | (1.21–1.75) | 4.89 × 10−5 | |
| 2* | rs2205960 | 171 458 098 | T<G | 0.331 | 0.227 | 1.69 | (1.39–2.06) | 1.95 × 10−7 | |
| 3* | rs844644 | 171 476 118 | C<A | 0.545 | 0.458 | 1.41 | (1.18–1.70) | 1.82 × 10−4 | |
| 4 | rs1012507 | 171 486 094 | T<G | 0.353 | 0.261 | 1.54 | (1.27–1.87) | 1.14 × 10−5 | |
| 5* | rs844648 | 171 490 486 | A<G | 0.546 | 0.465 | 1.39 | (1.16–1.66) | 4.18 × 10−4 | |
| 6 | rs704840 | 171 492 818 | G<T | 0.490 | 0.390 | 1.51 | (1.25–1.81) | 1.06 × 10−5 | |
| 7 | rs10489265 | 171 502 688 | G<T | 0.325 | 0.230 | 1.61 | (1.32–1.96) | 2.73 × 10−6 | |
| rs4916319 | 171 533 201 | G<A | 0.455 | 0.378 | 1.38 | (1.15–1.66) | 5.92 × 10−4 | ||
| rs1342032 | 171 537 272 | T<G | 0.451 | 0.375 | 1.37 | (1.14–1.65) | 7.42 × 10−4 |
Abbreviations: No., SNP number labeled in Figure 1; BP, base pair, MAF−case, minor allele frequency in cases; MAF−control, minor allele frequency in controls; SNPs marked by * were reported to be associated with SLE in Caucasian studies; SNP, single nucleotide polymorphism.
SNPs spanning chromosome 4: 102 830 159–103 307 514 (480 kb) including BANK1 and chromosome 1: 171 458 098–171 502 458 (440 kb) including TNFSF4 were scanned for association with SLE in GWAS. 21 SNPs in BANK1 region conferred the association, whereas 11 SNPs in TNFSF4 were associated (P<0.05). In addition to the top five SNPs with the most significant P-values for each of the genes, rs10516487 from BANK1 and rs844648 and rs844644 from TNFSF4 were also bolded because they have been reported to be associated with SLE in Caucasian studies. The bolded SNPs were also shown in Figure 1. BANK1 rs3733197 and rs17266594 were not included in Illumina Human 610-Quad arrays.
Replication experiment with increased samples
Next, to further verify the association of these two genes with SLE, a few SNPs were selected for further genotyping. From the reported study, three SNPs from BANK1, namely rs3733197, rs17266594 and rs10516487, have been found to be associated with the disease, but only rs10516487 was included in our Bead-chip data. To compare the effects of SNPs in Caucasians and Chinese, rs3733197 and rs17266594 were selected for further replication. SNP rs10516487 was not replicated as it has high linkage disequilibrium (LD) with rs17266594 (r2=1 in HapMap-HCB, r2=0.9 in the study of Kozyrev et al.4). In addition, rs4522865 showed the most significant association with the disease in our GWAS (Table 1) and was therefore also selected for replication (Table 2).
Table 2.
Case–control analysis in replication study
| SNP | Position | Allele | MAF−case | MAF−control | OR | (95% CI) | P | P* | |
|---|---|---|---|---|---|---|---|---|---|
| BANK1 | rs17266594 | 102 969 945 | C<T | 0.137 | 0.207 | 0.606 | (0.51–0.72) | 4.67 × 10−9 | 4.15 × 10−5 |
| BANK1 | rs3733197 | 103 058 310 | A<G | 0.204 | 0.234 | 0.838 | (0.72–0.97) | 0.021 | 0.048 |
| BANK1 | *rs4522865 | 102 934 911 | G<A | 0.378 | 0.456 | 0.725 | (0.64–0.82) | 2.93 × 10−3 | 9.12 × 10−3 |
| TNFSF4 | rs2205960 | 171 458 098 | T<G | 0.313 | 0.260 | 1.296 | (1.13–1.49) | 2.41 × 10−4 | 3.87 × 10−3 |
| TNFSF4 | rs844648 | 171 490 486 | A<G | 0.503 | 0.454 | 1.215 | (1.07–1.38) | 2.47 × 10−3 | 0.024 |
Abbreviations: P*, P-values adjusted for age and sex by logistic regression; SNP, single nucleotide polymorphism.
Note that in rs844648, ‘A' is the minor allele (frequency=0.454) in controls, but it becomes the major allele in cases (frequency=0.503); ‘*', all the SNPs were replicated by TaqMan in 949 SLE patients and 1042 controls, except rs4522865, which was replicated by Sequenom in 360 cases and 360 controls.
Independent contributions of the SNPs in TNFSF4 were difficult to disentangle as the study of Graham et al.5 was mainly conducted on trios. Based on the study of Graham et al.5 and our own GWAS, we have chosen rs2205960 and rs844648 for further replication. SNP rs2205960 was chosen because it had the most significant association in TNFSF4 region in our GWAS. Besides, based on LD analysis from both Caucasians (CEU) and Han Chinese in Beijing (HCB) in the HapMap data, rs2205960 is a good surrogate for its surrounding SNPs, such as rs1012507 and rs10489265 and so on; all of which have shown significant associations in either the reported study or our own GWAS. SNP rs844648 is a good proxy for rs844644 (r2=0.92 in our GWAS), whose minor allele (‘A') was found to tag the under-transmitted haplotype in SLE patients and was the only variant tested that showed significant P-value after conditional analysis in the reported study.5 Further, rs844648 also has high LD with the other SNPs in the TNFSF4 region and thus it can serve as a tag SNP. Altogether, rs3733197, rs17266594 from BANK1 and rs2205960, rs844648 from TNFSF4 were chosen for replication in 949 SLE cases (including the 314 cases in the GWAS) and 1042 healthy controls (independent from the 920 controls in the GWAS) by TaqMan. BANK1 rs4522865, which has the most significant association in GWAS, was separately genotyped in a non-duplicating set of 360 cases and 360 controls by Sequenom. All SNPs conferred associations with SLE, which remained significant after adjusting for age and sex (Table 2).
Independence test
To better define the relative contribution of each SNP in BANK1 and TNFSF4, conditional analyzes, including logistic regression and haplotype-based association test were performed (Table 3).
Table 3.
Conditional analysis of (a) BANK1 and (b) TNFSF4
| Logistic regression | Haplotype-based association test | ||||
|---|---|---|---|---|---|
| Controlled SNP | Effective SNP | P-values | Effective SNP | P-values | |
| (a) | |||||
| GWAS | rs4522865 | rs10516487 | 0.042 | Omnibus | 9.06 × 10−3 |
| *rs10516487 | rs4522865 | 1.03 × 10−3 | rs4522865 | 0.031 | |
| rs10516487 | 0.047 | ||||
| Replication | *rs17266594 | rs3733197 | 0.037 | Omnibus | 1.29 × 10−8 |
| rs3733197 | rs17266594 | 6.63 × 10−8 | rs3733197 | 0.028 | |
| rs17266594 | 2.01 × 10−8 | ||||
| (b) | |||||
| GWAS | rs2205960 | rs844648 | 0.467 | Omnibus | 3.12 × 10−5 |
| rs2205960 | rs844644 | 0.361 | rs2205960 | 3.21 × 10−3 | |
| rs844648 | 0.523 | ||||
| Replication | rs2205960 | rs844648 | 0.551 | Omnibus | 3.81 × 10−4 |
| rs844648 | rs2205960 | 6.26 × 10−3 | rs2205960 | 5.16 × 10−3 | |
| rs844648 | 0.489 | ||||
Abbreviations: GWAS, genome-wide association study; SNP, single nucleotide polymorphism.
Conditional analysis of SNPs from BANK1 and TNFSF4 in both GWAS and TaqMan replication data using PLINK.
‘*', rs10516487 and rs17266594 have strong LD in Chinese (r2=1 in HapMap-HCB).
SNPs chosen for analysis were the five top-rank SNPs from our GWAS data in the BANK1 and TNFSF4 regions, respectively, and SNPs with reported association with SLE in Caucasian studies, as bolded in Table 1 and labeled in Figure 1. In the section of ‘logistic regression', the column marked by ‘controlled SNP' lists the SNPs whose effects were controlled whereas the column marked by ‘effective SNPs' lists the SNPs in concern during pairwise logistic regression. P-value is for the individual effect of the effective SNP. Only SNPs with significant P-values during logistic regression were shown, except rs844648 and rs844644 in TNFSF4. In the section of ‘haplotype-based association test', haplotypes were constructed for BANK1 and TNFSF4 correspondingly with the selected SNPs bolded in Table 1. The column marker by ‘effective SNP' contains SNPs whose independent effects were in concern and ‘P' represents the P-values for the significance of ‘effective SNP' when the effects of the other SNPs within the same haplotype were controlled.
In BANK1 GWAS, five SNPs with the most significant P-values together with rs10516487 (bolded in Table 1 and labeled in Figure 1) were selected to undergo conditional analysis. Although rs10516487 was not the top-rank SNPs in our GWAS, it was included for analysis because it has been reported to be associated with SLEin the study of Kozyrev et al.4 Moreover, rs10516487 is a good surrogate for rs17266594, the branch-point site SNP that was not included in our Bead-chip data. Therefore, examining the independence of rs10516487 allowed an indirect look for the contribution of rs17266594. In logistic regression, after controlling the effect of rs4522865, the effects of the other SNPs disappeared except rs10516487. In the pairwise logistic regression, the effects of the two SNPs were independent to each other (rs4522865, P=1.03 × 10−3; rs10516487, P=0.042). Subsequently, haplotype was constructed with the six SNPs bolded in Table 1, which gave a significant overall effect to disease association (P=9.06 × 10−3). Global association was conditioned on rs4522865 and rs10516487, in turn, and the individual effects of the two SNPs remained marginally significant (rs4522865, P=0.031; rs10516487, P=0.047).
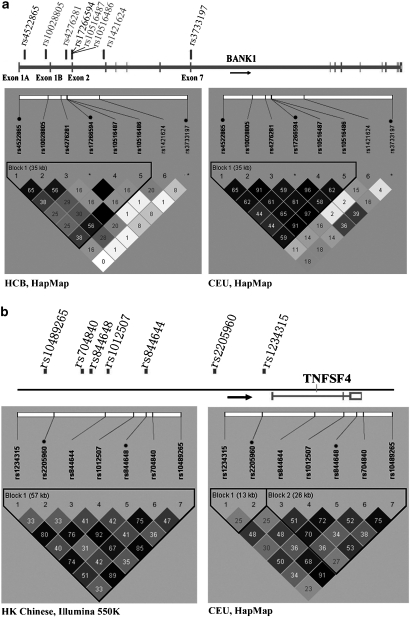
Figure 1.
Comparison of linkage disequilibrium of SNPs between Chinese and Caucasians. LD r2 prime charts from HaploView that summarize the LD patterns in Chinese (HCB) and Caucasians (CEU) are shown. (a) BANK1, (b) TNFSF4. Dark area represents regions of high-pairwise r2, whereas white color represents regions with low-pairwise r2. The numbers in the boxes are the pairwise r2 values and empty cells represent that pairwise r2=1. Haplotype-block is defined by ‘solid-spine' option in HaploView. Numbering of SNPs is consistent with that in Table 1. SNPs rs17266594 and rs3733197 were not included in GWAS but in replication experiment and thus are labeled as ‘*'. SNPs selected for replication study are marked by a black dot. In Figure 1a, rs17266594, rs10516487 and rs10516486 cluster together because of physical proximity.
In the replication experiment by TaqMan, rs3733197 and rs17266594 from BANK1 were significantly independent to each other in logistic regression (P=0.037 and 6.63 × 10−8, respectively), as well as haplotype-based association test (P=0.028 and 2.01 × 10−8, respectively). These results were consistent with the weak LD between them in Chinese (r2=0.08) (Figure 1a). On the whole, our data indicated that rs4522865 (a synonymous SNP within the first intron), rs10516487 (R61H) (or branch-site SNP rs17266594 because of the high LD between the two) and rs3733197 (A383T) were contributing to SLE risk independently in BANK1 (Table 3a).
In the case of TNFSF4, the effects of the other SNPs vanished after controlling rs2205960 by logistic regression in GWAS. This was consistent with the results of logistic regression using data from TaqMan replication, which showed that the effect of rs844648 became insignificant (P=0.551) after controlling the effect of rs2205960. In contrast, the association of rs2205960 was independent from rs844648 in logistic regression (P=6.26 × 10−3), indicating the presence of a major contribution from rs2205960 in the upstream region of TNFSF4. Haplotype-association test with SNPs bolded in Table 1 showed consistent results with logistic regression (Table 3b).
Sub-phenotype analysis and gene–gene interaction
The high heterogeneity of SLE manifestations may give clues on different pathogenesis of the disease. Therefore, case-only analysis was performed to determine potential genetic association with specific sub-phenotypes (for example, cases with arthritis versus cases without arthritis). Results showed that TNFSF4 rs2205960 was associated with the production of anti-Ro antibodies (odds ratio (OR)=1.25, P=0.043) and BANK1 rs3733197 was associated with malar rash (OR=0.67, P=0.003). Tests on other sub-phenotypes gave insignificant results (data not shown). However, these results should be interpreted with caution in the context of multiple testing effect and reduction in sample size in case-only analyzes. In addition to the individual contributions of genetic variants to the disease, interactions among them may also affect disease susceptibility. BANK1 and TNFSF4 encode a scaffold protein and a cytokine, respectively, and both of which can be found in the activated B cells. An interesting question to ask is whether interaction between the two, alters risk of SLE. However, there was no evidence to suggest such an interaction from our data by logistic regression (P>0.05).
Discussion
The recent advance in genetic association study has uncovered more and more susceptibility genes for complex diseases such as SLE. In populations of European ancestry, rs3733197 and rs17266594 in BANK1 were found to be associated with SLE (OR=1.23, P=4.67 × 10−5; OR=1.42, P=4.74 × 10−11, respectively).4 In our study, the contributions of rs3733197 and rs17266594 were verified (OR=1.19, P=0.021; OR=1.65, P=4.67 × 10−9), with a larger effect size for rs17266594 in Chinese (OR=1.65, CI=1.38–1.96) than that in the Caucasians (OR=1.42, CI=1.28–1.58).
Concerning the independent effects of SNPs, Kozyrev et al.4 reported that none of the SNPs studied in BANK1 (rs3733197, rs17266594 and rs10516487) were independent of each other in conditional logistic regression analysis as a result of the LD between them. In our study, the effects of rs3733197 and rs17266594 did not explain each other. This population difference could be a result of the larger effect size observed in Chinese, and thus a more readily detectable independence between the SNPs by statistical methods. In addition, the difference in LD patterns may also play a critical role. In Chinese, LD between rs3733197 and rs17266594 is much weaker than that in Caucasian (HapMap-HCB: r2=0.08; HapMap-CEU: r2=0.36, Figure 1a). In addition, rs3733197 also has a weak LD with the rest of the SNPs (Figure 1a) and therefore it is likely to confer SLE risk independently in the BANK1 region.
In addition to statistical analyzes of their independent effects, possible functional implications of the SNPs were also considered to better justify their roles in SLE. SNP rs3733197 causes a substitution in the ankyrin domain (A383T). The alanine residue at this position is well conserved in all the mammals examined, including Monodelphis domestica (Gray short-tailed opossum), suggesting a functional constraint on this position during evolutionary courses. The mutations in ankyrin motifs have also been shown to alter interactions with IP3R, thus affecting cytoplasmic calcium mobilization in cardiac arrhythmia and sudden cardiac death.13 These data supports the possible functional importance of rs3733197 and its independent contribution to the risk of SLE. On the other hand, although rs17266594 (the branch point-site polymorphism) confers a strong association with SLE, it is hard to distinguish a direct association at the functional level from an indirect association caused by its strong LD with rs10516487 (r2=1 in HapMap-HCB). The non-synonymous substitution because of rs10516487 (R61H) is at a highly variable position across species. On the same position in its orthologs, it is a histidine in dog, leucine in horse, proline in mouse and rat, cysteine in cow, and serine in Opossum. Variations at the position may indicate a relaxation of selection pressure on this amino acid residue, although it can also be explained by functional diversities among the orthologs. On the other hand, it has been shown that the expression of two isoforms of BANK1 transcripts are associated with the genotypes of rs17266594, the branch point-site SNP,4 which thus seems to be a more likely candidate responsible for the association of SLE.
It is noted that besides rs3733197 and rs17266594, rs4522865 also contributed to SLE susceptibility, which was not reported in the earlier study. SNP rs4522865 had the most significant P-value (P=8.49 × 10−4, Table 1) in our GWAS and was successfully replicated by further genotyping non-overlapping samples (P=2.93 × 10−3, Table 2). Furthermore, it was found that rs4522865 had a weak LD with rs10516487 (r2=0.25, HapMap-HCB, Figure 1a) and thus the effect of rs4522865 was unlikely to be dependent on rs10516487. This was consistent with the result of conditional analysis, in which it was found that after controlling the effect of rs4522865, the effects of most of the other SNPs in the BANK1 region were gone except rs10516487. Therefore, rs4522865 is likely to be the other independent contributor to SLE susceptibility besides rs3733197 and rs17266594. Locating at the first intron of BANK1, rs4522865 may have a role in the expression of BANK1. It is interesting to find out several candidates having independent contribution to SLE association within the same locus, however, further functional studies and deep sequencing are required to find out the genuine functional variants in this region.
In the case of TNFSF4, the minor alleles of rs844648 and rs2205960 tagged an over-transmitted upstream haplotype in SLE cases in Caucasians (MAF−case=0.485, MAF−control=0.439, P=0.03; MAF−case=0.276, MAF−control=0.233, P=7.0 × 10−3, respectively).5 In addition, rs844644, which has a high LD with rs844648 in our data and the Caucasians (r2=0.92 and 0.70, Figure 1b), had been shown to be the single genotyped variant that showed a marginal P-value after conditional analysis (P=0.015, Graham et al.,5 Supplementary Table 6). In our study, the association of rs844648 and rs2205960 were confirmed (MAF−case=0.50, MAF−control=0.45, P=2.47 × 10−3; MAF−case=0.31, MAF−control=0.26, P=2.41 × 10−4, respectively). However, after controlling the effect of rs2205960, the effect of rs844648 became insignificant in both of our GWAS and the replication data. The moderate independent effect of rs844644 in Caucasian study was also not observed by GWAS in our population. It is difficult to tell whether this is a genuine difference between populations or only inconsistent statistical results because of the moderate effect sizes of rs844644 and rs844648, which would reduce the power of the conditional analyzes. SNP rs2205960 is located 38.6 kb upstream of TNFSF4 and probably involved in a cis-acting factor of transcription regulation. Together with the results of conditional analysis, rs2205960 is therefore likely to be a major variant contributing to disease susceptibility in this locus.
In conclusion, this study evaluated SNPs from BANK1 and TNFSF4 to further define their roles in SLE risk. Their associations with SLE are confirmed in Chinese; nevertheless differences with the earlier studies are also showed, especially in terms of individual effects of SNPs. This suggests that multiple variants are probably present and affect the genes by different mechanisms. Further studies are needed to confirm or dissect out the genuine functional variants and to understand how these polymorphisms affect SLE.
Materials and methods
The study was approved by the Institutional Review Board of the University of Hong Kong and Hospital Authority, Hong Kong West Cluster, New Territory West Cluster and Hong Kong East Cluster. All patients gave informed consent.
Subjects
SLE patients were recruited from Queen Mary Hospital, Tuen Mun Hospital and Pamela Youde Nethersole Eastern Hospital, all of self-reported Chinese ethnicity living in Hong Kong. Their mean age was 31.8 and 93.1% were females. SLE in patients was diagnosed according to the criteria of the American College of Rheumatology.14 Clinical and serological data and autoantibody profile were recorded at the time of diagnosis. In the GWAS, 920 controls were obtained from a mixture of healthy subjects (35.5%) and patients with schizophrenia (43.2%) and chronic hepatitis B carriers (21.3%) with Chinese ethnicity. The minor allele frequencies of the SNPs in the two genes from the three control groups are similar to each other. Case–control analyzes were initially performed using the three control groups separately and the results were found to be consistent (Supplementary Table 1). In the replication study, healthy and ethnically matched controls were recruited from donors in Hong Kong Red Cross, with a mean age of 32.9 years old and 29.8% of females.
Genotyping
In GWAS, genotyping was carried out using Illumina Human 610-Quad arrays according to manufacturer's instructions. In the replication study, rs17266594 and rs3733197 from BANK1 and rs844648 and rs2205960 from TNFSF4 were genotyped by TaqMan SNP Assays (Applied Biosystems, Foster City, CA, USA). Genotyping accuracy was confirmed by direct sequencing of PCR products for 20 randomly selected samples, which showed 100% concordance. SNP rs4522865 from BANK1 was genotyped by Sequenom MassArray Genotyping according to the manufacturer's instructions.15 Concordance was checked against 48 positive controls out of 720 samples, and reached 100%.
Statistical analysis
GWAS phase
Preprocessing filters were applied for Illumina Human 610-Quad arrays data. Individuals were left out from further analysis if their relationship test failed, indicated by high proportion (>20% of diploid genome) of shared region identical by decent between two individuals calculated by PLINK. Further, individuals were also discarded because of failed heterozygosity test, probably because of sample mix-up. All together six individuals were filtered out and the final sample number is 314 in SLE cases and 920 in controls. In addition, 1172 SNPs and 73 338 SNPs were discarded because of low-genotyping call rate (<0.95) and low-minor allele frequency (<0.01), respectively. SNPs that failed the Hardy–Weinberg equilibrium test (P<0.01) were also excluded from further analysis. After filtering, 489 851 SNPs remained and the total-genotyping call rate for these SNPs in the remaining individuals was 99.9%. Possible population substructure was evaluated with EIGENSTRAT16 and no population substructure was observed among local cases and controls. Chi-square test was corrected by an genome inflation factor of 1.03 calculated by PLINK.17
Replication phase
The genotype frequencies of all SNPs were tested for Hardy–Weinberg equilibrium separately in cases and controls and all attained P>0.05. Disease associations were analyzed by basic allelic test, as well as logistic regression and other inheritance models. SNP effect models were tested by using SNPGWA (URL: http://www.phs.wfubmc.edu/public/bios/gene/downloads.cfm). Testing for deviation from additive model was not significant. Corresponding P-values were adjusted for age and sex by logistic regression. Independence test of SNPs in the same gene was performed by logistic regression, as well as haplotype-based association test. Sub-phenotype stratification analysis was performed by case-only approach, in which basic allelic test was performed by comparing minor allele frequency of patients with a specific sub-phenotype to patients without the specific sub-phenotype. Any interaction between BANK1 and TNFSF4 was tested by conditional analysis.
All statistical analyzes were performed by PLINK,17 version 1.05, unless otherwise specified. Linkage disequilibrium patterns and values were obtained by HaploView.18
Power calculation
Power calculations in this study considered allele frequency of SNPs from 0.05 to 0.5, a population prevalence of 0.06% for SLE,3 and a significance level of 0.05. This study has sufficient power (>80%) to detect association of OR of 1.2 or above with 900 cases and 1000 controls and OR of 1.3 or above with 350 cases and 350 controls. However, in sub-phenotype analysis using case-only approach, samples are reduced and the power of the study remarkably decreased.
Conflict of interest
The authors declare no conflict of interest.
Supplementary Material
Acknowledgments
This study is partially supported by the Shun Tak District Min Yuen Tong of Hong Kong. YKC thanks support from Mr and Mrs SH Wong Foundation Scholarship, BL Wong Scholarship and Award of Yu Chun Keung Memorial Scholarship for Master of Research in Medicine. MZ was supported by Edward Sai Kim Hotung Paediatric Education and Research Fund, and University Postgraduate Studentship. WY thanks support from UGC, UHK (200711159155).
Supplementary information accompanies the paper on Genes and Immunity website (http://www.nature.com/gene)
References
- Pao-Hsii F. Systemic lupus erythematosus. Annals of the New York Academy of Sciences. 2007;1108 Autoimmunity, Part D: Autoimmune Disease, Annus Mirabilis:114–120. [Google Scholar]
- Lau CS, Yin G, Mok MY. Ethnic and geographical differences in systemic lupus erythematosus: an overview. Lupus. 2006;15:715–719. doi: 10.1177/0961203306072311. [DOI] [PubMed] [Google Scholar]
- Mok CC, Lau CS. Lupus in Hong Kong Chinese. Lupus. 2003;12:717–722. doi: 10.1191/0961203303lu451xx. [DOI] [PubMed] [Google Scholar]
- Kozyrev SV, Abelson AK, Wojcik J, Zaghlool A, Linga Reddy MV, Sanchez E, et al. Functional variants in the B-cell gene BANK1 are associated with systemic lupus erythematosus. Nat Genet. 2008;40:211–216. doi: 10.1038/ng.79. [DOI] [PubMed] [Google Scholar]
- Graham DS, Graham RR, Manku H, Wong AK, Whittaker JC, Gaffney PM, et al. Polymorphism at the TNF superfamily gene TNFSF4 confers susceptibility to systemic lupus erythematosus. Nat Genet. 2008;40:83–89. doi: 10.1038/ng.2007.47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yokoyama K, Su Ih IH, Tezuka T, Yasuda T, Mikoshiba K, Tarakhovsky A, et al. BANK regulates BCR-induced calcium mobilization by promoting tyrosine phosphorylation of IP(3) receptor. EMBO J. 2002;21:83–92. doi: 10.1093/emboj/21.1.83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baum PR, Gayle RB, III, Ramsdell F, Srinivasan S, Sorensen RA, Watson ML, et al. Molecular characterization of murine and human OX40/OX40 ligand systems: identification of a human OX40 ligand as the HTLV-1-regulated protein gp34. EMBO J. 1994;13:3992–4001. doi: 10.1002/j.1460-2075.1994.tb06715.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito T, Wang YH, Duramad O, Hanabuchi S, Perng OA, Gilliet M, et al. OX40 ligand shuts down IL-10-producing regulatory T cells. Proc Natl Acad Sci USA. 2006;103:13138–13143. doi: 10.1073/pnas.0603107103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J, Li L, Shang X, Benson J, Merle Elloso M, Schantz A, et al. Negative regulation of IL-17 production by OX40/OX40L interaction. Cell Immunol. 2008;253:31–37. doi: 10.1016/j.cellimm.2008.04.010. [DOI] [PubMed] [Google Scholar]
- Cordell HJ, Clayton DG. Genetic association studies. Lancet. 2005;366:1121–1131. doi: 10.1016/S0140-6736(05)67424-7. [DOI] [PubMed] [Google Scholar]
- Lettre G, Rioux JD. Autoimmune diseases: insights from genome-wide association studies. Hum Mol Genet. 2008;17 R2:R116–R121. doi: 10.1093/hmg/ddn246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang W, Ng P, Zhao M, Hirankarn N, Lau CS, Mok CC, et al. Population differences in SLE susceptibility genes: STAT4 and BLK, but not PXK, are associated with systemic lupus erythematosus in Hong Kong Chinese Genes Immun 2009(E-pub ahead of print) doi:10.1038/gene.2009.1 [DOI] [PubMed]
- Mohler PJ, Schott JJ, Gramolini AO, Dilly KW, Guatimosim S, duBell WH, et al. Ankyrin-B mutation causes type 4 long-QT cardiac arrhythmia and sudden cardiac death Nature 2003421587589–590 [DOI] [PubMed] [Google Scholar]
- Tan EM, Cohen AS, Fries JF, Masi AT, McShane DJ, Rothfield NF, et al. The 1982 revised criteria for the classification of systemic lupus erythematosus. Arthritis Rheum. 1982;25:1271–1277. doi: 10.1002/art.1780251101. [DOI] [PubMed] [Google Scholar]
- Gabriel S, Ziaugra L, Tabbaa D. SNP Genotyping Using the Sequenom Mass ARRAY iPLEX Platform. Curr Protoc Hum Genet. 2009;Chapter 2:Unit 2.12. doi: 10.1002/0471142905.hg0212s60. [DOI] [PubMed] [Google Scholar]
- Price AL, Patterson NJ, Plenge RM, Weinblatt ME, Shadick NA, Reich D. Principal components analysis corrects for stratification in genome-wide association studies. Nat Genet. 2006;38:904–909. doi: 10.1038/ng1847. [DOI] [PubMed] [Google Scholar]
- Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MA, Bender D, et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet. 2007;81:559–575. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrett JC, Fry B, Maller J, Daly MJ. Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics. 2005;21:263–265. doi: 10.1093/bioinformatics/bth457. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.