Abstract
The life cycle of mammals begins when a sperm enters an egg. Immediately after fertilization, both the maternal and paternal genomes undergo dramatic reprogramming to prepare for the transition from germ cell to somatic cell transcription programs 1. One of the molecular events that takes place during this transition is the demethylation of the paternal genome 2,3. Despite extensive efforts, the factors responsible for paternal DNA demethylation have not been identified 4. To search for such factors, we developed a live cell imaging system that allows us to monitor the paternal DNA methylation state in zygotes. Through siRNA-mediated knockdown in zygotes, we identified Elp3/KAT9, a component of the elongator complex 5, to be important for paternal DNA demethylation. We demonstrate that knockdown of Elp3 impairs paternal DNA demethylation as indicated by reporter binding, immunostaining and bisulfite sequencing. Similar results were also obtained when other elongator components, Elp1 and Elp4, were knocked down. Importantly, injection of mRNA encoding the Elp3 radical SAM domain mutant, but not the HAT domain mutant, into MII oocytes before fertilization also impaired paternal DNA demethylation indicating that the SAM radical domain is involved in the demethylation process. Thus, our study not only establishes a critical role for the elongator in zygotic paternal genome demethylation, but also suggests that the demethylation process may be mediated through a reaction that requires an intact radical SAM domain.
Global removal of the methyl group from 5-methyl-CpG (5mC) of DNA has been observed in at least two stages of embryogenesis. One occurs in zygotes when the paternal genome is preferentially demethylated 2,3. However, imprinted genes are resistant to this wave of DNA demethylation 6. Instead, this group of genes is actively demethylated in primordial germ cells (PGCs) from E10.5 to E12.5 which results in the establishment of gender-specific methylation patterns 7. Given the importance of active DNA demethylation in embryogenesis, reprogramming, cloning, and stem cell biology, the identification of the putative demethylase has been one of the major focuses in the field 4.
The first molecule claimed to posses DNA demethylase activity is the methyl-CpG binding protein Mbd2 8. However, Mbd2 is not required for paternal genome demethylation as normal demethylation is still observed in Mbd2 deficient zygotes 9. Several recent studies in plants 10,11, zebrafish 12, and mammalian cells 13 have suggested that active DNA demethylation can occur through various DNA repair mechanisms. However, it is not known whether any of these proteins affect paternal genome demethylation.
Both Gadd45a and Gadd45b have been implicated in DNA demethylation in somatic cells 13,14, but the role of Gadd45a in DNA demethylation has been challenged by some recent studies 15,16. To determine whether Gadd45 proteins play a role in paternal DNA demethylation in zygotes, we performed RT-qPCR and found that Gadd45b is the most highly expressed gene among the Gadd45 family members in zygotes (Fig. S1a). Because Gadd45b has been shown to affect DNA demethylation in mature non-proliferating neurons 14, we examined whether loss of Gadd45b function affects zygotic paternal DNA demethylation. Immunostaining with the 5mC antibody indicates that paternal DNA demethylation is not affected by Gadd45b knockout suggesting that Gadd45b is not required for paternal DNA demethylation (Fig. S1b).
To facilitate the identification of factors involved in paternal DNA demethylation, we attempted to develop two molecular probes (Fig. S2a, b). The MBD domain of Mbd1 and the CxxC domain of Mll1 have high affinity towards methyl-CpG and non-methyl-CpG, respectively 17,18. Expectedly, EGFP-MBD exhibited a nuclear dotted pattern, while CxxC-EGFP exhibited diffuse nuclear staining in wild-type MEFs (Fig. S2c, d). In contrast, almost 100% of Dnmt1 null MEF cells that lack CpG methylation exhibited punctate nuclear localization of CxxC-EGFP. Unexpectedly, the nuclear dotted pattern of EGFP-MBD was still maintained in ~60% of the DKO cells (Fig. S2c, d). This result indicates that when compared to EGFP-MBD, CxxC-EGFP is the better probe whose subcellular localization pattern can reflect the DNA methylation state. We further confirmed the utility of the CxxC-EGFP reporter by demonstrating that 5-Aza-dC-mediated DNA demethylation resulted in a clear increase in the number and intensity of GFP bright dots in NIH 3T3 cells (Fig. S2e).
We next tested whether the CxxC-EGFP probe can accurately “report” paternal genome demethylation. Since injected plasmid DNA is transcriptionally inactive in 1-cell zygotes, we adapted an mRNA injection technique that allows for visualization of molecular events in the mammalian zygote as early as 3 hours after introduction 19. We generated poly(A) mRNAs for the CxxC-EGFP as well as H2B-mRFP1 (monomeric red fluorescent protein 1) by in vitro transcription 19 (Fig. S2b). Using the procedure outlined in Fig. S3a, the mRNAs were co-injected into the zygotes immediately after in vitro fertilization (IVF). Time-lapse imaging of the injected zygotes indicate that CxxC-EGFP is visible at the PN2 stage and accumulates throughout the PN3-4 and PN5 stages in the paternal pronucleus (Fig. S3b). The dynamics of the paternal PN CxxC-EGFP accumulation mimic paternal DNA demethylation dynamics reported previously 2,3. Based on this result, we conclude that paternal genome demethylation can be monitored by injection of CxxC-EGFP mRNA in zygotes.
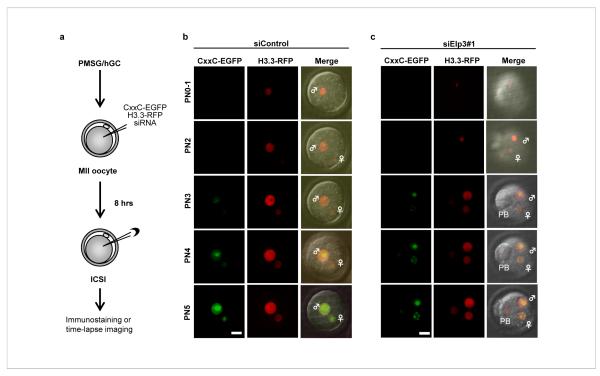
We next asked whether siRNA-mediated depletion of candidate mRNAs in the oocytes could affect paternal DNA demethylation in zygotes. We first determined the optimal siRNA concentration and the time needed for injected siRNA to become effective using siRNA against Lamin A/C. The tests established that effective knockdown required a minimum dose of 2 μM and incubation time of 8 hrs prior to intracytoplasmic sperm injection (ICSI). Based on the results from these tests, we established a modified experimental procedure (Fig. 1a). To facilitate early stage PN identification (PN0-2), we also replaced H2B-mRFP1 with H3.3-mRFP1 as H3.3 has been shown to be preferentially deposited in the paternal PN immediately following fertilization 20. This modified experimental scheme allows us to monitor H3.3 deposition and DNA demethylation simultaneously with time-lapse imaging. A representative snap shot of the various PN stages with the injection of a scrambled siRNA control is presented in Fig. 1b. This time-lapse imaging system coupled with siRNA knockdown, allowed us to test a dozen candidate genes selected based on several criteria that include: 1) their expression in zygotes; 2) the domain/structure motifs they contain; and 3) their potential in catalyzing DNA demethylation. Using these criteria, we designed siRNAs targeting a dozen candidate genes. We achieved a knockdown efficiency of at least 80% for six of the candidate genes that include Cyp11a1, Smc6-like, Brm, Alkbh5, NfuI, and Elp3 (Fig. S4a). We also attempted to knockdown the recently identified 5mC hydroxylase Tet1 21 but failed to achieve high efficiency. With the exception of Elp3 knockdown (Fig. 1c), none of the other knockdowns altered the preferential distribution of the reporter to the paternal pronucleus (Fig. S4b). To verify this preliminary observation, we performed immunostaining using the anti-5mC antibody. Results shown in Fig. 2a clearly demonstrate that knockdown of Elp3 prevent paternal DNA demethylation. Furthermore, a second siRNA targeting a different region of Elp3 also resulted in a similar result (Fig. 2a).
Figure 1. Knockdown of Elp3 prevents preferential incorporation of the CxxC-EGFP reporter into the paternal pronucleus.
(a) Scheme of the experimental procedure. (b, c) Time-lapse imaging of CxxC-EGFP (green; left column) and H3.3-mRFP1 (red; middle column) at various pronucleus stages of zygotic development in the absence (b) or presence (c) of siRNA that targets Elp3. ♂: male pronucleus, ♀: female pronucleus, PB: polar body. Bar = 25 μm.
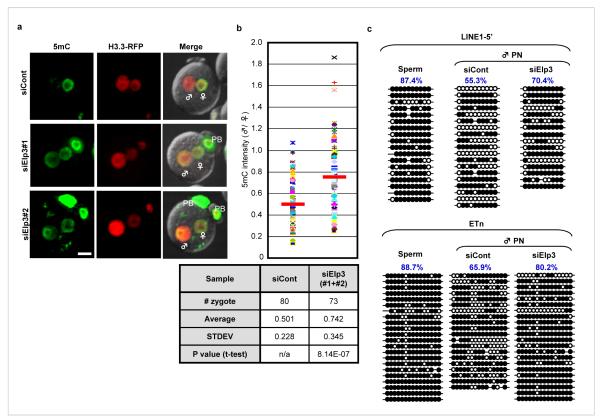
Figure 2. Knockdown of Elp3 impairs DNA demethylation in the paternal pronucleus.
(a) siRNA-mediated knockdown of Elp3 resulted in increased 5mC staining (green) in the PN5 paternal pronucleus. H3.3-mRFP1 (red) serves as a nuclear marker. H3.3-mRFP1 signal is more intense in male pronuclei than in female pronulcei due to preferential incoporation of H3.3 to the paternal genome. ♂: male pronucleus, ♀: female pronucleus, PB: polar body. Bar = 25 μm. (b) Quantification of the ratio (male/female) of 5mC intensity in Elp3 knockdown and control groups. Each dot represents a zygote. Red bars represent the average ratio of each group. The statistics of the injections are presented in the table. (c) Bisulfite sequencing of Line1-5′ and ETn indicates that knockdown of Elp3 impairs paternal DNA demethylation. Open circles and closed circles represent unmethylated and methylated CpG respectively. Each line represents an individual clone. 10 CpGs and 15 CpGs were analyzed for Line1-5′ and ETn respectively.
Although preferential demethylation of the paternal DNA in zygotes is a general phenomenon, the extent of demethylation of individual zygotes is variable (Fig. S5). Therefore, we quantified the effect of Elp3 knockdown on paternal DNA demethylation by analyzing a large number of zygotes. After determination of the highest 5mC intensity of the Z-section for both male and female pronucleus (Fig. S6a), a ratio of paternal over maternal 5mC intensity was determined for each zygote (Fig. S6b). Analysis of 80 PN4-5 stage zygotes with control injection results in an average ratio of 0.501. However, this ratio is significantly increased (average 0.742, p=8.14E-07) with injection of siRNAs that target Elp3 (Fig. 2b). These results indicate that Elp3 knockdown significantly impairs paternal DNA demethylation as judged by 5mC Ab staining.
To provide direct evidence that Elp3 knockdown affects paternal DNA demethylation, we evaluated DNA methylation levels by bisulfite sequencing. Previous studies indicated that the transposable elements Line1 and Etn (early retrotransposons) are actively demethylated in zygotes 22,23. We therefore asked whether knockdown of Elp3 would impair their demethylation. To this end, we injected siRNAs that target Elp3 prior to ICSI and isolated paternal pronuclei at the PN3-4 stages when DNA demethylation is at the beginning or is still occurring. We note that this is the latest time that we can still isolate the paternal pronuclei without co-isolating the maternal pronuclei as the two pronuclei become too close at the PN5 stage. Despite the fact that demethylation is far from completion at the PN3-4 stage, the effect of Elp3 knockdown on Line1 and Etn demethylation is still obvious (Fig. 3c). However, the methylation state of the imprinted H19 DMR or an unmethylated control Epha7 is not affected by Elp3 knockdown (Fig. S7) indicating the observed effect on Line1 and Etn is specific.
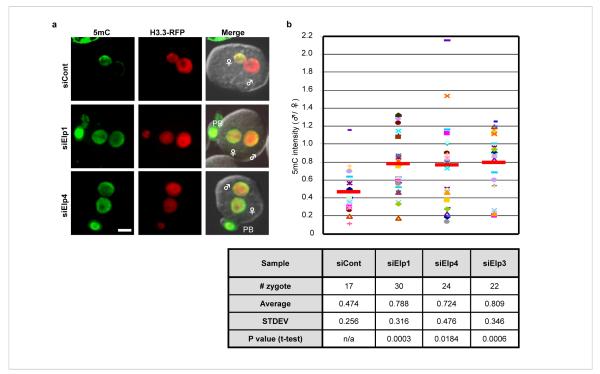
Figure 3. Knockdown of the elongator components Elp1 and Elp4 also impairs paternal DNA demethylation in zygotes.
(a) siRNA-mediated knockdown of Elp1 and Elp4 resulted in increased 5mC staining (green) in the PN5 paternal pronucleus. H3.3-mRFP1 (red) serves as a nuclear marker. ♂: male pronucleus, ♀: female pronucleus, PB: polar body. Bar = 25 μm. (b) Quantification of the ratio (male/female) of 5mC intensity in Elp1, Elp4, Elp3 knockdown and control groups. Each dot represents a zygote. Red bars represent the average ratio of each group. The statistics of the injections are presented in the table.
Elp3 is a component of the elongator complex that was initially identified based on its association with the RNA polymerase II holoenzyme involved in transcriptional elongation. Subsequent studies have revealed that the elongator complex has diverse functions that include cytoplasmic kinase signalling, exocytosis, and tRNA modification 5. The yeast elongator complex is composed of six subunits, Elp1-6, which includes the histone acetyltransferase (HAT) Elp3 24. The human elongator purified from HeLa is also composed of six subunits although the identities of Elp5 and Elp6 are unknown 25. To determine whether knockdown of other elongator subunits in oocytes also prevents paternal DNA from demethylation, we performed knockdown on two additional elongator subunits, Elp1 and Elp4. Results shown in Fig. 3 demonstrate that knockdown of these two subunits also significantly affects paternal genome demethylation. Collectively, these results suggest that paternal DNA demethylation in zygotes requires the elongator complex.
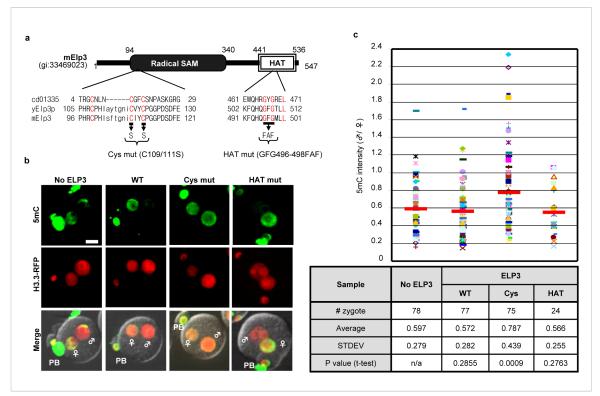
In addition to a HAT domain, Elp3 also contains a domain that shares significant sequence homology with the radical SAM domain present in the radical SAM superfamily (Fig. 4a). Members of this superfamily contain an iron-sulfur (Fe-S) cluster and use S-adenosylmethionine (SAM) to catalyze a variety of radical reactions 26. Interestingly, a recent study confirmed the presence of this Fe4S4 cluster in the bacteria Methanocaldococcus jannaschii Elp3 protein 27. To determine whether any of the two conserved domains of the mouse Elp3 is required for paternal DNA demethylation, we used a dominant negative approach and generated mRNAs that harbor mutations in the cysteine-rich motif and the HAT domain respectively (Fig. 4a). Injection of the cysteine mutant mRNA, but not the wild-type or HAT mutant mRNA, significantly impaired paternal DNA demethylation (Fig. 4b, c), indicating that the cysteine-rich motif, but not the HAT domain, is important for paternal DNA demethylation.
Figure 4. Mutation of the cysteine-rich radical SAM domain of Elp3 impairs paternal DNA demethylation.
Schematic representation of wild-type and mutant mElp3. Conserved domain (CD) of Elp3 protein sequences from NCBI are aligned with Elp3 sequences from budding yeast and mouse. Conserved amino acid residues are color coded in red. (b) Overexpression of the Cys mutant, but not the HAT mutant or wild-type, blocked paternal DNA demethylation. Representative images from PN5 stage were shown. ♂: male pronucleus, ♀: female pronucleus, PB: polar body. Bar = 25 μm. (c) Quantification of the ratio (male/female) of 5mC intensity in control and Elp3 (wild-type, Cyc mutant, or HAT mutant) mRNA injected groups. Each dot represents a zygote. Red bars represent the average ratio of each group. The statistics of the injections are presented in the table.
Using a live cell imaging reporter system coupled with siRNA knockdown, we uncovered a critical function of the elongator complex in paternal DNA demethylation. Several lines of evidence support our conclusion. First, three independent assays (reporter localization, 5mC staining, bisulfite sequencing) indicate that knockdown of Elp3 impairs paternal DNA demethylation (Figs. 1, 2). Second, knockdown of additional components of the elongator complex Elp1 and Elp4 also impaired paternal DNA demethylation (Fig. 3). Third, a dominant negative approach identified the radical SAM domain, but not the HAT domain, of Elp3 to be critical for the demethylation to occur (Fig. 4). Consistent with the involvement of the elongator complex in zygote demethylation, mRNA levels of Elp1-4 are up-regulated 3-9 fold in the PN1-2 stages prior to the start of paternal DNA demethylation at PN3 (Fig. S7). Considering the fact that it takes ~3 hrs for injected mRNA to be expressed, the upregulation of the Elp mRNA levels at PN1-2 stages is consistent with demethylation of the paternal DNA at PN3-5 stages. Although the exact molecular mechanism by which the elongator complex participates in the demethylation process awaits to be determined, the demonstration for the first time that a specific protein complex is involved in paternal genome demethylation in zygotes has set the stage for further studies. The fact that the radical SAM domain is required for demethylation to occur points to a potential mechanism that involves the generation of a powerful oxidizing agent, 5′-deoxyadenosyl radical, from SAM. 5′-deoxyadenosyl radical could then extract a hydrogen atom from the methyl group of 5mC to generate 5mC radical for subsequent reactions. Although this potential mechanism is attractive, currently we do not have evidence indicating that Elp3 directly acts upon DNA as a DNA demethylase. We note that reconstitution of the enzymatic activity may not be trivial as the maternal genome in the same zygote is not subject to demethylation indicating that certain features of the paternal genome might be required for the demethylation reaction to occur. Regardless of how elongator participates in the demethylation process, our studies not only uncover a novel function for the elongator complex, but also set the stage for understanding the functional significance of paternal genome demethylation.
METHOD SUMMARY
Oocyte/zygote preparation, 5mC staining, and time-lapse imaging
MII oocytes, collected from female BDF1 mice treated with PMSG (Harbor-UCLA) and hCG (Sigma Aldrich), were cultured in M16 medium (EmbryoMax, Millipore) or CZB medium at 37°C with 5% CO2 before being used in experiments. For 5mC staining, zygotes were fixed with 4% paraformaldehyde and then permeabilized with 0.4% TritonX-100 for 30 min at room temperature. Cells were then washed with PBS containing 0.05% Tween20 (PBST), treated with 4N HCl for 30 min, and then neutralized with 0.1M Tris-HCl (pH 8.5) for 10 min before being washed with PBST containing 0.5 M NaCl. After blocking with 1% BSA in PBST, cells were incubated with anti-5mC antibody (Eurogentic) for 0.5-1 hr at 37°C, and the signal is detected by FITC-conjugated donkey anti-mouse IgG (Jackson Immmunoresearch). Fluorescent images and time-lapse imaging were taken using a confocal microscope with a spinning disk (CSU-10, Yokogawa). Images were acquired as multiple 2μm-Z-axis intervals, and stacked images were reconstituted using Axiovision (Zeiss) or MetaMorph (Universal Imaging Co).
mRNA and siRNA injection, RT-qPCR, and bisulfite sequencing
About 3-5 pl of siRNAs (2μM) purchased from Ambion (Table S1) were co-injected with H3.3-mRFP1 (25 μg/ml) and CxxC-EGFP mRNAs (25 μg/ml). After 8 hrs of cultivation, cells were subjected to ICSI. To determine knockdown efficiency, RNA isolated from 10-20 zygotes at PN4-5 stage was used for RT-qPCR using SuperScript III cDNA synthesis kit and SYBR GreenER (Invitrogen). For bisulfite sequencing, Elp3 or control siRNA was co-injected with H3.3-mRFP1 mRNA to MII oocytes followed by ICSI. Male pronulei were harvested at PN3-4 stages. Forty-three and 47 male pronuclei from control or siElp3-injected zygotes were used for bisulfite conversion using EZ DNA Methylation-Direct Kit (Zymo Research), respectively.
FULL METHODS
DNA constructs
cDNA that encodes the CxxC domain (aa. 1144-1250) of mouse Mll-1 (NCBI# NP_005924) was cloned by RT-PCR. cDNA for H3.3 was provided by Dr. Nakatani 28. These cDNAs were subcloned into a pcDNA3.1-poly(A)83 vector with a C-terminal EGFP or mRFP1. pcDNA3.1-EGFP-MBD-poly(A)83 and pcDNA3.1-H2B-mRFP1-poly(A)83 were previously described 19. These plasmids were used for in vitro transcription using the RiboMAX Large Scale RNA production System T7 (Promega). Synthesized mRNAs were purified with Illustra MicroSpin G-25 columns (GE Healthcare) before being used for injection. The mouse Elp3 cDNA was amplified by RT-PCR and was subcloned into a pCDNA3.1-poly(A)83 vector with a Flag tag at the N-terminus. Both the cysteine and the HAT mutants of Elp3 were generated by PCR-based mutagenesis and confirmed by sequencing. The primers used for generation of these mutants were as follows: Cys-F) 5′ ACAGGGAATATATCTATATACTCCCCCGGAGGACCTG-3′, Cys-R) 5′-CAGGTCCTCCGGGGGAGTATATAGATATATTCCCTGT-3′, HAT-F) 5′-AATTTCAGCATCAGTTCGCCTTCATGCTGCTGATGG-3′, HAT-R) 5′-CCATCAGCAGCATGAAGGCGAACTGATGCTGAAATT-3′. The underlined nucleotides are substituted in the mutants.
Mice and oocyte/zygote preparation
All animal experiments were performed according to procedures approved by the Institutional Animal Care and Use Committee (IACUC protocol# 07-006) and the Animal Experiment Handbook at the Kobe Center for Developmental Biology (RIKEN). Four to twelve week old BDF1 mice (C57BL6 × DBA2, Charles River or Japan SLC) were used for all the experiments. MII oocytes, collected from female mice treated with PMSG (Harbor-UCLA) and hCG (Sigma Aldrich), were cultured in M16 medium (EmbryoMax, Millipore) or CZB medium at 37°C with 5% CO2 before being used in experiments. Gadd45b-deficient zygotes were obtained by mating of Gadd45b knockout mice pairs 14.
5mC staining and time-lapse imaging
Zygotes were fixed with 4% paraformaldehyde for at least 1.5 hrs at 4°C. After washing with PBS, the zygotes were permeabilized with 0.4% TritonX-100 for 30 min at room temperature. Cells were then washed with PBS containing 0.05% Tween20 (PBST) and treated with 4N HCl for 30 min at room temperature before being neutralized with 0.1M Tris-HCl (pH 8.5) for 10 min, and then washed with PBST containing 0.5 M NaCl. In the following procedure, all solutions and buffers contain 0.5 M NaCl. After blocking with 1% BSA in PBST, cells were incubated with anti-5mC antibody (1:100 dilution, Eurogentic) for 0.5-1 hr at 37°C, and the positive signal was detected by FITC-conjugated donkey anti-mouse IgG (Jackson Immmunoresearch). Fluorescent images were taken using a confocal microscope with a spinning disk (CSU-10, Yokogawa). The same confocal microscope system, combined with an on-stage incubation chamber, was used for time-lapse imaging. For both live and fixed zygotes, images were acquired as multiple 2μm-Z-axis intervals, and stacked images were reconstituted using Axiovision (Zeiss) or MetaMorph (Universal Imaging Co). The intensity of 5mC in each pronucleus was calculated by MetaMorph as shown in Fig. S6.
mRNA and siRNA injection, RT-qPCR, and bisulfite sequencing
About 3-5 pl of siRNAs (2μM) purchased from Ambion (Table S1) were co-injected with H3.3-mRFP1 (25 μg/ml) and CxxC-EGFP mRNAs (25 μg/ml) simultaneously. After 8 hrs of cultivation, cells were subjected to ICSI (Fig. 2a). For determination of knockdown efficiency, RNA isolated from 10-20 zygotes at PN4-5 stage were used for reverse transcription using SuperScript III Cell Direct cDNA synthesis kit (Invitrogen) followed by quantitative PCR (qPCR) using SYBR GreenER (Invitrogen). To determine the expression dynamics of Elp1-4 during pronuclear stages (Fig. S6), MII oocytes were subjected to in vitro fertilization. Groups of zygotes (100-120) were collected at 4 hours after insemination (PN1-2), and then every two hours (PN3, 4, 5, respectively) followed by acidic Tyrode’s treatment to remove cumulus cells. The extracted RNAs were subject to reverse transcription. Results were normalized with 18S rRNA as a standard. Primer sequences for qPCR are listed in Table S2.
For bisulfite sequencing, either Elp3 siRNA or control siRNA was co-injected with H3.3-mRFP1 mRNA to MII oocytes followed by ICSI after 6-8 hrs of siRNA/mRNA injection. Male pronulei, which were distinguished from female pronuclei based on their size, distance from polar bodies, and more intense H3.3-mRFP1 fluorescence, were harvested from zygotes of PN3-4 stages by breaking the zona and cytoplasm using Piezo drive (Prime Tech) and aspirating using a micromanipulator. Forty-three male pronuclei from control siRNA-injected zygotes and 47 male pronuclei from siElp3-injected zygotes were collected and subjected to bisulfite conversion using EZ DNA Methylation-Direct Kit (Zymo Research). Nested PCR was performed using Platinum Taq DNA polymerase (Invitrogen) or ExTaq HS (TaKaRa). The sequences of the PCR primers and PCR conditions are listed in Table S3 (ref29,30).
Cell culture and transfection
Immortalized p53 knockout (KO) and p53/Dnmt1 double knockout (DKO) mouse embryonic fibroblasts (MEFs) were previously described (Jackson-Grusby et al., 2001). The KO MEFs, DKO MEFs, and NIH3T3 cells were maintained in DMEM supplemented with 10% FBS. pcDNA3-EGFP-pA83 plasmids containing that MBD domain and CxxC motif were transfected using Fugene6 (Roche). NIH3T3 cells that stably express CxxC-EGFP were selected under 1 mg/ml G418. 5-Aza-2′deoxycytidine (Sigma Aldrich) was applied at the concentration of 5 μM for 72 hours.
Supplementary Material
Acknowledgements
We thank Dr. Hongjun Song (John’s Hopkins) for providing the Gadd45b null mice; Drs. Kazuhiro Aoki and Michiyuki Matsuda (Kyoto University) for helping with image quantification. We are grateful to Susan Wu for critical reading of the manuscript. Y.Z. is an investigator of the Howard Hughes Medical Institute.
Footnotes
Author Information Reprints and permissions information is available at www.nature.com/reprints.
Conflict of interest statement The authors declare no conflict of interests.
References
- 1.Reik W. Stability and flexibility of epigenetic gene regulation in mammalian development. Nature. 2007;447:425–432. doi: 10.1038/nature05918. [DOI] [PubMed] [Google Scholar]
- 2.Mayer W, Niveleau A, Walter J, Fundele R, Haaf T. Demethylation of the zygotic paternal genome. Nature. 2000;403:501–502. doi: 10.1038/35000656. [DOI] [PubMed] [Google Scholar]
- 3.Oswald J, et al. Active demethylation of the paternal genome in the mouse zygote. Curr Biol. 2000;10:475–478. doi: 10.1016/s0960-9822(00)00448-6. [DOI] [PubMed] [Google Scholar]
- 4.Ooi SK, Bestor TH. The colorful history of active DNA demethylation. Cell. 2008;133:1145–1148. doi: 10.1016/j.cell.2008.06.009. [DOI] [PubMed] [Google Scholar]
- 5.Svejstrup JQ. Elongator complex: how many roles does it play? Curr Opin Cell Biol. 2007;19:331–336. doi: 10.1016/j.ceb.2007.04.005. [DOI] [PubMed] [Google Scholar]
- 6.Howell CY, et al. Genomic imprinting disrupted by a maternal effect mutation in the Dnmt1 gene. Cell. 2001;104:829–838. doi: 10.1016/s0092-8674(01)00280-x. [DOI] [PubMed] [Google Scholar]
- 7.Hajkova P, et al. Epigenetic reprogramming in mouse primordial germ cells. Mech Dev. 2002;117:15–23. doi: 10.1016/s0925-4773(02)00181-8. [DOI] [PubMed] [Google Scholar]
- 8.Bhattacharya SK, Ramchandani S, Cervoni N, Szyf M. A mammalian protein with specific demethylase activity for mCpG DNA. Nature. 1999;397:579–583. doi: 10.1038/17533. [DOI] [PubMed] [Google Scholar]
- 9.Santos F, Hendrich B, Reik W, Dean W. Dynamic reprogramming of DNA methylation in the early mouse embryo. Dev Biol. 2002;241:172–182. doi: 10.1006/dbio.2001.0501. [DOI] [PubMed] [Google Scholar]
- 10.Choi Y, et al. DEMETER, a DNA glycosylase domain protein, is required for endosperm gene imprinting and seed viability in arabidopsis. Cell. 2002;110:33–42. doi: 10.1016/s0092-8674(02)00807-3. [DOI] [PubMed] [Google Scholar]
- 11.Gong Z, et al. ROS1, a repressor of transcriptional gene silencing in Arabidopsis, encodes a DNA glycosylase/lyase. Cell. 2002;111:803–814. doi: 10.1016/s0092-8674(02)01133-9. [DOI] [PubMed] [Google Scholar]
- 12.Rai K, et al. DNA demethylation in zebrafish involves the coupling of a deaminase, a glycosylase, and gadd45. Cell. 2008;135:1201–1212. doi: 10.1016/j.cell.2008.11.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Barreto G, et al. Gadd45a promotes epigenetic gene activation by repair-mediated DNA demethylation. Nature. 2007;445:671–675. doi: 10.1038/nature05515. [DOI] [PubMed] [Google Scholar]
- 14.Ma DK, et al. Neuronal activity-induced Gadd45b promotes epigenetic DNA demethylation and adult neurogenesis. Science. 2009;323:1074–1077. doi: 10.1126/science.1166859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Engel N, et al. Conserved DNA methylation in Gadd45a(-/-) mice. Epigenetics. 2009;4:98–99. doi: 10.4161/epi.4.2.7858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jin SG, Guo C, Pfeifer GP. GADD45A does not promote DNA demethylation. PLoS Genet. 2008;4:e1000013. doi: 10.1371/journal.pgen.1000013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Allen MD, et al. Solution structure of the nonmethyl-CpG-binding CXXC domain of the leukaemia-associated MLL histone methyltransferase. Embo J. 2006;25:4503–4512. doi: 10.1038/sj.emboj.7601340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jorgensen HF, Adie K, Chaubert P, Bird AP. Engineering a high-affinity methyl-CpG-binding protein. Nucleic Acids Res. 2006;34:e96. doi: 10.1093/nar/gkl527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yamagata K, Suetsugu R, Wakayama T. Long-term, six-dimensional live-cell imaging for the mouse preimplantation embryo that does not affect full-term development. J Reprod Dev. 2009;55:343–350. doi: 10.1262/jrd.20166. [DOI] [PubMed] [Google Scholar]
- 20.Torres-Padilla ME, Bannister AJ, Hurd PJ, Kouzarides T, Zernicka-Goetz M. Dynamic distribution of the replacement histone variant H3.3 in the mouse oocyte and preimplantation embryos. Int J Dev Biol. 2006;50:455–461. doi: 10.1387/ijdb.052073mt. [DOI] [PubMed] [Google Scholar]
- 21.Tahiliani M, et al. Conversion of 5-methylcytosine to 5-hydroxymethylcytosine in mammalian DNA by MLL partner TET1. Science. 2009;324:930–935. doi: 10.1126/science.1170116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kim SH, et al. Differential DNA methylation reprogramming of various repetitive sequences in mouse preimplantation embryos. Biochem Biophys Res Commun. 2004;324:58–63. doi: 10.1016/j.bbrc.2004.09.023. [DOI] [PubMed] [Google Scholar]
- 23.Lane N, et al. Resistance of IAPs to methylation reprogramming may provide a mechanism for epigenetic inheritance in the mouse. Genesis. 2003;35:88–93. doi: 10.1002/gene.10168. [DOI] [PubMed] [Google Scholar]
- 24.Wittschieben BO, et al. A novel histone acetyltransferase is an integral subunit of elongating RNA polymerase II holoenzyme. Mol Cell. 1999;4:123–128. doi: 10.1016/s1097-2765(00)80194-x. [DOI] [PubMed] [Google Scholar]
- 25.Hawkes NA, et al. Purification and characterization of the human elongator complex. J Biol Chem. 2002;277:3047–3052. doi: 10.1074/jbc.M110445200. [DOI] [PubMed] [Google Scholar]
- 26.Wang SC, Frey PA. S-adenosylmethionine as an oxidant: the radical SAM superfamily. Trends Biochem Sci. 2007;32:101–110. doi: 10.1016/j.tibs.2007.01.002. [DOI] [PubMed] [Google Scholar]
- 27.Paraskevopoulou C, Fairhurst SA, Lowe DJ, Brick P, Onesti S. The Elongator subunit Elp3 contains a Fe4S4 cluster and binds S-adenosylmethionine. Mol Microbiol. 2006;59:795–806. doi: 10.1111/j.1365-2958.2005.04989.x. [DOI] [PubMed] [Google Scholar]
- 28.Tagami H, Ray-Gallet D, Almouzni G, Nakatani Y. Histone H3.1 and H3.3 complexes mediate nucleosome assembly pathways dependent or independent of DNA synthesis. Cell. 2004;116:51–61. doi: 10.1016/s0092-8674(03)01064-x. [DOI] [PubMed] [Google Scholar]
- 29.Opavsky R, et al. CpG island methylation in a mouse model of lymphoma is driven by the genetic configuration of tumor cells. PLoS Genet. 2007;3:1757–1769. doi: 10.1371/journal.pgen.0030167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tremblay KD, Duran KL, Bartolomei MS. A 5′ 2-kilobase-pair region of the imprinted mouse H19 gene exhibits exclusive paternal methylation throughout development. Mol Cell Biol. 1997;17:4322–4329. doi: 10.1128/mcb.17.8.4322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ma DK, et al. Neuronal activity-induced Gadd45b promotes epigenetic DNA demethylation and adult neurogenesis. Science. 2009;323:1074–1077. doi: 10.1126/science.1166859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yamagata K, Suetsugu R, Wakayama T. Long-term, six-dimensional live-cell imaging for the mouse preimplantation embryo that does not affect full-term development. J Reprod Dev. 2009;55:343–350. doi: 10.1262/jrd.20166. [DOI] [PubMed] [Google Scholar]
- 33.Tagami H, Ray-Gallet D, Almouzni G, Nakatani Y. Histone H3.1 and H3.3 complexes mediate nucleosome assembly pathways dependent or independent of DNA synthesis. Cell. 2004;116:51–61. doi: 10.1016/s0092-8674(03)01064-x. [DOI] [PubMed] [Google Scholar]
- 34.Opavsky R, et al. CpG island methylation in a mouse model of lymphoma is driven by the genetic configuration of tumor cells. PLoS Genet. 2007;3:1757–1769. doi: 10.1371/journal.pgen.0030167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tremblay KD, Duran KL, Bartolomei MS. A 5′ 2-kilobase-pair region of the imprinted mouse H19 gene exhibits exclusive paternal methylation throughout development. Mol Cell Biol. 1997;17:4322–4329. doi: 10.1128/mcb.17.8.4322. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.