Abstract
Purpose
Hypoxia-inducible factor (HIF) controls the expression of genes in response to hypoxia, as well as a wide range of other cellular processes. We previously showed constitutive stabilization of HIF-1α in the majority of patients with diffuse large B-cell lymphoma (DLBCL). To our knowledge, the prognostic significance of HIF in lymphoma has never been investigated.
Patients and Methods
We studied the immunohistochemical protein expression of HIF-1α on tissue microarrays from 153 patients with DLBCL treated in sequential cohorts with cyclophosphamide, doxorubicin, oncovin, and prednisone (CHOP) or rituximab-CHOP (R-CHOP) from 1999 to 2002. Results were correlated with patient outcome.
Results
Median follow-up for all patients was 80 months. Among all patients, HIF-1α was expressed in 62% of germinal center and 59% of non–germinal center patients. With HIF-1α analyzed as a dependent variable, there were no survival differences in CHOP-treated patients. In the R-CHOP group, however, HIF-1α protein expression correlated with significantly improved progression-free survival (PFS) and overall survival (OS). Five-year PFS for HIF-1α–positive patients was 71% v 43% for HIF-1α–negative patients (P = .0187), whereas 5-year OS was 75% and 54%, respectively (P = .025). In multivariate analysis with International Prognostic Index criteria, HIF-1α remained a significant predictor for PFS (P = .026) and OS (P = .043). Compared with other biomarkers, HIF-1α correlated only with BCL6 (P = .004). In terms of gene expression, we found several common gene associations of HIF-1α and the stromal-1 signature with genes predominantly involved in regulation of the extracellular matrix (eg, BGN, COL1A2, COL5A1, and PLOD2).
Conclusion
The expression of HIF-1α protein is an important independent favorable prognostic factor for survival in patients with DLBCL treated with R-CHOP.
INTRODUCTION
Hypoxia-inducible factor (HIF) is a master regulator of gene transcription, as it controls the expression of more than 200 genes in response to cellular hypoxia.1–3 The genes regulated by HIF affect a wide range of cellular processes including glucose transport, cell metabolism, angiogenesis, erythropoiesis, vascular tone, cell proliferation, apoptosis, and extracellular matrix. Clinically, elevated HIF-1α protein has been correlated with poor prognosis in most solid tumor studies.4 However, relatively little is known about the role of HIF in lymphoma. We previously reported that there is constitutive stabilization of HIF-1α and HIF-2α in many non-Hodgkin's lymphoma cell lines, as well as among a significant fraction of patients with diffuse large B-cell lymphoma (DLBCL) and follicular lymphoma (FL),5 implicating a potential role of dysregulated HIF activation in non-Hodgkin's lymphoma.
DLBCL is curable with multiagent anthracycline-containing chemotherapy.6,7 Addition of rituximab to cyclophosphamide, doxorubicin, oncovin, and prednisone (R-CHOP) represented a significant clinical advance in the treatment of DLBCL.8,9 Several biologic factors have been examined in DLBCL, including TP53, FOXP1, BCL2, BCL6, and LMO2, in an effort to better understand lymphomagenesis and to potentially identify patients who might benefit from particular treatment approaches.10–14 To our knowledge, the prognostic significance of HIF has never been evaluated in non-Hodgkin's lymphoma. We examined whether HIF-1α protein expression, as assessed among 153 patients with DLBCL treated with anthracycline-based chemotherapy with and without rituximab (CHOP and R-CHOP), can predict outcome.
PATIENTS AND METHODS
Patient Selection
We retrospectively examined 153 patients who were treated with anthracycline-based chemotherapy in sequential cohorts from 1999 through 2002 (prerituximab [n = 75] and postrituximab era [n = 78]). All patients were treated through the British Columbia Cancer Agency (BCCA). Treatment was according to BCCA guidelines that instituted R-CHOP as the standard therapy for DLBCL as of March 1, 2001 (standard therapy before 2001 was CHOP), as previously described.15 Characteristics of all patients, based on HIF status, are shown in Table 1. The median follow-up for all living patients was 80 months (range, 7 to 106 months), with CHOP median follow-up of 95 months (range, 51 to 106 months) and R-CHOP median follow-up of 75 months (range, 7 to 90 months).
Table 1.
Patient Demographic and Clinical Characteristics
| Characteristic | HIF-1α Positive (n = 93) |
HIF-1α Negative (n = 60) |
P | ||
|---|---|---|---|---|---|
| No. | % | No. | % | ||
| Age | |||||
| Median | 59 | 65 | .163 | ||
| Mean | 59 | 62 | |||
| Range | 20-85 | 22-85 | |||
| Stage | |||||
| I-II | 37 | 39 | 18 | 30 | .218 |
| III-IV | 56 | 61 | 42 | 70 | |
| ECOG PS | |||||
| 0-1 | 58 | 62 | 40 | 67 | .588 |
| ≥ 2 | 35 | 38 | 20 | 33 | |
| LDH | |||||
| Normal | 36 | 39 | 25 | 42 | .715 |
| Elevated | 57 | 61 | 35 | 58 | |
| Extranodal sites | |||||
| 0-1 | 65 | 67 | 44 | 73 | .646 |
| ≥ 2 | 28 | 33 | 16 | 27 | |
| IPI score | |||||
| 0-2 | 48 | 52 | 29 | 48 | .692 |
| 3-5 | 45 | 48 | 31 | 52 | |
Abbreviations: HIF, hypoxia-inducible factor; ECOG, Eastern Cooperative Oncology Group; PS, performance status; LDH, lactate dehydrogenase; IPI, International Prognostic Index.
Tissue Microarray and Immunohistochemistry
Standard methods were used for tissue fixation (10% buffered formalin) and tissue processing. Tissue sections of 4 to 5 μm were cut from tissue microarrays (TMAs) and placed on glass slides. The TMAs consisted of duplicate 0.6-mm cores for all 153 patients. TMAs were shipped from the BCCA to Northwestern University. All TMAs were stained and scored without knowledge of treatment characteristics or patient outcome. Sections were deparaffinized, dehydrated, and stained with monoclonal antibody (anti–HIF-1α dilution 1:10,0000 Novus NB100-105; Novus Biologicals, Littleton, CO). HIF-1α expression was assessed with semiquantitative immunohistochemical analysis. Nuclear expression of HIF-1α within DLBCL malignant large cells was assessed and scored. Bright-field microscopy imaging coupled with advanced color detection software was used (Automated Cellular Imaging System; ChromaVision Medical Systems, San Juan Capistrano, CA)16–18 to detect and count stained cellular objects. The TMA was read according to the given sector map. Each core was scored individually, and the results were presented as the mean of three replicate core samples. TMA scoring was supervised and validated by pathology review. HIF-1α was dichotomized as either negative (no nuclear staining) or positive (any nuclear staining). The other immunostains (BCL-6, BCL2, CD10, MUM-1, and FOXP1) were determined by the methods described by Hans et al.19
Statistical Analysis
Progression-free survival (PFS) was defined as the interval between the date of initial diagnosis and the date of disease progression or death from lymphoma or treatment toxicity. Overall survival (OS) was defined as the time interval between the date of diagnoses and the date of death from any cause or last follow-up. Survival analyses were performed using Kaplan-Meier curves,20 which were compared using log-rank testing.21 Univariate analysis and multivariate regression according to the Cox proportional hazards regression model22 were used (for International Prognostic Index [IPI] and HIF-1α).
RESULTS
HIF Expression
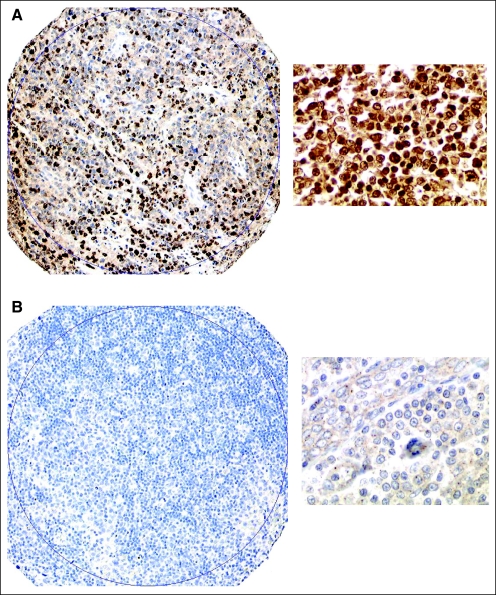
To ensure reproducibility between treatment groups, we assessed HIF-1α expression according to the two treatment eras of CHOP and R-CHOP. HIF-1α was expressed in 63% (49 of 78) of patients treated with R-CHOP compared with 59% (44 of 75) treated with CHOP (P = not significant). Figures 1A and 1B, respectively, show representative examples of increased HIF-1α protein expression (positive) and no evidence of HIF-1α expression (negative). Of patients who had increased HIF-1α protein expression (93 of 153), 87% (81 of 93) had moderate to high (> 10% malignant cells) HIF-1α staining.
Fig 1.
HIF-1α protein expression. Representative immunohistochemistry cases from the tissue microarray of increased HIF-1α protein expression (A) and a representative case with no evidence of HIF-1α protein expression (B); both cases shown at ×10 and ×40 magnification.
Patients
Sixty-four percent (98 of 153) of all patients had advanced-stage disease, whereas 50% had an IPI score of ≥ 3 (Table 1). HIF-1α–negative patients were older and had a higher incidence of stage III/IV disease compared with HIF-1α–positive patients (P = .22), whereas HIF-1α–positive patients had a slightly higher incidence of elevated lactate dehydrogenase, worse performance status, and increased frequency of at least two extranodal sites compared with the HIF-1α–negative group, although these differences were not statistically significant. We also determined HIF-1α expression status according to cell of origin, on the basis of the Hans algorithm.19 Among all patients (n = 153), HIF-1α was expressed in 62% of germinal center (GC) patients and 59% of non-GC patients (P = not significant).
HIF-1α Expression and Outcome
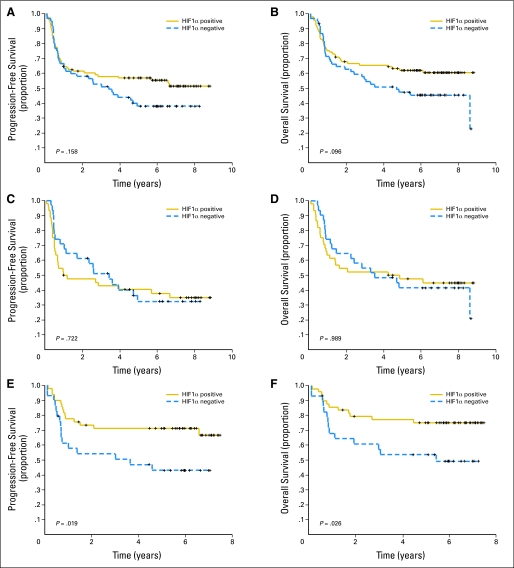
The relationship of HIF-1α protein expression with clinical outcome was examined. When HIF-1α protein was analyzed as a dependent variable (positive v negative) among all patients, there was a trend toward improved survival for HIF-1α–positive patients, as shown in Figures 2A and 2B (P = .158 and P = .096, respectively). When analyzed according to treatment group, there was no difference noted in PFS or OS among patients who received CHOP (Figs 2C and 2D). In the R-CHOP cohort of patients (Figs 2E and 2F), however, the 5-year PFS was 43% for HIF-1α–negative patients compared with 72% for HIF-1α–positive patients (log-rank P = .019), whereas the 5-year OS was 54% for HIF-1α–negative patients compared with 75% for HIF-1α–positive patients (log-rank P = .025). We examined other cutoffs of HIF-1α protein expression with different percentages of HIF-1α staining (eg, ≥ 10% v < 10%); no other HIF-1α expression level was more significant than using the dichotomization of any HIF-1α expression versus no HIF-1α expression (data not shown).
Fig 2.
Progression-free survival (PFS) and overall survival (OS) for all patients according to HIF-1α protein expression. Kaplan-Meier curves of (A) PFS and (B) OS in 153 patients with diffuse large B-cell lymphoma (DLBCL) showed a trend for prolonged OS (P = .096) with HIF-1α protein expression. Kaplan-Meier curves of (C) PFS and (D) OS in the 75 patients with DLBCL treated with cyclophosphamide, doxorubicin, oncovin, and prednisone (CHOP) grouped on the basis of HIF-1α protein expression showed no correlation with HIF-1α protein expression. Kaplan-Meier curves of (E) PFS and (F) OS in 78 patients with DLBCL grouped on the basis of HIF show that HIF-1α protein expression correlates with longer PFS (P = .019) and OS (P = .026).
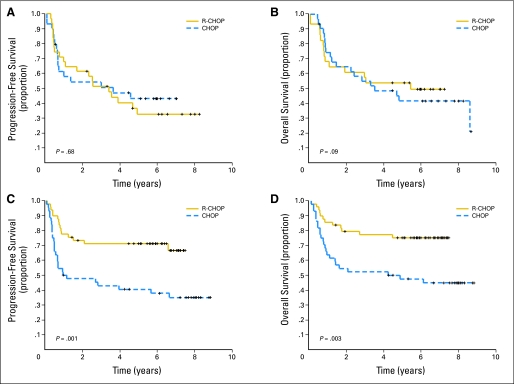
We subsequently analyzed survival according to induction therapy for HIF-1α–positive and HIF-1α–negative patients. For HIF-1α–negative patients, the 5-year PFS and OS did not appear to be significantly influenced by treatment (Figs 3A and 3B). Conversely, among patients with DLBCL who expressed HIF-1α protein, the outcomes were significantly improved with R-CHOP. As shown in Figures 3C and 3D, the 5-year PFS was 72% for R-CHOP versus 41% for CHOP (log-rank P = .001), whereas the 5-year OS was 77% for R-CHOP versus 48% for CHOP (log-rank P = .003).
Fig 3.
Progression-free survival (PFS) and overall survival (OS) according to induction therapy. Kaplan-Meier curves of (A) PFS and (B) OS in 60 patients with diffuse large B-cell lymphoma (DLBCL) with HIF-1α–negative status grouped on the basis of treatment received (cyclophosphamide, doxorubicin, oncovin, and prednisone [CHOP] v rituximab and CHOP [R-CHOP]). Kaplan-Meier curves of (C) PFS and (D) OS in 93 patients with DLBCL with HIF-1α protein expression grouped on the basis of treatment show that R-CHOP was associated highly increased PFS (P = .001) and OS (P = .003) compared with CHOP.
Using multivariate Cox regression analysis, we analyzed IPI and HIF-1α protein as dependent variables for correlation with PFS and OS among patients who received R-CHOP, with HIF dichotomized as positive versus negative and IPI as high (3 to 5) versus low (0 to 2), as shown in Table 2. In multivariate analysis, HIF-1α expression remained a significant independent factor predicting for improved PFS (P = .026) and OS (P = .043), whereas the IPI was significant for PFS and of borderline significance for OS. In terms of outcomes by HIF-1α based on cell of origin, the improvement in survival for HIF-1α–positive patients (v HIF-1α–negative patients) was seen primarily in non-GC (n = 77; OS P = .0015) compared with GC DLBCL (n = 61; OS P = .547). Furthermore, the improvement in survival among patients with HIF-1α–positive non-GC DLBCL (v HIF-1α negative) was more evident among R-CHOP–treated patients (OS P = .0027) than CHOP-treated patients (OS P = .117).
Table 2.
Multivariate Analysis of IPI and HIF-1α Protein Expression With PFS and OS in Patients With DLBCL Treated With R-CHOP
| Variable | Progression-Free Survival |
Overall Survival |
||||
|---|---|---|---|---|---|---|
| HR* | 95% CI | P | HR* | 95% CI | P | |
| IPI 3-5 | 2.41 | 1.17 to 4.99 | .018 | 2.21 | 0.99 to 4.89 | .051 |
| HIF1α-positive expression | 0.45 | 0.22 to 0.91 | .026 | 0.45 | 0.21 to 0.98 | .043 |
Abbreviations: IPI, International Prognostic Index; HIF, hypoxia-inducible factor; PFS, progression-free survival; OS, overall survival; DLBCL, diffuse large B-cell lymphoma; R-CHOP, rituximab with cyclophosphamide, doxorubicin, oncovin, and prednisone; HR, hazard ratio.
HR greater than 1 indicates a factor with poor prognosis, whereas a ratio of 1 or less indicates a factor with good prognosis.
HIF-1α Compared With Other Biomarkers
Among patients who received R-CHOP, we examined whether HIF-1α correlated with expression of other DLBCL prognostic markers. As shown in Table 3, HIF-1α did not correlate with BCL2, CD10, MUM-1, or FOXP1, whereas a correlation was detected between BCL6 protein (by the Hans method).19 Among HIF-1α–positive R-CHOP–treated patients, 94% (45 of 48) were BCL6 positive (P = .004), although among HIF-1α–negative patients, 66% (19/29) were also BCL6 positive. Among the CHOP-treated patient group (n = 75), BCL6 protein expression was associated with a superior PFS (P = .007) and OS (P = .009), whereas among the R-CHOP group (n = 78), there were no survival differences noted on the basis of BCL6 expression.
Table 3.
Comparison of HIF-1α Protein Expression With Other Biomarkers in R-CHOP-Treated Patients (n = 78)
| Biomarker | HIF-1α Expression |
P† | |
|---|---|---|---|
| Positive | Negative | ||
| BCL2* | |||
| Positive | 32 | 21 | .54 |
| Negative | 16 | 8 | |
| CD10 | |||
| Positive | 18 | 12 | .81 |
| Negative | 31 | 17 | |
| MUM-1* | |||
| Positive | 34 | 16 | .24 |
| Negative | 15 | 12 | |
| BCL6* | |||
| Positive | 45 | 19 | .004 |
| Negative | 3 | 10 | |
| FOXP1 | |||
| Positive | 14 | 10 | .59 |
| Negative | 35 | 19 | |
Abbreviations: HIF, hypoxia-inducible factor; R-CHOP, rituximab with cyclophosphamide, doxorubicin, oncovin, and prednisone.
One sample in each group could not be scored.
Two-sided P value.
Correlation With Gene Expression
We performed a survey of the gene signature that defines a list of HIF-1α target genes.23 Increased HIF-1α gene expression was associated with expression of BGN (biglycan), C8ORF4 (chromosome 8 open reading frame 4), COL1A2 (collagen, type I, α 2), COL5A1 (collagen, type V, α 1), HTRA1 (HtrA serine peptidase 1), LOX (lysyl oxidase), MAFF (v-maf musculoaponeurotic fibrosarcoma oncogene homolog F), PLOD2 (procollagen-lysine, 2-oxoglutarate 5-dioxygenase 2), and VEGFC.23 Interestingly, each of these genes are part of the favorable stromal-1 gene signature as defined by Lenz et al.24
DISCUSSION
We found that HIF-1α is an important and previously undiscovered independent favorable prognostic factor for PFS and OS in patients with DLBCL treated with R-CHOP. To our knowledge, this is the first report to examine the prognostic significance of HIF-1α in lymphoma. Although HIF is not an oncogene, a large amount of evidence has accumulated linking HIF-1α regulation with cancer pathogenesis.1,2,4,25,26 HIF is a dimer comprised of an α (HIF-α) and a β subunit (also known as ARNT).25 Under hypoxic conditions, its degradation is inhibited, allowing HIF-α to accumulate, dimerize with HIF-β, and translocate to the nucleus and activate transcription of HIF-1α target genes. Under normoxic conditions, minimal HIF-α protein is detected. Degradation of HIF-α in normoxia is initiated by the hydroxylation of two proline residues by prolyl hydroxylases.27,28 Proline hydroxylation permits interaction with von Hippel Lindau protein, thereby committing it to proteasome degradation.29 It should be noted that other factors, in addition to hypoxia, are able to independently increase the transcription or translation of HIF-1α. These include phosphatidylinositide 3-kinase-AKT-mammalian target of rapamycin, RAS-mitogen-activated protein kinase-kinase-extracellular signal regulated kinase, and erbB2 families.30–32
HIF-1α and HIF-2α represent the major isoforms regulated by hypoxia.33 Some overlap exists with respect to the genes they control, although the targets are not identical; HIF-1α has been more clearly described. We previously reported abnormal constitutive stabilization of HIF-1α and HIF-2α in Raji, Namwala, Ramos, SUDHL4, HF1, and Daudi NHL cell lines, and we demonstrated that HIF-1α was upregulated in lymph node biopsies obtained from patients with DLBCL and FL.5 We found that HIF-1α was more frequently expressed in DLBCL versus FL (65% v 47%, respectively; P = .05). Furthermore, the frequency of moderate to high HIF-1α expression was higher in DLBCL compared with FL (ie, ≥ 10% HIF-1α expression: 54% v 20%, respectively; P = .04).5 Giatromanolaki et al34recently confirmed the upregulation of HIF-1α in DLBCL, and they showed that normal lymph nodes had no detectable HIF-1α, HIF-2α, vascular endothelial growth factor (VEGF), or phosphorylated VEGF receptor-2 (pVEGFR-2/KDR) expression (0 of 42). They found HIF-1α protein expression (nuclear) in 53% of patients with DLBCL versus 39% of those with FL.
The majority of reports examining the relationship of HIF in solid tumors have found increased HIF-1α protein to be associated with an increased risk of metastasis and/or inferior survival,4,35 although the correlation of HIF-1α with improved outcome is not unprecedented. Two reports in epithelial tumors (renal cell and head/neck cancer), showed that increased HIF-1α protein expression correlated with improved survival.36,37 Although HIF is known to regulate genes that contribute to the growth of tumors, the overall balance of the effect of HIF may depend on the specific malignant phenotype and particular treatment modality. A recent report by Gratzinger et al38 showed that high VEGF and VEGFR-1 protein expression in DLBCL was associated with significantly improved PFS and OS. Gratzinger et al did not examine HIF, but the report by Giatromanolaki et al34 found a high concordance between increased tumor cell expression of HIF-1α protein with VEGF (P = .0005) and pVEGFR-2/KDR tumor cell expression (P < .0001).
We found in the present study that patients with DLBCL treated with R-CHOP had superior outcome if HIF-1α was expressed. This HIF-based survival difference was not apparent in CHOP-treated patients. The reason for this is not clear, but there are several possible explanations. We know that HIF-1α expression is regulated by reactive oxygen species (ROS). Gao et al39 recently found that treatment of mice bearing a MYC-inducible lymphoma with the antioxidant N-acetyl-l-cysteine resulted in inhibition of HIF-1α expression and inhibition of tumor growth. Similarly, Gupta et al40 showed that ROS increased CD20 expression in the Burkitt lymphoma cell lines, Daudi and Raji. Taken together, these data suggest that HIF-1α and CD20 expression are both regulated by ROS and that the HIF-1α expression in our current study may reflect an increase in CD20 expression, making those patients more likely to respond to a rituximab-containing regimen. We recently reported that density of CD20 expression correlates with outcome in R-CHOP–treated patients with DLBCL.41 Alternatively, there may be unappreciated interactions between rituximab and downstream targets of HIF-1α. Rituximab may interact with other oncogenic pathways that are known to stimulate HIF transcription or translation, as discussed before. A further explanation for inferior survival with HIF-negative DLBCL tumor cells may also be found in the data reported by Ginouves et al,42 in which chronic cellular hypoxia led to over-activation of prolyl hydroxylase, with associated HIF desensitization and subsequent degradation of HIF-1α protein as a mechanism of cell survival.
The results here also need to be reconciled with the study by Lenz et al,24 in which gene expression profiling of R-CHOP–treated DLBCL patients discovered two new signatures of the nonmalignant surrounding cells, termed stromal-1 and stromal-2. The unfavorable stromal-2 signature was associated with several angiogenic genes including von Willebrand factor and CD31 (PECAM1). Furthermore, cases with stromal-2 signature were associated with increased lymph node microvessel density (MVD). Gratzinger et al38 showed that increased MVD correlated with poor outcome in DLBCL, whereas among the same samples, increased cell expression of VEGF was associated with improved survival, suggesting an inverse relationship between tumor cell VEGF expression and MVD. It is important to note that the associations and interactions between the microenvironment and expression of cellular DLBCL HIF-1α are not known. In a survey of HIF-1α target genes,23 we found overlap with several of the genes included in the stromal-1 signature defined by Lenz et al.24 Several of the common gene associations are involved in regulation of the extracellular matrix (eg, BGN, COL1A2, COL5A1, and PLOD2). In addition to angiogenesis, HIF-1α regulates a wide array of genes involved in cancer, including those involved in cell metabolism, cell motility, inflammatory cell recruitment, pH regulation, and extracellular matrix function.3 On the basis of consistent upregulation of HIF-1α target genes within the stromal-1 signature, it could be hypothesized that increased gene expression of HIF-1α is also associated with a favorable survival in DLBCL treated with R-CHOP.
A correlation was detected between HIF-1α protein and Bcl-6 protein expression wherein 94% of HIF-1α–positive patients were Bcl-6 positive, although 30% of patients who expressed Bcl-6 were negative for HIF-1α. Carmeliet et al2 found a similar strong association between HIF-1α and Bcl-6 in breast cancer, whereas Bos et al43 correlated Bcl-6 expression with HIF-1α and p21. Further examination of the relationship of HIF-1α with Bcl-6 and other lymphoma biomarkers is warranted.
We conclude that expression of HIF-1α is an important and heretofore undiscovered independent favorable prognostic factor for PFS and OS in patients with DLBCL treated with R-CHOP, but not in patients treated with CHOP. These data suggest there may be an important biologic interaction between CD20 monoclonal antibody–based therapy and HIF-1α or downstream genes regulated by HIF. It is important to note that these findings should be validated and studied in prospective sample sets. Furthermore, the importance of HIF-1α gene expression in DLBCL, downstream targets of HIF, enzymes (eg, thioredoxins) and pathways that interact with HIF, and the relationship of malignant cell HIF-1α expression and the tumor microenvironment need to be studied.
Glossary Terms
- HIF (hypoxia-inducible factor):
HIF is a transcriptional factor that regulates the adaptive responses of mammalian cells to low oxygen (hypoxia). It is composed of HIF-1α, which is upregulated in conditions of hypoxia, and HIF-1β (or, aryl hydrocarbon receptor nuclear translocators), which is expressed constitutively. Dimerization of HIF-1α with HIF-1β leads to transcription of genes such as VEGF and PDGF.
- Rituximab:
A monoclonal antibody therapy that is indicated for relapsed or refractory low-grade or follicular, CD20+, B-cell non-Hodgkin's lymphoma.
- ROS (reactive oxygen species):
Molecules like hydrogen peroxide, hydroxyl radical, and superoxide anion are designated ROS, which are formed as a consequence of ionizing radiation with biologic molecules, cellular respiration (via the electron transport chain), and a metabolic byproduct of neutrophils and macrophages. The activity of ROS damages cellular molecules and structures. In phagocytic cells, ROS are essential to eliminate harmful agents like bacteria. Cellular enzymes such as catalase and superoxide dismutase, and antioxidants such as vitamins E and C dissipate ROS after they are formed.
- CD20:
Cell surface antigen present on lymphoid B cells, it is a widely used phenotypic marker for typing malignant lymphomas. It is involved in B-cell activation.
- Hypoxia:
Oxygen concentration below normal physiological limits in a specific tissue.
- Apoptosis:
Also called programmed cell death, it is a signaling pathway that leads to cellular suicide in an organized manner. Several factors and receptors are specific to the apoptotic pathway. The net result is that cells shrink, develop blebs on their surface, and their DNA undergoes fragmentation.
Footnotes
Supported in part by National Cancer Institute Grants No. K23 CA109613-A1 (A.M.E.); the National Cancer Institute of Canada, Terry Fox Program Project Grant No. 016003 (R.D.G.).
Terms in blue are defined in the glossary, found at the end of this article and online at www.jco.org.
Authors' disclosures of potential conflicts of interest and author contributions are found at the end of this article.
AUTHORS' DISCLOSURES OF POTENTIAL CONFLICTS OF INTEREST
Although all authors completed the disclosure declaration, the following author(s) indicated a financial or other interest that is relevant to the subject matter under consideration in this article. Certain relationships marked with a “U” are those for which no compensation was received; those relationships marked with a “C” were compensated. For a detailed description of the disclosure categories, or for more information about ASCO's conflict of interest policy, please refer to the Author Disclosure Declaration and the Disclosures of Potential Conflicts of Interest section in Information for Contributors.
Employment or Leadership Position: None Consultant or Advisory Role: Laurie H. Sehn, Roche (C); Randy D. Gascoyne, Roche Canada (C) Stock Ownership: None Honoraria: Laurie H. Sehn, Roche; Randy D. Gascoyne, Roche Canada Research Funding: Laurie H. Sehn, Roche; Randy D. Gascoyne, Roche Canada Expert Testimony: None Other Remuneration: None
AUTHOR CONTRIBUTIONS
Conception and design: Andrew M. Evens, Randy D. Gascoyne, Leo I. Gordon
Financial support: Andrew M. Evens, Randy D. Gascoyne, Leo I. Gordon
Administrative support: Andrew M. Evens, Randy D. Gascoyne, Leo I. Gordon
Provision of study materials or patients: Andrew M. Evens, Laurie H. Sehn, Pedro Farinha, Randy D. Gascoyne, Leo I. Gordon
Collection and assembly of data: Andrew M. Evens, Laurie H. Sehn, Beverly P. Nelson, Adekunle Raji, Yi Lu, Adam Brakman, Vamsi Parimi, Randy D. Gascoyne, Leo I. Gordon
Data analysis and interpretation: Andrew M. Evens, Laurie H. Sehn, Pedro Farinha, Beverly P. Nelson, Adekunle Raji, Yi Lu, Adam Brakman, Vamsi Parimi, Jane N. Winter, Paul T. Schumacker, Randy D. Gascoyne, Leo I. Gordon
Manuscript writing: Andrew M. Evens, Laurie H. Sehn, Pedro Farinha, Beverly P. Nelson, Jane N. Winter, Paul T. Schumacker, Randy D. Gascoyne, Leo I. Gordon
Final approval of manuscript: Andrew M. Evens, Laurie H. Sehn, Pedro Farinha, Beverly P. Nelson, Adekunle Raji, Yi Lu, Jane N. Winter, Paul T. Schumacker, Randy D. Gascoyne, Leo I. Gordon
REFERENCES
- 1.Semenza GL. HIF-1: Mediator of physiological and pathophysiological responses to hypoxia. J Appl Physiol. 2000;88:1474–1480. doi: 10.1152/jappl.2000.88.4.1474. [DOI] [PubMed] [Google Scholar]
- 2.Carmeliet P, Dor Y, Herbert JM, et al. Role of HIF-1alpha in hypoxia-mediated apoptosis, cell proliferation and tumour angiogenesis. Nature. 1998;394:485–490. doi: 10.1038/28867. [DOI] [PubMed] [Google Scholar]
- 3.Bertout JA, Patel SA, Simon MC. The impact of O2 availability on human cancer. Nat Rev Cancer. 2008;8:967–975. doi: 10.1038/nrc2540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zhong H, De Marzo AM, Laughner E, et al. Overexpression of hypoxia-inducible factor 1alpha in common human cancers and their metastases. Cancer Res. 1999;59:5830–5835. [PubMed] [Google Scholar]
- 5.Evens AM, Schumacker PT, Helenowski IB, et al. Hypoxia inducible factor-alpha activation in lymphoma and relationship to the thioredoxin family. Br J Haematol. 2008;141:676–680. doi: 10.1111/j.1365-2141.2008.07093.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fisher RI, Gaynor ER, Dahlberg S, et al. Comparison of a standard regimen (CHOP) with three intensive chemotherapy regimens for advanced non-Hodgkin's lymphoma. N Engl J Med. 1993;328:1002–1006. doi: 10.1056/NEJM199304083281404. [DOI] [PubMed] [Google Scholar]
- 7.Gordon LI, Harrington D, Andersen J, et al. Comparison of a second-generation combination chemotherapeutic regimen (m-BACOD) with a standard regimen (CHOP) for advanced diffuse non-Hodgkin's lymphoma. N Engl J Med. 1992;327:1342–1349. doi: 10.1056/NEJM199211053271903. [DOI] [PubMed] [Google Scholar]
- 8.Habermann TM, Weller EA, Morrison VA, et al. Rituximab-CHOP versus CHOP alone or with maintenance rituximab in older patients with diffuse large B-cell lymphoma. J Clin Oncol. 2006;24:3121–3127. doi: 10.1200/JCO.2005.05.1003. [DOI] [PubMed] [Google Scholar]
- 9.Feugier P, Van Hoof A, Sebban C, et al. Long-term results of the R-CHOP study in the treatment of elderly patients with diffuse large B-cell lymphoma: A study by the Groupe d'Etude des Lymphomes de l'Adulte. J Clin Oncol. 2005;23:4117–4126. doi: 10.1200/JCO.2005.09.131. [DOI] [PubMed] [Google Scholar]
- 10.Natkunam Y, Farinha P, Hsi ED, et al. LMO2 protein expression predicts survival in patients with diffuse large B-cell lymphoma treated with anthracycline-based chemotherapy with and without rituximab. J Clin Oncol. 2008;26:447–454. doi: 10.1200/JCO.2007.13.0690. [DOI] [PubMed] [Google Scholar]
- 11.Malumbres R, Chen J, Tibshirani R, et al. Paraffin-based 6-gene model predicts outcome in diffuse large B-cell lymphoma patients treated with R-CHOP. Blood. 2008;111:5509–5514. doi: 10.1182/blood-2008-02-136374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Winter JN, Weller EA, Horning SJ, et al. Prognostic significance of Bcl-6 protein expression in DLBCL treated with CHOP or R-CHOP: A prospective correlative study. Blood. 2006;107:4207–4213. doi: 10.1182/blood-2005-10-4222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mounier N, Briere J, Gisselbrecht C, et al. Rituximab plus CHOP (R-CHOP) overcomes bcl-2–associated resistance to chemotherapy in elderly patients with diffuse large B-cell lymphoma (DLBCL) Blood. 2003;101:4279–4284. doi: 10.1182/blood-2002-11-3442. [DOI] [PubMed] [Google Scholar]
- 14.Lenz G, Wright GW, Emre NC, et al. Molecular subtypes of diffuse large B-cell lymphoma arise by distinct genetic pathways. Proc Natl Acad Sci U S A. 2008;105:13520–13525. doi: 10.1073/pnas.0804295105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sehn LH, Berry B, Chhanabhai M, et al. The revised International Prognostic Index (R-IPI) is a better predictor of outcome than the standard IPI for patients with diffuse large B-cell lymphoma treated with R-CHOP. Blood. 2007;109:1857–1861. doi: 10.1182/blood-2006-08-038257. [DOI] [PubMed] [Google Scholar]
- 16.Giltnane JM, Rimm DL. Technology insight: Identification of biomarkers with tissue microarray technology. Nat Clin Pract Oncol. 2004;1:104–111. doi: 10.1038/ncponc0046. [DOI] [PubMed] [Google Scholar]
- 17.Tawfik OW, Kimler BF, Davis M, et al. Comparison of immunohistochemistry by automated cellular imaging system (ACIS) versus fluorescence in-situ hybridization in the evaluation of HER-2/neu expression in primary breast carcinoma. Histopathology. 2006;48:258–267. doi: 10.1111/j.1365-2559.2005.02322.x. [DOI] [PubMed] [Google Scholar]
- 18.Margulis V, Shariat SF, Ashfaq R, et al. Ki-67 is an independent predictor of bladder cancer outcome in patients treated with radical cystectomy for organ-confined disease. Clin Cancer Res. 2006;12:7369–7373. doi: 10.1158/1078-0432.CCR-06-1472. [DOI] [PubMed] [Google Scholar]
- 19.Hans CP, Weisenburger DD, Greiner TC, et al. Confirmation of the molecular classification of diffuse large B-cell lymphoma by immunohistochemistry using a tissue microarray. Blood. 2004;103:275–282. doi: 10.1182/blood-2003-05-1545. [DOI] [PubMed] [Google Scholar]
- 20.Kaplan EL, Meier M. Nonparametric estimation from incomplete observation. J Am Stat Assoc. 1958;53:457–481. [Google Scholar]
- 21.Mantel H, Haenszel W. Statistical aspects of the analysis of data from retrospective studies of disease. J Natl Cancer Inst. 1959;22:719–748. [PubMed] [Google Scholar]
- 22.Cox DR. Regression models and life tables. J R Stat Soc (B) 1972;34:187–220. [Google Scholar]
- 23.Shaffer AL, Wright G, Yang L, et al. A library of gene expression signatures to illuminate normal and pathological lymphoid biology. doi: 10.1111/j.0105-2896.2006.00373.x. http://lymphochip.nih.gov/signaturedb/ [DOI] [PubMed] [Google Scholar]
- 24.Lenz G, Wright G, Dave SS, et al. Stromal gene signatures in large-B-cell lymphomas. N Engl J Med. 2008;359:2313–2323. doi: 10.1056/NEJMoa0802885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Semenza GL. Hypoxia, clonal selection, and the role of HIF-1 in tumor progression. Crit Rev Biochem Mol Biol. 2000;35:71–103. doi: 10.1080/10409230091169186. [DOI] [PubMed] [Google Scholar]
- 26.Semenza GL, Roth PH, Fang HM, et al. Transcriptional regulation of genes encoding glycolytic enzymes by hypoxia-inducible factor 1. J Biol Chem. 1994;269:23757–23763. [PubMed] [Google Scholar]
- 27.Ivan M, Kondo K, Yang H, et al. HIFalpha targeted for VHL-mediated destruction by proline hydroxylation: Implications for O2 sensing. Science. 2001;292:464–468. doi: 10.1126/science.1059817. [DOI] [PubMed] [Google Scholar]
- 28.Jaakkola P, Mole DR, Tian YM, et al. Targeting of HIF-alpha to the von Hippel-Lindau ubiquitylation complex by O2-regulated prolyl hydroxylation. Science. 2001;292:468–472. doi: 10.1126/science.1059796. [DOI] [PubMed] [Google Scholar]
- 29.Maxwell PH, Wiesener MS, Chang GW, et al. The tumour suppressor protein VHL targets hypoxia-inducible factors for oxygen-dependent proteolysis. Nature. 1999;399:271–275. doi: 10.1038/20459. [DOI] [PubMed] [Google Scholar]
- 30.Majumder PK, Febbo PG, Bikoff R, et al. MTOR inhibition reverses Akt-dependent prostate intraepithelial neoplasia through regulation of apoptotic and HIF-1-dependent pathways. Nat Med. 2004;10:594–601. doi: 10.1038/nm1052. [DOI] [PubMed] [Google Scholar]
- 31.Laughner E, Taghavi P, Chiles K, et al. HER2 (neu) signaling increases the rate of hypoxia-inducible factor 1alpha (HIF-1alpha) synthesis: Novel mechanism for HIF-1-mediated vascular endothelial growth factor expression. Mol Cell Biol. 2001;21:3995–4004. doi: 10.1128/MCB.21.12.3995-4004.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Fukuda R, Hirota K, Fan F, et al. Insulin-like growth factor 1 induces hypoxia-inducible factor 1-mediated vascular endothelial growth factor expression, which is dependent on MAP kinase and phosphatidylinositol 3-kinase signaling in colon cancer cells. J Biol Chem. 2002;277:38205–38211. doi: 10.1074/jbc.M203781200. [DOI] [PubMed] [Google Scholar]
- 33.Hu CJ, Wang LY, Chodosh LA, et al. Differential roles of hypoxia-inducible factor 1alpha (HIF-1alpha) and HIF-2alpha in hypoxic gene regulation. Mol Cell Biol. 2003;23:9361–9374. doi: 10.1128/MCB.23.24.9361-9374.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Giatromanolaki A, Koukourakis MI, Pezzella F, et al. Phosphorylated VEGFR2/KDR receptors are widely expressed in B-cell non-Hodgkin's lymphomas and correlate with hypoxia inducible factor activation. Hematol Oncol. 2008;26:219–224. doi: 10.1002/hon.861. [DOI] [PubMed] [Google Scholar]
- 35.Semenza GL. Targeting HIF-1 for cancer therapy. Nat Rev Cancer. 2003;3:721–732. doi: 10.1038/nrc1187. [DOI] [PubMed] [Google Scholar]
- 36.Beasley NJ, Leek R, Alam M, et al. Hypoxia-inducible factors HIF-1alpha and HIF-2alpha in head and neck cancer: Relationship to tumor biology and treatment outcome in surgically resected patients. Cancer Res. 2002;62:2493–2497. [PubMed] [Google Scholar]
- 37.Lidgren A, Hedberg Y, Grankvist K, et al. The expression of hypoxia-inducible factor 1alpha is a favorable independent prognostic factor in renal cell carcinoma. Clin Cancer Res. 2005;11:1129–1135. [PubMed] [Google Scholar]
- 38.Gratzinger D, Zhao S, Tibshirani RJ, et al. Prognostic significance of VEGF, VEGF receptors, and microvessel density in diffuse large B cell lymphoma treated with anthracycline-based chemotherapy. Lab Invest. 2008;88:38–47. doi: 10.1038/labinvest.3700697. [DOI] [PubMed] [Google Scholar]
- 39.Gao P, Zhang H, Dinavahi R, et al. HIF-dependent antitumorigenic effect of antioxidants in vivo. Cancer Cell. 2007;12:230–238. doi: 10.1016/j.ccr.2007.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Gupta D, Crosby ME, Almasan A, et al. Regulation of CD20 expression by radiation-induced changes in intracellular redox status. Free Radic Biol Med. 2008;44:614–623. doi: 10.1016/j.freeradbiomed.2007.10.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Johnson NA, Boyle M, Bashashati A, et al. Diffuse large B cell lymphoma: Reduced CD20 expression is associated with an inferior survival. Blood. 2008;113:3773–3780. doi: 10.1182/blood-2008-09-177469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ginouves A, Ilc K, Macias N, et al. PHDs overactivation during chronic hypoxia “desensitizes” HIFalpha and protects cells from necrosis. Proc Natl Acad Sci U S A. 2008;105:4745–4750. doi: 10.1073/pnas.0705680105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bos R, van Diest PJ, van der Groep P, et al. Protein expression of B-cell lymphoma gene 6 (BCL-6) in invasive breast cancer is associated with cyclin D1 and hypoxia-inducible factor-1alpha (HIF-1alpha) Oncogene. 2003;22:8948–8951. doi: 10.1038/sj.onc.1206995. [DOI] [PubMed] [Google Scholar]